Abstract
Porous polymer monolithic (PPM) columns are employed to collect and concentrate neuronal release from invertebrate and vertebrate model systems, prior to their characterization with mass spectrometry. The monoliths are fabricated in fused silica capillaries from lauryl methacrylate (LMA) and ethylene glycol dimethacrylate (EDMA). The binding capacities for fluorescein and for fluorescently labeled peptides are on the order of nanomoles per millimeter length of monolith material for a 200 μm inner diameter capillary. To evaluate this strategy for collecting peptides from physiological solutions, angiotensin I and insulin in artificial seawater are loaded onto, and then released from the monoliths after a desalination rinse, resulting in femtomole limits of detection via matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Positioned in the extracellular media near Aplysia californica bag cell neurons, upon electrical stimulation, these LMA-EDMA monoliths are also used to collect and concentrate peptide release, with egg laying hormone and acidic peptide detected. In addition, the collection of several known peptides secreted from chemically stimulated mouse brain slices demonstrates their ability to collect releasates from a variety of neuronal tissues. When compared to collection approaches using individual beads placed on brain slices, the PPM capillaries offer greater binding capacity. Moreover, they maintain higher spatial resolution as compared to the larger-volume, solid phase extraction collection strategies.
Introduction
Porous polymer monoliths (PPMs) have generated a great deal of research interest since their introduction as alternative chromatographic media in the late 1980s and early 1990s.1–3 A single piece of porous material, the PPM, forms an interconnected network of channels that combines high permeability with efficient mass transfer to facilitate low back-pressure and high flow rates.4 PPM utility is further enhanced due to the ease of monolith preparation, as well as the controlled chemistry that can be achieved via monomer backbone and/or surface modifications. Such properties join a long list of analytical advantages that are available with monolithic media and have led to their ever-growing popularity.
Monolithic media maximize permeability and mass transfer rates, as well as separation abilities.4 In addition to conventional chromatographic approaches,5–8 PPMs have been successfully employed in other applications including proteomics,9, 10 thermally responsive valves,11 enzyme immobilization,12–14 static mixing,15 and assisted electrospray generation;16–20 many of these have focused on the incorporation of monoliths in microchips.9, 11, 14–17, 20 PPMs have also been used in solid-phase extraction (SPE), as recently reviewed by Svec and Fréchet.21 Since the pioneering report by Xie et al.,22 PPMs have been utilized in SPE coupled with preconcentration on-chip,23, 24 high performance liquid chromatography,25–27 protein digestion,28, 29 and molecularly imprinted monoliths.30, 31 Notwithstanding the increasing number of reports focused on the use of PPMs for many applications in a wide variety of fields, researchers continue to develop unique ways to employ these versatile materials. Here we sample the extracellular environment of two common neurobiological model systems, the invertebrate Aplysia californica abdominal ganglion and the vertebrate mouse cortical brain slice. Using PPMs created in 200 μm inner diameter silica capillaries, we collect and concentrate multiple peptides secreted from stimulated neurons for MS characterization.
Collection and detection of the chemical signals released from electrically or chemically stimulated neurons can be challenging. Within the chemically complex extracellular environment, the compounds are diluted by orders of magnitude upon release from their secretion site(s). The direct investigation of released peptides and neurohormones is generally achieved by sampling the media surrounding biological tissues, followed by offline radiochemical, amperometric,32, 33 or mass spectrometric34–36 detection. As opposed to the sensitive, yet selective, radiochemical and amperometric methods, the mass spectrometric approach delivers more information, with the detection of multiple peptides. We reported an electrophoretic separation-SPE collection technique to monitor the activity-dependent release of A. californica bag cell neurons using matrix-assisted laser desorption/ionization (MALDI) mass spectrometry (MS).34, 36 More recently, we demonstrated improved sample collection with the deposition of SPE beads directly onto the release sites of intact nervous tissues,35, 37 as well as using commercial SPE cartridges for improved collection efficiency of peptide release.37, 38 In addition to the spatial resolution afforded with the precise placement of beads, physiological salts were eliminated by rinsing the beads and cartridges prior to MS measurements. Although both of these approaches work well, there are some caveats. The bead approach offers higher spatial information, but limited peptide capacity (low fmole range per bead), while the commercial SPE cartridges offer orders of magnitude greater capacity, but reduced spatial resolution.
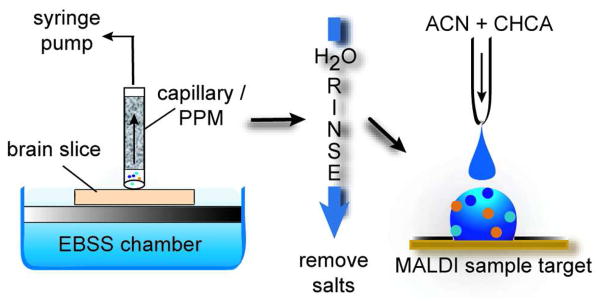
In an effort to address these limitations, we modify the previous approaches by employing PPMs to collect and desalt secreted peptides from extracellular media released upon stimulation. Our goal in using these size-adjustable PPM capillaries is to tailor the peptide collection process to meet the needs of a specific application. Specifically, capillaries containing monolithic columns, fabricated in-situ, are placed in close proximity to a bag cell cluster from the A. californica abdominal ganglion, as well as cortical regions of a mouse coronal brain slice (see Figure 1). Upon electrical or chemical stimulation, the placement of a PPM-containing capillary within a millimeter of the tissue region of interest allows the collection of releasate. The secreted chemicals are retained on the monolith, washed of excess salts, released from the PPM, and detected with MALDI time-of-flight (TOF) MS. The ability to custom design the micron-scale SPE column via photo-initiated polymerization offers a greater binding capacity in a shorter period of time than the direct bead approach. This also preserves greater spatial information than the larger diameter SPE cartridge approach, which gains its binding capacity at the cost of spatial information.37 Moreover, the 200 μm inner diameter of the capillary containing the monolith allows peptide collection from heterogeneous biological tissues with greater spatial differentiation than an SPE cartridge.
Figure 1.
Schematic showing the sampling process from a mouse brain slice using a capillary loaded with PPM; the individual steps shown include peptide collection, a monolith rinse and peptide deposition onto the MALDI target for MS characterization.
Experimental
Materials and Reagents
Lauryl methacrylate, 96% (LMA), ethylene glycol dimethacrylate, 98% (EDMA), benzoin methyl ether, 96%, 3-(trimethoxysilyl)propyl methacrylate, 98% (3-TMSM), L-leucine, fluorescein, angiotensin II, fluorescein isothiocyanate (FITC)-labeled insulin (from bovine pancreas), a-cyano-4-hydroxycinnamic acid (CHCA), and sodium hydroxide were all purchased from Sigma-Aldrich (St. Louis, MO). Reagent grade 2-propanol, methanol, ethanol, and acetonitrile (ACN) were obtained from Fisher Scientific (Fairlawn, NJ). Artificial seawater (ASW) for Aplysia experiments consisted of 460 mM NaCl, 10 mM KCl, 10 mM CaCl2, 22 mM MgCl2, 26 mM MgSO4, and 10 mM HEPES at pH 7.8. Polyimide-coated flexible fused silica capillary tubing (360 μm outer diameter × 200 μm inner diameter) was purchased from Polymicro Technologies (Phoenix, AZ). Ultrapure H2O was from a Millipore Milli-Q water purification system (Bedford, MA).
Porous Polymer Monolith Fabrication
LMA-co-EDMA porous polymer monoliths were fabricated in ~55 mm long pieces of fused silica capillary. Ultraviolet (UV) transparent windows, ~10 mm in length, were created by removing the polymer coating at the end of each fused silica capillary piece. The capillaries were connected to 0.01″ × 0.03″ tygon tubing (Cole-Parmer, Vernon Hills, IL) and filled with 1 M NaOH for 10 min to activate the silica surface, rinsed with ultrapure H2O for 10 min, coated with 20% 3-TMSM silanizing agent in ethanol, conditioned with porogenic solvent (50 wt% methanol, 50 wt% 2-propanol) for 10 min, and finally filled with the polymerization mixture (40 wt% monomer mix: 55 wt% LMA, 35 wt% EDMA, 10 wt% 3-TMSM, and 0.4% benzoin methyl ether with respect to monomer wt; 60 wt% porogenic solvent; adapted from Bedair and Oleschuk16). Here, the monomer mix included an addition of 3-TMSM39 to further promote adhesion during the polymerization and silanization. Once filled with the polymerization mixture, the capillaries with UV-transparent windows were exposed to 365 nm longwave UV radiation with a handheld lamp (UVP, Upland, CA) for 10 min at a distance of 3 cm. Upon fabrication, the PPMs remained untouched for approximately 5 min to eliminate any reactive species. The completed monolithic columns were then rinsed with 10 mL of porogenic solvent using a pressurized flow and stored in solvent until use.
Characterization of Monolithic Material
To observe the degree of polymerization and to characterize the porous material, a Philips XL30 environmental scanning electron microscope (FEI Company, Hillsboro, OR) in the backscattered electron mode was employed. The fused silica capillary piece was coated in gold using a Desk II turbo sputter coater (Denton Vacuum, Moorestown, NJ) and positioned vertically on the stage. Images were collected of the polymerized material at the capillary end.
The binding capacity of the LMA-co-EDMA monoliths was experimentally determined as a function of analyte size, as follows. Monolithic columns were first activated with 200 μL of ACN and then conditioned with 200 μL of ASW (high salt Aplysia bath solution) via pressure-induced flow delivered manually with a gastight syringe (Sigma-Aldrich) connected via fluid transfer fittings (Upchurch Scientific, Oak Harbor, WA). Using a PHD 22/2000 syringe pump (Harvard Apparatus, Holliston, MA) operating in the refill/withdraw mode at 0.2 μL/min, 1.0 mM of fluorescein in ASW was then loaded on the monolith. Fluorescein load amounts were determined by volume injection. Following loading, water was rinsed through the PPM material for 5 min at 0.05 mL/min using the syringe pump in the infuse mode. Bound analyte was then released with 5 μL of ACN delivered via the syringe pump operated in the infuse mode at 0.5 μL/min. The collected 5 μL sample was loaded via capillary action in a coating-free capillary piece and positioned in a custom-built holder (School of Chemical Sciences Machine Shop, University of Illinois at Urbana-Champaign, IL) adapted for mounting on a home-built laser-induced fluorescence system (previously described40) equipped with a 488 nm Ar+ ion laser (Melles Griot, Carlsbad, CA). The resulting fluorescence intensity was collected and averaged over a 60 s time period. This series of steps (from PPM activation to fluorescence detection) was repeated using the same monolith and varying load volumes in order to construct a binding curve. The same process was repeated to generate binding curves for 0.5 mM FITC-insulin in ASW, as well as for 0.5 mM FITC-insulin with 0.25 mM leucine in ASW. Curves were constructed for each analyte and/or analyte pair in triplicate with each experiment corresponding to the full range of analyte loading volumes on unique monolith columns. The final curve was the average of the triplicate curves. To ensure proper release of the analyte(s) between sequential runs, a loaded sample was collected in five 1 μL fractions and each fraction was diluted with 4 μL ACN for laser-induced fluorescence detection. The fluorescence signal was collected for each fraction over a 60 s time period and compared with that of background ACN.
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry
MALDI-TOF MS was performed with an Ultraflex II TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). Mass spectra were obtained with external calibration in reflectron mode with typical mass accuracies within 100 ppm. Spectra represent the accumulation of 300 shots and are presented as acquired signals without baseline correction or data filtering. Releasates were determined as stimulation-dependent analytes and were distinguished from the chemical background by comparing mass spectra from samples obtained before and during electrophysiological stimulation. Releasate analytes were initially identified by m/z. In cases where secreted peptides were collected in sufficient amounts, sequence identification was verified via tandem MS. MS analyses were performed with the accompanying Flex Analysis and BioTools software (Bruker Daltonics).
Limits of Detection
Limit of detection (LOD) studies for peptides collected with PPM capillaries were carried out using a standard solution containing 8.0 μM insulin (0.12 nmol) and 8.6 μM angiotensin I (0.13 nmol) in ASW and with successive 10-fold dilutions: 1/10, 1/100, and 1/1000. Monolithic columns were activated, conditioned, loaded, and rinsed as described above with a 15 μL load volume. To release the bound analyte, an aliquot (~4 μL) of ACN was spotted onto a MALDI target previously prepared with CHCA matrix. Less concentrated samples, 1/100 and 1/1000, are more easily observed when the bound sample is eluted with ~5 μL of ACN and collected in a sample vial prior to spotting the MALDI target. Using this spotting method, a more homogenous sample is prepared.
Invertebrate CNS Releasate Collection
A. californica animals (100–150 g) were obtained from Vivida Biological (Santa Barbara, CA). The abdominal ganglia and then the bag cell clusters were removed and placed in an ASW bath at room temperature (20–24 °C). Capillaries containing ~10 mm long monolithic columns were positioned directly above, but not in contact with, the surface of the bag cell region of the abdominal ganglion. The capillaries were connected to a syringe pump operating in refill/pull mode using 0.01″ × 0.03″ tygon tubing (Cole-Parmer, Vernon Hills, IL), typically with a collection rate of 200 nL/min.
The neuronal electrical potential of the A. californica bag cells was monitored and stimulated with extracellular suction electrodes as described previously.41 Electrophysiological data were stored digitally using a Digidata 1200 A/D converter and pCLAMP 7 software from Axon Instruments (Union City, CA). Extracellular sampling with the PPM capillaries was performed before and during electrical stimulation by switching on the syringe pump for 20 min collection intervals at a rate of 200 nL/min. Each PPM capillary was immediately disconnected following sample collection and rinsed with ultrapure H2O to remove saline salts. Collected releasates were eluted by rinsing the capillary with 100% ACN. Approximately 1 μL aliquots of releasate in ACN were spotted on five consecutive MALDI target sample regions previously prepared with CHCA matrix. The target was loaded in the mass spectrometer and mass spectra were generated.
EX VIVO Vertebrate Brain Slice Releasate Collection
Brain slices containing the cortical regions were prepared from mice bred from the FVB.129P2-fmr1tm1Cgr background and housed at the Beckman Institute animal facility (University of Illinois at Urbana-Champaign). Mice were sacrificed by rapid decapitation in accordance with protocols established by the University of Illinois Institutional Animal Care and Use Committee and in accordance with all state and federal regulations. Brains were removed, and 400 μm thick sections were prepared with a Vibratome (Vibratome 3000 Series; Ted Pella, Redding, CA). Sections were placed into a saline-perfused brain slice chamber (AutoMate Scientific, Inc., Berkeley, CA) equipped with a proportional temperature controller and gas perfusion bubbler. Tissues were perfused with typical Earle’s balanced salt solution (EBSS) without phenol red and supplemented with 24.6 mM of glucose, 26.2 mM of NaHCO3, and 2.5 mg/L of gentamycin. EBSS was maintained at saturated 95% O2/5% CO2 at 37 °C and pH 7.4.
Before collection, monolithic columns were first activated with 200 μL of ACN and then conditioned with 200 μL of EBSS via pressure-induced flow delivered manually with a gastight syringe (Sigma-Aldrich) and the appropriate fluid transfer fittings (Upchurch Scientific). Monoliths were connected to a syringe pump operating in refill/pull mode using 0.01″ × 0.03″ tygon tubing (Cole-Parmer) and placed over the region of interest until it was almost touching, within a millimeter, determined via visual inspection with a microscope. Slice regions were stimulated by dropwise application of elevated (55 mM) KCl-containing EBSS as described previously.37 As above, extracellular samples from the tissue surface were collected at a collection rate of 200 nL/min for 20 min collection intervals. Samples were prepared for MALDI TOF MS as above.
Results and Discussion
Porous Polymer Monolith Characteristics
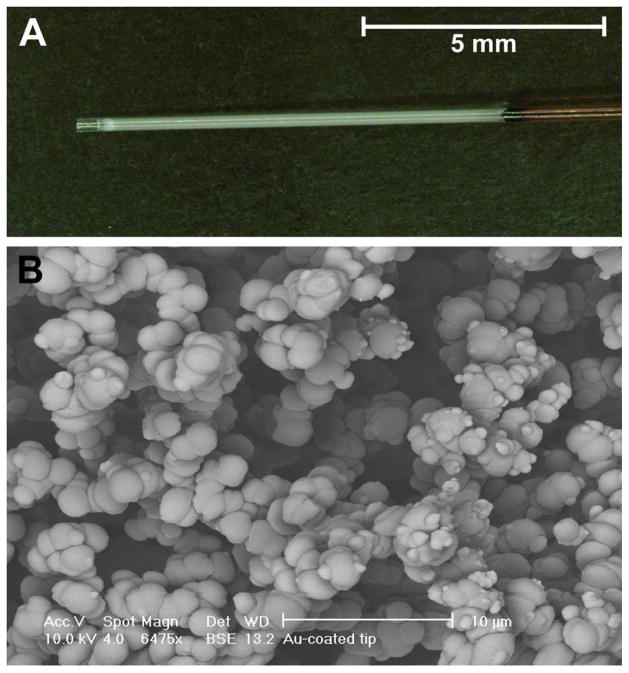
An LMA-EDMA block co-polymer, fabricated in a UV-transparent region at the end of a fused-silica capillary, is depicted in Figure 2A. An empty region at the exit of the capillary, ~0.5 mm in length, generally forms during polymerization due to a lack of applied pressure. This clear region is advantageous for the direct collection of biological samples, as the organic monolith material does not come in direct contact with the tissue, even if the capillary touches it. The SEM image, Figure 2B, shows the monolithic material at a capillary cross-section. The micron-sized through-pores made from the nanoporous polymer backbone are more easily observed with the use of a backscattered electron detector. Images collected using the more traditional secondary electron detector (not pictured) resemble those of LMA-EDMA monoliths described in previous reports.9, 42
Figure 2.
An LMA-EDMA block co-polymer fabricated in a UV-transparent region at the end of a fused-silica capillary. (A) White-light micrograph showing an ~10 mm long monolithic bed. (B) SEM image of the monolithic material at a capillary cross-section.
Prior to employing the hydrophobic PPMs for releasate collection, it is important to determine the binding capacity of different analytes in order to validate the technique for such mass/volume-limited samples, as well as to fabricate an optimal column length. Binding capacities were experimentally determined using the assembled monoliths via fluorescence spectroscopy. Due to the complex nature of the neuronal releasate, we present binding capacities for three distinct analyte types: small molecules, represented by 1.0 mM fluorescein in ASW; peptides and proteins, represented by 0.5 mM FITC-insulin in ASW; and a mixture, represented by 0.5 mM FITC-insulin with 0.25 mM leucine in ASW. The binding curve included in Figure 3 corresponds to the mixed sample and is representative of all sample types. As the volume of sample introduced into the PPM increases, there is an increase in fluorescence intensity upon its release. However, once the capacity to retain a compound is reached, no more is retained; thus, the fluorescence intensity of the collected materials levels off as the PPM region reaches its capacity. The binding capacity for 0.5 mM FITC-insulin in the presence of 0.25 mM leucine is 0.15 ± 0.03 nmol/mm of monolith. As expected, the single analyte systems exhibit higher binding capacities with 0.54 ± 0.03 nmol/mm for 1.0 mM fluorescein and 0.24 ± 0.02 nmol/mm for 0.05 mM FITC-insulin. As the binding capacity is a function of both binding affinity and saturation coverage, it is not surprising that the larger molecules have lower capacities. The PPMs collect a range of analytes as long as the amount collected is under the capacity of the monolith. The nmol/mm binding capacities observed are consistent with previously published binding capacities of similar materials.23, 43 Furthermore, as indicated in the binding curves, column-to-column variability is not as large for the single analyte systems—8.5% relative standard deviation (RSD) for FITC-insulin and 5.6% RSD for fluorescein—as compared to 20% RSD for the FITC-insulin + leucine system. For a given binding curve, the same PPM was used to collect each data point to ensure consistency. In results not shown here, the first three 1 μL fractions exhibited fluorescence signal, while fractions four and five displayed the same relative intensity as the ACN background, indicating that 3 μL should be sufficient to elute peptides.
Figure 3.
Binding curve demonstrating the capacity of the PPMs. The curve corresponds to a dual analyte sample containing 0.5 mM FITC-insulin and 0.25 mM leucine in ASW, but is representative of all tested sample types (n = 3). As the volume of the collected sample increases, there is an increase in fluorescence intensity. Eventually, the fluorescence intensity levels off as the column reaches capacity.
Limits of Detection
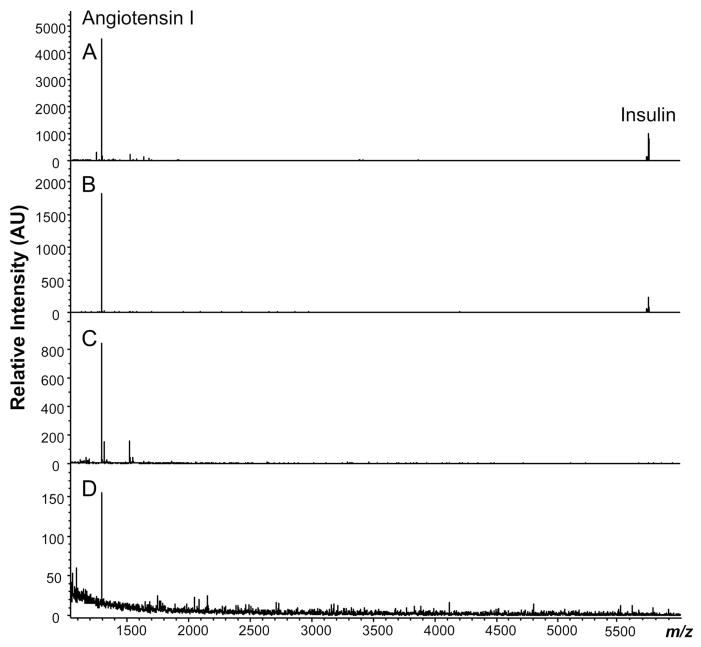
LOD studies, performed with insulin and angiotensin I peptide standards, were carried out to determine the overall ability of the PPM sampling and MALDI-TOF MS detection approach for two peptides with relatively low and high m/z ratios, respectively. Mass spectra corresponding to the collection of the standard peptide solution and three successive 10-fold dilutions are included in Figure 4A–D, respectively. The 1/1000 dilution contains 8.6 nM angiotensin I (0.17 ng) and the 1/10 dilution contains 803 nM insulin (69.7 ng) in the 15-μL load volume. These monoliths are, therefore, suitable for the collection of the nanogram amounts of peptides previously observed in bag cell releasates.44
Figure 4.
PPM-MALDI MS LODs. Mass spectra corresponding to the collection of a standard peptide solution: (A) 8.0 μM insulin (0.12 nmol) and 8.6 μM angiotensin I (0.13 nmol) in ASW; (B) 12 pmol insulin and 13 pmol angiotensin I; (C) 1.2 pmol insulin and 1.3 pmol angiotensin I; (D) 120 fmol insulin and 130 fmol of angiotensin I. Peaks corresponding to angiotensin I are observed at 1296.7 m/z and insulin at 5730.6 m/z.
The ability to characterize lower amounts of peptide is important for some applications. A longer monolith, for example, may be able to collect and concentrate less-abundant analytes in chemically heterogeneous biological samples when larger load volumes and longer collection times are employed. Alternatively, affinity collection columns could be utilized in such an application to preferentially collect a single analyte of interest from complex environments,45, 46 thereby eliminating the competition for binding among other components while increasing the collection efficiency of the analyte of interest. Thus, the fabrication of tailored monoliths could enable optimized release collections for a range of applications.
Invertebrate Releasate
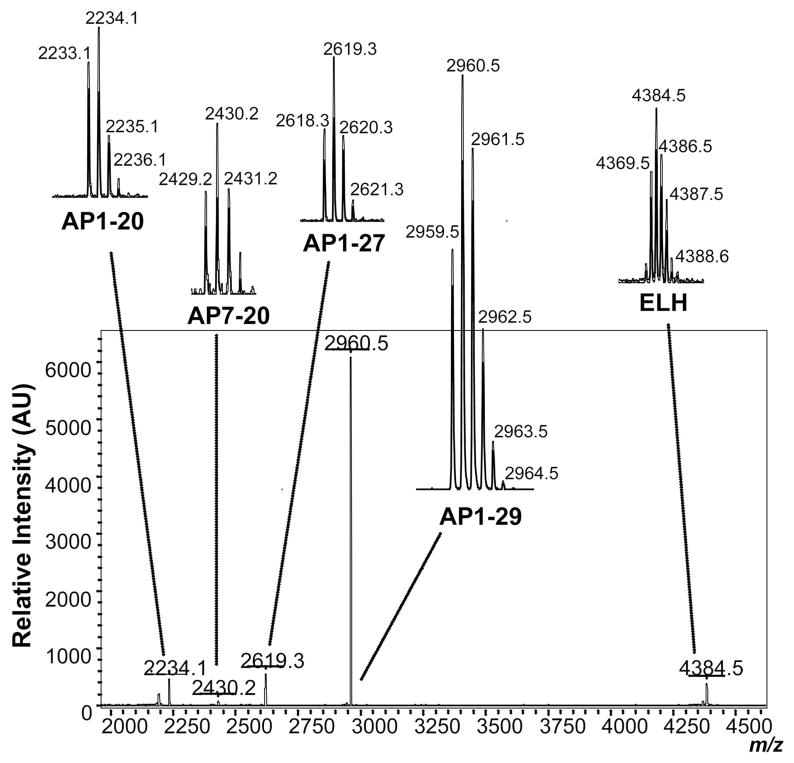
Release from the A. californica bag cell cluster served as a model system to evaluate the PPMs for collection and concentration of peptides. The released peptides collected during the first 20-min time period after stimulation are shown in Figure 5. The 20-min collection period was used as it is consistent with the length of peptide release from the bag cell neurons after electrical stimulation. Acidic peptide (AP)-related peptides are observed at 2234.1, 2430.2, 2619.3, and 2960.5 m/z, representing AP(1–20), AP(7–20), AP(1–27), and full length AP(1–29), respectively. In addition, egg laying hormone (ELH) is observed at 4384.5 m/z. Both AP and ELH have been characterized in the releasate of A. californica bag cell neurons.34, 36 These results also demonstrate that detectable levels of peptides are collected with the PPM from the extracellular media containing high levels of salts and other compounds present.
Figure 5.
Bag cell releasate. Mass spectra containing two known Aplysia californica bag cell peptides: acidic peptide (AP) at 2234.1, 2430.2, 2619.3, and 2960.5 m/z representing segments 1–20, 7–20, 1–27, and 1–29, respectively; and egg laying hormone (ELH) at 4384.5 m/z.
Mouse Brain Slice Releasate
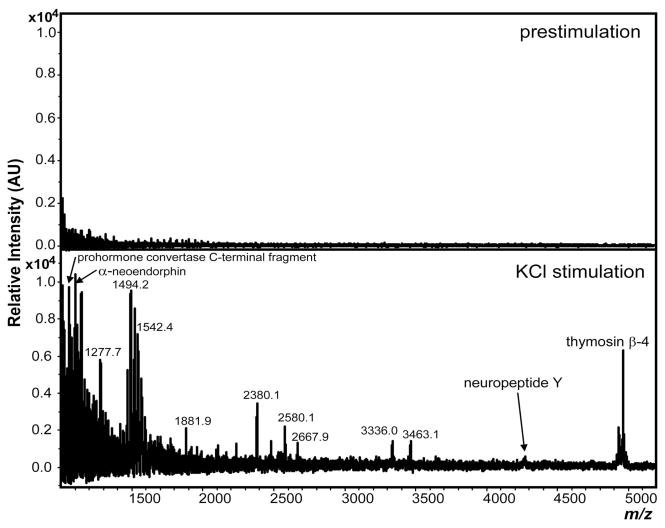
Release from the cortex region of the mouse brain slice demonstrates the application of this sampling scheme to the mammalian CNS. Figure 6 shows the mass spectra of collections obtained prior to and during cortex stimulation. Peaks were identified by matching the observed mass of the intact ion with the predicted masses of known peptides based on our group’s previous work38 as well as that from the Fricker group.47 Because many of the peaks are low intensity, the ability to accurately determine their m/z is hindered; here, the releasate peak assignments were within 100 ppm of the predicted mass. The peaks observed at 1057.7, 1099.6, 4269.1 and 4961.4 m/z match the predicted masses for C-terminal peptide from prohormone convertase 2, α-neoendorphin, neuropeptide Y and thymosin β-4, respectively.
Figure 6.
Mouse cortex releasate. MALDI-MS analysis of releasates collected from prestimulated (top spectrum) and stimulated (KCl, 55mM) (bottom spectrum) cortex from mice. In contrast to the relatively few analytes observed in the prestimulation samples, collections obtained during and after stimulation yielded an abundance of peptides. Using accurate mass match, several peaks were identified as peptides derived from C-terminal peptide from prohormone convertase 2, α-neoendorphin, neuropeptide Y and thymosin β-4, respectively.
Conclusions
PPM columns are well suited as collection, concentration, and treatment tools prior to MALDI-TOF MS detection. Fabricated at the end of a fused silica capillary piece, columns with nmol/mm capacities and sub-pmol LOD are used to collect and desalt A. californica bag cell and mouse brain peptide releasates. As it can be necessary to collect secreted chemicals from spatially defined release sites such as the SCN, the small (200 μm) inner diameter of the capillary has the advantage as it allows precise positioning and thus enables sample collection from such spatially defined areas of biological tissues. While individual beads placed over a release site offer higher spatial resolution, the amount of peptide that an individual bead can collect is relatively low, so that trace peptides are difficult to characterize and tandem MS approaches are hindered. The use of PPMs adds flexibility to the available neuronal release tool set; the material, including diameter and length, can be tailored to the specific application at hand, thereby expanding the ability to monitor peptide dynamics from ex vivo brain slices.
Acknowledgments
We gratefully acknowledge William T. Greenough for providing the mice for the brain slices, Agatha Luszpak for assistance with the brain slice preparation/techniques, and Sally Yoon for her laboratory assistance (all at the University of Illinois). Scanning electron microscopy was performed in the Imaging Technology Group at the Beckman Institute under the direction of Scott Robinson. This material is based upon work supported by the National Institute of Drug Abuse under Award No. DA017940 and Award No. DA018310 to the UIUC Neuroproteomics Center on Cell-Cell Signaling, and the National Institutes of Neurological Disease and Stroke under Award No. NS031609.
References
- 1.Hjerten S, Liao JL, Zhang R. J Chromatogr. 1989;473:273–275. [Google Scholar]
- 2.Svec F, Frechet JMJ. Anal Chem. 1992;64:820–822. doi: 10.1021/ac00035a008. [DOI] [PubMed] [Google Scholar]
- 3.Tennikova TB, Nahunek M, Svec F. J Liq Chromatogr. 1991;14:2621–2632. [Google Scholar]
- 4.Svec F, Huber CG. Anal Chem. 2006;78:2101–2107. doi: 10.1021/ac069383v. [DOI] [PubMed] [Google Scholar]
- 5.Jungbauer A, Hahn R. J Sep Sci. 2004;27:767–778. doi: 10.1002/jssc.200401812. [DOI] [PubMed] [Google Scholar]
- 6.Svec F. J Sep Sci. 2004;27:1419–1430. doi: 10.1002/jssc.200401825. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka N, Kobayashi H, Nakanishi K, Minakuchi H, Ishizuka N. Anal Chem. 2001;73:420A–429A. doi: 10.1021/ac012495w. [DOI] [PubMed] [Google Scholar]
- 8.Xie C, Fu H, Hu J, Zou H. Electrophoresis. 2004;25:4095–4109. doi: 10.1002/elps.200406155. [DOI] [PubMed] [Google Scholar]
- 9.Le Gac S, Carlier J, Camart JC, Cren-Olive C, Rolando C. J Chromatogr B Analyt Technol. 2004;808:3–14. doi: 10.1016/j.jchromb.2004.03.067. [DOI] [PubMed] [Google Scholar]
- 10.Chen HS, Rejtar T, Andreev V, Moskovets E, Karger BL. Anal Chem. 2005;77:2323–2331. doi: 10.1021/ac048322z. [DOI] [PubMed] [Google Scholar]
- 11.Yu C, Mutlu S, Selvaganapathy P, Mastrangelo CH, Svec F, Frechet JMJ. Anal Chem. 2003;75:1958–1961. doi: 10.1021/ac026455j. [DOI] [PubMed] [Google Scholar]
- 12.Luo Q, Zou H, Zhang Q, Xiao X, Ni J. Biotechnol Bioeng. 2002;80:481–489. doi: 10.1002/bit.10391. [DOI] [PubMed] [Google Scholar]
- 13.Pan Z, Zou H, Mo W, Huang X, Wu R. Anal Chim Acta. 2002;466:141–150. [Google Scholar]
- 14.Peterson DS, Rohr T, Svec F, Frechet JMJ. Anal Chem. 2002;74:4081–4088. doi: 10.1021/ac020180q. [DOI] [PubMed] [Google Scholar]
- 15.Rohr T, Yu C, Davey MH, Svec F, Frechet JMJ. Electrophoresis. 2001;22:3959–3967. doi: 10.1002/1522-2683(200110)22:18<3959::AID-ELPS3959>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 16.Bedair MF, Oleschuk RD. Anal Chem. 2006;78:1130–1138. doi: 10.1021/ac0514570. [DOI] [PubMed] [Google Scholar]
- 17.Koerner T, Oleschuk RD. Rapid Commun Mass Spectrom. 2005;19:3279–3286. doi: 10.1002/rcm.2181. [DOI] [PubMed] [Google Scholar]
- 18.Koerner T, Turck K, Brown L, Oleschuk RD. Anal Chem. 2004;76:6456–6460. doi: 10.1021/ac049438y. [DOI] [PubMed] [Google Scholar]
- 19.Lee SSH, Douma M, Koerner T, Oleschuk RD. Rapid Commun Mass Spectrom. 2005;19:2671–2680. doi: 10.1002/rcm.2109. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y, Li C, Kameoka J, Lee KH, Craighead HG. Lab Chip. 2005;5:869–876. doi: 10.1039/b503025k. [DOI] [PubMed] [Google Scholar]
- 21.Svec F. J Chromatogr B Analyt Technol. 2006;841:52–64. doi: 10.1016/j.jchromb.2006.03.055. [DOI] [PubMed] [Google Scholar]
- 22.Xie S, Svec F, Frechet JMJ. Chem Mater. 1998;10:4072–4078. [Google Scholar]
- 23.Yang Y, Li C, Lee KH, Craighead HG. Electrophoresis. 2005;26:3622–3630. doi: 10.1002/elps.200500121. [DOI] [PubMed] [Google Scholar]
- 24.Yu C, Davey MH, Svec F, Frechet JMJ. Anal Chem. 2001;73:5088–5096. doi: 10.1021/ac0106288. [DOI] [PubMed] [Google Scholar]
- 25.Fan Y, Feng YQ, Da SL, Shi ZG. Anal Chim Acta. 2004;523:251–258. [Google Scholar]
- 26.Kato K, Silva MJ, Needham LL, Calafat AM. Anal Chem. 2005;77:2985–2991. doi: 10.1021/ac0481248. [DOI] [PubMed] [Google Scholar]
- 27.Lim LW, Hirose K, Tatsumi S, Uzu H, Mizukami M, Takeuchi T. J Chromatogr A. 2004;1033:205–212. doi: 10.1016/j.chroma.2004.01.045. [DOI] [PubMed] [Google Scholar]
- 28.Zhang K, Wu S, Tang X, Kaiser NK, Bruce JE. J Chromatogr B Analyt Technol. 2007;849:223–230. doi: 10.1016/j.jchromb.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 29.Peterson DS, Rohr T, Svec F, Frechet JMJ. Anal Chem. 2003;75:5328–5335. doi: 10.1021/ac034108j. [DOI] [PubMed] [Google Scholar]
- 30.Courtois J, Fischer G, Sellergren B, Irgum K. J Chromatogr A. 2006;1109:92–99. doi: 10.1016/j.chroma.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 31.Zhang M, Xie J, Zhou Q, Chen G, Liu Z. J Chromatogr A. 2003;984:173–183. doi: 10.1016/s0021-9673(02)01847-2. [DOI] [PubMed] [Google Scholar]
- 32.Paras CD, Kennedy RT. Anal Chem. 1995;67:3633–3637. doi: 10.1021/ac00116a003. [DOI] [PubMed] [Google Scholar]
- 33.Paras CD, Qian W, Lakey JR, Tan W, Kennedy RT. Cell Biochem Biophys. 2000;33:227–240. doi: 10.1385/cbb:33:3:227. [DOI] [PubMed] [Google Scholar]
- 34.Garden RW, Moroz LL, Moroz TP, Shippy SA, Sweedler JV. J Mass Spectrom. 1996;31:1126–1130. doi: 10.1002/(SICI)1096-9888(199610)31:10<1126::AID-JMS403>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 35.Hatcher NG, Richmond TA, Rubakhin SS, Sweedler JV. Anal Chem. 2005;77:1580–1587. doi: 10.1021/ac0487909. [DOI] [PubMed] [Google Scholar]
- 36.Rubakhin SS, Page JS, Monroe BR, Sweedler JV. Electrophoresis. 2001;22:3752–3758. doi: 10.1002/1522-2683(200109)22:17<3752::AID-ELPS3752>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 37.Hatcher NG, Atkins N, Jr, Annangudi SP, Forbes AJ, Kelleher NL, Gillette MU, Sweedler JV. Proc Natl Acad Sci USA. 2008;105:12527–12532. doi: 10.1073/pnas.0804340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Annangudi SP, Kim SH, Weiler IJ, Rubakhin SS, Greenough WT, Sweedler JV. Proceedings of the 56th ASMS Conference on Mass Spectrometry and Allied Topics, Abstract #399; Denver, CO. 2008. [Google Scholar]
- 39.Delaunay-Bertoncini N, Demesmay C, Rocca JL. Electrophoresis. 2004;25:3204–3215. doi: 10.1002/elps.200305975. [DOI] [PubMed] [Google Scholar]
- 40.Tulock JJ, Shannon MA, Bohn PW, Sweedler JV. Anal Chem. 2004;76:6419–6425. doi: 10.1021/ac049601p. [DOI] [PubMed] [Google Scholar]
- 41.Hatcher NG, Sweedler JV. J Neurophysiol. 2008;99:333–343. doi: 10.1152/jn.00968.2007. [DOI] [PubMed] [Google Scholar]
- 42.Carlier J, Arscott S, Thomy V, Camart JC, Cren-Olive C, Le Gac S. J Chromatogr A. 2005;1071:213–222. doi: 10.1016/j.chroma.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 43.Baryla NE, Toltl NP. Analyst. 2003;128:1009–1012. [Google Scholar]
- 44.Wayne NL. J Endocrinol. 1995;147:1–4. doi: 10.1677/joe.0.1470001. [DOI] [PubMed] [Google Scholar]
- 45.Bedair M, El Rassi Z. J Chromatogr A. 2005;1079:236–245. doi: 10.1016/j.chroma.2005.02.084. [DOI] [PubMed] [Google Scholar]
- 46.Bedair M, El Rassi Z. J Chromatogr A. 2004;1044:177–186. doi: 10.1016/j.chroma.2004.03.080. [DOI] [PubMed] [Google Scholar]
- 47.Lim J, Berezniuk I, Che FY, Parikh R, Biswas R, Pan H, Fricker LD. J Neurochem. 2006;96:1169–1181. doi: 10.1111/j.1471-4159.2005.03614.x. [DOI] [PubMed] [Google Scholar]