Summary
Chlamydia is predicted to encode two alternative sigma factors that could provide a mechanism for the regulation of gene expression via alternative forms of RNA polymerase. We have demonstrated that σ28, one of these alternative sigma factors, is transcriptionally active. Chlamydial σ28 RNA polymerase was reconstituted from recombinant σ28 protein and core enzyme that was biochemically isolated from chlamydiae. In an in vitro transcription assay, σ28 RNA polymerase transcribed the hctB promoter in a σ28-dependent manner. Transcription by σ28 RNA polymerase was salt tolerant compared with transcription by σ66 RNA polymerase, the major form of chlamydial RNA polymerase. As hctB encodes a histone-like protein that is only expressed late in the developmental cycle, our results suggest that σ28 RNA polymerase has a role in the regulation of late gene expression in Chlamydia.
Introduction
Chlamydiae are pathogenic bacteria and obligate intracellular parasites. Chlamydia trachomatis is a leading cause of sexually transmitted disease in the developed world and of preventable blindness in the developing world (reviewed in Schachter, 1999). Chlamydiae have an unusual developmental cycle that requires growth and replication within a eukaryotic cell (reviewed in Hackstadt, 1999). During this cycle, there is conversion between two distinct morphological forms. The elementary body (EB) is a spore-like, infectious form that is metabolically inert. Upon entry into the host cell, the EB differentiates into a reticulate body (RB), which is larger and has de-condensed DNA. The RB is metabolically active and divides repeatedly by binary fission. Late in the developmental cycle, RBs convert back to EBs, which are released to infect new cells.
The mechanism of RB to EB conversion is incompletely defined, but a prominent feature of this process is the condensation of DNA to form a nucleoid. Two histone-like proteins, Hc1 and Hc2 are believed to mediate this change in DNA structure and both are first detectable late in the developmental cycle at the time of RB to EB conversion (Hackstadt et al., 1991; Tao et al., 1991; Perara et al., 1992; Brickman et al., 1993). Hc1 and Hc2 have been shown to condense DNA in vitro, to condense the Escherichia coli chromosome when expressed in E. coli and to repress transcription and translation in vitro and in vivo (Barry et al., 1992; 1993; Brickman et al., 1993; Pedersen et al., 1994; 1996). Hc1 and Hc2 are encoded by hctA and hctB, respectively, and both genes are only transcribed late in the developmental cycle (Brickman et al., 1993; Fahr et al., 1995). hctA, but not hctB, is transcribed by the major (σ66) RNA polymerase, as are several other late genes such as omcA, omcB, ltuA and ltuB (reviewed in Fahr et al., 1995), but the mechanism underlying the transcriptional regulation of late genes is unknown.
Gene expression in bacteria is often regulated at the transcriptional level by transcription factors and by alternative forms of RNA polymerase. Transcription factors modulate RNA polymerase activity while an alternative RNA polymerase contains an alternative sigma factor that recognizes a different promoter structure. The genomes of Chlamydia species sequenced to date (Stephens et al., 1998; Kalman et al., 1999; Read et al., 2000; Shirai et al., 2000; Read et al., 2003), each encode the major sigma factor, σ66, and two predicted alternative sigma factors, σ28 and σ54, that are homologous to known bacterial alternative sigma factors (Stephens et al., 1998). Transcripts for both predicted alternative sigma factors can be detected by RT-PCR (Mathews et al., 1999; Douglas and Hatch, 2000; Shaw et al., 2000), but neither sigma factor has been shown to transcribe chlamydial promoters. In fact, the genes that are regulated by σ28 or σ54 are not known, although some σ54 promoters have been predicted based on sequence similarity to bacterial σ54 promoters (Mathews and Timms, 2000).
A major impediment to studying gene regulation in Chlamydia has been the absence of an experimental genetic system, although in vitro assays have been used to define the promoter recognized by the major RNA polymerase (reviewed in Hatch, 1999; Schaumburg and Tan, 2003) and to reconstitute regulated transcription (Wilson and Tan, 2002). In this report, we describe the development of an assay for chlamydial σ28 activity that required the reconstitution of active chlamydial σ28 RNA polymerase and the identification of a σ28-dependent promoter. σ28 RNA polymerase was assembled from recombinant σ28 and core enzyme prepared from chlamydiae grown in tissue culture. We identified the hctB promoter as a candidate σ28 promoter because sequences upstream of its predicted transcription start site are similar to the consensus bacterial σ28 promoter structure. Using our in vitro assay, we demonstrated that the hctB promoter is transcribed by chlamydial σ28 RNA polymerase. As hctB is only transcribed late in the chlamydial developmental cycle, our results suggest that σ28 has a role in the developmental regulation of chlamydial gene expression.
Results
Overexpression and purification of chlamydial σ28
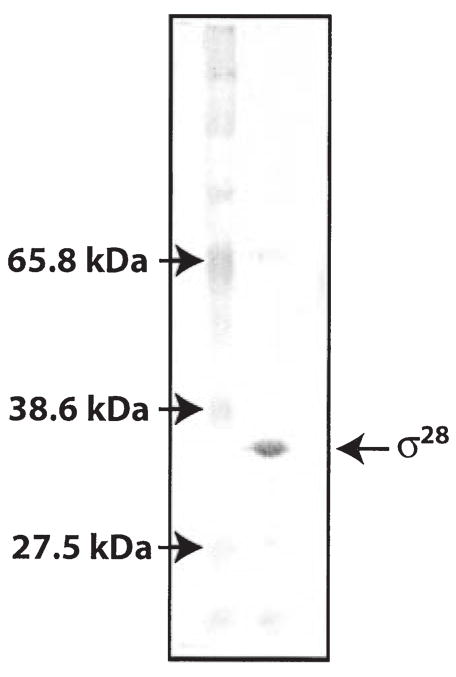
rpsD, the gene encoding a predicted σ28 factor, was cloned from C. trachomatis serovar L2 and σ28 protein was overexpressed in E. coli and purified as a six-histidine-tagged recombinant protein. The recombinant σ28 protein is highly purified and has a predicted size of 33 kDa (Fig. 1), from addition of a 4 kDa histidine tag to the 29 kDa σ28. To minimize co-purification of E. coli proteins that may have formed associations with recombinant chlamydial σ28, soluble protein was precipitated by ammonium sulphate and re-suspended in 6 M guanidine hydrochloride prior to purification with Ni-NTA agarose beads. Antibodies against E. coli σ28 did not recognize E. coli σ28 in our purified C. trachomatis σ28 protein preparation (immunoblot data not shown).
Fig. 1.
Silver stain of SDS–PAGE showing purified recombinant histidine-tagged chlamydial σ28 protein. Lane 1, marker; Lane 2, σ28 after purification with Ni-NTA beads.
Prediction that the hctB promoter is a σ28-regulated promoter
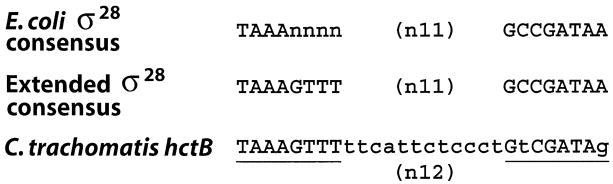
σ28 promoter structure is well conserved in bacteria and a consensus promoter structure [TAAAnnnn (n11) GCCGATAA] (reviewed in Helmann, 1991; Gross et al., 1992) and a more stringent, extended promoter structure [TAAAGTTT (n11) GCCGATAA] (Ide et al., 1999) have been proposed. By inspection, we noted that sequences upstream of the predicted transcription start site of C. trachomatis serovar L2 hctB (Brickman et al., 1993) closely resemble the extended σ28 promoter sequence but with a 12 bp spacer between promoter elements rather than the canonical 11 bp (Fig. 2).
Fig. 2.
Alignment of Chlamydia trachomatis serovar L2 hctB promoter (Brickman et al., 1993) with the σ28 consensus promoter structure (reviewed in Helmann, 1991; Gross et al., 1992) and σ28 extended promoter structure (Ide et al., 1999) derived from Escherichia coli and Salmonella sequences. Predicted promoter elements are underlined with the sequence differences from the extended promoter structure shown in lowercase.
Chlamydial σ28 RNA polymerase is active and specifically transcribes the hctB promoter
Chlamydial σ28 RNA polymerase was reconstituted by mixing recombinant σ28 with chlamydial core enzyme. As a source of core enzyme, we used RNA polymerase purified from reticulate bodies by heparin-agarose chromatography. We have previously demonstrated that heparin-agarose-purified RNA polymerase is transcriptionally active and contains core enzyme and σ66, the major sigma factor in Chlamydia (Tan and Engel, 1996). Using immunoblots with anti-chlamydial σ28 antibodies, we determined that σ28 protein was not detectable in the heparin-agarose-purified RNA polymerase (data not shown).
The reconstituted σ28 RNA polymerase was used to demonstrate that the hctB promoter is a σ28-regulated promoter. Recombinant σ28 was pre-incubated with heparin-agarose-purified RNA polymerase and an in vitro transcription reaction was performed with plasmid pMT1212, which contains the predicted hctB promoter upstream of a synthetic G-less cassette transcription template that does not encode any G residues (Fig. 3). The radiolabelled products of the transcription reaction were electrophoresed on a urea-polyacrylamide gel, which was exposed to a phosphorimaging screen for quantification of the amount of transcript present. There was no transcription with σ28 alone (Fig. 4A, lane 1), or with heparin-agarose-purified RNA polymerase alone (Fig. 4A, lane 2), which suggests that σ66 RNA polymerase was not able to recognize this hctB promoter. A readily detectable transcript of expected size (150 nt) was produced when the reaction was performed with σ28 RNA polymerase that had been reconstituted from recombinant σ28 and heparin-agarose-purified RNA polymerase (Fig. 4A, lane 3). There is almost a complete lack of background transcription because only the G-less cassette was transcribed in the absence of GTP in the reaction mix (Tan and Engel, 1996).
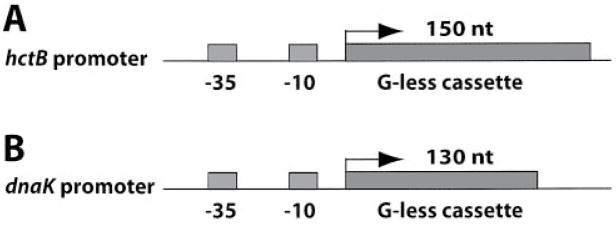
Fig. 3. Schematic diagram of the transcription plasmids.
A. The C. trachomatis hctB promoter precedes a 150 nt G-less cassette transcription template in pMT1212.
B. The C. trachomatis dnaK promoter precedes a 130 nt G-less cassette in pMT1211.
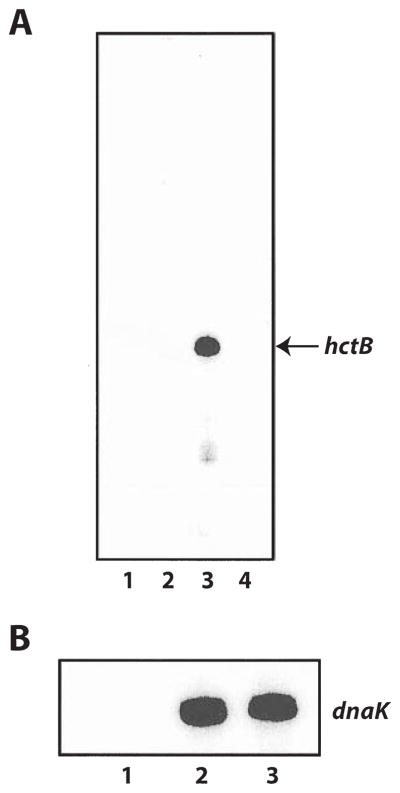
Fig. 4. In vitro transcription reactions with chlamydial σ28 RNA poly-merase.
A. Transcription from the hctB promoter with the following components present in the reaction: Lane 1, σ28 alone; Lane 2, heparin-agarose-purified RNA polymerase alone; Lane 3, chlamydial σ28 RNA polymerase reconstituted from σ28 and heparin-agarose-purified RNA polymerase; Lane 4, chlamydial σ28 RNA polymerase in the presence of anti-chlamydial σ28 antibodies.
B. σ66-dependent transcription from the dnaK promoter with the following components present in the reaction: Lane 1, σ28 alone; Lane 2, heparin-agarose-purified RNA polymerase alone; Lane 3, heparin-agarose-purified RNA polymerase in the presence of anti-chlamydial σ28 antibodies.
To confirm that the transcriptional activity of reconstituted σ28 RNA polymerase is dependent on chlamydial σ28, we pre-incubated the recombinant σ28-heparin-agarose-purified RNA polymerase mix with anti-chlamydial σ28 antibodies prior to in vitro transcription. Our results demonstrated that polyclonal antibodies against chlamydial σ28 completely inhibited transcription of the hctB promoter (Fig. 4A, lane 4). These antibodies had no effect on σ66-dependent transcription of the dnaK promoter contained in plasmid pMT1211 (Fig. 4B, compare lanes 2 and 3), which showed that inhibition of hctB transcription is σ28 specific. These experiments demonstrate that the transcriptional activity of reconstituted σ28 RNA polymerase is dependent on chlamydial σ28. Our antibodies against chlamydial σ28 do not recognize purified E. coli σ28 protein by immunoblot (data not shown), which rules out the formal possibility that the activity is due to contaminating E. coli σ28 in the chlamydial σ28 preparation.
σ28 associated with free core enzyme in the heparin-agarose-purified RNA polymerase preparations
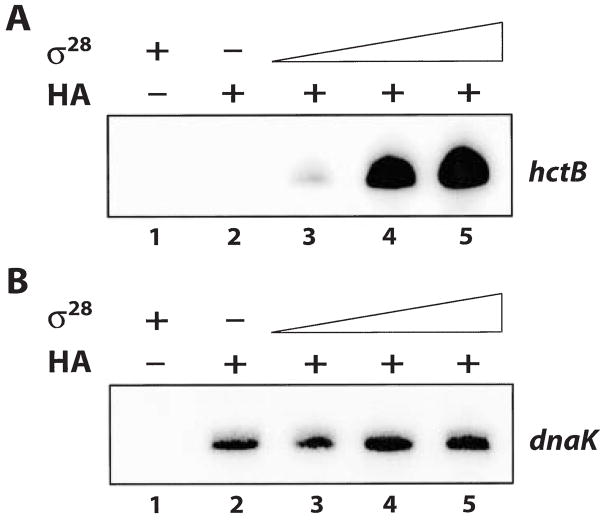
The preceding results demonstrated that σ28 RNA polymerase was successfully reconstituted from recombinant σ28 and heparin-agarose-purified RNA polymerase containing core enzyme and σ66. To examine the interactions between σ28, σ66 and core enzyme, we performed a titration experiment by adding increasing amounts of σ28 to the heparin-agarose-purified RNA polymerase. Each RNA polymerase mix was allowed to pre-incubate and then used in separate reactions containing either the hctB promoter or dnaK promoter transcription plasmid to assay for σ28-dependent or σ66-dependent activity respectively. There was increased transcription from the hctB promoter with increasing concentrations of σ28 (Fig. 5A, lanes 3–5) but transcription from the σ66-dependent dnaK promoter did not change (Fig. 5B, lanes 3–5). As the reactions were performed with non-saturating amounts of σ66 RNA polymerase (C. Schaumburg and M.T., unpubl. results), these results show that addition of σ28 did not affect the amount of σ66 RNA polymerase activity present. We infer from these results that σ28, in the concentrations added, associated with free core enzyme in the heparin-agarose-purified RNA polymerase and did not displace σ66 from intact holoenzyme.
Fig. 5. In vitro transcription reactions showing the effect of increasing amounts of σ28 on the reconstitution of σ28 RNA polymerase.
A. Transcription from the hctB promoter. Lane 1, σ28 alone; Lane 2, heparin-agarose-purified RNA polymerase (HA) alone; Lane 3–5, heparin-agarose-purified RNA polymerase (HA) with increasing amounts of σ28 (30, 120 and 480 nM final concentration).
B. Transcription from the dnaK promoter. Lane 1, σ28 alone; Lane 2, heparin-agarose-purified RNA polymerase (HA) alone; Lane 3–5, heparin-agarose-purified RNA polymerase (HA) with increasing amounts of σ28 (30, 120 and 480 nM final concentration).
Effect of salt concentration on transcription of the hctB promoter by σ28 RNA polymerase
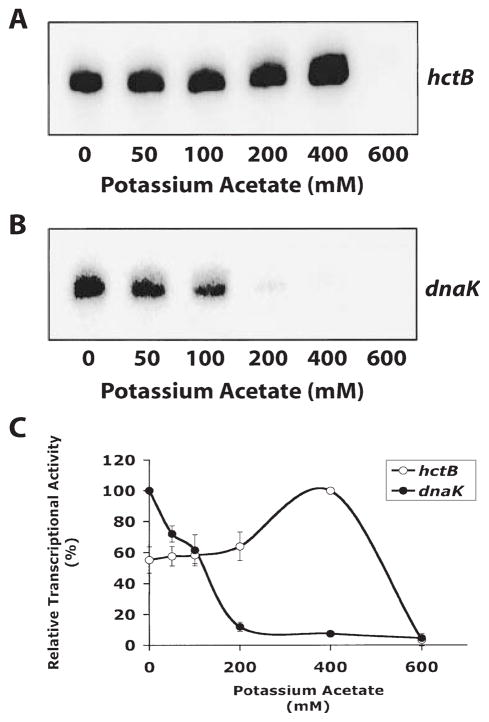
We performed in vitro transcription experiments with different salt concentrations to determine if the chlamydial σ28 RNA polymerase was salt tolerant relative to chlamydial σ66 RNA polymerase. Kundu et al. have shown that σ28 (σF)-regulated transcription in E. coli is highly salt tolerant compared with σ70-regulated transcription (Kundu et al., 1997). We tested potassium acetate concentrations from 0 to 600 mM and found that transcription of the hctB promoter by chlamydial σ28 RNA polymerase was salt tolerant, with peak transcriptional activity at 400 mM potassium acetate and a significant decrease at 600 mM potassium acetate (Fig. 6). In contrast, transcription of the dnaK promoter by σ66 RNA polymerase decreased to negligible levels at concentrations above 100 mM (Fig. 6).
Fig. 6. In vitro transcription reactions showing the effect of varying salt concentration.
A. Transcription from the hctB promoter by chlamydial σ28 RNA polymerase. Lane 1–6, increasing concentrations of potassium acetate (0, 50, 100, 200, 400 and 600 mM).
B. Transcription from the dnaK promoter by heparin-agarose-purified RNA polymerase. Lanes 1–6, increasing concentrations of potassium acetate (0, 50, 100, 200, 400 and 600 mM).
C. Quantification of transcriptions. Reactions were performed in triplicate and quantified by phosphorimager analysis. Results were normalized to 100% for the highest level of transcription for each promoter. Error bars are SD from the mean.
Discussion
This is the first report of the reconstitution of a transcriptionally active alternative chlamydial RNA polymerase. Our results indicate that rpsD encodes a functional alternative sigma factor, σ28, and that the hctB promoter is regulated by σ28 RNA polymerase. Shen and Zhang (2002) have presented preliminary work showing that recombinant chlamydial σ28 is active in a hybrid RNA polymerase with E. coli core enzyme and is able to transcribe a known E. coli σ28 promoter, fliC, in vitro. Our assay, which utilizes chlamydial σ28 RNA polymerase to transcribe a chlamydial promoter, has the advantage of being a purely chlamydial system.
As we purified recombinant chlamydial σ28 protein from E. coli, it was important to ensure that any transcriptional activity was not due to contaminating E. coli proteins. To prevent the co-purification of E. coli core enzyme that could potentially bind to chlamydial σ28, we purified chlamydial σ28 under denaturing conditions. In addition, two lines of evidence indicate that our chlamydial σ28 does not contain contaminating E. coli σ28. First, we did not detect E. coli σ28 in our chlamydial σ28 preparation with anti-E. coli σ28 antibodies. Second, the transcriptional activity of our reconstituted σ28 RNA polymerase was inhibited by anti-chlamydial σ28 antibodies, which do not cross-react with E. coli σ28.
We reconstituted σ28 RNA polymerase by adding recombinant σ28 protein to σ66 holoenzyme that had been purified from chlamydiae by heparin-agarose chromatography. While it has been noted that E. coli σ28 has a greater affinity for core enzyme compared with σ70 (the E. coli homologue of σ66) (Kundu et al., 1997), our results suggest that, under our reaction conditions, chlamydial σ28 was able to bind to free core enzyme without displacing σ66 from intact holoenzyme. The presence of free core enzyme in the heparin-agarose-purified RNA polymerase would not be surprising as E. coli RNA polymerase purified by this method is only 30% saturated with σ70 (Chamberlin et al., 1983).
The high salt tolerance of our reconstituted chlamydial σ28 RNA polymerase has also been described for E. coli σ28 (σF)-regulated transcription (Kundu et al., 1997). For E. coli, 200–300 mM potassium acetate was required for optimal in vitro transcription of two E. coli σ28-regulated promoters, fliC and fliD, while transcription of a σ70-regulated promoter decreased dramatically between 100 mM and 150 mM potassium acetate (Kundu et al., 1997). Our reconstituted σ28 RNA polymerase showed optimal transcription at 400 mM potassium acetate, a concentration at which σ66-dependent promoter activity is completely inhibited. These results provide further proof that the hctB promoter is only transcribed by σ28 RNA polymerase and not by σ66 holoenzyme from the heparin-agarose-purified RNA polymerase preparation.
While σ28 regulates the expression of genes involved in flagellar synthesis, chemotaxis and motility in other bacteria (reviewed in Helmann, 1991; Gross et al., 1992; Haldenwang, 1995), the genes that are regulated by σ28 in Chlamydia spp. are not known. Chlamydiae are non-motile organisms that do not possess flagella. σ28 regulates the expression of some type III secretion genes in Salmonella spp. (Eichelberg and Galan, 2000) and Chlamydia does encode homologues of a few flagellar-like genes that may be structural components of the type III secretion apparatus (reviewed in Kim, 2001). σ28 has also been proposed to have a role in developmental regulation. σ28-dependent transcription in Bacillus is highest during the transition from log phase to stationary phase growth and is dependent on many of the same spo0 gene products needed for the initiation of the sporulation cascade (Gilman and Chamberlin, 1983).
We predicted that the hctB promoter is a σ28-regulated promoter because sequences upstream of the mapped in vivo transcription start site showed strong conservation with the σ28 consensus promoter in other bacteria. This transcription start site had been mapped to a site 36 bp upstream of the predicted AUG translation start codon by Brickman et al. (1993). Fahr et al. (1995) also mapped this site as well as a second in vivo transcription start site 79 bp upstream of the predicted AUG. Neither site contains upstream sequences that resemble the optimal promoter structure recognized by the major sigma factor, σ66 (Tan et al., 1998; Schaumburg and Tan, 2003). In addition, neither predicted promoter produced a radiolabelled transcript in a σ66 transcription assay (Fahr et al., 1995). We have now shown that the promoter predicted from the transcription start site first mapped by Brickman et al. (1993) is recognized by σ28 RNA polymerase. The sequences upstream of the second transcription start site mapped by Fahr et al. (1995) do not resemble the σ28 consensus promoter, suggesting, perhaps, that a different form of RNA polymerase may recognize this putative promoter. Alternatively, this site may not be an actual transcription start site.
The hctB promoter strongly resembles the extended σ28 consensus promoter proposed by Ide et al. (1999). One small difference is that the hctB promoter has a 12 bp spacer while the extended promoter sequence and the majority of known bacterial σ28 promoters have an 11 bp spacer (reviewed in Helmann, 1991; Gross et al., 1992; Ide et al., 1999). Slight variability in spacer length of promoters is not without precedence. For example, E coli σ70 RNA polymerase prefers a 17 bp spacer, but a spacer range of 16–18 bp is not uncommon and σ70 promoters with a spacer between 15 and 21 bp have been identified (Mulligan et al., 1984).
Transcription of the hctB promoter by σ28 RNA polymerase suggests that this alternative form of chlamydial RNA polymerase is involved in the regulation of late gene expression. hctB is a prototypical late gene that is only transcribed late in the developmental cycle, at the time of RB to EB conversion (Brickman et al., 1993; Fahr et al., 1995). hctB encodes Hc2, a histone-like protein that is believed to be involved in this conversion through its ability to condense DNA. However, other late genes, such as omcB, hctA, ltuA, and ltuB have been shown to be transcribed by σ66 RNA polymerase (Fahr et al., 1995). Thus, it is likely that there is more than one mechanism for the regulation of late gene expression and that these mechanisms involve transcription by σ28 RNA polymerase and by regulation of σ66 RNA polymerase by an activator or repressor.
If σ28 is involved in transcriptional regulation of gene expression during the chlamydial developmental cycle, what regulates σ28 activity? The temporal pattern of chlamydial σ28 expression has been studied by measuring rpsD mRNA and σ28 protein expression levels during the developmental cycle. Mathews et al. (1999) measured peak rpsD mRNA levels early in the developmental cycle and speculated that σ28 RNA polymerase may be involved in EB to RB conversion. In contrast, Shaw et al. (2000) and Douglas and Hatch (2000) have shown that rpsD transcripts were undetectable at early times after infection, but present during the mid- and late period of infection. σ28 protein has been detected early and late in the developmental cycle (A. M. Douglas and T. Hatch, pers. comm.). While these results are not definitive, it does not appear that the presence of σ28 protein by itself is sufficient for the regulated transcription of hctB at late times in the developmental cycle.
It is known from other bacteria that σ28 activity is controlled by a partner switching mechanism that involves an anti-sigma factor (reviewed in Hughes and Mathee, 1998). In Bacillus spp., the anti-sigma factor, RsbW, binds to σ28 and prevents it from binding with core enzyme to form an active polymerase. RsbW is itself regulated by an anti-anti-sigma factor that is differentially phosphorylated in response to a signalling cascade. The sequenced chlamydial genomes all contain genes that encode homologues of RsbW, RsbV and RsbU, which are proposed members of this partner switching mechanism (Stephens et al., 1998; Kalman et al., 1999; Read et al., 2000; 2003; Shirai et al., 2000).
With the reconstitution of active chlamydial σ28 RNA polymerase, we now have a powerful assay for studying gene regulation by this alternative RNA polymerase and for identifying other σ28-regulated genes in addition to hctB. The hctB promoter strongly resembles the consensus bacterial σ28 promoter structure but we do not know if the sequences of other chlamydial σ28 promoters and the promoter specificity of chlamydial σ28 RNA polymerase are similarly conserved. Identification of additional σ28 promoters will allow us to gain insight into the role of σ28 RNA polymerase in the co-ordinate regulation of genes late in the developmental cycle and under other growth conditions in Chlamydia.
Experimental procedures
Reagents
The following products were obtained from the sources given and were used according to the manufacturers’ specifications: restriction enzymes, calf intestinal alkaline phosphatase, T4 DNA ligase, rRNasin and Taq DNA polymerase, Promega Biotech; T4 polynucleotide kinase and pRSET expression vector, Invitrogen; Sequenase kit, U.S. Biochemicals; E. coli BL21(DE3), Stratagene; nucleoside triphosphates, 3′-O-methylguanosine 5′-triphosphate, Amersham; Ni-NTA agarose beads, Qiagen; oligonucleotide primers, Sigma Genosys; 32P-labelled nucleoside triphosphates, ICN; Pwo DNA polymerase, Roche Diagnostics; protein assay reagent, Protein A agarose kit, Bio-Rad; E. coli σ28 protein, monoclonal anti-E. coli σ28 antibodies, Neoclone.
Cloning of chlamydial rpsD (σ28 gene)
All C. trachomatis sequences are based on information from the Chlamydia Genome Project (http://chlamydia-www.berkeley.edu:4231/). Plasmid pMT1104 contains the C. trachomatis serovar L2 σ28 gene (rpsD) cloned into a His-tagged expression vector pRSET-C. The insert (containing the entire rpsD gene with the exclusion of the start codon) was amplified by PCR from C. trachomatis L2 genomic DNA by Pwo DNA polymerase, using PCR primers T133 (5′ AAGACTCACGATCTCGCAGATACTTG) and T134 (5′ CCCGGTACCCTAAAGCAGACTG). As the sequence of C. trachomatis serovar L2 rpsD was not available, we compared the sequence of our clone with the sequence of serovar D rpsD. There were three nucleotide differences between our amplified L2 rpsD gene and the serovar D rpsD sequence, but none of them produced a change at the amino acid level. The PCR product was digested with KpnI and cloned into pRSET-C between KpnI and blunted BamHI sites.
Overexpression and purification of chlamydial σ28
σ28 was overexpressed in E. coli BL21(DE3) freshly transformed with pMT1104. Next, 250 ml of cells were grown at 37°C to an optical density at 600 nm of 0.5 and induced with 1 mM isopropyl-β-D-thiogalactosidase (IPTG). After 3 h, cells were collected by centrifugation, resuspended in 8 ml of Buffer N (10 mM Tris, pH 8.0, 0.3 M NaCl, 10 mM β-mercaptoethanol) containing 20 mM imidazole and disrupted with a Branson Sonifier 450 (30 s × 4) in the presence of 0.2% sarkosyl. Soluble protein was separated from cell debris by centrifugation at 10 000 r.p.m. for 10 min at 4°C (Beckman JA-17 rotor). 2% Triton X-100 was added to sequester the sarkosyl.
σ28 protein was then purified under denaturing conditions. The protein lysate was precipitated with 60% ammonium sulphate and the protein pellet was solubilized with 6 M guanidine hydrochloride. Proteins were allowed to bind to Ni-NTA agarose beads at 4°C for 1 h. Bound proteins were washed with Buffer N containing 30 mM imidazole. His-tagged protein was eluted with Buffer N containing 250 mM imidazole. Purified σ28 protein was dialysed overnight with two changes of storage buffer (50 mM Tris, pH 8.0, 200 mM KCl, 10 mM MgCl2, 10 μM ZnCl2, 1 mM EDTA, 5 mM 2-β-mercaptoethanol, 20% glycerol). The concentration of the purified σ28 protein was approximately 350 μg ml−1.
Production of polyclonal anti-σ28 antibodies
Recombinant σ28 was gel purified by SDS–PAGE and used to generate rabbit polyclonal antibodies (Harlan Bio-products for Science). Antibodies were purified by Protein-A agarose column according to the manufacturer’s instructions (Bio-Rad).
Purification of C. trachomatis RNA polymerase from chlamydiae grown in tissue culture
Chlamydia trachomatis LGV serovar L2 was grown in mouse L929 cells and harvested at 18 h post-infection. RNA polymerase was partially purified by heparin-agarose chromatography as previously described (Tan and Engel, 1996).
Construction of transcription plasmids
Plasmid pMT1212 contains the promoter region of C. trachomatis hctB (−135 to +5), which was amplified by PCR from C. trachomatis serovar L2 genomic DNA with primers T12 (5′-ATTTATTTGATCTATCGAC) and T13 (5′-GCCGAAT-TCAGGATTTGGTGTGC). Transcription from this plasmid produced a 150 nt transcript. Plasmid pMT1211 contains the C. trachomatis dnaK promoter (−180 to +5) amplified from C. trachomatis MoPn strain genomic DNA with primers dnaK2 (5′-AAGTTGGTGTCATTATAGGAAAACC) and dnaK180 (5′-AGGGAATTCGATCGAATCTCGATCTGG). Transcription from this plasmid produced a 130 nt transcript. For each promoter, the insert was cloned upstream of a promoter-less, G-less cassette transcription template in plasmid pMT1125 (Wilson and Tan, 2002).
In vitro transcription
The general transcription reaction was performed in a 10 μl final volume with the following components: 50 mM potassium acetate, 8.1 mM magnesium acetate, 50 mM Tris acetate pH 8.0, 27 mM ammonium acetate, 2 mM DTT, 400 μM ATP, 400 μM UTP, 1.2 μM CTP, 0.06 μM [α-32P]-CTP (3000 Ci mmol−1), 100 μM 3′-O-methylguanosine 5′-triphosphate Na salt, 18 units rRNasin, 10% glycerol and 25 nM supercoiled DNA template. In some experiments, the potassium acetate concentration was varied over a range from 0 to 600 mM. The transcription reaction was incubated at 37°C for 15 min and terminated by the addition of 10 μl stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol). Next, 7 μl of the sample was electrophoresed on an 8 M urea/6% polyacrylamide gel and quantified by phosphorimager analysis with a Bio-Rad Personal FX scanner and analysis with Bio-Rad Quantity One software, as previously described (Wilson and Tan, 2002).
In general, σ28 transcription reactions were performed with chlamydial σ28 RNA polymerase that had been reconstituted by mixing 0.5 μl purified His6-σ28 (final concentration of 480 nM) with 1 μl heparin-agarose-purified RNA polymerase (Schaumburg and Tan, 2000) at 4°C for 15 min, immediately prior to the transcription reaction. For the σ28 titration experiments, a range of σ28 concentrations (30, 120 or 480 nM final concentration) was used for the reconstitution. Next, 1 μl heparin-agarose-purified C. trachomatis RNA polymerase was used for the σ66 transcription reactions. For the antibody-inhibition reactions, anti-chlamydial σ28 antibodies were pre-incubated with each RNA polymerase for 20 min at room temperature prior to transcription.
Acknowledgments
We would like to thank Adam Wilson, Chris Schaumburg and Jamie Joyner for their support and Drs G. Wesley Hatfield, Bert Semler and Marian Waterman for critical review of the manuscript. This work is supported by a grant from the NIH (AI 44198). H.H.Y.Y. is supported by a pre-doctoral training grant from the NIH (National Research Service Award 1 T15 LM07443-01 from the National Library of Medicine).
References
- Barry C, Hayes S, Hackstadt T. Nucleoid condensation in Escherichia coli that express a chlamydial histone homolog. Science. 1992;256:377–379. doi: 10.1126/science.256.5055.377. [DOI] [PubMed] [Google Scholar]
- Barry CE, Brickman TJ, Hackstadt T. Hc1-mediated effects on DNA structure: a potential regulator of chlamydial development. Mol Microbiol. 1993;9:273–283. doi: 10.1111/j.1365-2958.1993.tb01689.x. [DOI] [PubMed] [Google Scholar]
- Brickman TJ, Barry CE, III, Hackstadt T. Molecular cloning and expression of hctB encoding a strain-variant chlamydial histone-like protein with DNA-binding activity. J Bacteriol. 1993;175:4274–4281. doi: 10.1128/jb.175.14.4274-4281.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin M, Kingston R, Gilman M, Wiggs J, deVera A. Isolation of bacterial and bacteriophage RNA polymerases and their use in synthesis of RNA in vitro. Methods Enzymol. 1983;101:540–569. doi: 10.1016/0076-6879(83)01037-x. [DOI] [PubMed] [Google Scholar]
- Douglas AL, Hatch TP. Expression of the transcripts of the sigma factors and putative sigma factor regulators of Chlamydia trachomatis L2. Gene. 2000;247:209–214. doi: 10.1016/s0378-1119(00)00094-9. [DOI] [PubMed] [Google Scholar]
- Eichelberg K, Galan JE. The flagellar sigma factor FliA (sigma 28) regulates the expression of Salmonella genes associated with the centisome 63 type III secretion system. Infect Immun. 2000;68:2735–2743. doi: 10.1128/iai.68.5.2735-2743.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahr MJ, Douglas AL, Xia W, Hatch TP. Characterization of late gene promoters of Chlamydia trachomatis. J Bacteriol. 1995;177:4252–4260. doi: 10.1128/jb.177.15.4252-4260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman MZ, Chamberlin MJ. Developmental and genetic regulation of Bacillus subtilis genes transcribed by sigma-28-RNA polymerase. Cell. 1983;35:285–293. doi: 10.1016/0092-8674(83)90231-3. [DOI] [PubMed] [Google Scholar]
- Gross CA, Lonetto M, Losick R. Bacterial sigma factors. In: McKnight SL, Yamamoto KR, editors. Transcriptional Regulation. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 129–176. [Google Scholar]
- Hackstadt T. Cell biology. In: Stephens RS, editor. Chlamydia: Intracellular Biology, Pathogenesis, and Immunity. Washington, DC: American Society for Microbiology Press; 1999. p. 321. [Google Scholar]
- Hackstadt T, Baehr W, Ying Y. Chlamydia trachomatis developmentally regulated protein is homologous to eukaryotic histone H1. Proc Natl Acad Sci USA. 1991;88:3937–3941. doi: 10.1073/pnas.88.9.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldenwang WG. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch TP. Developmental biology. In: Stephens RS, editor. Chlamydia: Intracellular Biology, Pathogenesis, and Immunity. Washington, DC: American Society for Microbiology Press; 1999. pp. 29–68. [Google Scholar]
- Helmann JD. Alternative sigma factors and the regulation of flagellar gene expression. Mol Microbiol. 1991;5:2875–2882. doi: 10.1111/j.1365-2958.1991.tb01847.x. [DOI] [PubMed] [Google Scholar]
- Hughes KT, Mathee K. The anti-sigma factors. Annu Rev Microbiol. 1998;52:231–286. doi: 10.1146/annurev.micro.52.1.231. [DOI] [PubMed] [Google Scholar]
- Ide N, Ikebe T, Kutsukake K. Reevaluation of the promoter structure of the class 3 flagellar operons of Escherichia coli and Salmonella. Genes Genet Syst. 1999;74:113–116. doi: 10.1266/ggs.74.113. [DOI] [PubMed] [Google Scholar]
- Kalman S, Mitchell W, Marathe R, Lammel C, Fan J, Hyman RW, et al. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nature Genet. 1999;21:385–389. doi: 10.1038/7716. [DOI] [PubMed] [Google Scholar]
- Kim JF. Revisiting the chlamydial type III protein secretion system: clues to the origin of type III protein secretion. Trends Genet. 2001;17:65–69. doi: 10.1016/s0168-9525(00)02175-2. [DOI] [PubMed] [Google Scholar]
- Kundu T, Kusano S, Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase sigmaF holoenzyme involved in transcription of flagellar and chemotaxis genes. J Bacteriol. 1997;179:4264–4269. doi: 10.1128/jb.179.13.4264-4269.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews SA, Timms P. Identification and mapping of sigma-54 promoters in Chlamydia trachomatis. J Bacteriol. 2000;182:6239–6242. doi: 10.1128/jb.182.21.6239-6242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews SA, Volp KM, Timms P. Development of a quantitative gene expression assay for Chlamydia trachomatis identified temporal expression of sigma factors. FEBS Microbiol Lett. 1999;458:354–358. doi: 10.1016/s0014-5793(99)01182-5. [DOI] [PubMed] [Google Scholar]
- Mulligan ME, Hawley DK, Entriken R, McClure WR. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984;12:789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen LB, Birkelund S, Christiansen G. Interaction of the Chlamydia trachomatis histone H1-like protein (Hc1) with DNA and RNA causes repression of transcription and translation in vitro. Mol Microbiol. 1994;11:1085–1098. doi: 10.1111/j.1365-2958.1994.tb00385.x. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Birkelund S, Christiansen G. Purification of recombinant Chlamydia trachomatis histone H1-like protein Hc2, and comparative functional analysis of Hc2 and Hc1. Mol Microbiol. 1996;20:295–311. doi: 10.1111/j.1365-2958.1996.tb02618.x. [DOI] [PubMed] [Google Scholar]
- Perara E, Ganem D, Engel JN. A developmentally regulated chlamydial gene with apparent homology to eukaryotic histone H1. Proc Natl Acad Sci USA. 1992;89:2125–2129. doi: 10.1073/pnas.89.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, et al. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read TD, Myers GS, Brunham RC, Nelson WC, Paulsen IT, Heidelberg J, et al. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 2003;31:2134–2147. doi: 10.1093/nar/gkg321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter J. Infection and disease epidemiology. In: Stephens RS, editor. Chlamydia: Intracellular Biology, Pathogenesis, and Immunity. Washington, DC: American Society for Microbiology Press; 1999. pp. 139–169. [Google Scholar]
- Schaumburg CS, Tan M. A positive cis-acting DNA element is required for high level transcription in Chlamydia. J Bacteriol. 2000;182:5167–5171. doi: 10.1128/jb.182.18.5167-5171.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaumburg CS, Tan M. Mutational analysis of the Chlamydia trachomatis dnaK promoter defines the optimal −35 promoter element. Nucleic Acids Res. 2003;31:551–555. doi: 10.1093/nar/gkg150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw EI, Dooley CA, Fischer ER, Scidmore MA, Fields KA, Hackstadt T. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol Microbiol. 2000;37:913–925. doi: 10.1046/j.1365-2958.2000.02057.x. [DOI] [PubMed] [Google Scholar]
- Shen L, Zhang Y-X. The fliA gene of chlamydia trachomatis encodes a novel heat shock responsive sigma factor. 102nd American Society for Microbiology General Meeting; Salt Lake City, UT: American Society for Microbiology; 2002. p. 233. [Google Scholar]
- Shirai M, Hirakawa H, Kimoto M, Tabuchi M, Kishi F, Ouchi K, et al. Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res. 2000;28:2311–2314. doi: 10.1093/nar/28.12.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- Tan M, Engel JN. Identification of sequences necessary for transcription in vitro from the Chlamydia trachomatis rRNA P1 promoter. J Bacteriol. 1996;178:6975–6982. doi: 10.1128/jb.178.23.6975-6982.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Gaal T, Gourse RL, Engel JN. Mutational analysis of the Chlamydia trachomatis rRNA P1 promoter defines four regions important for transcription in vitro. J Bacteriol. 1998;180:2359–2366. doi: 10.1128/jb.180.9.2359-2366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao S, Kaul R, Wenman WM. Identification and nucleotide sequence of a developmentally regulated gene encoding a eukaryotic histone H1-like protein from Chlamydia trachomatis. J Bacteriol. 1991;173:2818–2822. doi: 10.1128/jb.173.9.2818-2822.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Tan M. Functional analysis of the heat shock regulator HrcA of Chlamydia trachomatis. J Bacteriol. 2002;184:6566–6571. doi: 10.1128/JB.184.23.6566-6571.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]