Summary
We report here the NMR derived structure of the binary complex formed by the Itk SH3 and SH2 domains. The interaction is independent of both a phosphotyrosine motif and a proline-rich sequence; the classical targets of the SH2 and SH3 domain, respectively. The Itk SH3/SH2 structure reveals the molecular details of this non-classical interaction and provides a clear picture for how the previously described prolyl cis/trans isomerization present in the Itk SH2 domain mediates SH3 binding. The higher affinity cis SH2 conformer is pre-organized to form a hydrophobic interface with the SH3 domain. The structure also provides insight into how autophosphorylation in the Itk SH3 domain might increase the affinity of the intermolecular SH3/SH2 interaction. Finally, we can compare this Itk complex with other examples of SH3 and SH2 domains engaging their ligands in a non-classical manner. These small binding domains exhibit a surprising level of diversity in their binding repertoires.
Keywords: Itk, SH3, SH2, oligomerization, tyrosine kinase
Introduction
The non-receptor Interleukin-2 tyrosine kinase (Itk) is an important immunological protein involved in regulating T-cell receptor (TCR) response to antigen stimulation 1, actin cytoskeletal rearrangement 2; 3 and more recently, Itk has been implicated in the HIV replication pathway 4. Itk and the related Tec family kinases (Btk, Txk, Tec and Bmx) all contain the common SH3-SH2-Kinase domain structure and, with the exception of Txk, also contain an N-terminal Pleckstrin homology (PH) domain 1. The Src homology 2 and Src homology 3 (SH2 and SH3) domains are well characterized protein binding domains involved in protein regulation, substrate recognition, and cell signaling 5. Classical ligands for the SH3 domain consist of proline-rich sequences 6, and SH2 domains typically bind phosphotyrosine (pY)-containing motifs 5; 7; 8. While these canonical sequences constitute the most well understood targets of the SH3 and SH2 domains, alternative binding interactions for these modular domains are emerging 9; 10; 11; 12; 13; 14; 15; 16; 17; 18; 19; 20; 21; 22; 23; 24.
In this vein, the SH3 and SH2 domains of Itk participate in both classical ligand binding 25; 26 and non-classical interactions 27. We previously reported solution NMR work that defined a specific intermolecular interaction between the SH3 and SH2 domains of Itk 27. In that work, intermolecular self-association of the Itk dual domain fragment, SH3-SH2, is described and interaction surfaces on both domains were mapped using chemical shift perturbations and differential isotopic labeling. Chemical shift mapping indicated that the SH3/SH2 interaction involves the classical ligand-binding cleft of the Itk SH3 domain and a surface that only partially overlaps the canonical ligand-binding surface of the Itk SH2 domain. The interaction is independent of phosphotyrosine, and the proline-rich sequence that defines classical SH3 targets is not present within the Itk SH2 domain. Thus, the Itk SH3/SH2 interaction represents an alternative mode of binding for each of these well-studied domains.
Additional molecular details for the Itk SH3/SH2 interaction emerged following complete structure determination of the Itk SH2 domain 28. The Itk SH2 domain contains a rather unique residue, Pro 287, which interconverts between the cis and trans imide bond conformers in solution. This exchange process leads to two distinct structures of the Itk SH2 domain since cis/trans isomerization about the Asn 286-Pro 287 imide bond induces conformational differences across one-third of the domain surface. As a result, the cis and trans imide bond containing Itk SH2 conformers (also referred to as cis SH2 or trans SH2) exhibit different ligand binding properties. The classical phosphotyrosine ligand binds preferentially to the trans imide bond containing SH2 domain and the Itk SH3 domain binds with greater affinity to the cis imide bond containing SH2 domain 29. The structure of a classical phosphotyrosine containing peptide ligand bound to the trans Itk SH2 domain has been solved 26 providing insight into why this canonical SH2 ligand prefers the trans imide bond containing SH2 conformer over cis. The structure of the Itk SH3 domain bound to the cis imide bond containing conformer of the Itk SH2 domain reported here now provides a more complete picture of proline-driven molecular recognition within this small domain.
In due course, the intermolecular Itk SH3/SH2 interaction must be understood in the context of the full length Itk protein. In addition to the intermolecular interaction between SH3-SH2 fragments of Itk 27, the Itk PH domain also interacts with itself in an intermolecular fashion 30 and the structure of the Btk PH domain reveals a dimeric arrangement 31. Moreover, the other Tec family kinases; Btk, Tec and Txk, exhibit intermolecular self-association of their non-catalytic regulatory domains 32; 33; 34; 35. In the context of the intermolecular interactions reported for these domain fragments, it is notable that full length Itk readily co-immunoprecipitates with itself 30; 36 and Itk clustering at the T cell receptor has been observed upon T cell activation 37. We therefore hypothesize that the Itk regulatory domains mediate a functionally relevant self-association of full length Itk. The structure of the Itk SH3/SH2 complex reported here is a step toward gaining an understanding of the molecular details of Itk intermolecular self-association. Such information should ultimately translate into a better understanding of how these interactions mediate Itk signaling in the T cell.
Results
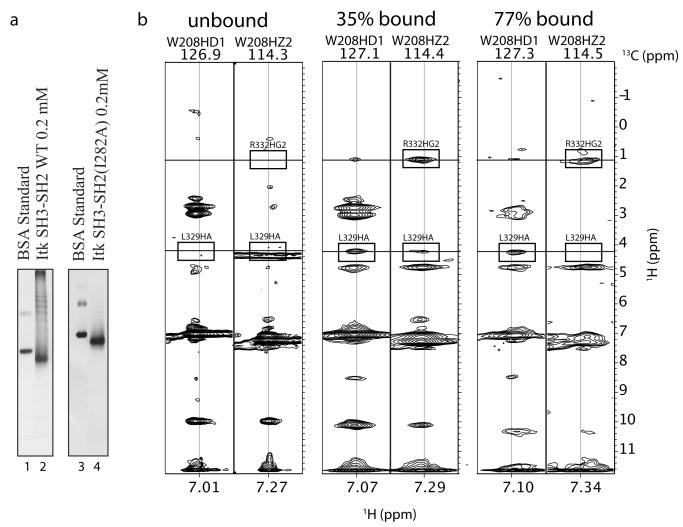
NMR solution studies on the dual domain SH3-SH2 fragment of Itk indicated an intermolecular interaction that is dependent on the aromatic binding cleft of the SH3 domain (mutation of the conserved W208 in the Itk SH3 domain abolishes the intermolecular interaction between SH3-SH2 fragments) 27. In this early work 27, the SH3-SH2 containing fragment of Itk was considered primarily dimeric. This conclusion was based on measured NMR linewidths and NMR diffusion data both of which would have been weighted toward the monomer and dimer species rather than larger aggregates given the inherent line broadening and disappearance of the NMR signal with increasing molecular weight. At the time, it was noted that the quality of NMR spectra acquired for samples containing the Itk SH3-SH2 dual domain was poor, likely due to exchange broadening. We have subsequently revisited the aggregation state of the Itk SH3-SH2 fragment using native gel electrophoresis. As shown in Figure 1a (lane 2), a native gel corresponding to the wild type Itk SH3-SH2 fragment exhibits multiple bands consistent with the notion that SH3-SH2 intermolecular self-association leads to formation of multiple aggregated species. In a similar manner, full length Itk also produces multiple bands on a native gel (data not shown) suggesting that the intermolecular oligomerization of the SH3-SH2 dual domain fragment is retained in the full-length molecule. Thus, formation of multiple oligomerized species is consistent with NMR data for the Itk SH3-SH2 dual domain fragment that are adversely affected by both exchange broadening and the presence of high molecular weight species. Indeed, while NMR spectra of SH3-SH2 are sufficient for chemical shift mapping, further NMR work to solve the structure of the Itk SH3-SH2 fragment posed significant challenges due to poor spectral quality.
Figure 1.
(a) Native gel electrophoresis was carried out to compare the Itk SH3-SH2 dual domain fragment (lane 2) and the same Itk SH3-SH2 fragment with Ile 282 mutated to alanine (Itk SH3-SH2(I282A), lane 4). Lanes 1 and 3 are Bovine Serum Albumin (BSA) standard. (b) Intermolecular NOEs are evident in 3D 13C-edited NOESY spectra. Two-dimensional slices corresponding to side chain protons of W208 are shown for free SH3 domain (unbound), 35% bound SH3 domain, and 77% bound SH3 domain. Intermolecular NOEs between protons on the SH3 and SH2 domains are boxed and labeled in the spectra of 35% and 77% bound SH3 domain. The corresponding regions of the spectrum of free SH3 domain (unbound) are also boxed showing the absence of these intermolecular NOEs for the isolated SH3 domain.
We found, however, that the intermolecular interaction can be recapitulated by expressing the single domains; purified Itk SH2 can be titrated into labeled Itk SH3 and chemical shift changes in the SH3 domain map the same surface as that of the dual domain interaction 27. Likewise, purified Itk SH3 can be titrated into labeled Itk SH2 to map the SH2 side of the interaction. The quality of the NMR spectra in these single domain titrations is superior to that acquired for the dual domain. We therefore proceeded with structure determination of the binary complex formed by the singly expressed Itk SH3 and SH2 domains.
Two different strategies were initially employed to solve the structure of the Itk SH3/SH2 complex. We began by screening conditions for crystallography yet found none that produced crystals of the complex; likely due to the relatively low affinity of the interaction (the Kd for the SH3/SH2 interaction is 670 μM 29). We therefore focused our efforts on structure determination by NMR. There has been great success in determining weak protein-protein interactions using conjoined rigid body/torsion angle simulated annealing 38; 39. In fact, the structure of the extremely weak (~3000 μM) protein-protein interaction between Nck-2 SH3-3 and PINCH-1 LIM4 was recently solved using this technique 21. This experimental method takes advantage of orientation restraints obtained from residual dipolar coupling (RDC) experiments and short distance restraints obtained from intermolecular NOEs. In addition, using starting structures for the individual domains that do not undergo a large conformational changes upon binding allows the calculation to be simplified by holding the atoms in the initial structures rigid but allowing for rotation and translation of the structures to minimize distance and orientation restraints. After this initial minimization, side chain residues at the interaction interface are given their full torsional degrees of freedom along with the rotational and translational degrees of freedom of the rigid bodies and the energy function is minimized to generate a final complex structure.
To solve the structure of the binary complex, distance restraints, orientation restraints, and starting structures of the individual Itk SH3 and SH2 domains were required. Distance restraints are obtained from the identification and assignment of intermolecular NOE restraints. Orientation restraints are obtained from RDC experiments that depend on the identification of one or several inert alignment media. Starting structures of the individual domains are obtained from selectively labeled NMR data on the bound binary complex. Additionally, we made use of the recently solved structure of the free Itk SH3 domain 40 and used RDCs to refine the previously solved structure of the unbound cis SH2 domain 28. These unbound domain structures permitted direct comparisons between free and bound domains upon completion of the SH3/SH2 complex structure.
Two NMR samples containing 15N/13C labeled SH2 alone or 15N/13C labeled SH3 alone were prepared (Table 1-1, 1-2). NMR samples containing the binary complex required special consideration during preparation. Based on previous experience with the Itk SH2 domain, a protein concentration above 5 mM is unstable due to precipitation over a short period of time. To avoid complications due to sample instability, two samples were generated containing 15N/13C labeled SH3 with reasonable excess unlabeled SH2 and 15N/13C labeled SH2 with the same excess unlabeled SH3 (Table 1-5, 1-6). This ratio of labeled to unlabeled protein resulted in a stable, labeled domain that was 77% bound as calculated following previously published methods 34.
Table 1.
Compositions of NMR Samples
| ID | NMR Sample | Percent bound |
|---|---|---|
| 1 | 1.5 mM 13C, 15N labeled Itk SH3 | 0% |
| 2 | 1.5 mM 13C, 15N labeled Itk SH2 | 0% |
| 3 | 3.4 mM 13C, 15N labeled Itk SH3 with 1.5 mM unlabeled Itk SH2 | 35% |
| 4 | 3.4 mM 13C, 15N labeled Itk SH2 with 1.5 mM unlabeled Itk SH3 | 35% |
| 5 | 1.5 mM 13C, 15N labeled Itk SH3 with 3.4 mM unlabeled Itk SH2 | 77% |
| 6 | 1.5 mM 13C, 15N labeled Itk SH2 with 3.4 mM unlabeled Itk SH3 | 77% |
To assign intermolecular NOE restraints, two additional NMR samples containing unlabeled SH2 with excess 15N/13C labeled SH3 and unlabeled SH3 with excess 15N/13C labeled SH2 were generated (Table 1-3, 1-4). For these samples, the ratio of labeled to unlabeled protein gave a labeled sample that was 35% bound. Thus, a total of six samples were generated for NMR data acquisition: unbound SH3 (15N/13C labeled), unbound SH2 (15N/13C labeled), and two samples each of 35% and 77% bound with either the SH3 or SH2 domain carrying the 15N/13C isotopic label (Table 1). Due to fast exchange the chemical shifts for a given atom differed in the 0%, 35% and 77% bound samples. The 35% and 77% bound samples provided complementary information that allowed NOESY cross peaks to be assigned. For example, the proton chemical shifts of unlabeled SH2 in sample 3 (Table 1-3) are identical to the proton chemical shifts of labeled SH2 in sample 6 (Table 1-6).
While the majority of resonances in the NMR spectra of the SH3/SH2 mixture are in the fast exchange regime, we find that exchange broadening for the samples containing 77% bound protein prohibited observation of many of the crosspeaks thus hampering efforts to identify and assign intermolecular NOEs. A direct comparison of the NOESY data of W208 for the unbound, 35% bound, and 77% bound samples (Figure 1b) show the extent of line broadening due to exchange. Exchange broadening is evident in the NOESY strip for W208HZ2: the NOE between L329HA and W208HZ2 is clearly visible in the 35% bound sample but not seen in the 77% bound sample. In addition, the NOE between W208HZ2 and R332HG2 is broadened yet still visible in the 77% bound sample. For some NOEs, we find little effect from exchange broadening; for example, the NOE between W208HD1 and L329HA is similar for the 35% and 77% bound samples. Overall, the NOESY data acquired for unbound and 35% bound samples showed sufficiently narrow linewidths to permit complete identification and resonance assignment of intra- and intermolecular NOEs. We therefore obtained distance restraints for structure calculations of the bound SH3 and SH2 domains as well as intermolecular NOE restraints using data from the 35% bound sample and, when linewidths permitted, the 77% bound sample. The NOESY data obtained from free SH3 or SH2 domain provided a useful comparative tool for the identification of intermolecular NOEs defining the SH3/SH2 interface since intermolecular NOEs will be absent in these datasets (Figure 1b).
To acquire orientation restraints from RDC experiments, a medium that induces weak alignment in the sample but does not otherwise interact with the protein complex of interest is needed. Due to differences in the isoelectric points for the two domains (4.3 versus 8.8, respectively for the Itk SH3 and Itk SH2 domains) and the large hydrophobic binding pocket on the SH3 domain, it was difficult to identify an alignment media that did not interact strongly with either the SH3 or SH2 domain. We found that despite its highly negative charge, Pf1 phage can be used in low concentration (8mg/ml) as an alignment media for the positively charged SH2 domain 41, the SH3 domain, and the binary complex.
To obtain orientation restraints, we used residual dipolar coupling (RDC) data acquired from the fractionally bound NMR samples described in Table 1 to extrapolate RDC values for the structure calculation. The dipolar coupling values, DCAHA and DNH, for all structure calculations were determined from the difference between one bond scalar couplings recorded in a phage containing weakly aligned sample (giving J+D) and a no-phage non-aligned sample (giving J). The couplings for the completely bound complex, Dbound, could not be measured directly and instead were calculated from the population weighted average couplings in the 35% and 77% bound samples using Equation 1 providing two estimates of the fully bound value42. The value of Dbound used in structural calculation was the average of the corresponding values calculated from the 35% and 77% bound datasets.
| 1) |
The binary complex structure was then calculated from the NOE-derived restraints and 58 orientation restraints derived from RDC experiments. The lowest energy structure obtained from conjoined rigid body/torsion angle simulated annealing was further refined using traditional non-rigid simulated annealing protocols 38; 43; 44; 45.
Structural overview of the Itk SH3/SH2 binary complex
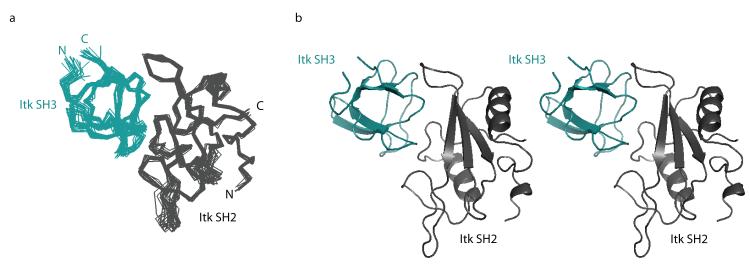
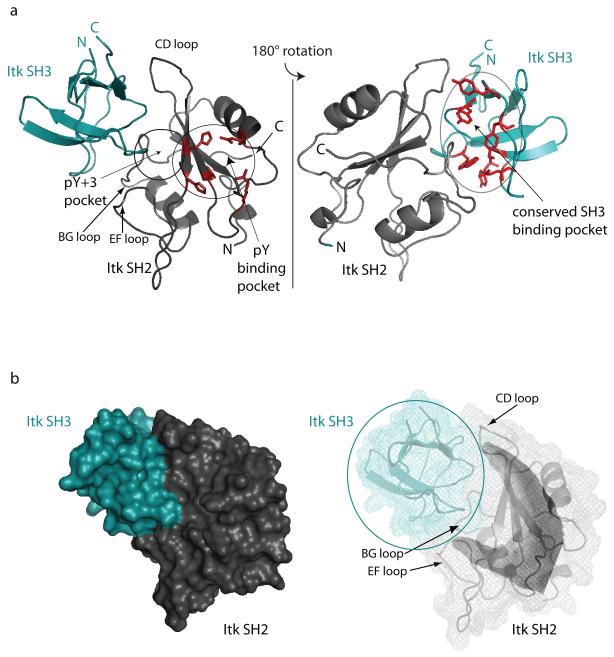
The structure of the binary SH3/SH2 complex is well defined (Figure 2 and Table 2). Backbone overlays of the 20 lowest energy structures show good convergence (Figure 2a) with a backbone RMSD (N, Cα, C’) of 0.68 Å and an overall heavy atom RMSD of 1.28 Å. The individual domains in the complex exhibit their characteristic folds (Figure 2b; Figure 1S in Supplementary Material). The bound Itk SH2 domain contains a central, three-strand anti-parallel beta sheet sandwiched between two alpha helices and has a backbone RMSD (including all loops) from the refined unbound cis SH2 structure of 1.28Å 28. The bound Itk SH3 domain contains two anti-parallel beta sheets that wrap around each other to form a beta barrel and has a backbone RMSD from the unbound SH3 structure of 0.98Å 40. In each case the RMSD was calculated between energy minimized average structures. The C-terminus of the SH3 domain and the N-terminus of the SH2 domain (the termini covalently linking each domain to its adjacent domain in the SH3-SH2 dual domain fragment) protrude from opposite sides of the SH3/SH2 intermolecular complex (Figure 3a). The structure reveals that the Itk SH2 domain binds to the conserved SH3 binding pocket and the Itk SH3 domain interacts with the pY+3 binding pocket and CD loop of the Itk SH2 domain, consistent with previously reported chemical shift mapping and ligand competition data 27. The interaction between Itk SH3 and SH2 domains does not involve the pY binding pocket on the SH2 domain (Figure 3a).
Figure 2.
(a) Superposition of Cα backbone traces of the 20 lowest energy structures calculated for the binary complex between the Itk SH3 and Itk SH2 domains. The SH3 domain is shown in teal and the SH2 domain is gray. The amino (N)- and carboxy (C)-termini of both the SH3 and SH2 domains are labeled. (b) Stereoview of the average SH3/SH2 binary complex structure.
Table 2.
Structural Statistics
| Experimental Restraints | SH2bound | SH3bound | SH3/SH2 | |
|---|---|---|---|---|
| NOEs | 2439 | 1351 | 3849+ | |
| CDIH | 191 | 110 | 301 | |
| RDC | NH | 33 | 25 | 58 |
| CAHA | 0 | 18 | 18 | |
| Total | 2663 | 1504 | 4226 | |
| Restraint Statistics | ||||
| RMSD from standard geometry |
||||
| Bonds(Å) | 0.004(0.001) | 0.004(0.001) | 0.005(0.001) | |
| Angles(deg) | 0.479(0.050) | 0.577(0.028) | 0.580(0.040) | |
| Improper(deg) | 0.405(0.042) | 0.466(0.041) | 0.501(0.026) | |
| RMSD from experimental restraints |
||||
| Distance(Å) | 0.046(0.003) | 0.051(0.003) | 0.058(0.003) | |
| cdih(deg) | 0.383(0.125) | 0.547(0.271) | .691(0.211) | |
| Ramachandran Plot++ | ||||
| (percent of residues) | ||||
| Core | 73.7 | 76.8 | 70.2 | |
| Allowed | 20 | 21.4 | 25.2 | |
| Generously Allowed | 3.2 | 1.8 | 2.6 | |
| Disallowed | 3.2 | 0 | 2.0 | |
| Average RMSD (Å) | ||||
| CA,C,N | 0.60 | 0.40 | 0.68 | |
| not hydrogen |
1.25 | 1.03 | 1.28 | |
includes 59 intermolecular NOEs
Determined from the average structure
Figure 3.
(a) Ribbon representation of the Itk SH3/SH2 complex structure showing the canonical ligand binding sites on each domain. (left) The pY and pY+3 binding pockets on the Itk SH2 domain are circled and labeled. The sidechains that comprise the pY pocket are shown in red. The loop regions of the Itk SH2 domain that contact the Itk SH3 domain in the complex are labeled CD, BG and EF using standard SH2 domain nomenclature. (right) The view in (a) has been rotated 180 degrees and the conserved SH3 biding pocket is circled and labeled. The aromatic residues that make up the proline-binding site on the SH3 domain are shown in red. In both views the amino (N)- and carboxy (C) termini of each domain are labeled. (b) Surface rendering of the Itk SH3/SH2 complex shows the fit between SH3 and SH2 domains in the complex. The conformation of the cis CD loop in the SH2 domain (labeled in the mesh representation on the right) contributes to the curved surface (or hand-like shape) presented to the SH3 domain. The color scheme for each domain is the same as in Figure 2.
In the surface rendering of the SH3/SH2 complex (Figure 3b) it is evident that the larger SH2 domain clasps the edges of the SH3 domain binding groove. The CD and EF loops of the SH2 domain resemble the fingers and thumb, respectively of a hand clasping the ball shaped SH3 domain (Figure 3b). The BG loop lies in the central, back part of the interface in the view shown in Figure 3b. The binding interface between the Itk SH3 and SH2 domains has a total buried surface area of 1430 Å2 ± 80. This is larger than the buried surface area reported for the ultra weak protein-protein interaction described by Vaynberg et al. (~480 Å2) and is similar to the mean buried surface area of nine previously reported weak protein complexes (1386 Å2 ± 214) 46 as determined by the protein-protein interface comparison server: PROTORP 47.
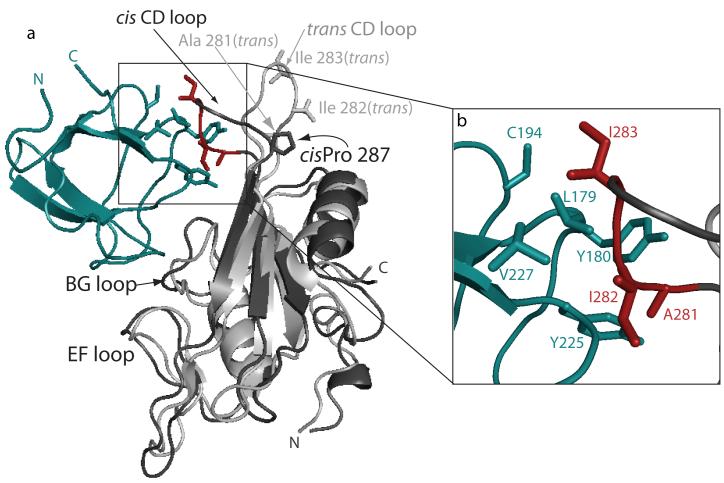
At the outset of this work, we wished to understand why the cis imide bond containing SH2 conformer exhibits a greater affinity for the Itk SH3 domain than does the trans conformer 29. Interestingly, the structure of the SH3/SH2 complex shows that Pro 287 itself does not make direct contact with the Itk SH3 domain (Figure 4a). The structural differences between the cis and trans conformers of the unbound Itk SH2 domain are largest in the CD loop and backbone 15N relaxation data for the cis and trans conformers showed that the CD loop in the cis conformer is more rigid than that in the trans conformer 28. The CD loop in the cis imide bond containing SH2 conformer appears pre-organized to make several contacts to a hydrophobic patch on the SH3 domain. Three hydrophobic side chains on the SH2 domain CD loop, A281, I282 and I283, pack into a hydrophobic cluster on the SH3 domain surface comprised of the conserved aromatic residues, Y180 and Y225, as well as L179, C194 and V227 (Figure 4b). Superposition of the trans imide bond containing SH2 conformer with the corresponding cis SH2 domain in the SH3/SH2 complex demonstrates how these contacts are lost on isomerization of the 286-287 imide bond to the trans configuration. The different CD loop conformation in the trans SH2 domain results in a decrease of approximately 25% of the total buried surface area of the SH3/SH2 complex and concomitant loss of hydrophobic contacts between the SH2 and SH3 domains; the side chains of A281, I282 and I283 are solvent exposed and considerably more flexible in the trans conformer28 (Figure 4).
Figure 4.
Cis/trans isomerization of the Itk SH2 domain affects contacts between SH2 and SH3 domains. (a) Ribbon representation of the complex between the Itk SH3 domain (teal) and Itk cis SH2 domain (darker gray). The structure of the trans imide bond containing SH2 conformer (light gray) is superimposed with the cis SH2 domain of the complex. The EF, BG and CD loops of the SH2 domain are labeled as is the cis Pro 287. Three residues of the CD loop (A281, I282 and I283) are shown in red (and labeled in (b)). For the trans CD loop, the same three side chains are shown in gray and labeled. (b) Expanded view of boxed region in (a) showing interface contacts between the CD loop of the cis SH2 domain and sidechains on the SH3 domain surface. SH3 domain side chains are teal and SH2 domain sidechains are red.
Of the SH2 residues at the SH3/SH2 interface, the CD loop residues (A281, I282 and I283) lie farthest from the canonical phospholigand binding pockets of the SH2 domain, make extensive contacts to the SH3 domain across the interface, and as previously reported, I282 in particular serves to stabilize the cis SH2 domain28. For these reasons we explored whether a mutation in this region of the Itk SH2 domain might effectively abolish the intermolecular SH3/SH2 interaction without adversely affecting phospholigand binding. Using the singly expressed domains, we find that the mutant Itk SH2(I282A) domain causes no chemical shift perturbations when titrated into an 15N labeled sample of Itk SH3 (data not shown) indicating no detectable interaction between SH3 and SH2(I282A). This result was then followed by incorporation of the I282A mutation into the Itk dual domain fragment (SH3-SH2(I282A)) and examination by native gel electrophoresis (Fig. 1a, lane 4). Unlike the wild type SH3-SH2 dual domain (Fig. 1a, lane 2), the single I282A point mutant in the Itk SH3/SH2 dual domain abolishes intermolecular self-association giving rise to a single band on the native gel.
For the I282A mutation to be a useful tool in disrupting Itk self-association for further functional studies, it must have a negligible effect on the phospholigand binding function of the Itk SH2 domain. We therefore measured the binding affinity of the Itk SH2(I282A) mutant to a classical phosphotyrosine containing ligand. Titration of a phosphopeptide derived from Slp76 (Ac-ADpYEPP-NH2) into 15N labeled SH2(I282A) was carried out to determine the dissociation constant for the mutant SH2/phosphopeptide interaction as previously described29. Unexpectedly, we found that mutation of isoleucine 282 to alanine in the SH2 domain caused a decrease in phosphopeptide binding affinity. The Kd for phosphopeptide binding to the SH2(I282A) mutant is 1.0 ± 0.2 mM compared to a Kd of 0.25 ± 0.2 mM for binding of the same phosphopeptide to the wild type SH2 domain 29.
The structure of the Itk SH2 domain bound to this phosphopeptide ligand has been solved previously 26 and Ile 282 is, as mentioned above, located far from the peptide binding site. We presume therefore that altered domain dynamics rather than loss of direct contacts to the ligand might be the source of the observed affinity loss. Whatever the cause, this result underscores an important conceptual point regarding functional probing of Itk self-association. Targeted functional experiments to elucidate the significance of full length Itk self-association will require a minimal set of mutations that selectively alter/abolish intermolecular Itk self-association but have no measurable effect on the classical ligand binding activities of each domain. Such mutations would allow for activity assays or in vivo experiments using Itk variants that do not exhibit intermolecular clustering but retain all other functions, in particular the protein-protein interactions mediated by the various domains of Itk during signaling. Thus, while the I282A mutation appears to abolish intermolecular self-association of the dual domain SH3-SH2 fragment (Fig. 1a), its negative effect on classical phospholigand binding means that this mutation cannot be incorporated into full length Itk to study self-association in a physiological context without also adversely affecting other signaling functions. The structure of the Itk SH3/SH2 complex solved here must therefore be probed more extensively to identify appropriately selective mutational tools for biochemical and cell biological experiments aimed at elucidating the functional significance of full length Itk self-association.
Comparison of Itk SH3/SH2 and the SH3/proline-rich ligand interfaces
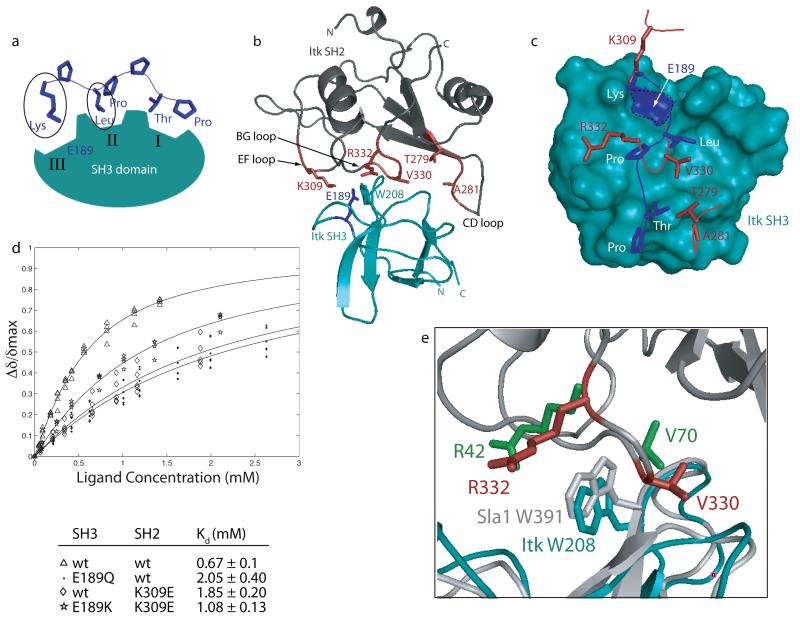
In addition to hydrophobic contacts between the CD loop of the cis SH2 domain at one end of the SH3 binding cleft, the structure of the Itk SH3/SH2 complex also reveals some similarities to classical SH3 mediated proline-rich ligand binding. The well-characterized SH3 binding surface consists of three shallow pockets (denoted I, II and III) that typically contact three distinct regions of a proline-rich peptide ligand 6. SH3 binding clefts I and II contact the two aliphatic-proline dipeptide units of the classical proline-rich ligand. In binding groove III, a conserved acidic site (E189 in Itk SH3) forms a salt bridge with a basic residue on the peptide ligand (Figure 5a).
Figure 5.
Comparing the Itk SH2 ligand with the classical proline-rich ligand for SH3 domains. (a) Schematic of classical SH3/ligand interaction. Proline-rich ligands (such as the KPLPPTP sequence shown here) make contacts to three distinct binding clefts on the SH3 domain, labeled I, II and III. The lysine and leucine ligand residues are circled since they appear to be most closely mimicked by residues of the Itk SH2 domain. (b) Structure of the Itk SH3/SH2 complex highlighting interface residues in the EF, BG and CD loops of the SH2 domain (side chains labeled and shown in red). Also shown in this structure are the sidechains of two highly conserved SH3 residues, W208 and E189, which contact both proline-rich ligands as well as the bound SH2 domain in this structure. (c) Structural comparison of the SH2 ligand and a proline-rich ligand bound to the Itk SH3 domain. The view in (b) is rotated to view straight into the SH3 binding surface and then the SH2 domain is removed for clarity leaving only five SH2 residues at the interface (K309, R332, V330, T279 and A281 in red). In addition, the structure of the Itk SH3 domain bound to a canonical ligand (pdbcode 1AWJ) is superimposed with the Itk SH3 domain in the SH3/SH2 complex. For clarity the Itk SH3 domain in the proline/SH3 structure is not shown and the proline-rich ligand is depicted in blue with side chains labeled: Lys, Leu, Pro, Thr, Pro. The conserved E189 is outlined with a dashed line and highlighted in blue on the teal surface of the Itk SH3 domain. (d) Increasing concentration of unlabeled Itk SH2 domain was titrated into 15N labeled Itk SH3 domain. 1H-15N HSQC spectra were obtained after addition of each aliquot of SH2 ligand. Binding curves were generated by plotting the normalized concentration dependence of amide chemical shifts for several different residues. Dissociation constants (Kd) were derived from binding curves shown as described in Methods. The pairs of SH3 and SH2 domains for each titration are: Δ wild type SH3/wild type SH2; · SH3 (E189Q)/wild type SH2; ◇ wild type SH3/SH2(K309E); SH3(E189K)/SH2(K309E). (e) Comparison of the Itk SH3/SH2 interface with the Sla-1 SH3/ubiquitin interface. Overlay of the Itk SH3(teal)/SH2 (dark gray) complex and the complex of Sla-1 SH3 (white)/ubiquitin complex. After alignment of only the SH3 portions of each complex, residues R332 and V330 of Itk SH2 (red) correspond very well with residues R42 and V70, respectively of ubiquitin (green) despite the very different tertiary structures of the Itk SH2 domain and ubiquitin. The conserved tryptophan in each SH3 domain are shown (Itk SH3 is teal, Sla1 SH3 is gray) and labeled.
Side chains on the BG and EF loops of the Itk SH2 domain point into two of the Itk SH3 binding clefts that are separated by W208 (Figure 5b). Contacts in the complex structure between Itk SH2 and the Itk SH3 domain in this region are consistent with earlier analysis showing the importance of the W208 side chain. Mutation of W208 in the Itk SH3 domain abolishes both proline ligand binding 25 and the interaction with the Itk SH2 domain 27. To compare the side chain contacts between SH3 and SH2 domain to classical proline-rich ligand recognition by the SH3 domain, the SH3 domain of the Itk SH3/SH2 complex is superimposed with the Itk SH3/peptide ligand complex solved previously25. To visualize the interface more clearly, the Itk SH2 domain is removed with the exception of the EF, BG and CD loop residues (K309, R332, V330, T279 and A281) that point toward the Itk SH3 domain. Using this superposition, the similarities and differences between the proline-rich ligand (KPLPPTP) and the SH2 domain ligand for the Itk SH3 domain are illustrated in Figure 5c.
The most notable similarity is the correspondence between SH2 residue K309 in the EF loop and the lysine residue of the proline-rich peptide ligand that contacts binding pocket III. In binding pocket II, V330 in the SH2 BG loop coincides in space to the similarly large, hydrophobic leucine of the proline-rich peptide. In a departure from the proline-rich peptide sequence, the other SH2 residue on the BG loop that makes contact to binding pocket II is not proline, as found in a classical proline-rich ligand, but rather R332. The long arginine sidechain appears to extend along the surface of the SH3 domain with the guanidinium headgroup of R332 possibly forming a hydrogen bond with the side chain amide group of Asn 185 on the SH3 domain. Indeed, the dissociation constant measured for the Itk SH2(R332A) mutant and wild type SH3 domain is 1.8 mM, three fold weaker than the wild type interaction. In binding pocket I, the SH2 ligand deviates from the proline-rich peptide as the CD loop (including residues T279 and A281) veers slightly out of the well-defined SH3 cleft to participate in the hydrophobic contacts depicted in Figure 4. Nevertheless, it is interesting to note that the SH2 ligand presents a threonine (T279) at a position that might mimic the threonine of the peptide ligand that contacts cleft I (Fig. 5c).
Since the electrostatic interaction between a basic residue of the prototypical proline-rich peptide and the conserved acidic site on the SH3 surface (E189 in Itk SH3) plays an established role in mediating proline ligand binding 48; 49; 50, we tested the contribution to binding affinity of both E189 and the basic side chain, K309 of SH2, that appears to mimic the canonical peptide. Curve fitting analysis of NMR data 34 obtained from the titration of unlabeled Itk SH2 domain into 15N labeled Itk SH3 domain was carried out to measure the affinities of several SH3 and SH2 mutants (Figure 5d). Three different combinations of mutants were tested and compared to the wild type SH3/SH2 interaction (Kd = 0.67 mM). First, E189 in the SH3 domain was mutated to glutamine (SH3(E189Q)) to test the putative role of this conserved negative charge on the SH3 domain surface. Consistent with the structure of the SH3/SH2 complex, we find that the affinity of the Itk SH2 domain for the SH3(E189Q) mutant drops significantly (Kd = 2.05 mM). Likewise, mutation of K309 in the SH2 domain, predicted from the structure to interact with E189, results in a similar loss of binding affinity for the wild type SH3 domain (Kd = 1.85 mM). Based on these data, we tested the interaction further by introducing a positive charge at position 189 in the SH3 domain (SH3(E189K)) and a corresponding negative charge at position 309 in the SH2 domain (SH2(K309E)). The complex formed by these swapped mutants exhibits a tighter binding affinity (Kd = 1.08 mM) than either of the single mutants alone. While the affinity does not reach wild type levels, the fact that these mutations improve binding compared to each of the single mutants is consistent with there being a stabilizing electrostatic interaction between E189 and K309 as suggested by the SH3/SH2 complex structure (Figure 5 c,d).
Of particular interest with respect to contacts between SH2 BG loop residues (R332 and V330) and cleft II of the Itk SH3 domain, are the striking similarities in this region with another SH3/protein complex. The Sla-1 SH3 domain binds ubiquitin and the structure of this protein-protein complex has been solved (pdbcode 2JT4)44. Superposition of the Itk and Sla1 SH3 domains shows that despite the completely different folds of their target protein ligands (Itk SH2 versus ubiquitin) side chain contacts between protein ligand (either Itk SH2 or ubiquitin) and SH3 domain binding site (either Itk SH3 or Sla1 SH3) are preserved (Figure 5e). The position and orientation of Itk SH2 residues R332 and V330 correspond quite well to ubiquitin side chains R42 and V70. This region of similarity between these two SH3 mediated protein complexes suggests some generality in the manner by which SH3 domains recognize their non-proline rich protein targets.
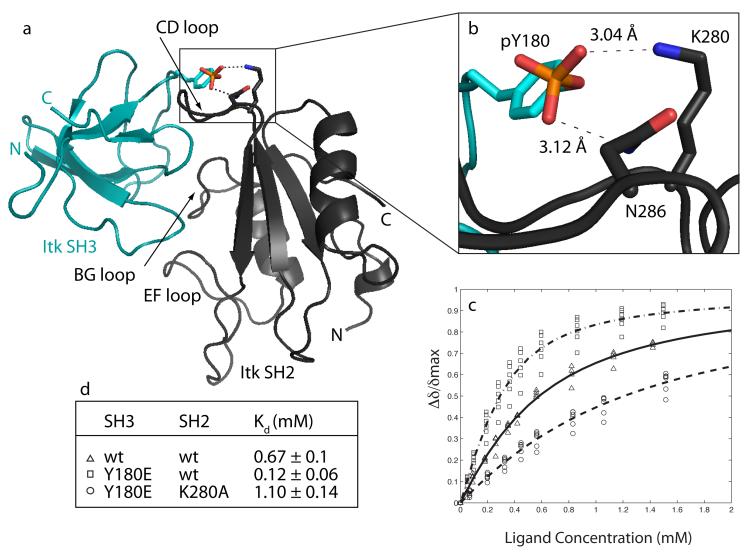
The Itk SH3/SH2 complex structure also provides a structural interpretation for previously reported mutational data51. Following T cell activation, Itk autophosphorylates Y180 in its own SH3 domain creating a highly negatively charged patch on the domain’s surface1. From the structure reported here, we can see that the Y180 side chain points directly at the SH2 domain in the binary complex and in particular wraps around the backside of the CD loop (Figure 4b). In earlier mutational analysis, we found that mutation of Y180 to glutamate, used to mimic a phosphotyrosine, leads to a six-fold increase in the affinity of the SH3 domain for Itk SH251. This result suggests that introducing a negative charge at this position of the SH3 domain stabilizes the SH3/SH2 interaction. We therefore modeled a phosphate group onto Y180 in the SH3 domain and find that in the Itk SH3/SH2 complex, the side chains of two basic SH2 residues, K280 and N286, lie within reasonable donor/acceptor distance from the phosphate oxygens on pY180 (Figure 6). We tested this model by further mutagenesis.
Figure 6.
Autophosphorylation of Y180 in the structural context of the Itk SH3/SH2 complex. (a) Structure of the Itk SH3/SH2 complex labeled as before. Y180 in the SH3 domain is shown with a phosphate group modeled as substituting at the hydroxyl group. (b) Enlarged view of boxed area in (a). Distances in the model between the phosphate oxygens and the side chain nitrogens of K280 and N286 are shown. (c & d) NMR titrations to determine dissociation constants as described in Figure 5d. The pairs of SH3 and SH2 domains for each titration are: Δ wild type SH3/wild type SH2; □ SH3(Y180E)/wild type SH2; ◯ SH3(Y180E)/SH2(K280A).
Based on the model shown in Figure 6, mutation of either K280 or N286 in the SH2 domain to alanine should reduce the affinity of the Y180E SH3 domain for the SH2 domain by removing potential hydrogen bond partners. Since the relative population of cis and trans SH2 domain also affects the affinity for the Itk SH3 domain29, we first examined the cis/trans ratio of the mutant and wild type Itk SH2 domains. Cis/trans ratio can be readily measured by comparing peak volume for cis versus trans crosspeaks in an HSQC spectrum of the SH2 domain28. The population of cis and trans wild type Itk SH2 domain is 40% and 60%, respectively. For the K280A mutant we find no change in the cis/trans ratio compared to wild type but the N286A SH2 mutant exhibits a reduction in the cis population to 17% cis and 83% trans (data not shown). We cannot test the N286A SH2 mutant for binding to SH3 (Y180E) without introducing another variable, namely reduction of cis population in the SH2 equilibrium ensemble and associated loss of affinity for SH3 by that virtue alone29. We therefore proceeded with the more straightforward K280A SH2 mutant and tested binding to the Itk SH3 (Y180E) phosphotyrosine mimic.
Binding affinity of the K280A SH2 mutant for the Itk SH3 Y180E mutant was measured as before34; 51 by titration of unlabeled Itk SH2 (K280A) mutant into 15N labeled Itk SH3 (Y180E) (Figure 6c). Consistent with a putative interaction between K280 and a negatively charged residue at position 180 in the SH3 domain (Figure 6b), loss of the basic lysine sidechain in the Itk SH2 domain results in a loss of binding affinity to the phosphomimic, Itk SH3(Y180E) (Figure 6 c&d). In fact, the affinity of SH3(Y180E) for the Itk SH2 domain drops ten fold upon mutation of K280 and is below that of the interaction between the wild type SH3 and SH2 domains (Figure 6d). The titration data and the structure of the Itk SH3/SH2 complex therefore support the idea that autophosphorylation within the Itk SH3 domain stabilizes the self-associated form of this protein by enhancing the affinity of the intermolecular SH3/SH2 interaction. These findings certainly need to be pursued in the context of the phosphorylated Itk SH3 domain, but suggest that pY180 might interact with K280 alone or with both K280 and N286 across the SH3/SH2 interface.
Discussion
The Itk SH3 and SH2 domains have been implicated in a number of different interactions 25; 27; 51; 52; 53; 54; 55; 56. Here we report the three dimensional structure of a direct intermolecular interaction between these two Itk regulatory domains. The structure of the SH3/SH2 complex reveals precisely how the cis prolyl imide bond at position 287 in the SH2 domain pre-organizes the large CD loop to contact the SH3 domain (Figure 4) and the structure suggests specific inter-domain contacts that might stabilize the SH3/SH2 interaction following autophosphorylation at Y180. As well, we have discerned several other specific contacts across the interface that contribute to binding. Comparison with other structures, such as classical proline ligand recognition and the Sla1/ubiquitin complex, suggests generalities in SH3 mediated protein interactions (Figure 5).
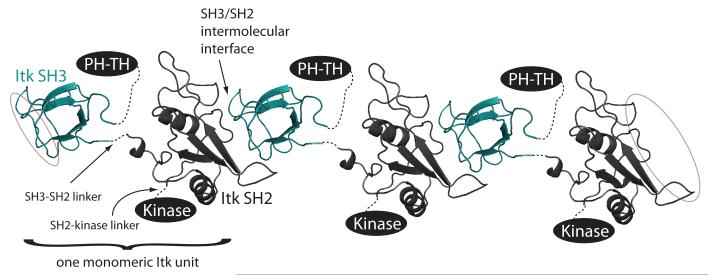
The structure of the binary SH3/SH2 complex, in conjunction with native gel electrophoresis, allows us to begin to visualize how larger fragments and even full length Itk might interact with itself intermolecularly. The Itk SH3-SH2 dual domain fragment does not form simple dimers but rather is a heterogeneous mixture of oligomers. This observation is consistent with the structure of the binary SH3/SH2 complex solved here. Grafting an SH3 domain onto the amino-terminus of the SH2 domain and likewise extending the carboxy-terminus of the SH3 domain to include the SH2 domain, suggests that a head-to-tail dimer arrangement of the SH3-SH2 dual domain fragment would not be sterically feasible (Fig. 7). Instead, the model of the dual domain that emerges from simply extending the binary SH3/SH2 structure suggests that each Src homology domain interacts with a target domain from a different Itk molecule (Fig. 7). This is consistent with the formation of different sized SH3-SH2 oligomers, or polymerization, as is evident on the native gel (Figure 1a).
Figure 7.
Model of Itk oligomerization. Two copies of the binary SH3/SH2 complex structure are shown using the same color scheme as in other figures. The amino-terminus of each SH2 domain is then connected (using a dotted line) to the carboxy-terminus of the Itk SH3 domain (labeled SH3-SH2 linker), the carboxy-terminus of each SH2 domain is extended toward the Kinase domain (labeled SH2-Kinase linker) and the amino-terminus of each SH3 domain is extended toward the PH-TH domains. Thus, one monomeric unit of Itk consists of PH-TH-SH3-SH2-Kinase. Three such Itk units are shown and the interaction interface between SH3 and SH2 domain is labeled. The SH2 and SH3 binding sites of the SH3 and SH2 domains, respectively, at each end of the oligomer are circled to suggest further interactions are possible.
Extending the model slightly we can place the other Itk domains (PH-TH and Kinase) onto this dual domain model based on the location of the amino-terminus of the SH3 domain and the carboxy-terminus of the SH2 domain. Albeit an incomplete picture of full length Itk at this point, it seems noteworthy that the PH domains of different Itk monomers could extend in the same direction from the N-termini of the SH3 domains in the context of the self-associated species. This is important since Itk clustering has thus far been observed in vivo at the membrane (rather than in the cytosol)37 and so the PH domain of each monomeric unit of the oligomer must be able to extend toward the membrane surface to engage its ligand. As well, PH domain intermolecular self-association has been reported30 and thus, the arrangement of the PH domain protruding on the same side of the SH3 domain in each Itk molecule would allow for PH/PH domain interactions.
The structure of the Itk SH3/SH2 complex does not by itself provide direct insight into the functional significance of Itk oligomerization. As already discussed, it does provide a valuable tool to begin to design targeted mutations that disrupt self-association in full length Itk for the purpose of understanding how these intermolecular interactions modulate Itk signaling. To this end, mutations must be identified that abolish the SH3/SH2 intermolecular interaction while maintaining classical SH3 and SH2 ligand binding properties. It is also likely that this approach will need to be extended to the other Itk domains (such as PH) that contribute to intermolecular self-association. Indeed, native gel electrophoresis suggests that disrupting the SH3/SH2 interaction (I282A mutation) in the context of full length Itk does not fully abolish Itk self-association (data not shown) and this is consistent with reports of the Itk PH domain interacting with itself in an intermolecular fashion 30.
In addition to setting the stage for targeted mutations to assess the functional outcome of Itk clustering in T cells, the structure reported here provides yet another example of the broader binding functions of the ubiquitous SH3 and SH2 domains. Add the Itk system to the many examples that have now been characterized demonstrating alternative SH3 and SH2 mediated binding 10; 11; 12; 13; 18; 19; 20; 57; 58; 59; 60; 61; 62 and we see that our original notions of simple target sequences for these ubiquitous Src homology domains, while correct, do not represent the complete story.
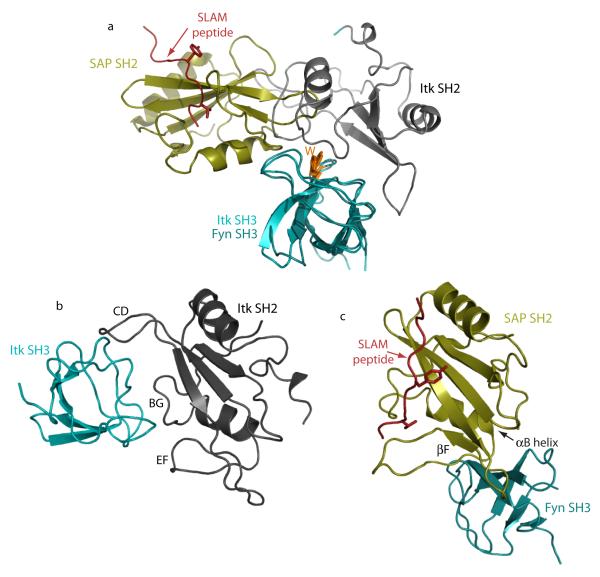
One comparison in particular stands out. A well-studied example of a direct regulatory interaction between an SH3 and SH2 domain is that of the SAP SH2 and Fyn SH3 domains mediating formation of the SLAM immune receptor complex10; 16. Like the Itk SH3/SH2 complex reported here, the SAP SH2/Fyn SH3 complex involves neither a proline-rich sequence nor a phosphotyrosine motif. In a manner similar to the Itk SH3/SH2 complex, the surface of the Fyn SH3 domain that binds SAP SH2 overlaps with the classical proline ligand-binding site. The SH3 interaction interfaces are not exactly the same; the Itk SH2 domain overlaps to a greater extent the canonical proline-binding site on its partner SH3 domain than does the SAP SH2 when binding to Fyn SH3 (Fig. 8a). The manner by which each SH2 binding partner is engaged by the Itk and Fyn SH3 domains is completely different (Fig. 8b,c). As discussed in detail above, the Itk SH3 domain contacts the CD, BG and EF loops of its cognate SH2 domain. In contrast, the Fyn SH3 domain contacts the βF strand, the base of the B helix and the loop connecting these two regions of secondary structure within the SAP SH2 domain. Thus, not only do SH3 and SH2 domains participate in a diverse set of non-classical interactions, even when these binding domains interact directly with each other there appears to be a significant degree of diversity.
Figure 8.
Comparison of Itk SH3/SH2 structure and Fyn SH3/SAP SH2 structure (pdbcode 1M27). (a) For both complex structures, the Itk SH3 domain and the Fyn SH3 domain (both shown in teal) are superimposed. The conserved tryptophan in the SH3 domain binding cleft is shown for both Fyn and Itk and labeled W. With superposition of the SH3 domains, it is clear that the bound SH2 domains do not contact precisely the same region of their cognate SH3 domain but both bind in a manner that would overlap with classical proline ligand binding. The Itk SH2 domain is gray and the SAP SH2 domain is gold. The SLAM derived phosphopeptide bound to the SAP SH2 domain is depicted in red. (b & c) The Itk SH3/SH2 complex (b) and the Fyn SH3/SAP SH2 complex (c) are shown separately with the SH2 domains in exactly the same orientation. The respective SH3 domains (shown in teal) interact with distinct regions of the different SH2 domains (SH2 secondary structural regions and loops involved in each complex are labeled). The SLAM peptide bound to the SAP SH2 domain is shown in red. It is clear in this comparison that the bound Itk SH3 domain partially occludes the classical ligand-binding site of the Itk SH2 domain.
The functional role of each of these interactions must be fully understood to appreciate the reasons for the observed diversity. For the Fyn/SAP complex, it has been clearly established that the interaction serves to activate the Fyn kinase while co-localizing Fyn to the SAP/SLAM complex10; 16. Thus, the mechanistic picture in this case is consistent with the fact that phospholigand binding to the SAP SH2 domain does not compete with the interaction between SAP SH2 and Fyn SH3. For the Itk SH3/SH2 interaction, phospholigand binding to the Itk SH2 domain is instead mutually exclusive with binding of Itk SH3 to the SH2 domain27. Hence, it seems likely that the functional significance of the intermolecular Itk interaction might coincide with states of the enzyme that do not require phospholigand association (i.e. not the activated state63). The structure of the binary Itk SH3/SH2 complex reported here is a first step toward deciphering precisely if and how intermolecular Itk association regulates activity and localization during T cell signaling.
Materials and Methods
NMR Sample Preparation
Protein expression and purification techniques were performed as described 27. NMR samples consisted of the free domain, 35% bound, and 77% bound for both labeled Itk SH2 and Itk SH3 domains (Table 1) in 50 mM NaPO4, 75 mM NaCl, 2 mM dithiothreitol (DTT), 5% D2O, and 0.02% (w/v) NaN3 at pH 7.4. The extinction coefficient (ε) at 280 nm and isoelectric point (pI) were calculated using ExPASy (Expert Protein Analysis System) for the Itk SH3 (pI=4.3, ε= 19940 mM-1 cm-1) and Itk SH2 (pI = 8.8, ε= 20400 mM-1 cm-1) domains.
NMR Spectroscopy
All NMR spectra were collected at 298K on a Bruker AVII 700 spectrometer equipped with a 5mm HCN z-gradient cryoprobe operating at a 1H frequency of 700.13 MHz. Chemical shift assignments and NOE correlations for the free and bound Itk SH3/Itk SH2 structures were analyzed using CARA 64 as previously reported 40. Chemical shift assignments were elucidated for all structures from double and triple resonance experiments: CBCA(CO)NH, HNCACB, HBHA(CO)NH, HBHANH, HNCO, (HB)CB(CGCDCE)HE, (HB)CB(CGCD)HD, and HCCH-TOCSY, along with a 3D 15N-edited TOCSY and 2D homonuclear TOCSY. Data were collected and independent assignments were made for the 35% and 77% bound samples (Table 1) since there were chemical shift variations among these samples due to fast exchange. NOE correlations were obtained from a 2D homonuclear NOESY, 3D 13C-edited aliphatic NOESY, 3D 13C-edited aromatic NOESY, and 3D 15N-edited NOESY spectra. All NOESY experiments were acquired with a mixing time of 100 ms. NOEs from each NOESY spectrum were binned into four categories: very weak, weak, medium, and strong with a total percent NOE restraint ratio of 20:30:30:20. The corresponding structural restraints have bounds of 1.8-6.0Å, 1.8-5.0Å, 1.8-3.3Å (3.5Å for amide groups), and 1.8-2.5Å (2.7Å for amide groups), respectively. To account for the higher intensity of NOEs involving methyl groups 0.5Å was added to each of these restraints with an upper bound no greater than the very weak upper bound limit of 6.0Å. IPAP 1H-15N correlation experiments and 3D HNCA E-COSY 15N-13C experiments were performed to measure residual dipolar coupling constants. Weak anisotropic alignment was achieved through the addition of 8 mg mL-1 of Pf1 phage (ASLA BIOTECH Ltd) 65; 66. The quality of all structures was evaluated using PROCHECK_NMR 67 and WHATCHECK 68.
Unbound Structure Calculations
The unbound Itk SH3 structure used for comparison to the bound complex was recently reported 40. The previously solved structure of the unbound cis imide bond containing SH2 conformer 28 was refined with the addition of RDC restraints from IPAP 1H-15N correlation experiments. RDC data from 3D HNCA E-COSY 15N-13C experiments could not be measured for the SH2 domain in both unbound and bound samples due to line broadening from exchange. Previously reported NOE restraints were verified and NOE restraints were generated in CARA from the newly acquired data. Hydrogen bond and torsion angle restraints from the previous unbound cis SH2 domain structure calculation were used unmodified. The refined unbound cis conformer of the Itk SH2 domain was calculated using simulated annealing in XPLOR-NIH with NOE, hydrogen bond, RDC, and torsion angle restraints. Ramachandran plot statistics in the core, allowed, generously allowed, and disallowed regions for residues in the average structure are 77.9%, 12.6%, 7.4%, and 2.1% respectively. The backbone (N, Ca, C’) and heavy atom RMSD is 0.89Å and 1.57Å respectively.
Individual Bound Structure Calculation
The individual bound structures of Itk SH2 and Itk SH3 were calculated independently using NOE, RDC, and torsion angle restraints. Initial estimates of alignment tensor terms Da (axial symmetry) and R (Rhombicity) for the bound structures were calculated via SVD (singular value decomposition) using the average minimized unbound structures of Itk SH2 and Itk SH3. The Da and R values of -11.00 and 0.3 respectively used in the bound structure calculations of SH2 and SH3 were calculated by a grid search of tensor terms around the initial estimate determined by SVD. Dihedral angle (φ, ψ) restraints were obtained from a large structural database using PREDITOR69. Chemical shifts from the more completely assigned 35% bound data set were used as input to PREDITOR. Bounds of ± 60° were chosen to allow for variability in calculated torsion angle restraints that may arise in chemical shift differences of the fully bound assignments and the 35% bound assignments.
Itk SH3/SH2 Complex Structure Calculation
The binary complex was determined from the bound domain structures in XPLOR-NIH using conjoined rigid body/torsion angle dynamics based on 59 intermolecular NOE distance constraints (depicted on the complex structure in Figure 2S; Supplementary Material) and 58 DNH RDC constraints. Intermolecular NOEs were obtained by comparing 3D 13C-edited aliphatic NOESY, 3D 13C-edited aromatic NOESY, and 3D 15N-edited NOESY spectra of bound and unbound samples. The intermolecular NOEs were manually assigned and binned. Distant restraints associated with the strong, medium and weak NOES were generated with upper bounds of 3.5, 4.7 and 5.5 Å, respectively. In this calculation, the bound structures are held rigid except for residues found at the interface. Interfacial residues were determined as those exhibiting all three of the following criteria: (a) significant chemical shift perturbation upon binding, (b) solvent exposure and (c) forming a contiguous surface 38. Criteria (b) and (c) were assessed by visual inspection of a space-filling representations of the domain structures. For Itk SH3 residues 179-185, 188-189, 205-208, 220, 222-225, and 227-228 and for Itk SH2 residues 258-260, 278-292, 326-332 comprised the flexible interfacial residues. A lowest energy structure from the calculation was chosen as an initial structure for further refinement using simulated annealing. Ramachandran plot statistics in the core, allowed, generously allowed, and disallowed regions for residues in the average structure are 70.2%, 25.2%, 2.6%, and 2.0%, respectively. The average backbone RMSD (N, Ca, C’) is 0.68 Å and average heavy atom RMSD is 1.28 Å (Table 1). The average buried interfacial surface area calculated from the twenty lowest energy structures of the SH3/SH2 binary complex was determined using NACCESS 70. The portion of the interaction surface that corresponds to the transient CD loop of the SH2 domain was calculated from the individual contributions of residues 279, 280, 281, 282, and 283 to the interaction surface.
Native Gel Analysis
Native gels were run as described previously71. Briefly, purified Itk wild-type or mutant SH3SH2 domain was mixed in a 1:1 ratio with native gel loading dye. The proteins were loaded on a 10 % discontinuous native gel and run at 180V with Tris Cl pH 8.8 as the running buffer. BSA was used as the standard. The gels were then stained with Coomassie stain.
Structural Comparison and Alignment
All alignments of unbound structures with the binary complex were performed in PyMol 72 between backbone atoms (C’, Cα, N) and residues 235-335 for the Itk SH2 domain and residues 173-229 for the Itk SH3, respectively. To ensure all backbone atoms are included, the align command was performed with zero cycles of refinement. Similar alignments were made between the Itk SH3 domain of the Itk SH3/SH2 complex with the Sla-1 SH3/ubiquitin complex and the SAP/Fyn complex.
To model pY180 in the Itk SH3 domain, the phosphotyrosine in the phosphopeptide ligand of Itk SH2/pY ligand complex structure was aligned with Y180 in the Itk SH3/SH2 complex structure. This aligned phosphotyrosine residue was used to represent the pY180 phosphorylated form of the SH3/SH2 complex structure.
NMR Titrations
Point mutations for the titrations were generated using the Quick change Site-Directed Mutagenesis kit (Stratagene). NMR titrations were carried out as described 29; 33; 34 where unlabeled protein was incrementally added to a 400 μM 15N-labeled protein sample. After each addition, the changes in chemical shift of resonances in a 1H-15N HSQC were recorded. Chemical shift perturbations were determined significant if the change in the position of a crosspeak was greater than the average of all crosspeak change plus one standard deviation. The 1H and 15N chemical shift difference were combined into an average chemical shift using the equation:
Dissociation constants (Kd) were determined using an in-house program written in Matlab (version 5.3.1, The Mathworks, Inc) that fits the following equation:
where Δδis the chemical shift difference, [P]0 is initial protein concentration, [L]0 is initial ligand concentration and Ka (= 1/Kd) is the association constant. Parameters Ka and δmax are both fitted in the analysis.
Percent Cis/Trans Calculation
Cis/trans isomerization of the Asn 286-Pro 287 imide bond is in the slow exchange NMR regime resulting in two distinct cross-peak resonances for approximately a third of the residues in a 1H/15N HSQC of the Itk SH2 domain. The ratio of cis/trans conformers for wild type Itk SH2, SH2(K309E), and SH2(K280A) was calculated from well resolved cross-peaks of the following residues: 256, 258, 260, 278, and 290. The fraction of SH2 domain that adopts the cis conformer expressed as percent is calculated by taking the intensity of a crosspeak that corresponds to the cis conformer, dividing by the sum of the crosspeak intensities of the cis and trans conformers, and multiplying by 100. The percent of SH2 domain containing the trans imide bond conformation is calculated in the same way.
Accession Codes
The RCSB PDB accession codes for 20 lowest energy structure ensemble and average minimized structure are 2K7A and 2K79, respectively. The BMRB accession number is 15912.
Supplementary Material
Figure 1S: Similarities between free and bound Src hmolgy domains are shown in the superposition of the structures of the isolated Itk SH3 and cis SH2 domains (shown in light grey) with the corresponding domains in the Itk SH3/SH2 complex.
Figure 2S: Superposition of the Itk SH3/SH2 complex depicting the 58 intermolecular NOE restraints used in the NMR structure calculation. The SH3 and SH2 domains are colored in cyan and gray, respectively. The intermolecular NOE restraints are primarily between the SH3 binding pocket and the three loops of the Itk SH2 domain (CD, BG and EF). The red and cyan lines are NOEs involving SH2 CD loop residues, magenta involves residues of the BG loop, orange indicates NOEs to the end of the βD strand, and green are NOEs to the EF loop. Side chains giving rise to NOEs are depicted but hydrogens are not included for clarity.
Acknowledgements
The authors would like to thank Dr. Robert J. Mallis for providing invaluable encouragement and guidance in the NMR structure determination process. Melissa Mayo and Patrick Breheny carried out preliminary work on SH2(I282A) mutant. This work is supported by a grant from the National Institutes of Health (National Institute of Allergy and Infectious Diseases, AI43957) to A.H.A and the generous support of a Roy J. Carver Charitable Trust training fellowship to A. Severin.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berg LJ, Finkelstein LD, Lucas JA, Schwartzberg PL. Tec Family Kinases in T Lymphocyte Development and Function. Annual Review of Immunology. 2005;23:549–600. doi: 10.1146/annurev.immunol.22.012703.104743. [DOI] [PubMed] [Google Scholar]
- 2.Dombroski D, Houghtling RA, Labno CM, Precht P, Takesono A, Caplen NJ, Billadeau DD, Wange RL, Burkhardt JK, Schwartzberg PL. Kinase-independent functions for Itk in TCR-induced regulation of Vav and the actin cytoskeleton. J Immunol. 2005;174:1385–92. doi: 10.4049/jimmunol.174.3.1385. [DOI] [PubMed] [Google Scholar]
- 3.Grasis JA, Browne CD, Tsoukas CD. Inducible T Cell Tyrosine Kinase Regulates Actin-Dependent Cytoskeletal Events Induced by the T Cell Antigen Receptor. The Journal of Immunology. 2003;170:3971–3976. doi: 10.4049/jimmunol.170.8.3971. [DOI] [PubMed] [Google Scholar]
- 4.Readinger JA, Schiralli GM, Jiang JK, Thomas CJ, August A, Henderson AJ, Schwartzberg PL. Selective targeting of ITK blocks multiple steps of HIV replication. Proc Natl Acad Sci USA. 2008;105:6684–9. doi: 10.1073/pnas.0709659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–52. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 6.Mayer BJ, Eck MJ. SH3 domains. Minding your p’s and q’s. Curr Biol. 1995;5:364–7. doi: 10.1016/s0960-9822(95)00073-x. [DOI] [PubMed] [Google Scholar]
- 7.Pawson T, Gish GD, Nash P. SH2 domains, interaction modules and cellular wiring. Trends Cell Biol. 2001;11:504–11. doi: 10.1016/s0962-8924(01)02154-7. [DOI] [PubMed] [Google Scholar]
- 8.Payne G, Stolz LA, Pei D, Band H, Shoelson SE, Walsh CT. The phosphopeptide-binding specificity of Src family SH2 domains. Chem Biol. 1994;1:99–105. doi: 10.1016/1074-5521(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 9.Barnett P, Bottger G, Klein AT, Tabak HF, Distel B. The peroxisomal membrane protein Pex13p shows a novel mode of SH3 interaction. EMBO J. 2000;19:6382–91. doi: 10.1093/emboj/19.23.6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan B, Lanyi A, Song HK, Griesbach J, Simarro-Grande M, Poy F, Howie D, Sumegi J, Terhorst C, Eck MJ. SAP couples Fyn to SLAM immune receptors. Nat Cell Biol. 2003;5:155–60. doi: 10.1038/ncb920. [DOI] [PubMed] [Google Scholar]
- 11.Cleghon V, Morrison DK. Raf-1 interacts with Fyn and Src in a non-phosphotyrosine-dependent manner. J Biol Chem. 1994;269:17749–55. [PubMed] [Google Scholar]
- 12.Harkiolaki M, Lewitzky M, Gilbert RJ, Jones EY, Bourette RP, Mouchiroud G, Sondermann H, Moarefi I, Feller SM. Structural basis for SH3 domain-mediated high-affinity binding between Mona/Gads and SLP-76. EMBO J. 2003;22:2571–82. doi: 10.1093/emboj/cdg258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia CY, Nie J, Wu C, Li C, Li SS. Novel Src homology 3 domain-binding motifs identified from proteomic screen of a Pro-rich region. Mol Cell Proteomics. 2005;4:1155–66. doi: 10.1074/mcp.M500108-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Kami K, Takeya R, Sumimoto H, Kohda D. Diverse recognition of non-PxxP peptide ligands by the SH3 domains from p67(phox), Grb2 and Pex13p. EMBO J. 2002;21:4268–76. doi: 10.1093/emboj/cdf428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang H, Freund C, Duke-Cohan JS, Musacchio A, Wagner G, Rudd CE. SH3 domain recognition of a proline-independent tyrosine-based RKxxYxxY motif in immune cell adaptor SKAP55. EMBO J. 2000;19:2889–99. doi: 10.1093/emboj/19.12.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Latour S, Roncagalli R, Chen R, Bakinowski M, Shi X, Schwartzberg PL, Davidson D, Veillette A. Binding of SAP SH2 domain to FynT SH3 domain reveals a novel mechanism of receptor signalling in immune regulation. Nat Cell Biol. 2003;5:149–54. doi: 10.1038/ncb919. [DOI] [PubMed] [Google Scholar]
- 17.Mayer BJ. SH3 domains: complexity in moderation. J Cell Sci. 2001;114:1253–63. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 18.Mongioví AM, Romano PR, Panni S, Mendoza M, Wong WT, Musacchio A, Cesareni G, Di Fiore PP. A novel peptide-SH3 interaction. EMBO J. 1999;18:5300–9. doi: 10.1093/emboj/18.19.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song YL, Peach ML, Roller PP, Qiu S, Wang S, Long YQ. Discovery of a novel nonphosphorylated pentapeptide motif displaying high affinity for Grb2-SH2 domain by the utilization of 3′-substituted tyrosine derivatives. J Med Chem. 2006;49:1585–96. doi: 10.1021/jm050910x. [DOI] [PubMed] [Google Scholar]
- 20.Tian L, Chen L, McClafferty H, Sailer CA, Ruth P, Knaus HG, Shipston MJ. A noncanonical SH3 domain binding motif links BK channels to the actin cytoskeleton via the SH3 adapter cortactin. FASEB J. 2006;20:2588–90. doi: 10.1096/fj.06-6152fje. [DOI] [PubMed] [Google Scholar]
- 21.Vaynberg J, Fukuda T, Chen K, Vinogradova O, Velyvis A, Tu Y, Ng L, Wu C, Qin J. Structure of an ultraweak protein-protein complex and its crucial role in regulation of cell morphology and motility. Mol Cell. 2005;17:513–23. doi: 10.1016/j.molcel.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 22.Hake MJ, Choowongkomon K, Kostenko O, Carlin CR, Sönnichsen FD. Specificity determinants of a novel Nck interaction with the juxtamembrane domain of the epidermal growth factor receptor. Biochemistry. 2008;47:3096–108. doi: 10.1021/bi701549a. [DOI] [PubMed] [Google Scholar]
- 23.Huang H, Li L, Wu C, Schibli D, Colwill K, Ma S, Li C, Roy P, Ho K, Songyang Z, Pawson T, Gao Y, Li SS. Defining the specificity space of the human SRC homology 2 domain. Mol Cell Proteomics. 2008;7:768–84. doi: 10.1074/mcp.M700312-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Li C, Schibli D, Li SS. The XLP syndrome protein SAP interacts with SH3 proteins to regulate T cell signaling and proliferation. Cell Signal. 2009;21:111–9. doi: 10.1016/j.cellsig.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 25.Andreotti AH, Bunnell SC, Feng S, Berg LJ, Schreiber SL. Regulatory intramolecular association in a tyrosine kinase of the Tec family. Nature. 1997;385:93–7. doi: 10.1038/385093a0. [DOI] [PubMed] [Google Scholar]
- 26.Pletneva EV, Sundd M, Fulton DB, Andreotti AH. Molecular details of Itk activation by prolyl isomerization and phospholigand binding: the NMR structure of the Itk SH2 domain bound to a phosphopeptide. J Mol Biol. 2006;357:550–61. doi: 10.1016/j.jmb.2005.12.073. [DOI] [PubMed] [Google Scholar]
- 27.Brazin KN, Fulton DB, Andreotti AH. A specific intermolecular association between the regulatory domains of a Tec family kinase. J Mol Biol. 2000;302:607–23. doi: 10.1006/jmbi.2000.4091. [DOI] [PubMed] [Google Scholar]
- 28.Mallis RJ, Brazin KN, Fulton DB, Andreotti AH. Structural characterization of a proline-driven conformational switch within the Itk SH 2 domain. Nature Structural Biology. 2002;9:900–905. doi: 10.1038/nsb864. [DOI] [PubMed] [Google Scholar]
- 29.Breheny PJ, Laederach A, Fulton DB, Andreotti AH. Ligand specificity modulated by prolyl imide bond Cis/Trans isomerization in the Itk SH2 domain: a quantitative NMR study. J Am Chem Soc. 2003;125:15706–7. doi: 10.1021/ja0375380. [DOI] [PubMed] [Google Scholar]
- 30.Huang YH, Grasis JA, Miller AT, Xu R, Soonthornvacharin S, Andreotti AH, Tsoukas CD, Cooke MP, Sauer K. Positive regulation of Itk PH domain function by soluble IP4. Science. 2007;316:886–9. doi: 10.1126/science.1138684. [DOI] [PubMed] [Google Scholar]
- 31.Hyvönen M, Saraste M. Structure of the PH domain and Btk motif from Bruton’s tyrosine kinase: molecular explanations for X-linked agammaglobulinaemia. EMBO J. 1997;16:3396–404. doi: 10.1093/emboj/16.12.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansson H, Mattsson PT, Allard P, Haapaniemi P, Vihinen M, Smith CI, Hard T. Solution structure of the SH3 domain from Bruton’s tyrosine kinase. Biochemistry. 1998;37:2912–24. doi: 10.1021/bi972409f. [DOI] [PubMed] [Google Scholar]
- 33.Laederach A, Cradic K, Brazin KN, Zamoon J, Fulton DB, Huang XY, Andreotti AH. Competing modes of self-association in the regulatory domains of Bruton’s tyrosine kinase: …. Protein Sci. 2002;11:36–57. doi: 10.1110/ps.26702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laederach A, Cradic KW, Fulton DB, Andreotti AH. Determinants of intra versus intermolecular self-association within the regulatory domains of Rlk and Itk. J Mol Biol. 2003;329:1011–20. doi: 10.1016/s0022-2836(03)00531-x. [DOI] [PubMed] [Google Scholar]
- 35.Pursglove SE, Mulhern TD, Mackay JP, Hinds MG, Booker GW. The solution structure and intramolecular associations of the Tec kinase SRC homology 3 domain. J Biol Chem. 2002;277:755–62. doi: 10.1074/jbc.M108318200. [DOI] [PubMed] [Google Scholar]
- 36.Colgan J, Asmal M, Neagu M, Yu B, Schneidkraut J, Lee Y, Sokolskaja E, Andreotti A, Luban J. Cyclophilin A regulates TCR signal strength in CD4+ T cells via a proline-directed conformational switch in Itk. Immunity. 2004;21:189–201. doi: 10.1016/j.immuni.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 37.Qi Q, Sahu N, August A. Tec kinase Itk forms membrane clusters specifically in the vicinity of recruiting receptors. J Biol Chem. 2006;281:38529–34. doi: 10.1074/jbc.M609180200. [DOI] [PubMed] [Google Scholar]
- 38.Clore GM, Schwieters CD. Docking of protein-protein complexes on the basis of highly ambiguous intermolecular distance restraints derived from 1H/15N chemical shift mapping and backbone 15N-1H residual dipolar couplings using conjoined rigid body/torsion angle dynamics. J Am Chem Soc. 2003;125:2902–12. doi: 10.1021/ja028893d. [DOI] [PubMed] [Google Scholar]
- 39.McCoy MA, Wyss DF. Structures of protein-protein complexes are docked using only NMR restraints from residual dipolar coupling and chemical shift perturbations. J Am Chem Soc. 2002;124:2104–5. doi: 10.1021/ja017242z. [DOI] [PubMed] [Google Scholar]
- 40.Severin A, Fulton DB, Andreotti AH. Murine Itk SH3 domain. J Biomol NMR. 2008;40:285–290. doi: 10.1007/s10858-008-9231-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ojennus D. Dahlke, Mitton-Fry RM, Wuttke DS. Induced alignment and measurement of dipolar couplings of an SH2 domain through direct binding with Pf1 phage. J Biomol NMR. 1999;14 doi: 10.1023/A:1008304332574. [DOI] [PubMed] [Google Scholar]
- 42.Lipsitz RS, Tjandra N. Residual dipolar couplings in NMR structure analysis. Annual review of biophysics and biomolecular structure. 2004;33:387–413. doi: 10.1146/annurev.biophys.33.110502.140306. [DOI] [PubMed] [Google Scholar]
- 43.Garrett DS, Seok YJ, Peterkofsky A, Gronenborn AM, Clore GM. Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr. Nat Struct Biol. 1999;6:166–73. doi: 10.1038/5854. [DOI] [PubMed] [Google Scholar]
- 44.He Y, Hicke L, Radhakrishnan I. Structural Basis for Ubiquitin Recognition by SH3 Domains. J Mol Biol. 2007;373:190–6. doi: 10.1016/j.jmb.2007.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 46.Nooren IM, Thornton JM. Diversity of protein-protein interactions. EMBO J. 2003;22:3486–92. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reynolds C, Damerell D, Jones S. ProtorP: A Protein-Protein Interaction Analysis Server. Bioinformatics. 2008:1–2. doi: 10.1093/bioinformatics/btn584. online. [DOI] [PubMed] [Google Scholar]
- 48.Feng S, Chen JK, Yu H, Simon JA, Schreiber SL. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science. 1994;266:1241–7. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 49.Feng S, Kasahara C, Rickles RJ, Schreiber SL. Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands. Proc Natl Acad Sci US A. 1995;92:12408–12415. doi: 10.1073/pnas.92.26.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–45. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 51.Joseph RE, Fulton DB, Andreotti AH. Mechanism and functional significance of Itk autophosphorylation. J Mol Biol. 2007;373:1281–92. doi: 10.1016/j.jmb.2007.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bunnell SC, Diehn M, Yaffe MB, Findell PR, Cantley LC, Berg LJ. Biochemical Interactions Integrating Itk with the T Cell Receptor-initiated Signaling Cascade. Journal of Biological Chemistry. 2000;275:2219–2230. doi: 10.1074/jbc.275.3.2219. [DOI] [PubMed] [Google Scholar]
- 53.Bunnell SC, Henry PA, Kolluri R, Kirchhausen T, Rickles RJ, Berg LJ. Identification of Itk/Tsk Src Homology 3 Domain Ligands. Journal of Biological Chemistry. 2005;271:25646–25656. doi: 10.1074/jbc.271.41.25646. [DOI] [PubMed] [Google Scholar]
- 54.Joseph RE, Min L, Andreotti AH. The Linker between SH2 and Kinase Domains Positively Regulates Catalysis of the Tec Family Kinases. Biochemistry. 2007;46:5455–62. doi: 10.1021/bi602512e. [DOI] [PubMed] [Google Scholar]
- 55.Joseph RE, Min L, Xu R, Musselman ED, Andreotti AH. A remote substrate docking mechanism for the tec family tyrosine kinases. Biochemistry. 2007;46:5595–603. doi: 10.1021/bi700127c. [DOI] [PubMed] [Google Scholar]
- 56.Su YW, Zhang Y, Schweikert J, Koretzky GA, Reth M, Wienands J. Interaction of SLP adaptors with the SH2 domain of Tec family kinases. Eur J Immunol. 1999;29:3702–11. doi: 10.1002/(SICI)1521-4141(199911)29:11<3702::AID-IMMU3702>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 57.Dutartre H, Harris M, Olive D, Collette Y. The human immunodeficiency virus type 1 Nef protein binds the Src-related tyrosine kinase Lck SH2 domain through a novel phosphotyrosine independent mechanism. Virology. 1998;247:200–11. doi: 10.1006/viro.1998.9244. [DOI] [PubMed] [Google Scholar]
- 58.Joung I, Strominger JL, Shin J. Molecular cloning of a phosphotyrosine-independent ligand of the p56lck SH2 domain. Proc Natl Acad Sci USA. 1996;93:5991–5. doi: 10.1073/pnas.93.12.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malek SN, Desiderio S. A cyclin-dependent kinase homologue, p130PITSLRE is a phosphotyrosine-independent SH2 ligand. Journal of Biological Chemistry. 1994;269:33009–33020. [PubMed] [Google Scholar]
- 60.Nantel A, Mohammad-Ali K, Sherk J, Posner BI, Thomas DY. Interaction of the Grb10 adapter protein with the Raf1 and MEK1 kinases. J Biol Chem. 1998;273:10475–84. doi: 10.1074/jbc.273.17.10475. [DOI] [PubMed] [Google Scholar]
- 61.Pendergast AM, Muller AJ, Havlik MH, Maru Y, Witte ON. BCR sequences essential for transformation by the BCR-ABL oncogene bind to the ABL SH2 regulatory domain in a non-phosphotyrosine-dependent manner. Cell. 1991;66:161–71. doi: 10.1016/0092-8674(91)90148-r. [DOI] [PubMed] [Google Scholar]
- 62.Schmandt R, Liu SK, McGlade CJ. Cloning and characterization of mPAL, a novel Shc SH2 domain-binding protein expressed in proliferating cells. Oncogene. 1999;18:1867–79. doi: 10.1038/sj.onc.1202507. [DOI] [PubMed] [Google Scholar]
- 63.Ching KA, Grasis JA, Tailor P, Kawakami Y, Kawakami T, Tsoukas CD. TCR/CD3-Induced activation and binding of Emt/Itk to linker of activated T cell complexes: requirement for the Src homology 2 domain. J Immunol. 2000;165:256–62. doi: 10.4049/jimmunol.165.1.256. [DOI] [PubMed] [Google Scholar]
- 64.Keller R. Optimizing the Process of Nuclear Magnetic Resonance Spectrum Analysis and Computer Aided Resonance Assignment. Swiss Federal Institute of Technology; Zurich: 2007. [Google Scholar]
- 65.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat Struct Biol. 1998;5:1065–74. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 66.Zweckstetter M, Bax A. Characterization of molecular alignment in aqueous suspensions of Pf1 bacteriophage. J Biomol NMR. 2001;20:365–77. doi: 10.1023/a:1011263920003. [DOI] [PubMed] [Google Scholar]
- 67.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 68.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 69.Berjanskii MV, Neal S, Wishart DS. PREDITOR: a web server for predicting protein torsion angle restraints. Nucleic Acids Res. 2006;34:W63–9. doi: 10.1093/nar/gkl341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.SJ H, JM T. NACCESS, Computer Program. Department of Biochemistry and Molecular Biology, University College; London: 1993. [Google Scholar]
- 71.SR. G. One-Dimensional Electrophoresis Using Non-Denaturing Conditions. Current Protocols in Protein Science. 2001:10.3.1–10.3.11. doi: 10.1002/0471140864.ps1003s00. Chapter 10. [DOI] [PubMed] [Google Scholar]
- 72.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific LLC; San Carlos, CA, USA: http://www.pymol.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1S: Similarities between free and bound Src hmolgy domains are shown in the superposition of the structures of the isolated Itk SH3 and cis SH2 domains (shown in light grey) with the corresponding domains in the Itk SH3/SH2 complex.
Figure 2S: Superposition of the Itk SH3/SH2 complex depicting the 58 intermolecular NOE restraints used in the NMR structure calculation. The SH3 and SH2 domains are colored in cyan and gray, respectively. The intermolecular NOE restraints are primarily between the SH3 binding pocket and the three loops of the Itk SH2 domain (CD, BG and EF). The red and cyan lines are NOEs involving SH2 CD loop residues, magenta involves residues of the BG loop, orange indicates NOEs to the end of the βD strand, and green are NOEs to the EF loop. Side chains giving rise to NOEs are depicted but hydrogens are not included for clarity.