Abstract
A method that enables metabolomic profiling of single cells and subcellular structures is described using capillary electrophoresis coupled to electrospray ionization time-of-flight mass spectrometry. A nebulizer-free coaxial sheath-flow interface completes the circuit and provides a stable electrospray, yielding a signal with a relative standard deviation of under 5% for the total ion electropherogram. Detection limits are in the low nanomolar range (i.e., < 50 nM (< 300 amol)) for a number of cell-to-cell signaling molecules, including acetylcholine (ACh), histamine, dopamine, and serotonin. The instrument also yields high efficiency separations, e.g., ~600,000 for eluting ACh bands. The utility of this setup for single cell metabolomic profiling is demonstrated with identified neurons from Aplysia californica—the R2 neuron and metacerebral cell (MCC). Single cell electropherograms are reproducible, with a large number of metabolites detected; more than 100 compounds yield signals of over 104 counts from the injection of only 0.1% of the total content from a single MCC. Expected neurotransmitters are detected within the cells (ACh in R2 and serotonin in MCC), as are compounds that have molecular masses consistent with all of the naturally-occurring amino acids (except cysteine). Tandem MS using a quadrupole time-of-flight tandem mass spectrometer distinguishes ACh from isobaric compounds in the R2 neuron and demonstrates the ability of this method to characterize and identify metabolites present within single cells.
Introduction
The field of comparative metabolomics is especially useful in medical and life science research. More specifically, the ability to uncover and evaluate biochemical differences within healthy and diseased organisms provides information as to the underlying cause(s) of disease, which in turn suggests targets for pharmacological intervention.1–6 A variety of analytical platforms have been developed to facilitate these and other types of metabolomics experiments.5, 7–12 For instance, capillary electrophoresis (CE), gas chromatography and liquid chromatography (LC) have been coupled to MS detection methods in order to perform metabolomic profiling of a variety of biological samples.3, 7, 9, 10, 13–17
There are a number of investigative queries for which comparative metabolomics experiments on a single selected cell would be particularly advantageous. For example, within the nervous system, neighboring cells often exhibit different chemical compositions and functional roles (see http://www.brainmap.org/). Thus, in order to characterize even the simplest neural circuit, it is necessary to determine not only the connectivity between the relevant cells, but also the signaling molecules used by each individual neuron (and perhaps the neighboring supporting cells) within a neuronal network. Furthermore, although even adjacent neurons have distinct signaling molecule and protein complements,18–21 differences in the cellular metabolome are less well studied. Clearly, an analytical platform capable of assaying the small molecule content of single cells would be an invaluable tool for gaining insights into these areas.
Both CE21–24 and LC23, 25, 26 are well-suited for single cell measurements. Using various detectors with CE has been effective for characterizing chemical content and even release from individual cells.22–24, 27–31 There are a number of features of CE that make it a method of choice for such analyses. For example, it is possible to simultaneously concentrate and separate analytes.32–34 In addition, because the scaling laws of CE make it amenable to small-volume sampling, it has been used extensively for single cell and even subcellular analyses, and is well-suited as a separation method for use in metabolomics experiments.35–37
MS has also been used to characterize the contents of individual neurons, with considerable efforts devoted to characterizing their peptide complements. Many of these studies employed direct MS profiling; however, pairing CE or LC to MS often leads to better analyte coverage. For example, several studies have used off-line fractionation and matrix assisted laser desorption/ionization in order to profile the peptide content of individual cells.18, 19, 38, 39
Electrospray ionization (ESI)-MS offers a number of capabilities that make it appropriate for single cell applications. It is amenable to low flow-rate applications,40–44 and enables the detection of analytes of interest not readily detected by other methods. One such example is the neurotransmitter acetylcholine (ACh), which is not directly electroactive, does not absorb light appreciably, and cannot readily be derivatized by the fluorogenic reagents commonly used in conjunction with laser-induced fluorescence detection methods. Furthermore, ESI coupled with tandem MS (MS/MS) allows for the identification and/or characterization of unknown or unexpected compounds, such as metabolites, via their mass-to-charge ratio (m/z) and MS/MS fragmentation pattern, and complements the other detection schemes used for single cell measurements.
CE-ESI-MS interface development has been an active area of investigation for over 20 years.45–47 However, completing the electrical circuit required for CE in a manner that results in a stable electrospray and suitable detection limits has been a challenge. System stability and sensitivity are both essential in single cell metabolomic investigations. High sensitivity can be the key to success, as sample preparation for single cell analysis inevitably involves some dilution of the femtoliter to nanoliter cell volumes to levels that can be manipulated in the laboratory. CE-ESI-MS analysis has been demonstrated with sufficient sensitivity to detect proteins from within single cells.25, 48, 49 In addition to adequate sensitivity, a robust system is required, so that run-to-run variations reflect real differences between samples and not instabilities in instrument performance. Here, we describe a sensitive and robust CE-ESI-MS system, and demonstrate its potential for single cell and subcellular metabolomics applications using an ESI-TOF mass spectrometer and an ESI-Qq-TOF-tandem mass spectrometer, both from Bruker Daltonics.
EXPERIMENTAL SECTION
Chemicals
Sigma-Aldrich (St. Louis, MO) was the source for all chemicals, unless stated otherwise. Ultrapure deionized water was used for all solution preparation, and was obtained from an Elga Purelab Ultra water system (U.S. Filter, Lowell, MA).
Animals
Adult Aplysia californica weighing 175–250 g were obtained from Charles M. Hollahan (Santa Barbara Marine Bio., Santa Barbara, CA). Animals were maintained in constantly circulated, aerated artificial seawater (Instant Ocean; Aquarium Systems, Mentor, OH), chilled to 14 °C.
Sample Preparation
A. californica were anesthetized by injection into the vascular cavity of a solution containing 390 mM MgCl2 dissolved in water, equal by mass to one-third of each animal’s body weight. Ganglia and adjacent nerves were surgically dissected and placed in artificial seawater (ASW) containing (in mM) 460 NaCl, 10 KCl, 10 CaCl2, 22 MgCl2, 26 MgSO4, and 10 HEPES (pH 7.7) supplemented with antibiotics (100 units/mL penicillin G, 100 μg/mL streptomycin, and 100 μg/mL gentamicin). A 1% protease type IX in ASW-antibiotics solution treatment for 60–120 min (depending on animal size) at 34 °C was used to reduce adherence between cells. This treatment also helps to remove connective tissue surrounding the ganglia and nerves and improves the success of individual neuron isolation. After the protease treatment, ganglia were washed in ASW and stored at 14 °C in the ASW-antibiotics solution until use. Isolation of individual neurons was performed manually using sharpened tungsten needles under visual control, assisted by a Leica MZ 7.5 high-performance stereomicroscope with a 7.9:1 zoom (Leica Microsystems Inc., Bannockburn, IL). While the neuron size depends on the animal, typical cell diameters for animals of this size are ~150 μm for MCC neurons and ~300 μm for R2 neurons.
Capillary Electrophoresis
Aliquots (100–500 nL) of standard solutions or cellular extracts were placed into a custom-fabricated stainless steel nanovial. Approximately 6 nL of an aliquot was then injected into the capillary hydrodynamically by maintaining a height difference of 15 cm between the capillary inlet and outlet for 60 s. After sample injection, the CE inlet was placed into a stainless steel buffer vial; the background electrolyte was 1% formic acid in water. A voltage of 20 kV was applied across the capillary for CE separations using a Bertan high voltage power supply (Valhalla, NY). The voltage was applied gradually, by manually ramping the voltage from 0 V to 20 kV over ~30 s. The injection equipment was enclosed in an acrylic box equipped with safety interlocks. A 100 cm long, 40 μm inner-diameter (ID), 105 μm outer-diameter (OD) fused silica capillary (Polymicro Technologies, Phoenix, AZ) was used for all separations.
Sheath Flow Interface
The capillary outlet was secured into a PEEK tee by using an FEP sleeve and a 10–32 one-piece fitting; this PEEK tee served as the body of the sheath flow interface. Fluidics components, unless otherwise noted, were obtained from Upchurch Scientific (Oak Harbor, WA). A 1″ long segment of HTX-33X stainless steel hypodermic tubing (0.0083″ OD, 0.0065 ID; Small Parts, Inc., Miramar, FL), which served as the sheath flow tube and electrospray needle, was secured to the port of the PEEK tee opposite the port in which the CE capillary was secured, also using an FEP sleeve and 10–32 one-piece fitting. The tip of the ESI needle was polished using 12 μm diamond lapping paper (3M, St. Paul, MN). The sheath liquid, which consisted of 50/50 (v/v) methanol/water with 0.1% (by volume) formic acid, was supplied through the remaining port of the tee. A PHD 22/2000 syringe pump (Harvard Apparatus, Holliston, MA), in combination with a 2.5 mL Gastight syringe (Hamilton, Reno, NV), delivered the sheath liquid. The luer-lock-tipped syringe was connected to a ~1 meter length of PEEK tubing (1/16″ OD, 0.005″ ID) via a PEEK luer-to-fitting adapter and a 10–32 one-piece fitting. This tubing was then secured into the final port of the PEEK tee via another 10–32 one-piece fitting.
Mass Spectrometry
Two mass spectrometers were used—a micrOTOF ESI-TOF-MS and a maXis ESI-Qq-TOF-MS/MS (Bruker Daltonics, Billerica, MA). Both instruments used the same nanospray interface and feature a grounded needle design; the ESI needle (the outlet end of the coaxial sheath-flow sprayer) was connected to ground using a copper wire. The metal inlet orifice of the mass spectrometer was held at −1700 V and 180 °C. A 3-axis translation stage (Model H RH, Line Tool Co., Allentown, PA) was used to optimize the position of the ESI needle in order to provide a stable electrospray, and to optimize the amount of the plume sampled by the mass spectrometer. The ion optics were tuned for the mass range of 50–500 m/z using lithium formate clusters from a direct infusion of 100 μg/mL lithium formate in water at 1 μL/min. The lithium formate solution was also infused after completed CE-ESI-MS runs and used for mass calibration. MS/MS analysis was carried out using the maXis, where argon was used as a collision gas to fragment precursor ions via collision-induced dissociation prior to mass analysis. A suitable collision energy for small molecule fragmentation was found by maximizing intensity of the 87 m/z fragment generated from the dissociation of ACh. This was accomplished by spiking the sheath liquid with 7.5 μM ACh and infusing it through the CE-ESI-MS interface at 1 μL/minute.
RESULTS AND DISCUSSION
CE-ESI-MS Interface Design and Characterization
Several strategies for coupling CE and ESI-MS have emerged—the direct coupling or sheathless interface, and the coaxial sheath-flow interface.50, 51 Each of these approaches has an associated set of benefits and challenges. We selected the sheath-flow interface because it allows the independent optimization of the background electrolyte in the CE capillary and the sheath liquid; typically, the solution characteristics that provide optimal separation and electrospray performance are quite different from one another. Moreover, the use of a sheath flow allows the flow rate of the system to be optimized in order to provide a stable electrospray, which is beneficial since the flow rate through the CE capillary due to electroosmotic flow alone is often too low to maintain a stable Taylor cone.
Even with the inherent benefits of the sheath flow interface, achieving spray stability is, in practice, still difficult at flow rates that are low enough to provide acceptable detection limits. This is due in part to the fact that the dimensions of commonly-used CE capillaries require that the diameter of the sheath-flow needle be much larger than is optimal for stable ESI plume generation at sufficiently low flow rates. Confronting this challenge, we developed a sheath-flow sprayer that accommodates a capillary with a reduced OD (105 μm versus the more typical 365 μm).
Unfortunately, the performance of the initial scaled-down interface was negatively affected by bubbles that formed at the needle tip due to solvent boiling and the generation of gaseous electrolysis products. To prevent bubble formation, several further modifications were made. The tip of the sprayer needle was polished with 12 μm diamond lapping paper in order to reduce the number of available nucleation sites for bubbles. In addition, the CE current was restricted by reducing the ID of the capillary from 50 μm to 40 μm and operating the CE system at a relatively low voltage (20 kV); it was also found that gradually ramping the voltage over 30 s to its final value was helpful in preventing initiation of bubbling. Finally, a heated desolvation chamber was omitted in our interface design. As a result of these modifications, a stable electrospray can be achieved (defined as a relative standard deviation of the baseline under 5%) with flow rates of 250 nL/min up to ~2 μL/min (versus 4–10 μL/min with commercial interfaces); 1 μL/min is typically used, and represents a compromise between enhanced sensitivity at lower flow rates and the ability to use higher electric fields without bubble formation at increased flow rates.
The use of a nanospray interface also enhances sensitivity in that it enables the sprayer position to be optimized so as to maximally sample analytes from the ESI plume. As a result, limits of detection on the order of < 50 nM (< 300 amol) were achieved for 6 nL hydrodynamic injections of a variety of analytes, including dopamine (20 nM), serotonin (5-HT) (35 nM), ACh (5 nM), and histamine (15 nM). Figure 1 shows the CE-ESI-MS analysis of a 6 nL injection of a standard solution containing 75 nM each of histamine, ACh, DA, and 5-HT (450 amol of each injected), and demonstrates the sensitivity of the method. These detection limits are sufficient to carry out metabolomic profiling at the single cell level. A linear MS detector response is observed as a function of analyte concentration (R2~ 0.99) from low nM to tens of μM concentrations, thus allowing quantitative measurements to be made.
Figure 1.
Sensitivity of the CE-ESI-MS method. Analysis of a 6 nL injection of a standard solution containing 75 nM each of histamine, acetylcholine, dopamine, and serotonin (450 amol of each injected). Extracted ion electropherograms with m/z ranges corresponding to the masses of the expected compounds show clear peaks for each analyte, demonstrating the low nM detection limits that can be achieved with the method. Data was processed using a Gaussian smoothing algorithm with a 3-point window.
One issue associated with the use of a sheath-flow interface for CE-ESI-MS is the potential for a reduction in separation quality. Many sheath-flow interfaces use a nebulizer gas in order to facilitate droplet formation and subsequent evaporation. This nebulizer gas can induce siphoning, resulting in a parabolic flow profile within the separation capillary, which leads to band broadening. Our sprayer does not require a nebulizer for operation, as it relies on the formation of a stable Taylor cone to generate an electrospray plume, making high column efficiencies possible. For example, the peak efficiency is ~600,000 for ACh peaks.
Single Cell and Subcellular Analysis
In order to evaluate the suitability of the CE-ESI-MS system for performing single cell metabolomic profiling, several identified neurons from the well-studied neurobiological model A. californica were selected for analysis. The R2 is an identifiable cholinergic neuron, and the extensively studied metacerebral cell (MCC) uses 5-HT as its neurotransmitter.52–54 After isolation, cells were quickly rinsed with a 1 μL aliquot of ultrapure water to remove excess extracellular inorganic salts, and then placed at the bottom of a 200 μL vial. This water rinse facilitates online analyte enrichment via sample stacking, which is dependent on maintaining low conductivity in the sample plug.32 A 5 μL aliquot of an extraction and preservation solution consisting of 49/50/1 (v/v/v) water/methanol/acetic acid was then added to each cell. The acid content of this solution also served to protonate chargeable metabolites, thereby facilitating CE separation and ESI-MS detection. The cells were disrupted using a sharp tungsten needle in order to enable metabolite extraction.
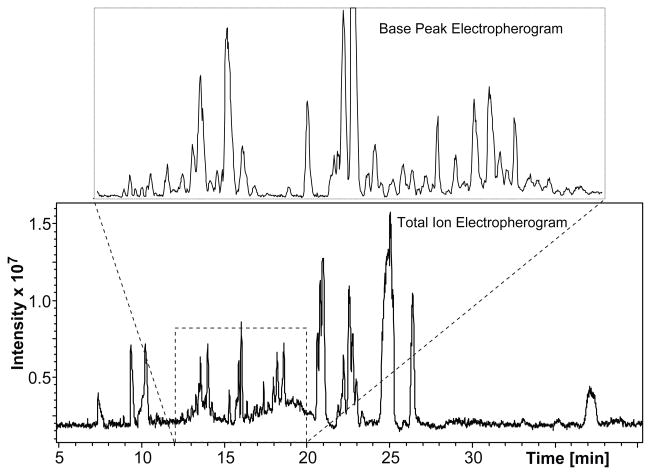
Figure 2 shows the TIE resulting from the injection of 6 nL of the cellular extract, corresponding to 0.1% of the total content, of a single R2 neuron (in this case, using the micrOTOF mass spectrometer). In the base peak electropherogram (Figure 2, inset), a larger number of peaks are evident, as is the quality of the CE separation—many peaks are either baseline resolved or nearly so. As a preliminary evaluation of the capability of the system to perform metabolomic profiling, the R2 CE-ESI-MS data was screened for signals corresponding to ACh and the naturally occurring amino acids. A putative ACh signal was detected, and peaks with m/z consistent with all of the amino acids except cysteine were observed.
Figure 2.
Metabolomic profiling of a single R2 neuron using CE-ESI-MS. An injection of 0.1% of a single R2 neuron yields a TIE with numerous well-defined peaks. The base peak electropherogram also shows a high-quality separation, with a number of peaks that are either base-line separated or nearly so.
In order for variations in the data to be correlated with differences in the samples being analyzed, it is important for the method to be reproducible. Figure 3 compares a subset of extracted ion electropherograms (XIE) generated from two different MCC neurons. Although the time scales have been normalized in order to account for slight differences in the migration times due, in part, to manually ramping the voltage when the separation is initiated, the overall qualitative features of the analyses are similar. The presence of peaks with m/z consistent with 5-HT and all of the naturally-occurring amino acids (except cysteine) was confirmed in the MCC data as well. In addition, over 100 substances gave rise to signals greater than 104 counts, which is encouraging, given that only 0.1% of each cell was injected, and that the MCC neurons are ~4 fold smaller by volume than the R2 neurons.
Figure 3.
Repeatability of single cell CE-ESI-MS metabolomic profiling. Composite extracted ion electropherograms from two different Aplysia californica MCC neurons are highly similar. Time axes have been normalized to account for variations in migration time caused, in part, by manually ramping the CE voltage at the beginning of CE runs.
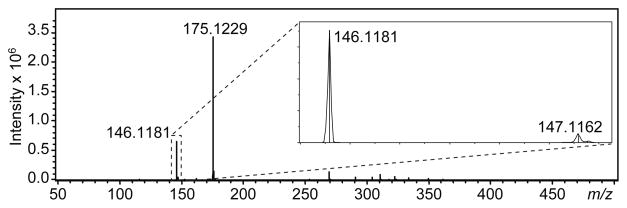
The use of a Bruker MicrOTOF mass spectrometer allows for the generation of high quality mass spectral data. For example, Figure 4 shows the mass spectrum obtained by summing the scans that were performed as the ACh band from an R2 neuron eluted. The method generates relatively clean mass spectra, with background ions present at or below the 104 level (the peak at 175 m/z is a co-eluting compound, and not present in the background). A mass resolution of over 11,000 is achieved for ACh, with a signal-to-noise ratio of over 1300. Preliminary data acquired using a Bruker maXis mass spectrometer displays even higher resolving power, >20,000 for ACh. The measured m/z for ACh in the R2 data presented in Figure 4 is 146.1181 (m/ztheory = 146.1181). The observed mass accuracy for metabolites varies (average < 25 ppm) and depends on a number of variables, including peak intensity, data acquisition rate, m/z, quality of calibration, and presence of background interferences. Nonetheless, low-ppm mass accuracies are possible with these systems.
Figure 4.
Generation of high quality mass spectra from single cell samples. Mass spectrum showing ACh from a single R2 neuron. The ACh peak at m/z = 146 has a signal-to-noise ratio of ~1300 and a mass resolution of >11,000.
Comparative metabolomic profiling of subcellular structures is also made possible by this method, and is dependent on the ability to isolate these structures. Figure 5 compares the XIEs obtained for 146 m/z from a single R2 cell soma with that obtained from the same R2 neurite. It is evident from the trace that the different subcellular regions contain different relative amounts of several metabolites.
Figure 5.
Different subcellular regions of the R2 neuron (neurite versus soma) yield different metabolite profiles. In this case, compounds with m/z = 146 +/− 0.5 Da are compared in the extracted ion electropherograms shown. Inset: Image of an isolated Aplysia R2 neuron and neurite.
Analyte Identification
There are a number of approaches that can be used to confirm the identity of an analyte, including the use of analytical standards for a comparison of molecular mass and migration time information. Figure 6(a) shows a comparison of an XIE generated at 177 m/z from the analysis of an MCC neuron to that generated from a 7.5 μM 5-HT standard solution. The high degree of similarity of these parameters between the MCC sample and the 5-HT standard confirms the presence of 5-HT in the neuron. However, if the XIE has several isobaric peaks, and/or there is enough of the single cell sample remaining, then the sample can be spiked with the putative analyte in order to increase the confidence of the identification; this has been done in the case of ACh in the R2 neurons (data not shown), as several isobaric compounds appear in the 146 m/z XIE from the R2.
Figure 6.
Analyte characterization and identification by single cell CE-ESI-MS. (a) Confirmation of 5-HT in a single MCC neuron. Migration time and mass information for the putative 5-HT peak in an MCC neuron matches the information obtained from analyzing a 5-HT standard solution. (b) Metabolite characterization via MS/MS. Different isobaric compounds (m/z = 146.1) present in a single R2 neuron are subjected to MS/MS analysis as they elute from the CE capillary using a Bruker maXis mass spectrometer. The peak at 15.4 min yields a fragment at 87 m/z, which is consistent with ACh, while other compounds either do not fragment appreciably, or yield fragments that are inconsistent with ACh. The peak corresponding to ACh was also confirmed by sample spiking (data not shown).
MS/MS can also be used to characterize or identify metabolites
The Bruker maXis tandem mass spectrometer provides sufficient sensitivity to enable MS/MS characterization of analytes present in single cells. The identification of ACh within an R2 demonstrates this capability. As mentioned above, there are a number of compounds in the R2 with m/z similar to ACh that appear in the electropherograms at different migration times. CE-ESI-MS/MS analysis enables the fragmentation spectra of these isobaric compounds to be compared; several representative examples are presented in Figure 6(b). The first peak in the XIE yields an MS/MS spectrum with a fragment at 87 m/z, consistent with the fragmentation profile of ACh. However, the other compounds selected for MS/MS analysis either do not fragment appreciably, or give rise to fragments that are inconsistent with the fragmentation pattern of ACh. Thus, MS/MS data supports the identity of the 1st peak in the XIE as ACh—a conclusion that has been confirmed with spiking experiments (data not shown).
CONCLUSIONS
The CE-ESI-MS/MS-based approach described herein enables a new volume regime to be probed, allowing investigation of the metabolome of individual cells and subcellular structures. The sensitivity of the ESI-TOF-MS and an ESI-Qq-TOF-MS/MS systems, when combined with the CE interface presented here, allows an important set of the cell metabolome to be characterized. The mass and concentration LODs reported here for neurotransmitters and other small molecules analyzed via a CE-MS interface are among the best reported to date for this platform and enables single cell measurements.55, 56 Improved CE sampling interfaces, such as using an optical trap or smaller nanovials, will allow a larger fraction of individual cell contents to be used for these measurements, therefore increasing the depth of metabolome coverage and/or enabling smaller structures to be characterized. Future work involves determining the intercellular signaling molecules at work in feeding, defensive, and reproductive neuronal networks, as well as identifying changes in the metabolome of these neurons in an activity-dependent manner. The combination of this analytical platform with the study of well-defined functional networks will allow changes in the cellular metabolome to be related to cell function.
Acknowledgments
This work is based upon work supported by the National Institutes of Health under Award Nos. DE018866 and NS031609. The authors would like to thank Elena Romanova for her assistance and advice regarding the maXis mass spectrometer and single cell sampling approaches, Stephanie Baker for her assistance in the preparation of this manuscript, the UIUC School of Chemical Sciences machine shop for fabrication of various auxiliary system components, and Christine Cecala for comments and suggestions regarding this manuscript.
References
- 1.Chen C, Shah YM, Morimura K, Krausz KW, Miyazaki M, Richardson TA, Morgan ET, Ntambi JM, Idle JR, Gonzalez FJ. Cell Metab. 2008;7:135–147. doi: 10.1016/j.cmet.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ippolito JE, Xu J, Jain S, Moulder K, Mennerick S, Crowley JR, Townsend RR, Gordon JI. Proc Natl Acad Sci U S A. 2005;102:9901–9906. doi: 10.1073/pnas.0500756102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiehn O. Plant Mol Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- 4.Kell DB. Drug Discov Today. 2006;11:1085–1092. doi: 10.1016/j.drudis.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Nicholson JK, Connelly J, Lindon JC, Holmes E. Nat Rev Drug Discov. 2002;1:153–161. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson JK, Lindon JC. Nature. 2008;455:1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 7.Dettmer K, Aronov PA, Hammock BD. Mass Spectrom Rev. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn WB, Ellis DI. TrAC, Trends Anal Chem. 2005;24:285–294. [Google Scholar]
- 9.Denkert C, Budczies J, Kind T, Weichert W, Tablack P, Sehouli J, Niesporek S, Konsgen D, Dietel M, Fiehn O. Cancer Res. 2006;66:10795–10804. doi: 10.1158/0008-5472.CAN-06-0755. [DOI] [PubMed] [Google Scholar]
- 10.Groger T, Welthagen W, Mitschke S, Schaffer M, Zimmermann R. J Sep Sci. 2008;31:3366–3374. doi: 10.1002/jssc.200800340. [DOI] [PubMed] [Google Scholar]
- 11.Hollywood K, Brison DR, Goodacre R. Proteomics. 2006;6:4716–4723. doi: 10.1002/pmic.200600106. [DOI] [PubMed] [Google Scholar]
- 12.Lindon JC, Holmes E, Nicholson JK. Pharm Res. 2006;23:1075–1088. doi: 10.1007/s11095-006-0025-z. [DOI] [PubMed] [Google Scholar]
- 13.Halket JM, Przyborowska A, Stein SE, Mallard WG, Down S, Chalmers RA. Rapid Commun Mass Spectrom. 1999;13:279–284. doi: 10.1002/(SICI)1097-0231(19990228)13:4<279::AID-RCM478>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 14.Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, Nishioka T. J Proteome Res. 2003;2:488–494. doi: 10.1021/pr034020m. [DOI] [PubMed] [Google Scholar]
- 15.Tolstikov VV, Lommen A, Nakanishi K, Tanaka N, Fiehn O. Anal Chem. 2003;75:6737–6740. doi: 10.1021/ac034716z. [DOI] [PubMed] [Google Scholar]
- 16.Tolstikov VV, Fiehn O, Tanaka N. Methods Mol Biol. 2007;358:141–155. doi: 10.1007/978-1-59745-244-1_9. [DOI] [PubMed] [Google Scholar]
- 17.Welthagen W, Schnelle-Kreis J, Zimmermann R. J Chromatogr A. 2003;1019:233–249. doi: 10.1016/j.chroma.2003.08.053. [DOI] [PubMed] [Google Scholar]
- 18.Hummon AB, Amare A, Sweedler JV. Mass Spectrom Rev. 2006;25:77–98. doi: 10.1002/mas.20055. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Garden RW, Sweedler JV. Trends Biotechnol. 2000;18:151–160. doi: 10.1016/s0167-7799(00)01427-x. [DOI] [PubMed] [Google Scholar]
- 20.Moroz LL, Edwards JR, Puthanveettil SV, Kohn AB, Ha T, Heyland A, Knudsen B, Sahni A, Yu F, Liu L, Jezzini S, Lovell P, Iannucculli W, Chen M, Nguyen T, Sheng H, Shaw R, Kalachikov S, Panchin YV, Farmerie W, Russo JJ, Ju J, Kandel ER. Cell. 2006;127:1453–1467. doi: 10.1016/j.cell.2006.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stuart JN, Hummon AB, Sweedler JV. Anal Chem. 2004;76:121A–128A. [PubMed] [Google Scholar]
- 22.Fuller RR, Moroz LL, Gillette R, Sweedler JV. Neuron. 1998;20:173–181. doi: 10.1016/s0896-6273(00)80446-8. [DOI] [PubMed] [Google Scholar]
- 23.Kennedy RT, Oates MD, Cooper BR, Nickerson B, Jorgenson JW. Science. 1989;246:57–63. doi: 10.1126/science.2675314. [DOI] [PubMed] [Google Scholar]
- 24.Stuart JN, Sweedler JV. Anal Bioanal Chem. 2003;375:28–29. doi: 10.1007/s00216-002-1609-z. [DOI] [PubMed] [Google Scholar]
- 25.Hu S, Le Z, Newitt R, Aebersold R, Kraly JR, Jones M, Dovichi NJ. Anal Chem. 2003;75:3502–3505. doi: 10.1021/ac034154j. [DOI] [PubMed] [Google Scholar]
- 26.Shen Y, Tolic N, Masselon C, Pasa-Tolic L, Camp DG, 2nd, Hixson KK, Zhao R, Anderson GA, Smith RD. Anal Chem. 2004;76:144–154. doi: 10.1021/ac030096q. [DOI] [PubMed] [Google Scholar]
- 27.Arcibal IG, Santillo MF, Ewing AG. Anal Bioanal Chem. 2007;387:51–57. doi: 10.1007/s00216-006-0690-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olefirowicz TM, Ewing AG. J Neurosci Methods. 1990;34:11–15. doi: 10.1016/0165-0270(90)90036-f. [DOI] [PubMed] [Google Scholar]
- 29.Tong W, Yeung ES. J Chromatogr, B: Anal Technol Biomed Life Sci. 1997;689:321–325. doi: 10.1016/s0378-4347(96)00317-9. [DOI] [PubMed] [Google Scholar]
- 30.Miao H, Rubakhin SS, Scanlan CR, Wang L, Sweedler JV. J Neurochem. 2006;97:595–606. doi: 10.1111/j.1471-4159.2006.03791.x. [DOI] [PubMed] [Google Scholar]
- 31.Liu YM, Moroz T, Sweedler JV. Anal Chem. 1999;71:28–33. doi: 10.1021/ac9808977. [DOI] [PubMed] [Google Scholar]
- 32.Burgi DS, Chien RL. Anal Chem. 1991;63:2042–2047. [Google Scholar]
- 33.Quirino JP, Terabe S. Anal Chem. 1999;71:1638–1644. [Google Scholar]
- 34.Osbourn DM, Weiss DJ, Lunte CE. Electrophoresis. 2000;21:2768–2779. doi: 10.1002/1522-2683(20000801)21:14<2768::AID-ELPS2768>3.0.CO;2-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiu DT, Lillard SJ, Scheller RH, Zare RN, Rodriguez-Cruz SE, Williams ER, Orwar O, Sandberg M, Lundqvist JA. Science. 1998;279:1190–1193. doi: 10.1126/science.279.5354.1190. [DOI] [PubMed] [Google Scholar]
- 36.Johnson RD, Navratil M, Poe BG, Xiong G, Olson KJ, Ahmadzadeh H, Andreyev D, Duffy CF, Arriaga EA. Anal Bioanal Chem. 2007;387:107–118. doi: 10.1007/s00216-006-0689-6. [DOI] [PubMed] [Google Scholar]
- 37.Monton MRN, Soga T. J Chromatogr, A. 2007;1168:237–246. doi: 10.1016/j.chroma.2007.02.065. [DOI] [PubMed] [Google Scholar]
- 38.Li L, Sweedler JV. Ann Rev Anal Chem. 2008;1:451–483. doi: 10.1146/annurev.anchem.1.031207.113053. [DOI] [PubMed] [Google Scholar]
- 39.Rubakhin SS, Sweedler JV. Nat Protoc. 2007;2:1987–1997. doi: 10.1038/nprot.2007.277. [DOI] [PubMed] [Google Scholar]
- 40.Wilm MS, Mann M. Int J Mass Spectrom Ion Processes. 1994;136:167–180. [Google Scholar]
- 41.Gale DC, Smith RD. Rapid Commun Mass Spectrom. 1993;7:1017–1021. [Google Scholar]
- 42.Wood TD, Moy MA, Dolan AR, Bigwarfe PM, Jr, White TP, Smith DR, Higbee DJ. Appl Spectrosc Rev. 2003;38:187–244. [Google Scholar]
- 43.Koster S, Verpoorte E. Lab Chip. 2007;7:1394–1412. doi: 10.1039/b709706a. [DOI] [PubMed] [Google Scholar]
- 44.Manisali I, Chen DDY, Schneider BB. TrAC, Trends Anal Chem. 2006;25:243–256. [Google Scholar]
- 45.Olivares JA, Nguyen NT, Yonker CR, Smith RD. Anal Chem. 1987;59:1230–1232. [Google Scholar]
- 46.Maxwell EJ, Chen DDY. Anal Chim Acta. 2008;627:25–33. doi: 10.1016/j.aca.2008.06.034. [DOI] [PubMed] [Google Scholar]
- 47.Schmitt-Kopplin P, Frommberger M. Electrophoresis. 2003;24:3837–3867. doi: 10.1002/elps.200305659. [DOI] [PubMed] [Google Scholar]
- 48.Valaskovic GA, Kelleher NL, McLafferty FW. Science. 1996;273:1199–1202. doi: 10.1126/science.273.5279.1199. [DOI] [PubMed] [Google Scholar]
- 49.Hofstadler SA, Severs JC, Smith RD, Swanek FD, Ewing AG. Rapid Commun Mass Spectrom. 1996;10:919–922. doi: 10.1002/(SICI)1097-0231(19960610)10:8<919::AID-RCM597>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 50.Smith RD, Barinaga CJ, Udseth HR. Anal Chem. 1988;60:1948–1952. [Google Scholar]
- 51.Smith RD, Olivares JA, Nguyen NT, Udseth HR. Anal Chem. 1988;60:436–441. [Google Scholar]
- 52.Eisenstadt M, Goldman JE, Kandel ER. Proc Natl Acad Sci U S A. 1973;70:3371–3375. doi: 10.1073/pnas.70.12.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weinreich D, McCaman MW, McCaman RE, Vaughn JE. J Neurochem. 1973;20:969–976. doi: 10.1111/j.1471-4159.1973.tb00067.x. [DOI] [PubMed] [Google Scholar]
- 54.Hatcher NG, Zhang X, Stuart JN, Moroz LL, Sweedler JV, Gillette R. J Neurochem. 2008;104:1358–1363. doi: 10.1111/j.1471-4159.2007.05084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mayboroda OA, Neusüβ C, Pelzing M, Zurek G, Derks R, Meulenbelt I, Kloppenburg M, Slagboom EP, Deelder AM. J Chromatogr, A. 2007;1159:149–153. doi: 10.1016/j.chroma.2007.04.055. [DOI] [PubMed] [Google Scholar]
- 56.Soga T, Heiger DN. Anal Chem. 2000;72:1236–1241. doi: 10.1021/ac990976y. [DOI] [PubMed] [Google Scholar]