Abstract
There is a significant need for new therapeutics to treat infections caused by the biodefense agent Bacillus anthracis. In pursuit of drug discovery against this organism, we have developed novel propargyl-linked inhibitors that target the essential enzyme dihydrofolate reductase (DHFR) from B. anthracis. Previously, we reported an initial series of these inhibitors and a high-resolution crystal structure of the ternary complex of the enzyme bound to its cofactor and one of the most potent inhibitors, UCP120B [Beierlein, J., Frey, K., Bolstad, D., Pelphrey, P., Joska, T., Smith, A., Priestley, N., Wright, D., and Anderson, A. (2008) J. Med. Chem. 51, 7532–7540]. Herein, we describe a three-dimensional solution structure of the ternary complex as determined by NMR. A comparison of this solution structure to the crystal structure reveals a general conservation of the DHFR fold and cofactor interactions as well as differences in the location of an active site helix and specific ligand interactions. In addition to data for the fully assigned ternary complex, data for the binary (enzyme–cofactor) complex were collected, providing chemical shift comparisons and revealing perturbations in residues that accommodate ligand binding. Dynamics of the protein, measured using 15N T1 and T2 relaxation times and {1H}–15N heteronuclear NOEs, reveal residue flexibility at the active site that explains enzyme inhibition and structure–activity relationships for two different series of these propargyl-linked inhibitors. The information obtained from the solution structure regarding active site flexibility will be especially valuable in the design of inhibitors with increased potency.
Bacillus anthracis, the causative agent of anthrax, is a well-known bioterrorism threat. The limited number of approved therapeutics exhibit serious drawbacks including indication spectrum, resistance, and expense, especially in a case of large-scale exposure. The development of stable, effective new therapeutics is certainly warranted.
Dihydrofolate reductase (DHFR),1 an essential enzyme in cellular metabolism, catalyzes the reduction of di-hydrofolate to form tetrahydrofolate using the cofactor NADPH. Over the past five decades, DHFR has been recognized as a validated drug target for both human malignancies and infections by pathogenic organisms. Building on the interest in this enzyme, structures of DHFR from several species have been determined by both X-ray crystallography and NMR methods. These structures include apo, binary (NADPH only), and ternary forms (NADPH and substrate or NADPH and inhibitor). Three complete ternary structures of DHFR determined using experimental NMR methods have been previously reported: Lactobacillus casei DHFR (LcDHFR) bound to NADPH and trimethoprim ((1); PDB ID 1LUD), an antibacterial DHFR inhibitor, LcDHFR bound to NADPH and methotrexate ((2); PDB ID 1AO8), and human DHFR (hDHFR) bound to NADPH and trimethoprim ((3); PDB ID 1YHO). In general, the solution structures are similar to the related X-ray crystal structures and yield additional details regarding the cooperativity of cofactor and ligand binding. Specifically, the solution structures of LcDHFR and hDHFR bound to NADPH and trimethoprim reveal that NADPH and trimethoprim are within an optimal distance for interaction. The hDHFR structure further identifies a hydrophobic interface formed by Trp 24 and Leu 22 that interacts with both trimethoprim and NADPH (3).
Previous NMR studies have also examined some of the conformational changes in active site loops that occur upon ligand binding. The majority of chemical shift differences observed between the binary and ternary complexes of E. coli DHFR (EcDHFR) (4, 5) are predictably located near the substrate and cofactor binding sites. Results from these studies showed that the enzyme predominantly populates two structural states in solution: one in which the loop containing residues Ala 9–Asn 23 (often called the Met 20 loop; this loop corresponds with residues Asp 10–Arg 24 in BaDHFR) closes over the active site when NADPH is bound and one in which the loop occludes the site when only folate is bound.
We have initiated a program to develop novel inhibitors of DHFR from pathogenic organisms, including B. anthracis (6). Through this program, we screened a number of propargyl-based DHFR inhibitors and identified 2,4-diamino-5-(3-(2,5-dimethoxyphenyl)prop-1-ynyl)-6-ethyl-pyrimidine (referred to here as UCP120B) as an efficient inhibitor of B. anthracis DHFR (BaDHFR). UCP120B is characterized by a 2,4-diaminopyrimidine ring with an ethyl substitution at the C6 position, a propargylic linker, and a 2′,5′-dimethoxyphenyl ring (Figure 1). It has a 50% inhibition concentration (IC50)of 0.89 μM for BaDHFR and modestly inhibits the growth of the bacteria. In order to design more potent and selective analogues of UCP120B, we previously determined a crystal structure of the complex of BaDHFR–NADPH and UCP120B (6). Herein, we describe a companion solution structure of the ternary complex that yields informative results regarding residue flexibility. This complete solution structure of BaDHFR represents the third species of DHFR to be determined by NMR methods. Overall, the solution structure resembles the crystal structure with a few notable exceptions, including the location of a helix that interacts with the ligand. Chemical shift differences that are evident between the binary (BaDHFR–NADPH) and ternary (BaDHFR–NADPH–UCP120B) complexes correspond with those that have been previously identified in EcDHFR and indicate that BaDHFR undergoes a concerted network of conformational changes to accommodate ligand binding. In addition, the solution structure also provides details of residue flexibility in the active site, explaining some of the structure–activity relationships evident in the biochemical assays.
Figure 1.
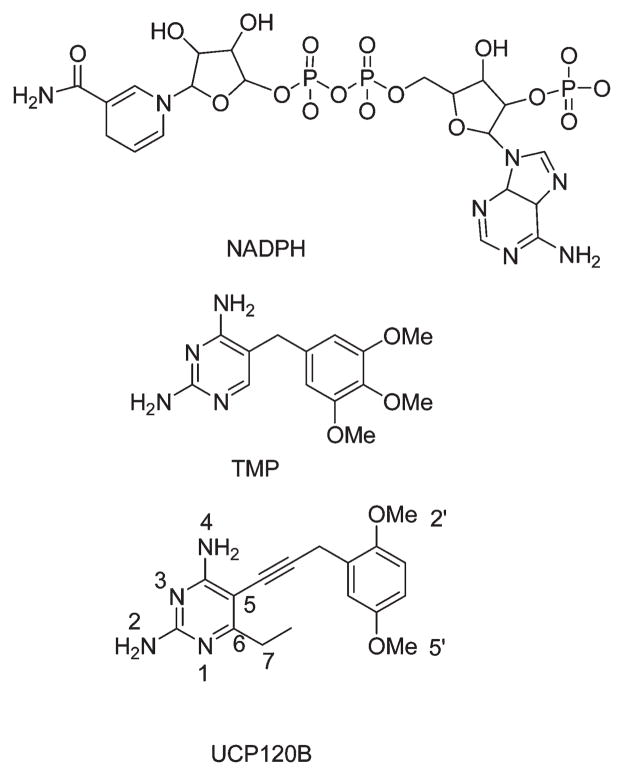
Schematic of NADPH, trimethoprim, and UCP120B with atom numbers.
MATERIALS AND METHODS
Protein Expression, Labeling, and Purification
Cloning methods for the BaDHFR pET41 construct were previously reported (6). The BaDHFR construct containing a C-terminal 8× histidine tag was used for NMR experiments and transformed into Escherichia coli BL21(DE3) expression cells. Cells were grown in M9 minimal media containing [13C]glucose and/or [15N]ammonium chloride (Cambridge Isotope Laboratories) and induced at midlog phase with 1 mM IPTG. Protein expression was extended for an additional 6 h at 37 °C and harvested by centrifugation.
Half-liter pellets were chemically lysed with 1× Bugbuster (Novagen) and DNase and then incubated for 20 min at room temperature. Supernatant was collected by high-speed centrifugation and loaded onto a preequilibrated nickel affinity column. Size exclusion chromatography was used to ensure homogeneity and desalt the labeled protein into a final buffer of 20 mM TES, 50 mM KCl, 10 mM DTT, and 0.5 mM EDTA. Samples were concentrated to 0.25 mM and incubated with 2 mM NADPH (for binary complex samples) or 2 mM NADPH and 2 mM UCP120B (for ternary complex samples) for 1 h. After incubation, ligand-bound 15N/13C-labeled protein was concentrated to 1 mM and used for NMR data collection.
Enzyme Inhibition Assays
Enzyme activity assays were performed at 25 °C by monitoring the rate of enzyme-dependent NADPH oxidation at an absorbance of 340 nm over several minutes (6). Reactions were performed in a buffer containing 20 mM TES, pH 7.0, 50 mM KCl, 10 mM 2-mercaptoethanol, 0.5 mM EDTA, and 1 mg/mL bovine serum albumin. All enzyme assays were performed with a single, limiting concentration of enzyme and saturating concentrations of NADPH and dihydrofolate. IC50 values were calculated as the average of three independent experiments.
Data Collection
All heteronuclear NMR experiments were performed on 1 mM 15N and/or 13C samples of BaDHFR (20 mM TES, 50 mM KCl, 10 mM DTT, 0.5 mM EDTA, 2 mM NADPH, and 2 mM UCP120B adjusted to pH 7.0 and 5% D2O) as described in Clore and Gronenborn (7) on Varian Inova 500 MHz or 600 MHz spectrometers equipped with inverse triple-resonance cold probes. All of the spectra were recorded at 25 °C. The pulse sequences used to record 15N T1/T2 and steady-state heteronuclear 1H–15N NOE values are from the last version of BioPack (Varian, Inc.). Relaxation data were collected on a Varian Inova 600 MHz spectrometer using the scheme adopted from Kay (8). 15N T1 values were measured from the spectra recorded with 14 different durations of the delay T: T = 0, 60, 150, 250, 370, 530, 760, 1150, 1350, 1550, 1750, 2500, 3000, and 4000 ms. T2 values were determined from spectra recorded with 8 different durations of the delay T: T = 10, 30, 50, 70, 90, 110, 130, and 150 ms. The data were processed and analyzed with CCPN (9). Steady-state heteronuclear 1H–15N NOE values were determined from spectra recorded in the presence and absence of a proton presaturation period of 5s. In order to define exchanged and protected regions, the lyophilized DHFR ternary complex was dissolved in D2O, and a series of 2D 1H–15NHSQC spectra were collected, starting 20 min after the dissolution and continuing over a period of 13 days.
Data Processing
The resonance assignments of 15N/13 C-labeled BaDHFR were determined using standard triple resonance experiments at 25 °C. Briefly, HNCACB and HNCA experiments were used for the backbone and HBHA(CBCACO)HN, and (H)CC(CO)HN and HCCH-TOCSY experiments were used for side chain assignments. All spectra were processed with nmrPipe (10)and visualized with PIPP (10) and/or CARA (11).
Structure Calculations
Dihedral angle restraints were obtained from TALOS (12). Initial protein structure calculations were performed using CYANA 2.1 (13). Twenty best conformers with the lowest target energy functions were used for ternary complex calculations. BaDHFR– NADPH–UCP120B and BaDHFR–NADPH NOEs were identified from 3D 15N and 13C NOESY spectra. Two hydrogen bond restraints for the ternary complex were inferred from the crystal structure(6). The parameter and topology files for NADPH and UCP120B were generated using the Dundee PRODRG2 server (14). Ternary complex calculations were performed using the sa_cross_tor.inp protocol from XPLOR-NIH (15). Fifteen lowest energy structures were further subjected to simulated annealing refinement (refine.inp) from XPLOR-NIH (15). Structures were validated by PSVS (http://psvs-1_3.nesg.org) and visualized by PyMol (16).
RESULTS AND DISCUSSION
Determination of the Solution Structure
We determined the three-dimensional solution structure of BaDHFR by modern triple resonance NMR methods as described in Materials and Methods. Twenty best conformers with lowest target energy functions, as defined by CYANA 2.1 (13), were used for the ternary complex calculations. In order to place the ligands, BaDHFR–NADPH–UCP120B and BaDHFR–NADPH NOEs were identified from 3D 15N and 13C NOESY experiments. Additionally, two conserved hydrogen bond restraints for the ternary complex, between the protonated N1 and 2-amino group of the ligand and the conserved acidic residue Glu 28, were inferred from the crystal structure (6). While these interactions were not observed experimentally, their conservation in every DHFR structure with a 2,4-diaminopyrimidine ring justifies their inclusion. Ternary complex calculations were performed, and 15 lowest energy structures were further subjected to simulated annealing refinement (Figure 2A). Statistics of the ensemble are presented in Table 1 and demonstrate the high quality of the structure.
Figure 2.
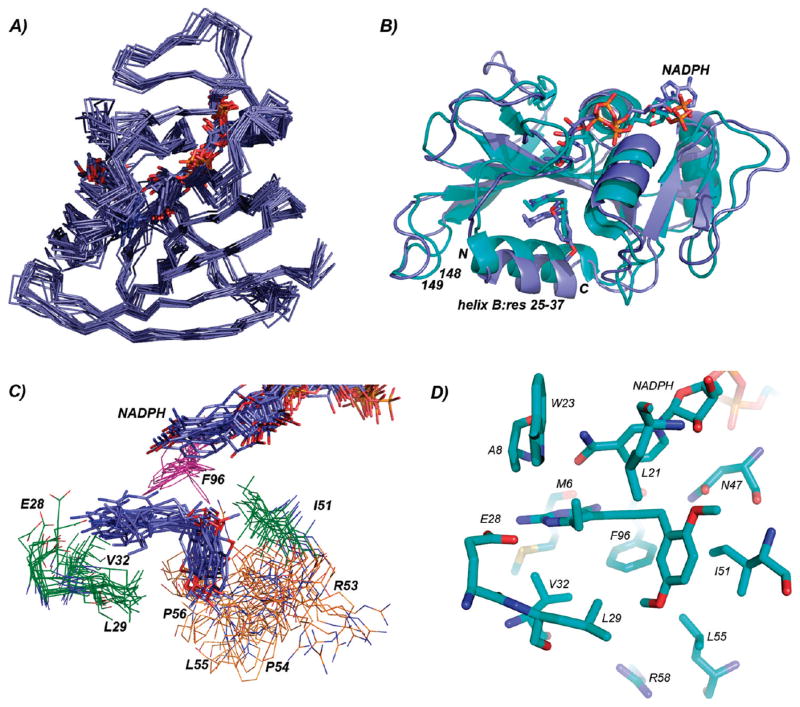
(A) Superposition of the members of the solution structure ensemble. (B) Superposition of the solution (dark blue) and crystal (cyan) structures of BaDHFR bound to ligands. Helix B (including the N-and C-termini of the helix) is labeled. (C) Superposition of the models of active site residues in the solution structure. Residues in the active site are shown as lines; residues within the 53–56 loop are colored orange; ligands are shown in stick form. (D) Active site residues and ligands in the crystal structure.
Table 1.
Structural Statistics for BaDHFR–NADPH–UCP120B
| distance restraints | |
| short range (|i − j| ≤ 1) | 2139 |
| medium range (1 < |i − j| < 5) | 230 |
| long range (|i − j| ≥ 5) | 416 |
| DHFR–NADPH | 10a |
| DHFR–NADPH–UCP120B | 5a |
| dihedral angle restraints | |
| phi (ϕ) | 178b |
| psi (ψ) | 178b |
| hydrogen bond restraints | |
| intra-DHFR | 78c |
| 13d | |
| DHFR—NADPH—UCP120B | 2d |
| violations | |
| NOE (0.5 Å) | 0 |
| dihedral (5°) | 0 |
| rmsd | |
| average backbone rmsd to mean (Å) | 1.0,e 0.7f |
| average heavy atom rmsd to mean (Å) | 1.7,e 1.3f |
| van der Waals energy (kcal mol−1)g | −661.2 ± 25.6 |
| rmsd from idealized geometry | |
| bonds (Å) | 0.0015 ± 0.0001 |
| angles (deg) | 0.444 ± 0.01 |
| impropers (deg) | 0.355 ± 0.006 |
| Ramachandran ploth | |
| residues in most favored regions (%) | 82.5 |
| residues in additional allowed regions (%) | 15.4 |
| residues in generously allowed regions (%) | 2.1 |
| residues in disallowed regions (%) | 0.1 |
Based upon 15N-edited and 13C-edited NOESY.
Generated by TALOS (12).
Intra-DHFR hydrogen bonds were introduced at the last stage of structure calculations.
Inferred from crystal structure (6).
All residues; calculated using PSVS (http://psvs-1_3.nesg.org).
Ordered residues (2–15, 17–20, 26–37, 40–51, 60–121, 123–128, 135–160); calculated using PSVS (http://psvs-1_3.nesg.org).
After energy refinement using XPLOR-NIH (15).
Calculated using PSVS (http://psvs-1_3.nesg.org).
Comparison of the Solution and Crystal Structures of BaDHFR–NADPH–UCP120B
A direct comparison of the ternary solution (Figure 2) and crystal structures of BaDHFR (6), both bound to NADPH and UCP120B, reveals that the general fold of DHFR and the binding of NADPH are conserved while there appear to be two different binding modes for UCP120B. Overall, the Cα atoms of the solution and crystal structures superimpose with an rmsd of 2.3 Å (Figure 2B). The central β-sheets of both structures superimpose particularly well. The crystal structure assigns two short additional β-strands (residues 13–16 and 124–126) relative to the solution structure. The most noticeable difference between the two structures is the translation of a helix (residues 25–37; often called helix B in DHFR nomenclature) near the active site (Figure 2B). A survey of each of the models in the solution structure ensemble reveals that the N-terminus of this helix is displaced from the crystal structure by 1.2–3.6 Å and the C-terminus is displaced by 2.7–5.3 Å. The conformation of helix B in this solution structure is supported by 11 long-range NOE signals (a full list of the NOE signals in this region is found in Supporting Information) arising from interactions between two residues in helix B (Pro 26 and Ser 27) and two residues in the remainder of the protein (Lys 148 and Asn 149). Additionally, an NOE is observed between the HD1 proton from Leu 29 and the C7 atom of the ligand.
NADPH is bound in the same extended conformation in the crystal and solution structures. The nicotinamide ring in the solution structure appears to be translated between 0.9 and 2.1 Å away from the active site, relative to the molecule in the crystal structure. The adenine ring, on the surface of the protein, appears to be flexible.
The ligand, UCP120B, exhibits two different binding modes in the two structures (Figure 2B–D). In the crystal structure, the diaminopyrimidine ring forms hydrogen bonds with Glu 28 as well as the backbone carbonyl atoms of Met 6 and Phe 96. The propargyl linker extends over the phenyl ring of Phe 96, and the dimethoxyphenyl ring forms van der Waals interactions with Ile 51, Leu 55, and Leu 21. In the solution structure, the hydrogen bonds between the protonated N1 and 2-amino group of the ligand were constrained, shifting the pyrimidine ring toward helix B. A measured NOE signal between the C7 atom and Leu 29 provides experimental evidence for the interaction of the pyrimidine ring and the displaced helix. The propargyl linker of the ligand extends under Phe 96 (as opposed to “over” the residue in the crystal structure). NOE signals between the two protons on the 4-amino group and Phe 96 provide experimental evidence of the interaction. The dimethoxyphenyl ring generally superimposes well with the same ring in the crystal structure and exhibits NOE signals with Ile 51. It also appears to form interactions with the loop containing Pro 54-Leu 55-Pro 56, supported by an NOE signal between Leu 55 and the dimethoxyphenyl ring.
Previously published ternary solution structures with LcDHFR and hDHFR show agreement with related crystal structures. Human DHFR TMP–NADPH solution and crystal structures have a root mean squared deviation of 1.26 Å and validate that there are no significant conformational changes observed between the structures (3). Similar solution and crystal structure comparisons have been observed in the ternary complex of LcDHFR (1). For hDHFR and LcDHFR, the position of helix B varies between the solution and crystal structures by approximately 1–2.5 Å.
While the extended conformation of NADPH dominates in most DHFR structures, both human and BaDHFR ensembles confirm an overall flexibility in the NADPH cofactor. Specifically, the pyrophosphate groups show the most flexibility and influence the distances between the nicotinamide ring and trimethoprim in hDHFR. The pyrophosphate is also variable in the BaDHFR structure, conferring flexibility to the deoxyribose sugar moiety.
Dynamics of BaDHFR
In order to better understand the backbone conformational flexibility of BaDHFR, we measured values of the relaxation parameters, T1, T2, and heteronuclear 1H–15N NOEs for the ternary complex (Figure 3). The average value of T1 is 926 ms (with a standard deviation of 92); the average value of T2 is 47 ms (with a standard deviation of 9). This results in an average T1/T2 ratio of approximately 20 (with a standard deviation of 3) that can be used for a rough estimate, using the related spectral density function, of the overall correlation time of the molecule, τm (17). The average NOE value is 0.67 (with a standard deviation of 0.1). For most of the residues, T1 and T2 values do not deviate significantly beyond experimental error from the average numbers. However, several regions (14–18, 65–73, 85–89, 124–133, 160–162) demonstrate some tendency to be involved in fast internal motion with larger T2 and smaller NOE values. These regions of fast internal motion correlate well with the general notion that regions of secondary structure show lower mobility while turns, loops, and N-/C-termini exhibit higher mobility. However, one amino acid, Val 32, located in the center of helix B, behaves very differently. The T1 and T2 values of Val 32 are 544 and 90 ms, respectively, resulting in a ratio of only 6. Its NOE value is also reduced to 0.25 as compared to an average NOE value of 0.67 or compared to the NOE value of the neighboring Lys 33 (1.0). All of these large deviations suggest that Val 32 is involved in fast internal motion (17), which is a striking feature since this residue is making strong contacts with UCP120B. In addition to the motion of Val 32, the majority of the residues in helix B (residues 25–32) exhibit fast exchange in hydrogen/deuterium exchange experiments (see Figure 3). There are also several regions such as residues 20–25 that lack backbone assignments due to an unfavorable conformational exchange and residues 89–102 and 117–120 that show evidence of conformational exchange with reduced T2 values.
Figure 3.
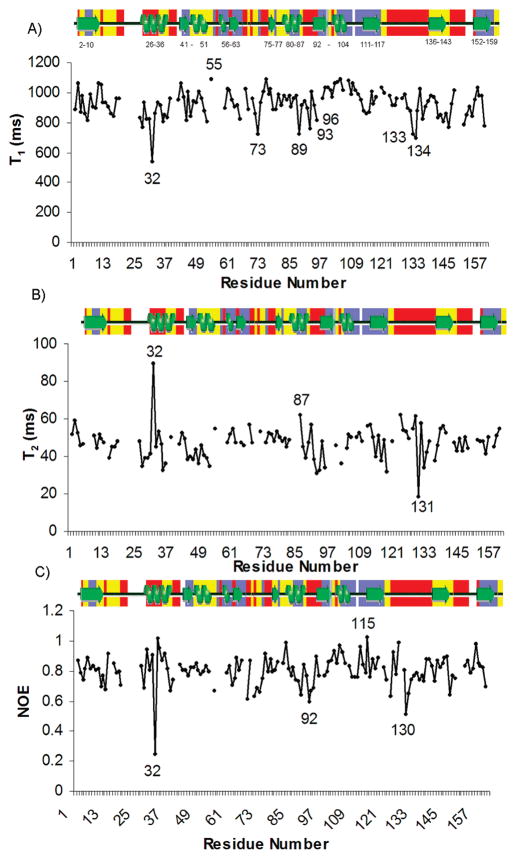
Relaxation data plotted for the 137 measured amide residues. Labeled secondary structure elements are shown above each plot and colored according to the rate of hydrogen/deuterium exchange (red = fast exchange in 20 min, yellow = intermediate exchange in 1 day, and blue = protected). (A) T1 values, (B) T2 values, and (C) 1H–15N NOE values.
Comparison between Binary and Ternary Enzyme Complexes
In order to optimize conditions for data collection and to observe the effect of the ligand on the complex, we acquired 15N HSQC spectra during a titration experiment in which the molar ratios of protein to ligand included no ligands (apoprotein) (Figure 4a), the binary protein–NADPH complex, a 1:1 mix of protein–NADPH:ligand and a 1:4 mix of protein–NADPH:ligand (the last two represent ternary complexes) (Figure 4b). Both the binary and ternary complexes were significantly more resolved than the apo form, reflecting results from previous experiments showing that the apoenzyme exists in at least two different isoforms (18–21). In addition, notable differences were observed between the 15N HSQC spectra. Using the complete assignments of the ternary HSQC and a superposition with the unassigned binary form, we analyzed regions of the protein that significantly shifted upon ligand binding.
Figure 4.
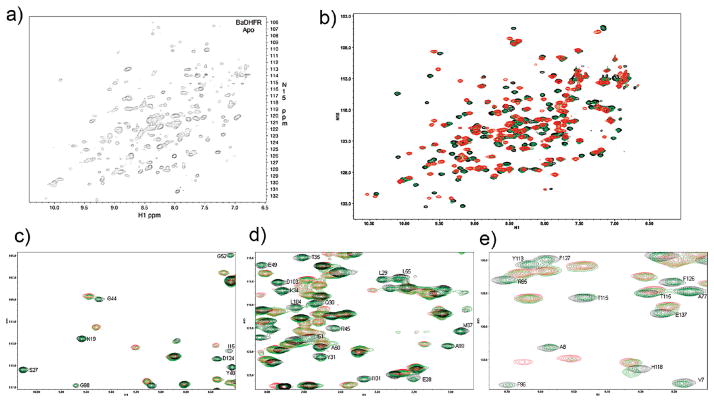
15N HSQC spectra of (a) apo, (b) binary (red), 1:1 protein–NADPH:ligand (green), and 1:4 protein–NADPH:ligand (black) complexes of BaDHFR. Expanded views of the superposition of binary and ternary spectra with labeled assignments for residues in the ternary complex are shown in panels c–e.
Examination of the shifted residues shows that the majority of these residues predictably lie near the active site. Figure 5 highlights the regions of the protein that shift between the two complexes. The acidic residue Glu 28 that makes conserved interactions to the diaminopyrimidine ring, along with the remainder of the residues in helix B, shows significant shift differences between the binary and ternary complexes. Another helix located near the NADPH cofactor containing residues Ile 95–Leu 104 and the β-strand containing F5–A8, which interacts with the ligand and NADPH, also show distinct shift differences. Interestingly, it appears that sections of the core canonical eight-stranded twisted β-sheet also undergo some slight to significant perturbations.
Figure 5.
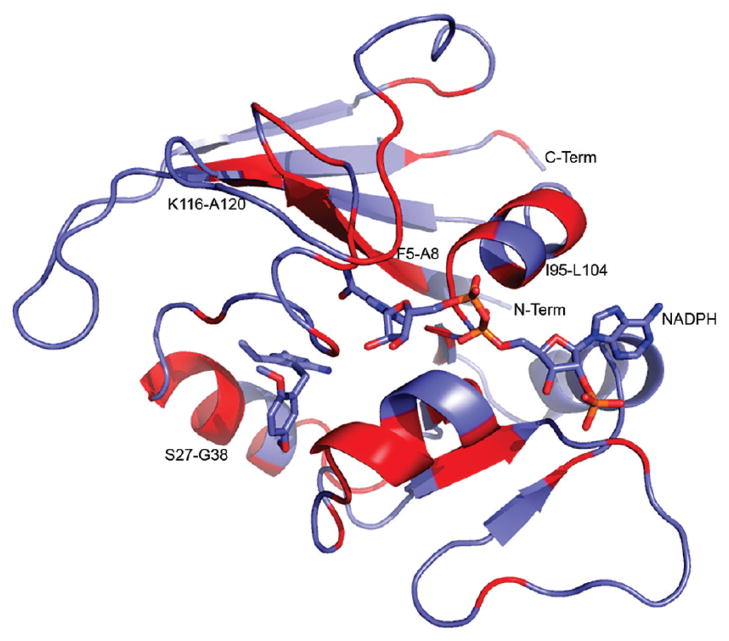
Cartoon depiction of the solution structure of BaDHFR–NADPH–UCP120B (UCP120B and NADPH are shown in blue sticks). Regions that show significant chemical shift differences between the binary (BaDHFR–NADPH) and ternary (BaDHFR–NADPH–UCP120B) complexes are colored in red. Figure generated in PyMol.
Movement of the β-sheet region in response to the addition of ligand has been observed in previous NMR studies. In EcDHFR, differences between the NADPH binary complex and the ternary complex with folate have been analyzed by NMR (4, 5). The analysis reveals shifts in many of the same regions as observed in BaDHFR, including Ala 8 in the first β-strand and helix B containing the conserved acidic residue. Solution structures of the binary and ternary LcDHFR enzyme complexes are also available (1, 2, 22, 23), though no analysis of shift differences between binary and ternary enzyme complexes has been previously reported. Comparison of the NADPH-bound binary complex (PDB ID 2HQP) and the trimethoprim – NADPH-bound ternary complex (PDB ID 1LUD (1)) shows the greatest structural differences in regions already indicated by the EcDHFR NMR studies (4, 5) and our own BaDHFR NMR studies (Figure S1 in Supporting Information shows a superposition of the binary and ternary LcDHFR structures and notes the structural differences). A superposition of the three structures (BaDHFR, EcDHFR, and LcDHFR) yields a structural alignment (Figure 6) and facilitates the interpretation of residue characteristics at equivalent positions.
Figure 6.
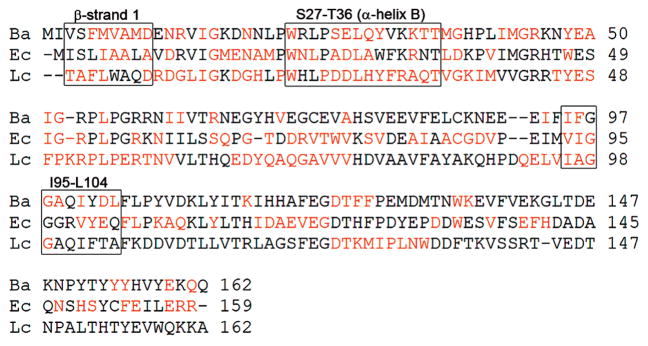
Structural alignment of BaDHFR, EcDHFR, and LcDHFR. Regions of difference between binary and ternary complexes are colored in red. For BaDHFR, differences were interpreted from the titration HSQC data shown in Figure 4. For EcDHFR, differences were taken from refs 4 and 5. For LcDHFR, differences were interpreted from comparisons between the reported binary and ternary structures.
Analyses of these comparisons are useful to illustrate the network of intramolecular interactions that are involved in the conformational transitions between the binary and ternary enzyme states. Helix B containing the conserved acidic residue and several other residues critical to ligand binding shows significant movement between the binary and ternary complexes. The core β-sheet region seems to shift upon ligand binding, ostensibly due to the fact that the initial β-strand (comprising residues 4–9 in BaDHFR) at the N-terminus contains residues critical to both ligand and cofactor binding. With this strand perturbed, the other strands must adjust to maintain the intramolecular interactions required for the twisted β-sheet conformation. Also of interest is the movement observed in the helix containing residues 99–104 in BaDHFR. The residues in the loop directly before the helix, including Phe 96 in BaDHFR, play an important role in ligand binding, sometimes including the formation of a hydrogen bond between the ligand and the protein backbone. This helix shows significant chemical shift displacement in BaDHFR and some displacement in EcDHFR or LcDHFR. In EcDHFR and LcDHFR, the corresponding residues to Phe 96 are smaller (Ile and Ala, respectively) and may not shift as dramatically to accommodate a ligand. It is also important to note that the ternary enzyme states are bound to three different ligands: UCP120B (Ba), folate (Ec), and TMP (Lc). The ligands have their own individual effects on enzyme conformation and account for some of the differences observed between species. Specifically in the 99–104 helix, UCP120B might play a role in the degree of shift since Phe 96 forms π-stacking interactions with the propargyl linker, which is unique to this inhibitor.
Structure–Activity Relationship Analysis of Previously Designed Ligands
We have previously designed, synthesized, and evaluated several inhibitors for many species of DHFR (6,24–26). However, none of the currently designed inhibitors has shown remarkable potency against BaDHFR (Table 2). To date, the most active inhibitor (UCP120B), the same ligand bound in the crystal structure and bound to the solution structure reported here, has an IC50 of 0.89 μM. Features observed in the solution structure of BaDHFR could assist in explaining the lack of potency in this series.
Table 2.
IC50 Values for the Trimethoxyphenyl and Biphenyl Inhibitor Seriesa
| Compound No. | Inhibitor | IC50 (μM) |
|---|---|---|
| 1 | 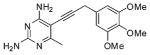 |
3.7 ± 0.2 |
| 2 |  |
30.3 ± 0.4 |
| 3 | 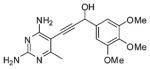 |
14.5 ± 0.5 |
| 4 | 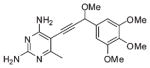 |
29.1 ± 0.06 |
| 5 |  |
10.5 ± 0.5 |
| 6 |  |
12.7 ± 1.0 |
| 7 | 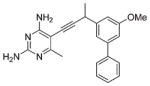 |
7.9 ± 0.8 |
| 8 | 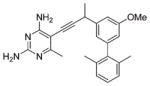 |
5.5 ± 1.0 |
| 9 | 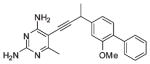 |
17.3 ± 1.4 |
| 10 |  |
49.1 ± 1.0 |
IC50 values for compounds 1–4 in Table 2 were also previously reported in ref 6 and are included here for comparison.
The first series of inhibitors contain the core scaffold of the propargyl-linked compounds with the 3,4,5-tri-methoxyphenyl ring of TMP and a series of substituents at the bridge position of the propargyl linker (compounds 1–6 in Table 2). Enzyme inhibition assays reveal that the addition of any group at the propargylic position lowers potency. The addition of a methyl group or methoxy group in particular causes nearly a 10-fold loss in activity.
The solution structure shows that the phenyl ring of Phe 96 is positioned near this propargyl linker (see Figure 2C). Relaxation data for Phe 96 show that the residue is undergoing significant conformational exchange, even in the presence of saturating ligand concentrations. These data, in combination with the significant line broadening observed for Phe 96, suggest its flexibility. While the steric interference of Phe 96 was suspected based on the crystal structure, the solution structure shows nicely how the flexibility of the phenyl ring may interfere with this linker region.
After the initial series of trimethoxy inhibitors were evaluated, a series of biphenyl compounds were designed and synthesized (compounds 7–10 in Table 2). The rationale for this series was to better occupy the hydrophobic pocket containing residues Leu 29, Ile 51, and Leu 55, normally occupied by the p-aminobenzoic acid moiety of the substrate. However, the expected increase in activity was not observed for these compounds.
An analysis of the solution structure of BaDHFR provides a possible explanation for the lack of improved affinity. The distal phenyl ring of the compound (relative to the pyrimidine) is predicted to be located next to the loop containing residues 52–59 (Figure 2C). This loop contains the flexible motif GRPLPGRR. Leu 55, a critical residue in this predicted biphenyl binding pocket, has a T2 value of 54.5 ms, slightly greater than the average T2 value (47 ms). Signals for the preceding residues in the loop (51 – 53) are broadened, and those for the following residues (57 – 59) are not assigned, most likely reflecting an unfavorable exchange rate. Leu 55 also appears to be undergoing fast (20 min) hydrogen/ deuterium exchange (Figure 3). Taken together, these results suggest significant flexibility in this loop region, which has the potential to cause steric interference with the biphenyl compounds. It is also possible that the phenyl ring simply does not create strong enough interactions to overcome the entropic cost that is incurred by ordering this loop region to prevent steric interference. The direct cause of the lower potency for the biphenyl compounds still needs further investigation; however, the information gained about this loop region from the solution structure will be very useful in the design of future inhibitors. Some potential avenues of future investigation include increasing the size of substituents and varying the substitution pattern of substituents around the aryl ring. Primarily, the ortho and meta positions have been investigated most extensively, and varying the substituents on the para position could prove fruitful.
CONCLUSIONS
Given the significance of identifying new therapeutics for infections caused by B. anthracis, we have begun a drug discovery program to identify inhibitors of the essential enzyme dihydrofolate reductase (DHFR). In this work, we present a three-dimensional solution structure of DHFR from B. anthracis bound to its cofactor, NADPH, and UCP120B, the most potent of our inhibitors to date. The ternary solution structure is the third species of DHFR to be fully determined by NMR methods and reveals the conserved DHFR fold with an eight-stranded β-sheet and four flanking α-helices. Differences with our previously determined ternary crystal structure are located primarily in the position of an α-helix at the active site and specific interactions formed by UCP120B and the active site residues. Comparisons of chemical shifts between the binary enzyme – NADPH complex and the ternary complex yield insight into the residues significantly affected by ligand binding. Specifically, Phe 96, which interacts with the propargyl group of UCP120B, displays significant chemical shift differences between the binary and ternary complex and appears to be flexible in the active site. Significantly for our drug design effort, the solution structure reveals a possible basis for observed structure–activity relationships. The flexibility of Phe 96 may interfere with propargyl-substituted compounds, and the flexibility of Leu 55 in a loop at the active site may affect binding of a biphenyl series of these inhibitors. Understanding the flexibility of the active site residues of BaDHFR will be important in designing more potent inhibitors of the enzyme.
Supplementary Material
Acknowledgments
We thank Martha Morton for assistance with NADPH resonance assignments.
Footnotes
Coordinates have been deposited in the Protein Data Bank with accession code 2KGK.
Abbreviations: DHFR, dihydrofolate reductase; NMR, nuclear magnetic resonance; NADPH, nicotinamide adenine dinucleotide phosphate (reduced); LcDHFR, DHFR from Lactobacillus casei; hDHFR, human DHFR; EcDHFR, DHFR from Escherichia coli; BaDHFR, DHFR from Bacillus anthracis; UCP120B, 2,4-diamino-5-(3-(2,5-dimethoxyphenyl)prop-1-ynyl)-6-ethylpyrimidine; NOE, nuclear Overhauser effect; rmsd, root mean squared deviation; HSQC, heteronuclear single-quantum coherence.
Funding provided by NIH Grant R01AI073375 (to A.C.A.).
SUPPORTING INFORMATION AVAILABLE
Figure S1 showing a superposition of the binary and ternary LcDHFR structures with structural differences noted and tables of chemical shift assignments for NADPH (Table S1) and UCP120B (Table S2) as well as a table of NOE signals involving helix B and the ligand (Table S3). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Polshakov V, Smirnov E, Birdsall B, Kelly G, Feeney J. NMR-based solution structure of the complex of Lactobacillus casei dihydrofolate reductase wtih trimethoprim and NADPH. J Biomol NMR. 2002;24:67–70. doi: 10.1023/a:1020659713373. [DOI] [PubMed] [Google Scholar]
- 2.Gargaro A, Soteriou A, Frenkiel T, Bauer C, Birdsall B, Polshakov V, Barsukov I, Roberts G, Feeney J. The solution structure of the complex of Lactobacillus casei dihydrofolate reductase with methotrexate. J Mol Biol. 1998;277:119–134. doi: 10.1006/jmbi.1997.1560. [DOI] [PubMed] [Google Scholar]
- 3.Kovalevskaya N, Smurnyy Y, Polshakov V, Birdsall B, Bradbury A, Frenkiel T, Feeney J. Solution structure of human dihydrofolate reductase in its complex with trimethoprim and NADPH. J Biomol NMR. 2005;33:69–72. doi: 10.1007/s10858-005-1475-z. [DOI] [PubMed] [Google Scholar]
- 4.Osborne M, Venkitakrishnan R, Dyson J, Wright P. Diagnostic chemical shift markers for loop conformation and substrate and cofactor binding in dihydrofolate reductase complexes. Protein Sci. 2003;12:2230–2238. doi: 10.1110/ps.03219603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venkitakrishnan R, Zabrowski E, McElheny D, Benkovic S, Dyson J, Wright P. Conformational changes in the active site loops of dihydrofolate reductase during the catalytic cycle. Biochemistry. 2004;43:16046–16055. doi: 10.1021/bi048119y. [DOI] [PubMed] [Google Scholar]
- 6.Beierlein J, Frey K, Bolstad D, Pelphrey P, Joska T, Smith A, Priestley N, Wright D, Anderson A. Synthetic and crystallographic studies of a new inhibitor series targeting Bacillus anthracis dihydrofolate reductase. J Med Chem. 2008;51:7532–7540. doi: 10.1021/jm800776a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clore GM, Gronenborn AM. NMR structure determination of proteins and protein complexes larger than 20 kDa. Curr Opin Chem Biol. 1998;2:564–570. doi: 10.1016/s1367-5931(98)80084-7. [DOI] [PubMed] [Google Scholar]
- 8.Farrow N, Muhandiram R, Singer A, Pascal S, Kay C, Gish G, Shoelson S, Pawson T, Forman-Kay J, Kay L. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 9.Fogh R, Ionides J, Ulrich E, Boucher W, Vranken W, Linge J, Habeck M, Rieping W, Bhat T, Westbrook J, Henrick K, Gilliland G, Berman H, Thorton J, Nilges M, Markley J, Laue E. The CCPN project: an interim report on a data model for the NMR community. Nat Struct Biol. 2002;9:416–418. doi: 10.1038/nsb0602-416. [DOI] [PubMed] [Google Scholar]
- 10.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 11.Keller R. 2004 http://www.cara.nmr-software.org/downloads/3-85600-112-3.pdf.
- 12.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 13.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 14.Schuettelkopf A, van Aalten D. PRODRG—a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. 2004;D60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 15.Schwieters C, Kuszewski J, Tjandra N, Clore G. The XPLOR-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:66–74. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 16.DeLano W. DeLano Scientific LLC. Palo Alto, CA: 2008. [Google Scholar]
- 17.Clore G, Driscoll P, Wingfield P, Gronenborn A. Analysis of the backbone dynamics of interleukin-1 beta using two-dimensional inverse detected heteronuclear 15N-1H NMR spectroscopy. Biochemistry. 1990;29:7387–7401. doi: 10.1021/bi00484a006. [DOI] [PubMed] [Google Scholar]
- 18.Epstein D, Benkovic S, Wright P. Dynamics of the dihydrofolate reductase-folate complex: catalytic sites and regions known to undergo conformational change exhibit diverse dynamical features. Biochemistry. 1995;34:11037–11048. doi: 10.1021/bi00035a009. [DOI] [PubMed] [Google Scholar]
- 19.Falzone C, Wright P, Benkovic S. Evidence for two interconverting protein isomers in the methotrexate complex of dihydrofolate reductase from Escherichia coli. Biochemistry. 1991;30:2184–2191. doi: 10.1021/bi00222a023. [DOI] [PubMed] [Google Scholar]
- 20.Osborne M, Schnell J, Benkovic S, Dyson J, Wright P. Backbone dynamics in dihydrofolate reductase complexes: role of loop flexibility in catalytic mechanism. Biochemistry. 2001;40:9846–9859. doi: 10.1021/bi010621k. [DOI] [PubMed] [Google Scholar]
- 21.Schnell J, Dyson J, Wright P. Structure, dynamics and catalytic function of dihydrofolate reductase. Annu Rev Biophys Biomol Struct. 2004;33:119–140. doi: 10.1146/annurev.biophys.33.110502.133613. [DOI] [PubMed] [Google Scholar]
- 22.Polshakov V, Birdsall B, Feeney J. Effects of co-operative ligand binding on protein amide NH hydrogen exchange. J Mol Biol. 2006;356:886–903. doi: 10.1016/j.jmb.2005.11.084. [DOI] [PubMed] [Google Scholar]
- 23.Polshakov V, Birdsall B, Frenkiel T, Gargaro A, Feeney J. Structure and dynamics in solution of the complex of Lactobacillus casei dihydrofolate reductase with the new lipophilic antifolate drug trimetrexate. Protein Sci. 1999;8:467–481. doi: 10.1110/ps.8.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolstad D, Bolstad E, Frey K, Wright D, Anderson A. A structure-based approach to the development of potent and selective inhibitors of dihydrofolate reductase from Cryptosporidium. J Med Chem. 2008;51:6839–6852. doi: 10.1021/jm8009124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Bolstad D, Smith A, Priestley N, Wright D, Anderson A. Structure-guided development of efficacious antifungal agents targeting Candida glabrata dihydrofolate reductase. Chem Biol. 2008;15:990–996. doi: 10.1016/j.chembiol.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelphrey P, Popov V, Joska T, Beierlein J, Bolstad E, Fillingham Y, Wright D, Anderson A. Highly efficient ligands for DHFR from Cryptosporidium hominis and Toxoplasma gondii inspired by structural analysis. J Med Chem. 2007;50:940–950. doi: 10.1021/jm061027h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


