Abstract
The present work studies the influence of hydrolytic enzymes (α-amylase or lipase) on the degradation of fiber mesh scaffolds based on a blend of starch and poly(ɛ-caprolactone) (SPCL) and the osteogenic differentiation of osteogenic medium–expanded rat bone marrow stromal cells (MSCs) and subsequent formation of extracellular matrix on these scaffolds under static culture conditions. The biodegradation profile of SPCL fiber meshes was investigated using enzymes that are specifically responsible for the enzymatic hydrolysis of SPCL using concentrations similar to those found in human serum. These degradation studies were performed under static and dynamic conditions. After several degradation periods (3, 7, 14, 21, and 30 days), weight loss measurements and micro-computed tomography analysis (specifically porosity, interconnectivity, mean pore size, and fiber thickness) were performed. The SPCL scaffolds were seeded with rat MSCs and cultured for 8 and 16 days using complete osteogenic media with and without enzymes (α-amylase or lipase). Results indicate that culture medium supplemented with enzymes enhanced cell proliferation after 16 days of culture, whereas culture medium without enzymes did not. No calcium was detected in groups cultured with α-amylase or without enzymes after each time period, although groups cultured with lipase presented calcium deposition after the eighth day, showing a significant increase at the sixteenth day. Lipase appears to positively influence osteoblastic differentiation of rat MSCs and to enhance matrix mineralization. Furthermore, scanning electron microscopy images showed that the enzymes did not have a deleterious effect on the three-dimensional structure of SPCL fiber meshes, meaning that the scaffolds did not lose their structural integrity after 16 days. Confocal micrographs have shown cells to be evenly distributed and infiltrated within the SPCL fiber meshes up to 410 μm from the surface. This study demonstrates that supplementation of culture media with lipase holds great potential for the generation of bone tissue engineering constructs from MSCs seeded onto SPCL fiber meshes, because lipase enhances the osteoblastic differentiation of the seeded MSCs and promotes matrix mineralization without harming the structural integrity of the meshes over 16 days of culture.
Introduction
Tissue engineering, as defined by Langer and Vacanti1 in 1993, “is an interdisciplinary field of research that applies the principles of engineering and the life sciences towards the development of biological substitutes that restore, maintain or improve tissue function.” Specific tissue-engineering applications, such as bone regeneration, often need a temporary scaffold with a three-dimensional (3D) architecture to support tissue growth. The ideal scaffold should be biodegradable and bioresorbable to support the growth of new bone. The scaffold degradation rate should complement the growth rate of the new bone in a manner that, by the time the defect or injury site is completely regenerated, the scaffold is totally degraded.1,2 Biodegradable materials have the capacity to temporarily mimic the original structural function of the tissue and to degrade by controlled mechanisms into products easily eliminated by the body's metabolic pathways.3 In this perspective, we used enzymes present in human serum that are responsible for the enzymatic degradation of starch and poly(ɛ-caprolactone) (PCL) (α-amylase and lipase, respectively) to simulate conditions found in vivo. In this study, we used a blend of starch and poly(ɛ-caprolactone) (SPCL). Starch itself is one of the most abundant naturally occurring polymers, presenting a combination of attractive properties that is being increasingly used in a variety of applications. Starch-based biomaterials are biocompatible4,5 and biodegradable.6,7 Studies with starch-based scaffolds have shown that their degradation products do not have a negative effect on human osteosarcoma cells (SaOS-2 cell line).8 Previous studies with SPCL fiber meshes have also shown that these scaffolds are able to support the attachment, proliferation, and differentiation of bone marrow stromal cells (MSCs).9,10 α-amylase degrades starch-based biomaterials. In principle, the in vivo resorption rate of blends based on starch can be regulated by controlling the percentage of starch in the material, preferentially attacked by α-amylase, an enzyme present in the human body.
PCL is a biodegradable aliphatic polyester currently used in an array of biomedical applications. Several studies have reported that lipase degrades PCL.11–14 Lipases are water-soluble enzymes that hydrolyze ester bonds of water-soluble substrates such as triglycerides, phospholipids, and cholesteryl esters.15,16 The lipase gene family consists of multiple lipases that share a similar sequence structure at the genetic and protein level, thus indicating a common ancestral origin. However, these lipases have disparate and organ-specific expression, indicating that they may have evolved relatively specific roles. The human lipases include the pre-duodenal lingual and gastric lipase and the extra-duodenal pancreatic, hepatic, lipoprotein, and endothelial lipase.17 The pancreatic, hepatic, lipoprotein, and endothelial lipases are members of the lipase gene family.17 Lipoprotein lipase has been described as a marker of adipogenic differentiation.18–20 Serum lipase is mainly derived from pancreatic cells; other sources in the human body are the digestive tract, adipose tissue, lung, milk, and leucocytes.3,21 Several authors have described different enzymatic effects of lipase on PCL using species of lipase obtained from diverse sources.12,13 Enzyme characteristics such as source and specificity seem to influence the experimental results obtained with lipase. Some studies have also reported that the biodegradation rate of PCL depends on the shape and inherent surface area:volume ratio of the PCL substrate, with films being slowly degraded, whereas nano- and microparticles are quickly degraded.13
Previous studies using static and dynamic (flow perfusion bioreactor) culture conditions for rat marrow stromal cells on SPCL fiber meshes have shown that flow perfusion conditions enhance differentiation of marrow stromal cells with subsequent formation of a bone-like extracellular matrix (ECM).9,10 Holtorf et al. 22 showed that flow perfusion culture induced osteoblastic differentiation of rat marrow stromal cells seeded on scaffolds, even in the absence of dexamethasone (a glucocorticoid medium supplement that promotes osteoblastic differentiation of MSCs in culture).
In this work, we propose a novel approach, using static cultures and complete osteogenic media supplemented with enzymes, to study simultaneously the influence of α-amylase and lipase on the degradation of SPCL fiber meshes as a function of immersion time and on the osteodifferentiation of rat marrow stromal cells.
This work was intended to mimic in vitro the in vivo degradation of the SPCL fiber meshes using enzymes responsible for their degradation at the same concentrations as those found in human serum. In addition to characterizing the susceptibility of SPCL fiber meshes to enzymatic degradation, this study aimed also to investigate the osteogenic differentiation of osteogenic medium–expanded rat MSCs and subsequent formation of ECM under static conditions in the presence of hydrolytic enzymes (lipase or α-amylase). By seeding rat MSCs onto SPCL fiber meshes and culturing them under static conditions using osteogenic media supplemented with lipase and α-amylase enzymes, we aimed to answer the following questions. What is the role of lipase and α-amylase in the degradation of SPCL fiber mesh scaffolds? Is the integrity of SPCL fiber mesh scaffolds rapidly compromised by enzymatic degradation, and if so, which enzyme is the most influential? Is there any effect of α-amylase and lipase on the osteogenic differentiation of rat MSCs?
Materials and Methods
Scaffold preparation and characterization
Porous scaffolds used in this study were based on a blend of corn starch and poly(ɛ-caprolactone) (SPCL, 30/70%) prepared using a fiber bonding technique.9 Scaffolds were cylinders with 8-mm diameters and thickness of 1.6 ± 0.1 mm. Before cell seeding, scaffolds were sterilized using ethylene oxide gas for 14 h. All chemical reagents were purchased from Sigma (St. Louis, MO) unless otherwise specified.
Degradation tests
Serum α-amylase and serum lipase in healthy adults are in the range of 46 to 244 U/L3,23 and 30 to 190 U/L,3,7,24 respectively. Degradation studies were carried out by incubating samples in phosphate buffered saline (PBS) solution (0.1 M, pH 7.4) containing α-amylase (150 U/L)3,23 from Bacillus sp. or lipase (110 U/L)3,24 from Aspergillus niger, both concentrations similar to those found in human serum, and sodium azide (0.2%), to avoid contamination, at 37ºC up to 30 days under static and dynamic conditions (shaker at 70 rpm). Based upon reports from the literature, the inclusion of sodium azide in the buffer solution was not expected to affect PCL degradation.25 The studies were performed in triplicate. The enzymatic degradation solutions were changed every 7 days to restore the original level of enzymatic activity. At the end of the degradation period, the samples were removed, washed thoroughly with distilled water, and dried in a graded series of ethanol for later calculation of weight loss.1
 |
Micro-computed tomography
The scaffolds (n = 3) were analyzed using micro-computed tomography (μ-CT) before and after degradation with both enzymes (lipase or α-amylase) for the different periods of immersion (0, 3, 7, 14, 21, and 30 days) under static and dynamic conditions. μ-CT was carried out with a high-resolution μ-CT Skyscan 1072 scanner (Skyscan, Kontich, Belgium) using a resolution pixel size of 8.79 μm and integration time of 1.9 ms. The X-ray source was set at 40 keV of energy and 250 μA of current. Approximately 400 projections were acquired over a rotation range of 180° with a rotation step of 0.45°. Data sets were reconstructed using standardized cone-beam reconstruction software (NRecon v1.4.3, SkyScan). The output format for each sample was 300 serial 1024 × 1024 bitmap images. Representative data sets of 200 slices were segmented into binary images with a dynamic threshold of 40 to 255 (grey values) and were used for morphometric analysis (CT Analyser, v1.5.1.5, SkyScan) and to build 3D models (ANT 3D creator, v2.4, SkyScan). The morphometric analysis included porosity, pore interconnectivity, mean pore size, and mean fiber thickness. Three-dimensional virtual models of representative regions in the bulk of the scaffolds were created, visualized, and registered using both image processing software packages (CT Analyser and ANT 3D creator).
Isolation and expansion of rat MSCs
Rat MSCs were harvested from the tibiae and femora of 41- to 44-day-old (151-175 g) male Wistar rats (Charles River Laboratories, Wilmington, MA) using established methods.26 Briefly, rats were euthanized using 4% isoflurane in carbon dioxide (CO2). The leg bones were excised, the soft tissue was removed, and the leg bones were placed in Dulbecco's modified Eagle medium supplemented with penicillin streptomycin Fungizone antibiotic cocktail. This concentration of antibiotics is three times as great as the normal concentration used in cell culture to avoid contamination during the harvest process. The proximal end of each femur and distal end of each tibia were clipped. An 18-gauge needle was inserted into the hole in the knee joint in each bone, and the bone was flushed with 5 mL of complete osteogenic medium (alpha minimum essential medium supplemented with 10% fetal bovine serum (FBS; Cambrex/BioWhittaker, Walkersville, MD), 50 μg/mL gentamicin, 100 μg/mL ampicillin, 10 mM Fungizone, 50 μg/mL L-ascorbic acid, 0.01 M β-glycerophosphate, and 10−8 M dexamethasone). The resulting marrow pellet was broken up by trituration, and cell suspensions from all bones were combined in a centrifuge tube.
The cells were plated in 75-cm2 flasks and cultured for 6 days in complete osteogenic media at 37ºC in a humidified atmosphere of 95% air, 5% CO2 to allow the expansion of the cells. Medium was changed at 1 and 3 days to remove the non-adherent cell population. After 6 days, the cells were trypsinized, centrifuged, and resuspended in a known amount of medium and 10% dimethyl sulfoxide to obtain a final concentration of 3 × 106 cells/mL. Cells were aliquoted into cryotubes for storage (∼1 mL cell suspension/cryotube). Each cryotube of cells was stored at −80ºC overnight and then stored in liquid nitrogen.
Cell seeding
Cryopreserved cells were thawed, and 5 mL of osteogenic medium was added per cryotube. Then, 2 mL of cell suspension (∼1×106 cells) was plated per 75-cm2 culture flask and cultured until confluence to have the necessary number of cells for seeding (500,000 cells/scaffold). At the end of this primary culture period, cells were lifted with trypsin solution (0.25% trypsin/0.02% ethylenediaminetetraacetic acid), centrifuged, and resuspended in a known amount of medium.
SPCL fiber mesh scaffolds were immersed in complete osteogenic medium overnight and press-fit into 8-mm-diameter cassettes on the following day. Cassettes were placed into 6-well plates, and then SPCL scaffolds were seeded with 500,000 cells in 300 μL of complete osteogenic medium. Cells were allowed to attach for 3 h before 10 mL of osteogenic medium was added to each well of a 6-well plate. After 24 h of attachment, the scaffolds were placed in new 6-well plates for 7 and 15 days with three different medium conditions (complete osteogenic medium with (1) lipase (110 U/L) or (2) α-amylase (150 U/L) or (3) without enzymes) to study the degradation of SPCL fiber meshes under conditions simulating those found in vivo. Media were changed every 3 days. Each group consisted of six samples.
DNA analysis
The cells in the cell–scaffold constructs were lysed by freeze-thaw in double distilled water (ddH2O). The DNA content of the lysates was quantified using a DNA quantification kit (PicoGreen, Molecular Probes, Eugene, OR) and correlating to a known amount of rat MSCs. Frozen scaffolds were thawed at room temperature, sonicated for 10 min, and vortexed to allow cellular DNA into solution. Buffer provided with the kit was diluted with ddH2O and placed in each well of a 96-well plate at 100 μL/well. DNA standard solutions in ddH2O in concentrations from 0 to 6 μg/mL were prepared, and 50 μL of standards/sample was added to each well of the 96-well plate. PicoGreen dye solutions were prepared according the instructions of the manufacturer using reagents provided in the kit and added at 150 μL/well. All of these procedures were performed in triplicate. After 10 min of incubation in a dark room, the fluorescence at 545 nm was read on a plate reader (FLx800, Bio-Tek Instruments Inc., Winooski, VT) to determine DNA concentrations per scaffold. A conversion factor of 2.7 pg of DNA/cell was used to calculate cell number based on MSC standards.
Alkaline phosphatase activity
Alkaline phosphatase (ALP) activity was determined using an established protocol27 by measuring the conversion of p-nitrophenylphosphate to p-nitrophenol. All reagents were purchased from Sigma. For the assay, 96-well plates were used. In each well. 80 μL of sample or standard, 20 μL of 1.5 M alkali buffer solution (Sigma, cat. no. A9226), and 100 μL of phosphatase substrate solution (Sigma, cat. no. P5869) were added, and the plate was incubated for 1 h at 37ºC. Each sample and standard was prepared in triplicate. The reaction was stopped by adding 100 μL of stop solution (0.3 N sodium hydroxide). The standard curve was generated by preparing dilutions of p-nitrophenol standard solution (0-250 μM, Sigma, cat. no. N7660). Absorbance was read at 405 nm on a plate reader to determine enzyme concentration per construct.
Calcium deposition measurement
Calcium content in the SPCL fiber mesh scaffolds was measured using a Calcium Kit (Diagnostic Chemical Limited, Oxford, CT). After the ALP and DNA assays, a volume of 1 N acetic acid equal to the volume remaining in each well was added to each well (final concentration of 0.5 N), and the plate was placed on an orbital shaker overnight. A volume of 300 μL of calcium assay reagent was added to 20 μL of sample solution in triplicate in 96-well plates. To generate a standard curve, serial dilutions of calcium chloride were prepared (0-50 μg/mL). Absorbance was read at 650 nm in a spectrophotometric plate reader. The calcium deposition in each scaffold is reported as μg of Ca2+.
Microscopic analysis
After each time period, SPCL fiber mesh scaffolds were removed from the cassettes, washed with PBS, and stained with calcein-acetoxymethyl ester (calcein-AM; Molecular Probes) according to the manufacturer's instructions. To visualize the distribution of cells within the scaffolds, after 45 min of immersion in PBS with calcein-AM, images were recorded using a laser scanning confocal microscope with a 10 × APO objective (Zeiss LSM 510 META, Carl Zeiss, Oberkochen, Germany). Depth projections of the surface (up to 410 μm) were obtained, and the cells were colored as a function of their distance from the surface.
Scanning electron microscopy (SEM) was used to evaluate the morphological appearance of the rat bone marrow cells. Samples for SEM analysis were washed twice in PBS and fixed with 2.5% glutaraldehyde for 30 min. Then the scaffolds were washed with PBS, dehydrated in a graded series of ethanol, and dried using hexamethyldisilazane.
Fourier transform infrared spectroscopy with attenuated total reflectance
The cell–scaffold constructs were analyzed after 16 days of culture using Fourier transform infrared spectroscopy with attenuated total reflectance FTIR-ATR. The scaffolds were fixed in 2.5% glutaraldehyde, dehydrated in a graded series of ethanol, and dried at room temperature. All spectra were recorded using 64 scans and 4 cm−1 in a FTIR spectrophotometer (IR Prestige-21 FTIR spectrometer, Shimadzu).
Statistics
Results of porosity, pore interconnectivity, pores size, fibers thickness, DNA, ALP, and calcium assay are expressed as means ± standard deviations, with n = 3 for each group. Statistical significance of differences was determined using the Student t-test multiple comparison procedure with a 95% confidence interval (p < 0.05).
Results and Discussion
Degradation studies
The main aims of the degradation studies were to simulate physiological conditions using enzymes present in human serum that would be responsible for enzymatic hydrolysis of SPCL fiber mesh scaffolds in vivo, to control and tailor the degradation rate of the SPCL scaffolds, and finally to evaluate the possibility of increasing their porosity and pore interconnectivity through degradation.
SPCL is a blend of starch and poly(ɛ-caprolactone). In this study, scaffolds based on SPCL were prepared using a fiber bonding process that uses heat to promote the bonding of the fibers that had previously been obtained using melt-spinning.9
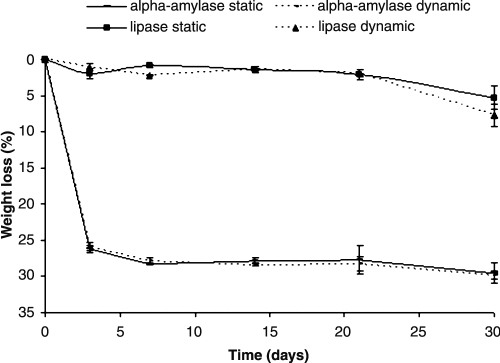
When analyzing weight loss profiles using α-amylase and lipase, few differences were observed between static and dynamic conditions for each enzyme (Fig. 1). One of the enzymes responsible for starch hydrolysis is α-amylase. The scaffolds immersed in α-amylase presented greater weight loss than the scaffolds immersed in lipase. A weight loss of approximately 25% was observed after 3 days of immersion in α-amylase, which increased slightly to 30% weight loss after 30 days of immersion (Fig. 1). Nevertheless, immersion periods up to 30 days did not cause the scaffolds to lose their structural integrity. These observed results are in agreement with previous studies using α-amylase.3,7
FIG. 1.
Weight loss profile (pH 7.4, 37ºC) of fiber mesh scaffolds made of a blend of starch and poly(ɛ-caprolactone) using α-amylase (150 U/L) and lipase (110 U/L) under static and dynamic conditions up to 30 days.
Lipase is an enzyme responsible for hydrolysis of ester bonds in polyesters, and some studies have shown that lipase enhances PCL degradation.7,28,29 PCL is a hydrophobic crystalline polymer that degrades slowly in vitro in the absence of enzymes and in vivo as well. The in vitro degradation can be enhanced in the presence of the enzyme lipase.29 The values of weight loss in the presence of lipase were lower than those obtained in the presence of α-amylase, but these values are in agreement with other studies.28 Weight loss remains constant up to 21 days, increasing slightly afterward (Fig. 1). Several studies have reported the enzymatic degradation of PCL by lipase.11–14,30 It is apparent from Figure 1 that the degradation of SPCL occurs in the presence of lipase, this enzyme being responsible for only 7.7 ± 1.6% weight loss after 30 days of immersion under dynamic conditions. These results are in accordance with several published studies.12,13 Hoshino et al.12 showed the influence of different commercially available lipases on degradation of PCL. They presented the results of biodegradability of PCL films after 100 days incubation at 37ºC, and some commercial lipase species were able to degrade PCL completely, namely from alcaligenes sp., Rhizopus niveus, Rhizopus oryzae, Chromobacterium viscosum, and Candida rugosa. However, they presented different results with other lipases that showed values of weight loss of less than 5%, as when using the same lipase employed in this study—Aspergillus niger. These results are in accordance with the weight loss after 30 days reported here. In contrast to its slow hydrolytic degradation, other studies with PCL showed high values of weight loss during enzymatic degradation when using Pseudomonas cepacia lipase, which has a specific activity on PCL segment degradation.14
α-amylase and lipase contributed to different degrees to the degradation of the SPCL fiber mesh scaffolds, and the studies presented here with α-amylase show that this enzyme is responsible for higher values of weight loss than the ones obtained with lipase (Fig. 1).
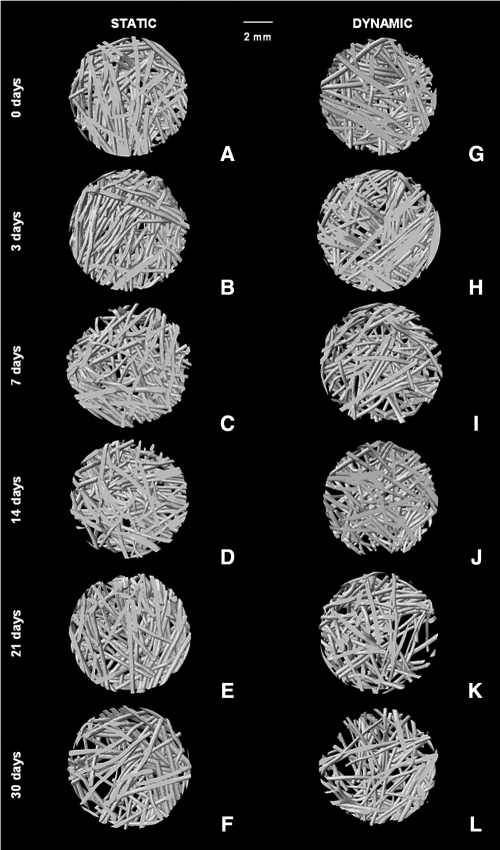
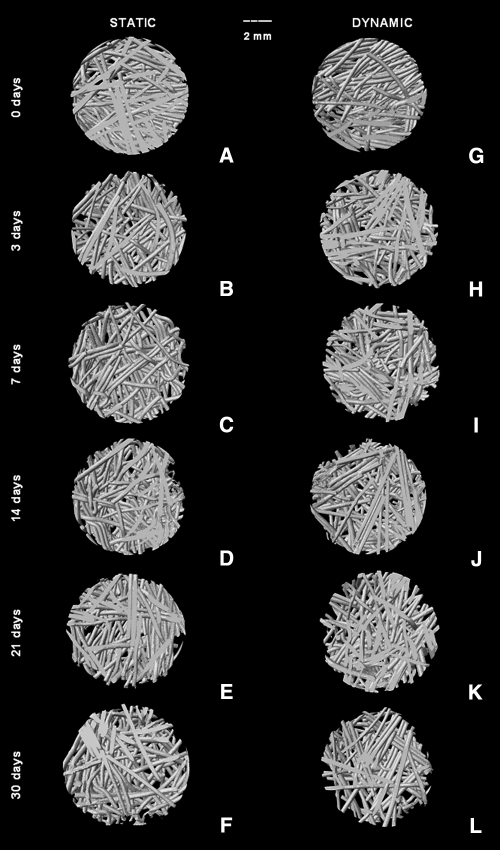
The morphological changes induced by degradation with these enzymes can be further confirmed using virtual 3D models obtained using μ-CT, where it can be seen that α-amylase and lipase affected SPCL fiber mesh scaffolds (Fig. 2). However, as confirmed by weight loss results, α-amylase had a great effect in SPCL fiber meshes mainly at day 21 (Fig. 2K) and day 30 (Fig. 2F, L). In Figure 3, it is possible to observe the 3D virtual models obtained using μ-CT as a function of immersion time in PBS using lipase solution. The effect of lipase is not as pronounced as images obtained after degradation with α-amylase; nevertheless, slight changes in SPCL fiber meshes were observed as a function of immersion time (Fig. 3).
FIG. 2.
Starch and poly(ɛ-caprolactone) fiber mesh images obtained using micro-computed tomography before (A, G) and after degradation with α-amylase (150 U/L) under static (B–F) and dynamic (H–L) conditions up to 30 days. The scale bar is 2 mm and applies to all images.
FIG. 3.
Starch and poly(ɛ-caprolactone) fiber mesh images obtained using micro-computed tomography before (A, G) and after degradation with lipase (110 U/L) under static (B–F) and dynamic (H–L) conditions up to 30 days. The scale bar is 2 mm and applies to all images.
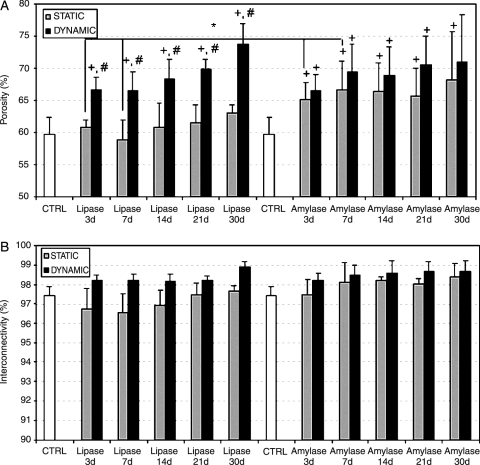
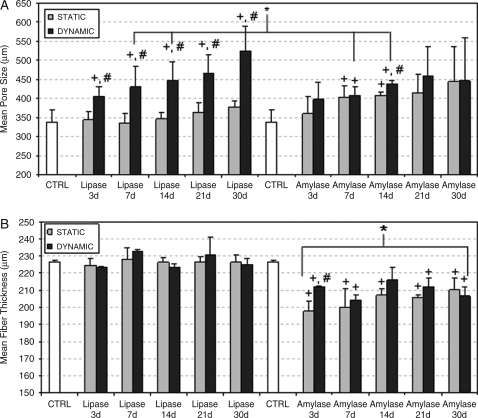
SPCL fiber mesh scaffolds before degradation presented an average porosity of 59.8 ± 2.6% (Fig. 4A), with approximately 97% pore interconnectivity (Fig. 4B). Figure 4A shows that both degradation conditions (lipase and α-amylase) enhanced the porosity of the scaffolds. Porosity results (Fig. 4A) show that both enzymes (lipase and α-amylase) have a significant effect on SPCL degradation as a function of degradation time. When lipase was used, significantly greater increase porosity was observed as a function of immersion time using dynamic conditions than with control samples. Furthermore, a significant difference in porosity was obtained using lipase and under dynamic conditions from results obtained with the same enzyme under static conditions. Agitation seems to influence lipase (dynamic conditions), as seen in Figure 4A; the porosity is greater than in control samples and under static conditions. In addition, no effect of lipase upon SPCL porosity using static conditions was observed.
FIG. 4.
Porosity (A) and pore interconnectivity (B) of starch and poly(ɛ-caprolactone) fiber meshes after different degradation periods with lipase and α-amylase under static and dynamic conditions. Results are expressed as means ± standard deviation with n = 3 for each bar. +Significant difference (p < 0.05) between conditions as a function of time compared with control samples. #Significant difference (p < 0.05) between static and dynamic conditions (using the same enzyme) at the same time point. *Significant difference (p < 0.05) between enzymes (lipase and α-amylase) under static conditions at the same time point. No significant differences were observed between dynamic conditions.
In contrast to results obtained with lipase, α-amylase has a significant effect on SPCL degradation under static or dynamic conditions (Fig. 4A). Moreover, results when α-amylase was used show that all time periods (except day 30 under dynamic conditions) under static and dynamic conditions presented significantly greater porosity than in control samples (Fig. 4A). The effect of α-amylase on porosity seems to be independent of using static or dynamic conditions, as can be seen in Figure 4A. When comparing results obtained after degradation with lipase or α-amylase for the same period of time, significant greater porosity was observed only for the third and seventh days using α-amylase under static conditions. Some differences can be observed between static and dynamic conditions for both enzymes; the positive effect of lipase on the porosity of the scaffolds is higher for dynamic conditions, whereas the effect of α-amylase on porosity appears to be less dependent on the circulation (static/dynamic) of the degrading medium (Fig. 4A).
The interconnectivity of SPCL scaffolds before degradation is high (∼97%), and for all periods, the interconnectivity increases under static and dynamic conditions with α-amylase (Fig. 4B), although such differences are not easily perceptible through visual inspection because of the high range of interconnectivity. The same trend was observed for lipase (Fig. 4B), and no significant differences were observed. However, dynamic conditions seemed to influence the interconnectivity when α-amylase or lipase was used.
Average pore size also increased upon degradation with lipase. According to porosity results (Fig. 4A), the same trend was observed for pore size when lipase was used (Fig. 5A). The pore size increase upon degradation with lipase was more pronounced for dynamic conditions (Fig. 5A), being significantly different from control samples or static conditions (Fig. 5A). In this respect, dynamic conditions enhance the size of the pores (Fig. 5A) relative to static conditions. As observed in Figure 4A, static conditions seemed not to interfere with the size of pores when lipase was used (Fig. 5A). The pore size of the scaffolds was greater upon degradation with α-amylase than in control samples, although this difference wais only significant for the seventh and fourteenth days using static or dynamic conditions. A significantly greater pore size was also observed when α-amylase was used under static conditions than when lipase was used (7th and 14th day).
FIG. 5.
Pores size (A) and fibers thickness (B) of starch and poly(ɛ-caprolactone) fiber meshes after degradation with lipase and α-amylase as a function of immersion time under static and dynamic conditions. Results are expressed as means ± standard deviation, with n = 3 for each bar. +Significant difference (p < 0.05) between conditions as a function of time compared with control samples. #Significant difference (p < 0.05) between static and dynamic conditions (using the same enzyme) at the same time point. *Significant difference (p < 0.05) between enzymes (lipase and α-amylase) under static or dynamic conditions at the same time point. In (B), *α-amylase has a significant difference at the same time point under static or dynamic conditions compared with lipase (except at day 21 under static conditions).
The thickness of the fibers remained more or less constant using lipase (Fig. 5B), which supports the low weight loss observed for lipase degradation media (Fig. 1). No significant differences were observed between static or dynamic conditions. α-amylase caused thinner fibers under static and dynamic conditions than control samples as a function of time. Fiber diameter decreased upon degradation with α-amylase, although differences between static and dynamic conditions were not significant, except for the third day of degradation under dynamic conditions. Comparing the action of lipase and α-amylase on fiber thickness, it is possible to observe significantly thinner fibers when lipase was used under static or dynamic conditions.
Assays
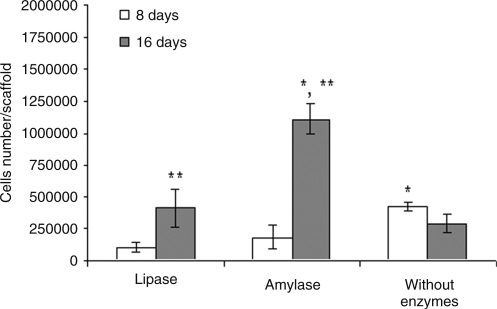
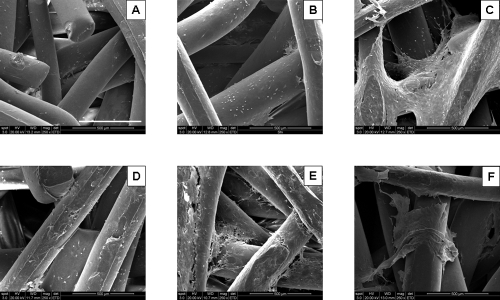
The results of scaffold cellularity are presented in Figure 6. The DNA assay showed a significant increase (p < 0.05) in late cell proliferation for SPCL fiber meshes that were cultured with osteogenic medium with lipase or α-amylase. After 16 days, the number of cells from scaffolds cultured with enzymes was much higher than the scaffolds cultured without enzymes; their cellularity increased three to five times between day 8 and day 16 of culture. In contrast, the DNA results showed a non-significant decrease in cell proliferation after 16 days for SPCL scaffolds cultured in static cultures without enzymes, in agreement with previous experiments.9,10 SEM images confirm these results. A significant improvement in cell proliferation on the surface of SPCL fiber meshes cultured with enzymes is apparent (Fig. 7).
FIG. 6.
Cellularity of starch and poly(ɛ-caprolactone) fiber meshes cultured in vitro with lipase or α-amylase or without enzymes for 8 and 16 days. Results are expressed as means ±standard deviation with n = 3 for each bar. *Significant difference (p < 0.05) between conditions at the same time point. **Significant difference (p < 0.05) between 8 and 16 days for each condition.
FIG. 7.
Scanning electron microscipy images of the surfaces of starch and poly(ɛ-caprolactone) fiber mesh scaffolds cultured with lipase (A, D) or α-amylase (B, E) and without enzymes (C, F) after day 8 (A–C) and day 16 (D–F). The scale bar is 500 μm and applies to all images.
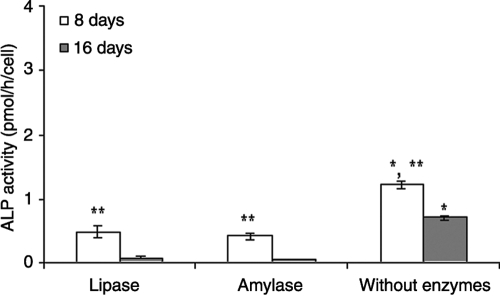
ALP is an enzyme expressed by cells and is an early marker of osteoblastic differentiation. ALP expression was highest values at day 8 of culture for all conditions (Fig. 8). According to Figure 8, ALP activity without enzymes (control) was significantly higher than in groups with enzymes. Usually a maximum value of ALP coincides with early osteoblastic differentiation of marrow stromal cells. There was a significant difference between the scaffolds cultured with and without enzymes for each time period. The presence of each enzyme caused lower ALP activity with than in the control. All conditions presented a significant decrease in ALP activity between day 8 and day 16, which was expected, because after this period, ALP activity usually decreases, and mineralization starts to take place.9
FIG. 8.
Alkaline phosphatase activity of rat marrow stromal cells cultured in vitro on starch and poly(ɛ-caprolactone) fiber mesh scaffolds with lipase or α-amylase or without enzymes for up to 16 days. Results are expressed as means ± standard deviations with n = 3 for each bar. *Significant difference (p < 0.05) between conditions at the same time point. **Significant difference (p < 0.05) between 8 and 16 days for each condition.
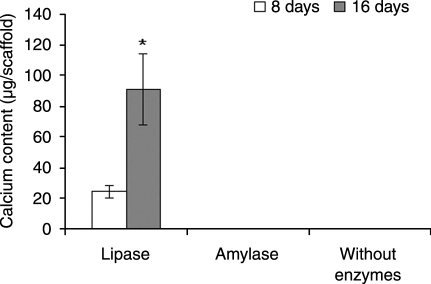
The decrease in ALP expression when lipase was added to the culture medium (Fig. 8) was related to an increase in calcium deposition (Fig. 9). The observed ALP activity levels corresponded with the observed calcium content only for the group exposed to lipase. However, ALP activity is a transient, early marker of osteoblastic differentiation, and peaks in ALP activity may have occurred at time points other than the ones assessed (days 8 and 16) for some groups. Because the production of a mineralized matrix is considered the endpoint in the osteoblastic differentiation of MSCs, calcium deposition results definitively mark the osteoblastic differentiation and expression of the cells. Previous studies showed low levels of calcium deposition in static cultures.9,10 After calcium deposition measurements, no calcium was detected for groups cultured with α-amylase or without enzymes after both time periods. However, constructs from the SPCL fiber mesh group cultured with lipase showed calcium deposition after day 8, with a significant increase after 16 days (p < 0.05). These values obtained for calcium deposition were similar to22,31 or higher than32,33 in previous studies conducted in static cultures using different fiber meshes. Lipase seems to positively influence osteoblastic differentiation of rat MSCs and enhance matrix mineralization.
FIG. 9.
Calcium content of starch and poly(ɛ-caprolactone) fiber mesh scaffolds cultured in vitro with lipase or α-amylase or without enzymes for 8 and 16 days expressed as total calcium per scaffold. Results are expressed as means ±standard deviations with n = 3 for each bar. *Significant difference (p < 0.05) between conditions after 16 days of culture.
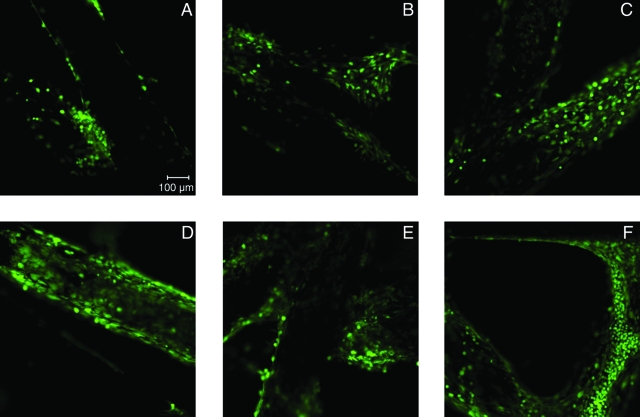
Figure 10 presents different images obtained using laser scanning confocal microscopy showing that cells are viable and well spread and cover the entire surface of the fibers at day 8 and 16.
FIG. 10.
Laser scanning confocal microscope images of rat marrow stromal cells stained with calcein-acetoxymethyl ester at the top surface of starch and poly(ɛ-caprolactone) fiber meshes with lipase (A, D) or α-amylase (B, E) or without enzymes (C, F) at 8 days (A–C) and 16 days (C, F). All images obtained using a 10 × APO objective. The scale bar is 100 μm and applies to all images.
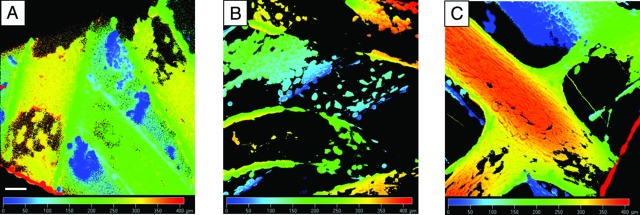
The images presented in Figure 11 were obtained from depth projections of SPCL fiber meshes after 16 days and show the infiltration of cells to a depth of 410 μm for all culture conditions and the presence of monolayers of cells covering the fibers.
FIG. 11.
Laser scanning confocal microscopy images of starch and poly(ɛ-caprolactone) fiber meshes cultured for 16 days with lipase (A) or α-amylase (B) or without enzymes (C) obtained from depth projections. The scale bar is 100 μm and applies to all images.
FIG. 12.
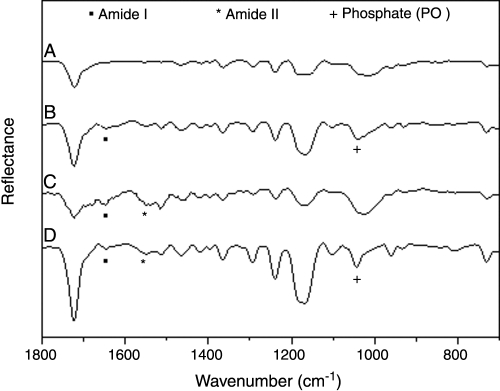
Fourier transform infrared spectroscopy with attenuated total reflectance spectra of starch and poly(ɛ-caprolactone) fiber meshes before (A) and after 16 days of culture without enzymes (B), with α-amylase (C), and with lipase (D).
Fourier transformed infrared spectroscopy with attenuated total reflectance
FTIR-ATR spectra were obtained from SPCL cultured for 16 days under static conditions with and without enzymes (α-amylase or lipase). This method was used as a complement to the calcium assay. When α-amylase or lipase was added to osteogenic media, it was possible to see bands centered at 1650 and 1542 cm−1 that can be attributed to amide I and amide II. The amide I band represents the stretching vibrations of C=O bonds in the backbone of the proteins.34 Furthermore, the amide II band arises from the combination of C-N stretching and N-H bending vibrations of the protein backbone.34 Analyzing the spectra when enzymes were added to culture media, it is possible to detect amide I and amide II bands, which confirms that these enzymes were able to adsorb to the SPCL fiber mesh surfaces. In addition, these bands can be correlated to the protein matrix formation. Moreover, phosphate bands located at 1020 cm−1 similar to the ones found in carbonate apatite were observed in SPCL fiber meshes cultured with lipase, clearly suggesting the presence of mineralized ECM and corroborating the results obtained from the calcium assay.
Conclusions
This study demonstrates the effect of lipase and α-amylase on degradation of SPCL fiber mesh scaffolds, showing that, after 30 days, these enzymes did not have a negative effect on the structural integrity of SPCL fiber mesh scaffolds. The positive effect of lipase on the porosity and size of pores was clearly visible under dynamic conditions. α-amylase also enhanced porosity, pore size, and fiber thickness. However, no significant differences were observed under static and dynamic conditions when α-amylase solution was used. SPCL fiber meshes cultured with media supplemented with α-amylase or lipase revealed that these enzymes promoted the proliferation of rat MSCs. Furthermore, this study confirms by calcium content measurement and FTIR analysis that lipase has a superior effect in osteodifferentiation of rat MSCs and promotes ECM mineralization better when added to osteogenic culture medium than results obtained without enzymes or with α-amylase. These results demonstrate that supplementation of culture media with lipase holds great potential for the generation of bone tissue-engineering constructs from MSCs seeded onto SPCL fiber mesh scaffolds, because lipase enhances the osteoblastic differentiation of the seeded MSCs and promotes matrix mineralization without harming the structural integrity of the meshes over 16 days of culture.
Acknowledgments
The authors would like to acknowledge Dr. Helena Azevedo and Dr. Serena Danti. This work was supported by the European NoE EXPERTISSUES (NMP3-CT-2004-500283), the European STREP HIPPOCRATES (NMP3-CT-2003-505758), and the Portuguese Foundation for Science and Technology (FCT) through POCTI and/or FEDER programmes. This work was also supported by grants from the National Institutes of Health (NIH R01 DE15164 and R01 DE17441) (AGM) and a Bioengineering Research Partnership with the Baylor College of Medicine through the National Institute of Biomedical Imaging and Bioengineering (NIH Grant 5 R01 EB005173-02). FKK is supported by a training fellowship from the Keck Center Nanobiology Training Program of the Gulf Coast Consortia (NIH Grant 5 T90 DK070121-03).
References
- 1.Langer R. Vacanti J.P. Tissue engineering. Science. 1993;260:920. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 2.Hutmacher D.W. Schantz J.T. Lam C.X.F. Tam K.C. Lim T.C. State of the art and future directions of scaffold-based bone engineering from a biomaterials perspective. J Tissue Eng Regen Med. 2007;1:245. doi: 10.1002/term.24. [DOI] [PubMed] [Google Scholar]
- 3.Azevedo H.S. Reis R.L. Understanding the enzymatic degradation of biodegradable polymers and strategies to control their degradation rate. In: Reis R.L., editor; Roman J.S., editor. Biodegradable Systems in Tissue Engineering and Regenerative Medicine. Boca Raton, FL: CRC Press; 2005. pp. 177–201. [Google Scholar]
- 4.Mendes S.C. Reis R.L. Bovell Y.P. Cunha A.M. van Blitterswijk C.A. de Bruijn J.D. Biocompatibility testing of novel starch-based materials with potential application in orthopaedic surgery: a preliminary study. Biomaterials. 2001;22:2057. doi: 10.1016/s0142-9612(00)00395-1. [DOI] [PubMed] [Google Scholar]
- 5.Marques A.P. Reis R.L. Hunt J.A. The biocompatibility of novel starch-based polymers and composites: in vitro studies. Biomaterials. 2002;23:1471. doi: 10.1016/s0142-9612(01)00272-1. [DOI] [PubMed] [Google Scholar]
- 6.Gomes M.E. Ribeiro A.S. Malafaya P.B. Reis R.L. Cunha A.M. A new approach based on injection moulding to produce biodegradable starch-based polymeric scaffolds: morphology, mechanical and degradation behaviour. Biomaterials. 2001;22:883. doi: 10.1016/s0142-9612(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 7.Azevedo H.S. Gama F.M. Reis R.L. In vitro assessment of the enzymatic degradation of several starch based biomaterials. Biomacromolecules. 2003;4:1703. doi: 10.1021/bm0300397. [DOI] [PubMed] [Google Scholar]
- 8.Salgado A.J. Coutinho O.P. Reis R.L. Novel starch-based scaffolds for bone tissue engineering: cytotoxicity, cell culture, and protein expression. Tissue Eng. 2004;10:465. doi: 10.1089/107632704323061825. [DOI] [PubMed] [Google Scholar]
- 9.Gomes M.E. Sikavitsas V.I. Behravesh E. Reis R.L. Mikos A.G. Effect of flow perfusion on the osteogenic differentiation of bone marrow stromal cells cultured on starch-based three-dimensional scaffolds. J Biomed Mater Res A. 2003;67:87. doi: 10.1002/jbm.a.10075. [DOI] [PubMed] [Google Scholar]
- 10.Gomes M.E. Holtorf H.L. Reis R.L. Mikos A.G. Influence of the porosity of starch-based fiber mesh scaffolds on the proliferation and osteogenic differentiation of bone marrow stromal cells cultured in a flow perfusion bioreactor. Tissue Eng. 2006;12:801. doi: 10.1089/ten.2006.12.801. [DOI] [PubMed] [Google Scholar]
- 11.Mochizuki M. Hirano M. Kanmuri Y. Kudo K. Tokiwa Y. Hydrolysis of polycaprolactone fibers by lipase—effects of draw ratio on enzymatic degradation. J Appl Polym Sci. 1995;55:289. [Google Scholar]
- 12.Hoshino A. Isono Y. Degradation of aliphatic polyester films by commercially available lipases with special reference to rapid and complete degradation of poly(L-lactide) film by lipase PL derived from Alcaligenes sp. Biodegradation. 2002;13:141. doi: 10.1023/a:1020450326301. [DOI] [PubMed] [Google Scholar]
- 13.Zeng J. Chen X. Liang Q. Xu X. Jing X. Enzymatic degradation of poly(L-lactide) and poly(epsilon-caprolactone) electrospun fibers. Macromol Biosci. 2004;4:1118. doi: 10.1002/mabi.200400092. [DOI] [PubMed] [Google Scholar]
- 14.Miao Z.M. Cheng S.X. Zhang X.Z. Wang Q.R. Zhuo R.X. Degradation and drug release property of star poly(epsilon-caprolactone)s with dendritic cores. J Biomed Mater Res B Appl Biomater. 2007;81:40. doi: 10.1002/jbm.b.30634. [DOI] [PubMed] [Google Scholar]
- 15.Wong H. Schotz M.C. The lipase gene family. J Lipid Res. 2002;43:993. doi: 10.1194/jlr.r200007-jlr200. [DOI] [PubMed] [Google Scholar]
- 16.Hasham S.N. Pillarisetti S. Vascular lipases, inflammation and atherosclerosis. Clin Chim Acta. 2006;372:179. doi: 10.1016/j.cca.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 17.Mukherjee M. Human digestive and metabolic lipases—a brief review. J Mol Catal, B Enzym. 2003;22:369. [Google Scholar]
- 18.Schoonjans K. PeinadoOnsurbe J. Lefebvre A.M. Heyman R.A. Briggs M. Deeb S. Staels B. Auwerx J. PPAR alpha and PPAR gamma activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. Embo J. 1996;15:5336. [PMC free article] [PubMed] [Google Scholar]
- 19.Palacios V.G. Morita I. Murota S. Expression of adipogenesis markers in a murine stromal cell line treated with 15-deoxy Delta(12,14)-prostaglandin J(2), interleukin-11,9-cis retinoic acid and vitamin K-2. Prostaglandins Leukot Essent Fatty Acids. 2001;65:215. doi: 10.1054/plef.2001.0314. [DOI] [PubMed] [Google Scholar]
- 20.De Coppi P. Bartsch G. Siddiqui M.M. Xu T. Santos C.C. Perin L. Mostoslavsky G. Serre A.C. Snyder E.Y. Yoo J.J. Furth M.E. Soker S. Atala A. Isolation of amniotic stem cell lines with potential for therapy. Nat Biotechnol. 2007;25:100. doi: 10.1038/nbt1274. [DOI] [PubMed] [Google Scholar]
- 21.Tietz N.W. Shuey D.F. Lipase in serum—the elusive enzyme: an overview. Clin Chem. 1993;39:746. [PubMed] [Google Scholar]
- 22.Holtorf H.L. Jansen J.A. Mikos A.G. Flow perfusion culture induces the osteoblastic differentiation of marrow stroma cell-scaffold constructs in the absence of dexamethasone. J Biomed Mater Res A. 2005;72:326. doi: 10.1002/jbm.a.30251. [DOI] [PubMed] [Google Scholar]
- 23.Junge W. Troge B. Klein G. Poppe W. Gerber M. Evaluation of a new assay for pancreatic amylase: performance characteristics and estimation of reference intervals. Clin Biochem. 1989;22:109. doi: 10.1016/s0009-9120(89)80007-4. [DOI] [PubMed] [Google Scholar]
- 24.Chawla J.S. Amiji M.M. Biodegradable poly(epsilon-caprolactone) nanoparticles for tumor-targeted delivery of tamoxifen. Int J Pharm. 2002;249:127. doi: 10.1016/s0378-5173(02)00483-0. [DOI] [PubMed] [Google Scholar]
- 25.Rutkowska M. Krasowska K. Heimowska A. Steinka I. Janik H. Haponiuk J. Karlsson S. Biodegradation of modified poly(e-caprolactone) in different environments. Pol J Environ Stud. 2002;11:413. [Google Scholar]
- 26.Maniatopoulos C. Sodek J. Melcher A.H. Bone formation in vitro by stromal cells obtained from bone marrow of young adult rats. Cell Tissue Res. 1988;254:317. doi: 10.1007/BF00225804. [DOI] [PubMed] [Google Scholar]
- 27.Datta N. Holtorf H.L. Sikavitsas V.I. Jansen J.A. Mikos A.G. Effect of bone extracellular matrix synthesized in vitro on the osteoblastic differentiation of marrow stromal cells. Biomaterials. 2005;26:971. doi: 10.1016/j.biomaterials.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Zeng J. Chen X.S. Liang Q.Z. Xu X.L. Jing X.B. Enzymatic degradation of poly(L-lactide) and poly (epsilon-caprolactone) electrospun fibers. Macromol Biosci. 2004;4:1118. doi: 10.1002/mabi.200400092. [DOI] [PubMed] [Google Scholar]
- 29.Chawla J.S. Amiji M.M. Biodegradable poly(epsilon-caprolactone) nanoparticles for tumor-targeted delivery of tamoxifen. Int J Pharm. 2002;249:127. doi: 10.1016/s0378-5173(02)00483-0. [DOI] [PubMed] [Google Scholar]
- 30.Pulkkinen M. Malin M. Tarvainen T. Saarimaki T. Seppala J. Jarvinen K. Effects of block length on the enzymatic degradation and erosion of oxazoline linked poly-epsilon-caprolactone. Eur J Pharm Sci. 2007;31:119. doi: 10.1016/j.ejps.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Sikavitsas V.I. Bancroft G.N. Holtorf H.L. Jansen J.A. Mikos A.G. Mineralized matrix deposition by marrow stromal osteoblasts in 3D perfusion culture increases with increasing fluid shear forces. Proc Natl Acad Sci U S A. 2003;100:14683. doi: 10.1073/pnas.2434367100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sikavitsas V.I. Bancroft G.N. Lemoine J.J. Liebschner M.A. Dauner M. Mikos A.G. Flow perfusion enhances the calcified matrix deposition of marrow stromal cells in biodegradable nonwoven fiber mesh scaffolds. Ann Biomed Eng. 2005;33:63. doi: 10.1007/s10439-005-8963-x. [DOI] [PubMed] [Google Scholar]
- 33.Holtorf H.L. Datta N. Jansen J.A. Mikos A.G. Scaffold mesh size affects the osteoblastic differentiation of seeded marrow stromal cells cultured in a flow perfusion bioreactor. J Biomed Mater Res A. 2005;74:171. doi: 10.1002/jbm.a.30330. [DOI] [PubMed] [Google Scholar]
- 34.Xie J. Riley C. Kumar M. Chittur K. FTIR/ATR study of protein adsorption and brushite transformation to hydroxyapatite. Biomaterials. 2002;23:3609. doi: 10.1016/s0142-9612(02)00090-x. [DOI] [PubMed] [Google Scholar]