Abstract
The gene regulatory network (GRN) established experimentally for the pre-gastrular sea urchin embryo provides causal explanations of the biological functions required for spatial specification of embryonic regulatory states. Here we focus on the structure of the GRN which controls the progressive increase in complexity of territorial regulatory states during embryogenesis; and on the types of modular subcircuits of which the GRN is composed. Each of these subcircuit topologies executes a particular operation of spatial information processing. The GRN architecture reflects the particular mode of embryogenesis represented by sea urchin development. Network structure not only specifies the linkages constituting the genomic regulatory code for development, but also indicates the various regulatory requirements of regional developmental processes.
Keywords: Subcircuit structure/function, Spatial transcriptional regulation, Embryonic specification
Introduction: GRNs for development
In the development of animals the establishment of spatial domains of specific gene expression underlies the formation of morphology and the diversification of function. The specification of regulatory states, the basic process that organizes development, is continuous, progressive and irreversible. This requires an entirely different mode of control as compared to the stable parallel activation of structural genes in terminally differentiated cells. As it turns out, the key informational transactions in developmental control systems depend on higher level functional interactions between different parts of the genome, rather than on the biochemical properties of any single genes. Two types of genomic sequence are essential for the control of genomic activity: sequences encoding transcription factors which read the genomic sequence by binding their DNA target sites in a sequence-dependent way, and the cis-regulatory sequences which control the expression of the regulatory genes. Gene regulatory networks (GRNs) contain both, and most importantly, they specify the functional interactions between them. By definition, structural genes do not possess this kind of regulative capacity. These genes represent a “dead end” output of GRNs. Their expression is controlled by the GRN, but they do not contribute to the GRN. Regulatory genes, however, do both at the same time: their expression is dependent on the GRN and they contribute to the flow of regulatory information by changing the activity of the genome.
System level analysis of genomic developmental control mechanisms requires the identification of the relevant drivers, the regulatory molecules which are expressed in the right cells and at the right stage to possibly contribute to a specific developmental process. Solving the regulatory interactions which functionally connect these factors reveals the process program from the genomic perspective. A number of GRNs have been described so far (for review see [1 Ch. 2]). Thus we are able to compare the topology of different GRNs. Recurrent network modules have been identified, in which the same topological constellations of regulatory interactions are used to solve comparable biological processes, even though connecting a different set of regulatory genes [2]. Examples from the GRN driving endoderm and mesoderm specification in embryos of the sea urchin Strongylocentrotus purpuratus demonstrate how the architecture of regulatory interactions relates to biological function, as we discuss in the following. First, however, we address some of the experimental approaches used to solve GRN structure.
1. Experimental approaches to GRN analysis
1.1. Identification of GRN nodes
The nodes of GRNs represent regulatory genes and their cis-regulatory control systems. The identification of specifically expressed regulatory factors has been facilitated by databases of annotated gene expression patterns which are available for many model systems. However, regulatory factors have the inconvenient property that it is not necessary for them to be present in vast amounts to be major effectors in a process. For example, pmar1, a gene which is crucial for specification of skeletogenic cells in sea urchin embryos, is never expressed at more than about 100 mRNA copies per cell and is present only in a small fraction of cells. Commonly used array-based technologies to detect gene expression levels have a much lower sensitivity than PCR-based approaches [3], which limits their usefulness for identifying low-abundance regulatory gene transcripts. Regulatory genes in the genome of the purple sea urchin S. purpuratus were identified based on orthologous sequences from other species [4-8]. The spatial and temporal gene expression patterns of zygotically expressed transcription factor genes were systematically analyzed at early embryonic stages to generate a map of regulatory states present in the sea urchin embryo. The results indicate that the majority of regulatory genes are expressed simultaneously in multiple territories, or dynamically first in one and later in another territory, or first in a broad set of precursor cells and later restricted to a subset of these. Many transcription factors are therefore employed in a number of possibly independent processes, which needs to be kept in mind when addressing their function. Regulatory genes that are ubiquitously expressed are generally not included in GRN models, unless their contribution to the spatial control of target gene expression was demonstrated. The outcome of this analysis is the identification of the complete regulatory toolkit underlying a specific biological process, the prerequisite to analysis of the regulatory system. The analysis of regulatory gene expression patterns might not be the most exciting part in GRN analysis. However, the more complete our knowledge of network components, the higher the predictive value of the resulting GRN model.
1.2. Identification of regulatory linkages
Regulatory interactions and thus the architecture of GRNs are encoded in cis-regulatory control regions. The network structure of regulatory interactions is a consequence of the fact that transcription factors act in a combinatorial way. The cis-regulatory apparatus of every gene thus contains information on type and number of regulatory proteins required for precise transcriptional output. This information is, however, difficult to access, since cis-regulatory control regions are modular, each module containing clusters of transcription factor binding sites which can be located in great distance either 5′ or 3′ to the transcriptional start site. Furthermore, DNA sequences recognized by DNA binding factors are short and usually contain ambiguous positions. Purely computational prediction of regulatory interactions and of network topology is therefore impossible at our present level of understanding of the exact sequence requirements for gene regulation. Many current approaches aim at mapping the physical binding of a transcription factor to specific sites in the genome by CHIP-based experiments. However, this approach appears to be of limited predictive value for the identification of functional enhancers due to a substantial level of non-specific interactions between transcription factors and DNA. The chances of identifying regulatory sequences are much improved by accurate prior prediction of enhancer functions, however. These predictions should include temporal and spatial expression profiles as well as regulatory inputs.
Regulatory interactions which are required for the correct expression of a gene are most efficiently identified by perturbation approaches. In sea urchin embryos, injection of morpholino antisense oligonucleotides specifically blocking the translation of proteins has proven most efficient for interfering with transcription factor expression. Expression of every transcription factor in the endomesoderm GRN model was perturbed in this way, and its putative target genes were identified using quantitative methods like QPCR or the Ncounter system, measuring the gene expression levels of all other regulatory genes which are included in the network [9-12]. Spatial changes in transcript distribution upon perturbation of a putative upstream regulatory factor are analyzed by in situ hybridizations on morpholino-injected embryos. Perturbation approaches are invaluable for determining the necessity of regulatory interactions, but they do not provide information on whether this interaction occurs directly or not. If changes in gene expression levels are observed long after the first apparent regulatory function of a transcription factor, they are very likely to be indirect consequences of earlier events. Once spatial and temporal gene expression patterns as well as predicted regulatory inputs of an endogenous gene are known, conserved sequence fragments surrounding the gene can be tested in functional assays. cis-Regulatory modules which are both necessary and sufficient to drive the expected expression pattern and responding to the perturbation of predicted regulatory inputs must contain the relevant information. Functional transcription factor binding sites are then identified by mutation of candidate sites.
In summary, a validated regulatory linkage between an upstream transcription factor and a downstream cis-regulatory control region requires the following experimental evidence: (i) transcription factor and target gene are at least partially co-expressed; (ii) the expression of the target gene is affected by the perturbation of the upstream regulatory gene; (iii) identification of a DNA fragment in the vicinity of the target gene which drives an expression pattern overlapping with the pattern of the endogenous gene and responds to perturbation of upstream transcription factor; and (iv) mutation of predicted binding site(s) of the upstream regulatory factor within the cis-regulatory fragment recapitulates the effect observed in (iii).
1.3. Assembly of a GRN model
As soon as interactions between more than two molecules are considered, it becomes very useful to work with models. They not only help in identifying missing parts in a network and in the design of future experiments, but they are also a tool for communicating one's results. A number of properties of developmental GRNs must be represented in models in order to reflect their functionality: (i) Most processes involve several territories which are specified by separate GRNs; (ii) these territories usually communicate by intercellular signaling; (iii) linkages in GRNs represent functional interactions which are directional and either activating or repressing; (iv) GRNs are hierarchical and regulatory interactions occur in a temporal sequence.
The GRN models shown in Figs. 2 and 3 were generated using BioTapestry (http://www.biotapestry.org). In these models, different regulatory territories are represented as colored rectangles with signaling events between them indicated as linkages between a gene encoding a signaling ligand in one domain and a gene encoding the corresponding receptor in the other domain. Each gene in the GRN is represented as a combination of a horizontal line, which represents its cis-regulatory control region, and as an outgoing arrow which represents the protein product of this gene. The interaction of a regulatory factor with its target gene is shown as horizontal bar if it causes repression or as arrow if it results in transcriptional activation. Each of the arrows and bars going into a gene model therefore represents a hypothetical interaction between a transcription factor and its cognate DNA binding site which can be tested by cis-regulatory analysis. Diamonds shown below modeled cis-regulatory regions are used to indicate different levels of experimental evidence for each linkage. Interactions shown as thick line have been validated by cis-regulatory experiments. In addition, online versions of BioTapestry models can be interactive, linking gene expression and perturbation data to each gene model. These models can also incorporate the time parameter and using a time-slider, the sequence of regulatory interaction and activation of gene expression can be observed, revealing the hierarchical structure of the GRN.
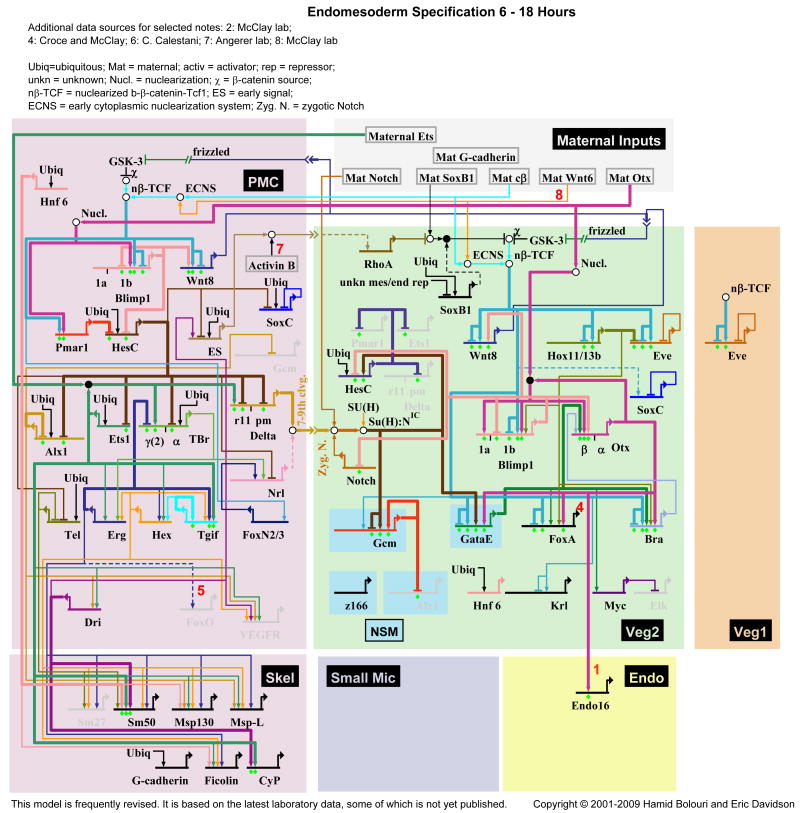
Fig. 2.
GRN model for the endomesoderm 6-18 h of development. This is a “view from the genome” in which all regulatory interactions occurring through time are portrayed in the various domains (cf. Fig. 1). For data and temporal and spatial regulatory views see http://sugp.caltech.edu/endomes/.
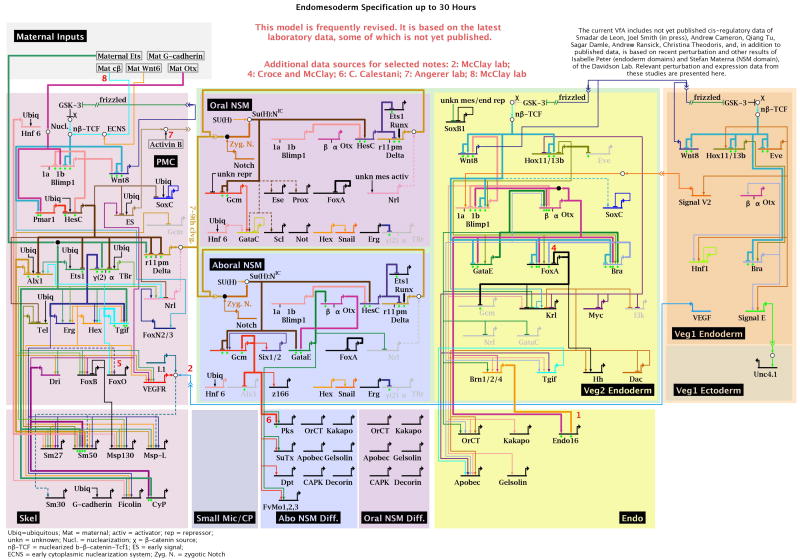
Fig. 3.
GRN model for the endomesoderm up to 30 h of development, as in Fig. 2.
2. The GRN for endomesoderm specification in Strongylocentrotus purpuratus embryos
Developmental GRNs ultimately control a biological process according to the structure of its regulatory interactions. Network architecture becomes apparent once a GRN model is generated. However, to relate the structural features to their biological functions requires thorough understanding of these functions, in our case of the development of the endomesodermal cell lineages.
3.1 The biological process
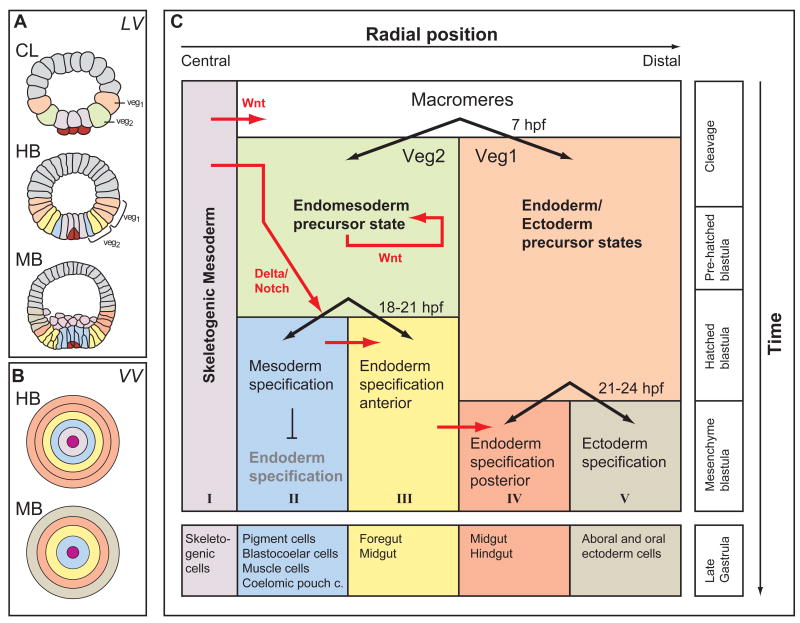
The process which is driven by the sea urchin endomesoderm GRN is summarized in Fig. 1. By 6th cleavage stage, about seven hours post fertilization (hpf), the vegetal half of the embryo consists of four cell lineages: the small micromeres, the skeletogenic micromeres, the veg2 and veg1 cell lineages. The latter three of these cell lineages constitute all the endodermal and mesodermal cell types of the larva, whereas the small micromeres are set-aside cells used at larval stages to form the adult organism [13]. As the small micromeres have no apparent function in embryonic development, they are not further considered here [14]. Skeletogenic micromeres, shown in light purple in Fig. 1, are located at the vegetal pole, a position with highest levels of certain maternal determinants, which turn out to be crucial for their specification. The cis-regulatory apparatus of a regulatory gene functioning at the top of the hierarchy of the skeletogenic GRN, the pmar1 gene, requires these maternal inputs in order to drive expression specifically in the skeletogenic micromeres [11,15,16]. Due to localization of these maternal determinants, specification of skeletogenic micromeres is the first such process in sea urchin embryos, and it requires no signaling inputs until much later, after the cells have differentiated and migrated into the interior of the embryo. Other specification processes in vegetal cells are dependent on signaling ligands. These are expressed by skeletogenic micromeres as a consequence of the skeletogenic GRN. Surrounding the skeletogenic micromeres are cells of the veg2 lineage, shown in green in Fig. 1. The veg2 lineage gives rise to mesodermal (Fig. 1, blue) as well as endodermal (Fig. 1, yellow) cells. Mesodermal pigment cells are specified depending on a Delta/Notch signaling input from the skeletogenic micromeres [17-19]. To transmit the Delta/Notch signal, cell membranes of signal sending and signal receiving cells must be in contact. The pigment specification program is therefore only induced in cells located adjacent to the skeletogenic micromeres expressing the Delta ligand [12,17]. Endoderm specification, on the other hand, is not dependent on Delta/Notch signaling and occurs in cells located more distally from the skeletogenic micromeres. Two sister cell lineages give rise to the future endoderm, but specification of these lineages depends on partially different sets of regulatory genes. Endoderm specification in veg2-derived cells is dependent on signals emitted from the micromeres [20-23]. Endoderm specification in veg1-derived cells (Fig. 1, orange) is accomplished by a different GRN architecture. Veg1-derived cells mainly contribute to posterior compartments of the larval gut ([12,24], unpublished results). The decision between ectodermal and endodermal cell fates in veg1-derived cells occurs slightly later than the mesoderm-endoderm distinction in the veg2 lineage, indicating that specification events occur in a spatio-temporal way, starting with the most central cells at the vegetal pole and spreading outward to the distal veg1-derived ectodermal cells (Fig. 1C).
Fig. 1.
Process Diagram indicating regulatory state domains in the sea urchin embryo up to gastrulation. (A) Optical cross sections of embryos at cleavage, 8 h (CL), hatching blastula, 18 h (HB), and mesenchyme blastula, 24 h (MB). Embryos are viewed from the side (lateral view, LV) and color coded to indicate territorial regulatory states. (B) Diagrammatic image of the concentric arrangement of regulatory states viewed from the vegetal pole (vegetal view, VV). (C) Process Diagram: the colored rectangles represent the territories of the endomesoderm. Subdivision of the embryos proceeds in a progressive manner so that just before gastrulation there are five domains indicated at the bottom of the diagram. Modified from Peter and Davidson [12].
3.2. Current status of the endomesoderm GRN
The GRNs driving the specification processes in the endomesoderm up to 30 hpf are modeled in Figs. 2 and. 3. Since the landscape of regulatory territories changes quite dramatically during the first 30 hours after fertilization, two different layouts are used to model regulatory interactions between 6-18 hpf (Fig. 2) and 21-30 hpf (Fig. 3), respectively. These layouts accommodate the different regulatory states which are present in the vegetal cell lineages. By the end of the time period covered by the GRN, which extends till just before the beginning of gastrulation, the embryo has become partitioned into regulatory territories in which specification of most larval cell types is at least initiated. Morphologically however, there is almost no difference between cells of the different territories at this stage (30 hpf), and these cells have not yet acquired their functionally differentiated state. The GRN model shown in Fig. 2 represents regulatory interactions which establish the regulatory states in three cell lineages, the skeletogenic micromeres (purple), the veg2 (green) and veg1 (orange) cell lineages. At early stages (15 hpf), each cell lineage therefore represents a specific regulatory state. After 21 hpf, however, the cell lineages have been subdivided so that cells originally deriving from a given lineage have now activated different and exclusive GRNs. Thus, in the veg2 cell lineage, GRNs underlying endoderm and mesoderm specification are at first initiated in the same cells, but are subsequently maintained in a completely exclusive set of cells (by 21 hpf; Fig. 3). Mesoderm precursor cells are further subdivided into oral and aboral territories, which will acquire different cell fates. A few hours later, some of the veg1 descendant cells have activated an endodermal GRN. Not all the domains of the endomesoderm GRN model have been analyzed to an equal extent. The GRNs underlying the specification of skeletogenic cells and endoderm cells are in the most complete state, which means that perturbation experiments have been performed for all the regulatory factors expressed specifically in these domains [11,12]. Many of the linkages in these domains have been tested by cis-regulatory analysis. In its current state, the oral and aboral mesoderm domains of the GRN, even though including the complete set of regulatory genes specifically expressed there, are still missing many of the interactions which functionally link these genes, but this is being remedied in current work. The whole of endomesoderm development up to the gastrula stage of the S. purpuratus embryo will soon be encompassed in a relatively complete network structure.
4. Modular GRN subcircuits
Though the GRNs of Figs.2 and 3 at first glance resemble a featureless maze of wiring connections, it might be suspected on first principles that their structure is actually modular, and such indeed is the case. The lens that best resolves the modularity of GRNs is that which detects function, though of course, ex post facto, given structural features can be associated with each type of function that these modules, the GRN subcircuits, mediate.
4.1 Subcircuits and their biological “jobs”
A basic argument that has emerged from the sea urchin developmental GRN is that if the phenomenological developmental biology is well enough understood, the processes of specification and subsequent territorial diversification can be broken down into individual “biological jobs” [1 Ch. 4]. As examples of such “jobs”, particular regulatory states must be established in the cells of an embryonic territory; these states must be made stable; regulatory states must be made exclusive; signals must be emitted; signals received must be interpreted functionally so as to produce a change in regulatory state; territories must be spatially subdivided; and so forth. The concept of the substituent biological jobs that constitute a process of development underlies the idea of the Process Diagram (e.g., Fig. 1), as the first step in construction of a developmental GRN. But how can we transform the fuzzy concept of “biological jobs”, such as the above, into an incisive tool for identification of GRN modules? The answer lies in the specific topologies of the subcircuits that execute these jobs. There are three simple principles here: first, that animal embryonic development invariably requires progressive installation of new spatial and temporal regulatory states; second, that given GRN subcircuit topologies are utilized to accomplish given kinds of spatial specification of regulatory state; third that other kinds of subcircuit topologies are utilized to accomplish given temporal projects. There turns out to be a one-to-one correspondence between subcircuit topology and the function it performs. This is a nice simplification, but an obvious one, since the subcircuits are composed of genes that regulate one another, and the output of each type of subcircuit is directly predictable from the linkages of which it is composed. Another valuable simplification is that the function of the subcircuits of various topologies do not depend in a unique way on the biochemical properties of the transcription factors that execute its interactions. Thus there can be found even within thesea urchin embryo subcircuits of identical or very similar topologies that execute the same jobs, but that are built of entirely different, non-overlapping sets of regulatory genes and entirely non-homologous signaling systems [25].
To illustrate this in detail, in the following we consider a set of seven different canonical subcircuit topologies extracted from the GRNs of Figs. 2 and 3, each of which effects a particular aspect of developmental regulatory state specification. Not only do these subcircuit topologies occur elsewhere composed of different genes, but as given topologies, they are also repeatedly deployed to accomplish the same developmental job whenever required. These same statements apply to subcircuits that execute temporal rather than spatial functions (e.g., transformation of a transient to a stable regulatory state by installation of positive feedback loops [10,11,25]). Thus the modular subcircuits of the GRN are in a sense the “building blocks” of the developmental regulatory system.
4.2 Topological structure and spatial logic processing functions: a repertoire of developmental subcircuits
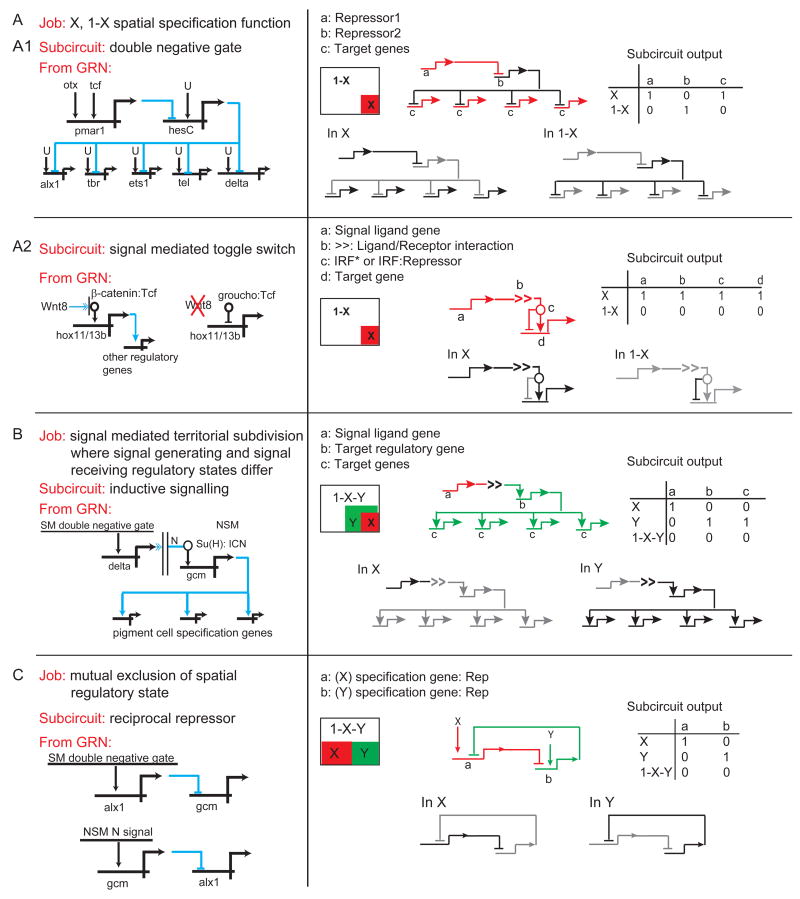
Our examples are shown in Fig. 4. In each section of this Figure the biological job is given at the top and immediately below is the name of the type of subcircuit that executes that job. There follows an excerpt from the GRN shown in Figs. 2 and/or 3; a geometrical diagram of the spatial domains affected by the subcircuit; and then a slightly more abstract, redrawn version of the subcircuit indicating the “on” or “off” regulatory states generated by the subcircuit in each spatial domain.
Fig. 4.
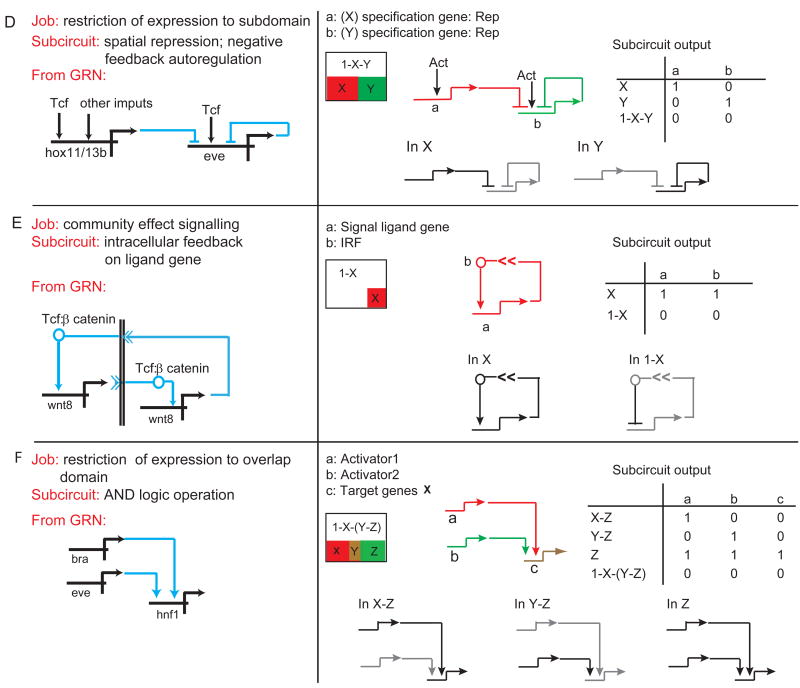
Canonical subcircuit from the endomesoderm GRN utilized to execute particular spatial specification processes. Each section of the figure (A-F) describes a particular spatial specification function, the “job” to be done, and the GRN subcircuit by which this job is executed. Following appear the subcircuit as excerpted from the GRNs in Figs. 2 and 3; a spatial expression cartoon; examples of the activity state of the subcircuit in different domains of the embryo; and a Boolean activity matrix for all relevant embryonic domains.
Figure 4A presents two different kinds of subcircuit that share the ultimate function of dividing embryonic space into two regulatory states, which we shall term “X” and “1-X”, such that a transcriptional state is established in X and specifically prohibited everywhere else. The subcircuit in Fig. 4A1 which accomplishes this function is the double negative gate subcircuit. Its definitive features are that the initial specification function activates a gene encoding a repressor, which transcriptionally prevents expression of a second repressor. This occurs only in the specific domain X. The second repressor gene of the gate is driven by widespread activators (here ubiquitous), and its targets are the initial, immediately downstream, regulatory genes constituting the territory-specific regulatory state. The result is that the second repressor specifically turns off these genes in 1-X, though they too respond to widespread activators, while specifically allowing their expression in X. There is a great deal of experimental evidence as to the details of operation of this gate in the skeletogenic domain of the sea urchin embryo, and for its cis-regulatory basis [11,16,26], and using entirely different genes it is also deployed in another domain of this embryo, the oral ectoderm [E. Li & E. Davidson, unpublished work]. The second type of “X, 1-X” subcircuit (Fig. 4A2), is what we call the signal mediated toggle switch subcircuit. A number of the commonly used developmental signaling systems have the feature that the ubiquitously present immediate response transcription factor (IRF), that is, the pre-existent factor altered in some way by ligand reception in the process of signal transduction, is a janus factor [27]. That is, in cells receiving the ligand the activated IRF* is permissive for transcription of its target genes, or actively promotes it, but in all other cells the IRF binds a dominant transcriptional silencer such as Groucho and becomes a repressor. The example from the GRN is Wnt8 signaling, which is required for spatially confined expression of a number of target genes in exactly this way. The IRF for the Wnt ligand is the Tcf transcription factor, and in the absence of this ligand Tcf forms a dominant repressive complex with Groucho. Thus, mutation of Tcf sites in the target gene cis-regulatory sequence causes ectopic expression.
The next function, considered in Fig. 4B, is signal-mediated subdivision of a prior regulatory state to set up a new state subdomain where different genes are expressed, by use of an inductive signaling subcircuit. Here the cells emitting the signal do that as a function of their prior regulatory state, and the cells receiving it are thereby caused to express a new regulatory state, different from their former one.
Our example from the GRN is Notch (N) signaling. This is a special case because N is activated by a cell bound ligand, so that the receiving cells are exclusively those in immediate contact with the sending cells. Their location thus determines the spatial location of the induced regulatory state (Y domain in the diagram, where the ligand emitting cells are in X domain). Here we have to consider the state of the target genes in X, Y, and the remainder of the embryo, 1-X-Y.
Like the other subcircuits and functions considered here, that in Fig. 4C is very widespread, perhaps almost universally to be found wherever developmental specification is occurring. This is the mutual exclusion of regulatory state, executed by reciprocal repression subcircuits [28]. When a given state of specification is installed, one of the constituent regulatory genes has the explicit function of repressing the possible expression of a regulatory gene that is a required component of an alternative regulatory state. Reciprocally, in the domain where this alternative regulatory state obtains, a repressive function targeting the first regulatory state, and sometimes the same gene therein, is activated. The example in Fig. 4C, one among many that could have been chosen, is reciprocal repression between a gene high up in the regulatory hierarchy of the skeletogenic lineage, alx1, and a gene high up in the regulatory hierarchy of the adjacent non-skeletogenic mesoderm, gcm [[28,29]; S. Damle & E. Davidson, unpublished work]. Spatially, the reciprocal repression subcircuit is usually deployed between domains of cells that could have deviated into one another's specification states, e.g., descendants of former sister cells, or as in the present case, of cells exposed to the same signaling ligands. Here again we have to consider the regulatory state of the target genes in the two mutually excluded domains and in the rest of the embryo as well.
Figure 4D shows another genomically encoded strategy for spatial regulatory state subdivision, executed by repression. Two kinds of repressive circuits are found: spatial repression subcircuits in which the boundary of a domain of expression of a given gene is set by institution of a repressive function for that gene within the area where another gene is active; and negative feedback autoregulation subcircuits. In the example shown, both are applied to the same target gene, here eve [12]. The autoregulation cuts the level of expression down and at a certain time in development, the external repressor, hox11/13b, then prohibits eve expression in an inner ring of cells where the former gene is expressed [12,30]. Boundary formation, by spatial repression subcircuits activated immediately across the future boundary is probably the most common mechanism for accomplishing this “job”, an essential and universally deployed aspect of spatial regulatory state subdivision.
Less well known is what we have termed “community effect signaling”, following an early study of John Gurdon [31]. This term is applied to signaling within a territory, in which each cell expresses the same regulatory state and in order to maintain this condition quantitatively, the cells signal to one another. Fig. 4E shows the canonical topology, the intercellular feedback on the ligand gene subcircuit. This subcircuit underlies the known examples of community effect signaling. The key topological feature, in several cases from the sea urchin GRN as well as elsewhere [32-35], is that the gene encoding the ligand responds to the same signal transduction system as it activates: thus each cell of the domain both receives and expresses the signal, and within the domain each cell is locked into a positive, signal mediated embrace with each other cell. A model calculation [32] shows that community effect signaling may account for the homogeneity of gene expression within multicellular territories.
Finally, and also a widespread development spatial specification device, is the use of cis-regulatory AND logic to establish novel spatial domains of regulatory state. Many examples are reviewed in a variety of systems in ref [1 Ch. 2]. Here two prior regulatory state domains overlap, and due to the operational constraints of the relevant cis-regulatory systems, only where both inputs are available are target genes defining a new regulatory state expressed. The example from the GRN is a regulatory gene of the endoderm, hnf1, which requires inputs from two genes, brachyury and eve, which at the relevant time overlap in part of their domains. The AND logic operation subcircuit defines the states of expression of target genes in the region of input overlap and also everwhere else, i.e., in the remainder of the expression domain of each input as well as elsewhere in the embryo.
Knowledge of developmental GRNs is still new and undoubtedly there will emerge further canonical spatial regulatory state subcircuits utilized in embryonic development, particularly when diverse kinds of developmental process are considered. As we consider in the next Section, sea urchins develop in a particular way which involves particular strategies and particular network architecture.
But the general point can be made that the modular structure of developmental GRNs, as here exemplified, at the same time illuminates the repertoire of topological subcircuits used by the Bilateria to build their embryos. Thus this repertoire is an ancient and definitive property of the bilaterian genomic regulatory system.
5. GRN design and the process of embryonic development
5.1 Different GRN designs underlie developmental strategies: comparisons within the endomesoderm GRN
For two specification processes within the sea urchin endomesoderm lineages, the specification of skeletogenic micromeres and of veg2 endoderm, we have highly elaborated GRN models, as noted above. The function of all these regulatory factors has been analyzed by perturbation experiments and many of the predicted interactions have been tested by cis-regulatory analyses. This, and the fact that both processes run at the same embryonic stages and in a similar time window, permit direct comparison of the general architecture of these two networks. Regulatory interactions in the skeletogenic and the veg2 endoderm territories were analyzed over a time period of about 18 hours. An obvious difference in the biology of these two lineages is that all the progeny of the skeletogenic micromere lineage execute skeletogenic cell fate, while the veg2 lineage gives rise to both endoderm and to various mesodermal cell types. This would suggest that the network architecture controlling the two processes might be fundamentally different. Surprisingly, however, it seems that the regulatory interactions initiating endoderm specification in veg2 cells run fairly independently of those controlling specification of mesodermal cells [12].
The similarity between the skeletogenic and veg2 endoderm GRNs relies on the fact that both are initiated by regulatory interactions which function in the way of an ON/OFF switch. The result is that these GRNs are turned ON in a restricted number of cells, and turned OFF by active repression in all other cells of the embryo. The control processes underlying this regulatory function are, however, solved very differently in the two GRNs. In skeletogenic precursor cells, this function is executed by the double negative gate (Fig. 4A1). In the endoderm GRN this very same function is executed by a direct positive gate function mediated by Tcf, the transcription factor which controls target gene expression in response to the Wnt signaling pathway. In cells which do not receive Wnt signaling, β-catenin is absent from the nuclei and Tcf interacts with the co-repressor Groucho, mediating repression of exactly the same genes (the signal-mediated toggle switch, Fig. 4A2 [36]).
Both systems are elegantly designed for the initiation of specific GRNs in the very early embryo. The employment of the double negative gate and the signal mediated toggle switch have in common that they rely on only one transcription factor to initiate spatially restricted expression. Even though the double negative gate consists of two transcriptional repressors, the cis-regulatory regions of the target genes require only one type of binding site to respond to this mode of control, specific for HesC (skeletogenic GRN) or Tcf (endodermal GRN). In both systems, the transcriptional activities of the target genes are binary readouts of the presence or activity of this transcription factor in all cells of the embryo (both the hesC and the tcf genes are driven by ubiquitous activators). This system is most useful at early embryonic stages when specifically expressed transcription factors are relatively rare. Surprisingly, in both GRNs multiple regulatory genes are directly controlled by this initiation function. However, most of these target genes also contain binding sites for other regulatory factors present in the corresponding domain. It remains to be seen whether these observations turn out to be general features of GRN wiring for the earliest embryonic specification functions.
5.2 Network design and the embryonic process
Changing focus, subcircuits which we would predict to be a general feature of bilaterian developmental GRNs, must be embedded in an organization which represents specifically the mode of development of the embryo. Sea urchin embryos accomplish specification and differentiation according to what has been termed a “Type 1” developmental process [1 Ch. 3,37]. This is a very widespread form of embryogenesis in invertebrate animals which produce relatively small eggs, and which generate free-living larvae after only about 10-12 cell divisions. The essential features of type I embryonic process are as follows: (i) The cleavage stage cell lineage is fixed and more or less canonical for given clades, so that in each individual embryo of the species cells of given fate occupy the same position in respect to the three-dimensional embryonic coordinates, and to the primordial polarities of the egg. (ii) The embryo nuclei become transcriptionally active immediately after fertilization, and development is controlled zygotically from early cleavage on, though (as in all forms of development) extensive use is also made of maternally deposited transcripts and proteins. (iii) The embryo assigns regulatory states to spatial territories that can be defined in terms of the cell lineage, and within each territory every cell expresses the same regulatory state (until the territory is subdivided), while each territory gives rise to a given part of the later embryo. (iv) The initial zygotic regulatory states are always set up by reference to maternal anisotropies that are interpreted in such a way as to spatially localize early regional zygotic regulatory state(s) within the confines of given cell lineage components. (v) All subsequent spatial territorial regulatory states, and progressive territorial subdivision, depend on inter- and intra-territorial signaling between cells, beginning at once during cleavage. (vi) Differentiation gene batteries begin to be activated even before gastrulation. (vii) There is no net growth during embryogenesis, and no significant cell migration or “salt and pepper” mixing of embryonic lineages until after territorial regulatory states have been established all over the embryo, only following which do gastrular movements ensue. None of these process characteristics are true of early vertebrate or Drosophila development, though (v) pertains to Drosophila development after cellularization, and eventually, following the delayed activation of the blastomere genomes at the end of cleavage, to the remainder of vertebrate development as well. Type 1 embryogenesis is an evolutionarily ancient mode of building the “body plan” of an animal embryo, since it is found in branches of animal phylogeny so distant that their last common ancestor was the last common bilaterian ancestor. Now that we have for the first time a reasonably complete and explanatory GRN for a significant portion of the specification processes in a Type 1 embryo, can we identify the components of this GRN that mediate the canonical features of this form of embryogenesis? If so we might then predict the essential modular features that should apply to any Type 1 embryonic process.
The basic output of any developmental GRN is establishment of regulatory states in the appropriate cells, or in a Type 1 embryo, in the appropriate cell lineages and polyclonal territories. Thus those of the above developmental process criteria that are directly concerned with establishment and deployment of spatial regulatory states should be directly controlled by the GRN. On the other hand, those features that depend on other than direct zygotic transcriptional control must be excluded from the discussion. In the case at hand, little is known about the mechanism by which canonical cleavage patterns are loaded into the egg (i), except that this is a property of the egg cytoplasmic organization that is established during oogenesis and in the earliest cleavages, and it is clear that cleavage plane localization is not controlled by zygotic gene expression [38 Ch. 6]. Similarly, the timing of zygotic gene activation (ii) is a function of egg size and nucleo-cytoplasmic ratio early in development [38 Ch. 2]. The issue we address is whether the endomesoderm GRN can explain, for this case, how those features of Type 1 embryogenesis that are zygotically controlled are genomically encoded. These are points (iii)-(vi) above.
The key mechanism by which this form of embryogenesis is initiated is the use of the invariant lineage, the result of the fixed geometry of the cleavage planes ((i) above), in setting up the initial zygotic regulatory states. The initial genes of the regulatory state are thus supposed to be turned on in response to localized factors that affect gene expression, and that have been segregated into the lineage founder cells. This theory was inferred from a vast amount of phenomenological evidence accrued from classical and modern experimental embryology and developmental molecular biology [1 Ch. 3,37,38 Ch. 6], but now we can see exactly how the transcriptional part of the process is genomically encoded. With respect to point (iii) above, the GRN in Fig. 2 affords three examples: activation of the specific regulatory states in the founder cells of the skeletogenic lineage (lavender area of Fig.2) and of the veg2 and veg1 endomesodermal lineages (green and tan areas). In the skeletogenic lineage, the maternal transcription factors Otx and β-catenin/Tcf are localized in the four (4th cleavage) skeletogenic founder cell nuclei and these inputs are used to activate the double negative gate discussed above, by turning on the pmar1 gene exclusively in these cells (point (iv) above [11]). This response is genomically encoded in the cis-regulatory target sites of the pmar1 gene(s) [15,16]. Additional community effect circuitry (see above) ensures the continuance of the β-catenin/Tcf feed in these cells by driving expression of Wnt8 [33,39]. Maternal β-catenin is also nuclearized in the eight veg2 and eight veg1 (6th cleavage) founder cells when they are born (point (iv); [40]), and the Wnt8 community effect thereafter runs in these cells as well. The β-catenin/Tcf input activates both the key early veg2 activator hox11/13b (see Fig. 2) and the key early veg1 activator eve, the cis-regulatory systems of which respond sharply to this input [12,30,41].
The GRN also affords a classical example of inductive signaling between cleavage stage blastomeres, another canonical hallmark of Type 1 embryonic process (point v above) also well known to occur in early cleavage C. elegans [42,43] and Ciona [44] embryos, for example. Though there is widespread evidence of cleavage stage inductive signaling in Type 1 embryos (for reviews [1 Ch. 3,21,37,44]), neither the genomic program by which this is caused to occur, nor the genomic program that determines its particular consequences is usually evident. Here again the sea urchin embryo GRN provides an exact solution. In these embryos, non-skeletogenic mesoderm specification depends on inductive Delta-Notch signaling from the skeletogenic lineage to the adjacent cells of the veg2 lineage ([18,19]; Fig.1). The GRN explains why the delta gene is expressed in the skeletogenic lineage: it is one of the targets of the double negative gate that sets up the regulatory state in this lineage, and is directly under transcriptional control of the second repressor of this gate, HesC (Figs. 2 and 4; [26,45]). The GRN also explains at the regulatory DNA sequence level why Notch signaling in the veg2 recipients is necessary and sufficient for the specification of the mesodermal pigment cell type. The target of the signal transduction system is a cis-regulatory module of the gcm gene [17]. As the GRN shows (Figs. 2 and 3), gcm is a regulator of pigment cell genes, including both genes encoding other transcription factors and differentiation genes. Thus we have in the GRN a comprehensive causal explanation of both the incidence of the inductive signal and its developmental output, couched in the required terms, the regulatory DNA sequence. For though the effects are conditional on signal reception, the role of inductive signaling in territorial specification of the Type 1 embryo is ultimately just as hardwired as any other aspect of the transcriptional developmental control system.
Finally we come to the activation of differentiation gene batteries, which occurs at the periphery of the GRN, as far downstream in any given phase of the developmental process as transcriptional control goes (cf. Fig. 1). Type 1 embryos characteristically display direct cell type specific activation of differentiation genes (point vi above; [1, Ch. 3,37]), often precociously with respect to the morphological generation of differentiated structures. For example, differentiation genes are likely to be activated in Type 1 embryos during blastula stages. By this point the embryo is already territorially specified, as it consists of a spatial mosaic formed by its diverse regulatory states, but morphologically is yet of simple form, lacking the terminally differentiated cell types that will appear only later. Here again the sea urchin GRN shows us how this phenomenon occurs. In the skeletogenic domain, for example (Figs. 2 and 3), a number of differentiation genes are indeed activated in the blastula stage, and network analysis shows that the regulatory genes downstream of the double negative gate all contribute feeds into various differentiation genes (only a small fraction of which are yet included in the GRN). cis-Regulatory studies have confirmed, for example that the inputs that drive the cyclophylin gene, a cytoskeletal gene expressed only in these cells, are the factors Deadringer (Dri) and Ets, which are generated as components of this specific lineage regulatory state ([46]; Fig.2). These factors plus Hnf6 also provide inputs into the sm50 biomineralization gene [47-49]. Similarly, in the mesodermal domain the aforementioned gcm gene provides a direct input into the polyketide synthase gene, which produces an enzyme utilized in pigment synthesis (C. Calestani, unpublished results). Why do these differentiation genes go on as early as they do? Because, as the GRN shows, there are no other intermediate steps to be traversed, and as soon as the regulators to which they respond become available, their target genes are activated. The timing of their activation follows the same general dynamics as does the rest of the GRN, being controlled essentially by the time it takes for the successive steps of transcription of regulatory genes, processing and translation of the mRNAs, and activation of transcription of the next target downstream. In these sea urchin embryos, which live at 15°C, the time separating any two such immediately sequential steps is 2-3 h [50].
In summary we here show how the definitive canonical process characteristics of Type 1 embryogenesis, which were formulated much more than a decade prior to the GRN, can be explained in terms of the network regulatory code. Of course this is only one example and the argument will be strengthened when equally comprehensive GRNs are available for other species of Type 1 embryos. But this exercise already provides us with specific predictions of the structures that should emerge from these networks to come. The more general implication is important: a specific form of network architecture should underlie each form of developmental process.
6. Concluding Comment
The sea urchin endomesoderm GRN successfully confirms one of the fundamental precepts of systems biology, viz. that to obtain a comprehensive causal explanation of a process, all (or most) of its component parts must be included in the analysis; but conversely, if they are, and the analysis is appropriately based, then the outcome should indeed provide the answers as to why the biology operates as it does. We note in this connection two fundamental epistemological features. First, the GRN structure was formulated on the results of system wide perturbation experiments, not on the basis of system wide measurements of the unperturbed state. No such measurements, spatial, kinetic, genomic or biochemical, could alone or in combination, ever have revealed the specific topologies of the subcircuits of Fig. 4, which as we see provide the heart of the functional explanations emergent from the GRN. Second, since the ultimate locus of control of the developmental process must lie in the genomic regulatory sequence, the system analysis must be couched in, and be able to be validated, in terms of functional significance of the genomic DNA sequence. This is the fundamental reason why the representation of the GRN in the BioTapestry platform has been important: it reveals directly, without further deconvolution or separation of one kind of interaction from another, not only the circuit topology but also the expected features of the network nodes that are subject to systematic test at the cis-regulatory level. Furthermore, it does so while preserving the territorial components of the biological process. But this is only the first stage at which we have arrived. What lies ahead are extensions of developmental GRNs in multiple directions: their extension to more and more advanced embryonic territories, to more embryonic and postembryonic systems, so that the immense resolving power of comparative meta-analysis can be brought to bear on the functional meaning of the bilaterian regulatory genome at the system level.
Acknowledgments
Research was supported by NIH grant HD37105 and the Lucille P. Markey Charitable Trust. I. P. was supported by a fellowship from the Swiss National Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Davidson EH. The Regulatory Genome Gene Regulatory Networks in Development and Evolution. Academic Press/Elsevier; San Diego, CA: 2006. [Google Scholar]
- 2.Davidson EH, Levine MS. Properties of developmental gene regulatory networks. Proc Natl Acad Sci USA. 2008;105:20063–20066. doi: 10.1073/pnas.0806007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, et al. Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics. 2006;7:59. doi: 10.1186/1471-2164-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howard-Ashby M, Materna SC, Brown CT, Chen L, Cameron RA, Davidson EH. Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus, and their expression in embryonic development. Dev Biol. 2006;300:74–89. doi: 10.1016/j.ydbio.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 5.Howard-Ashby M, Materna SC, Brown CT, Chen L, Cameron RA, Davidson EH. Gene families encoding transcription factors expressed in early development of Strongylocentrotus purpuratus. Dev Biol. 2006;300:90–107. doi: 10.1016/j.ydbio.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 6.Materna SC, Howard-Ashby M, Gray RF, Davidson EH. The C2H2 zinc finger genes of Strongylocentrotus purpuratus and their expression in embryonic development. Dev Biol. 2006;300:108–120. doi: 10.1016/j.ydbio.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 7.Rizzo F, Fernandez-Serra M, Squarzoni P, Archimandritis A, Arnone MI. Identification and developmental expression of the ets gene family in the sea urchin (Strongylocentrotus purpuratus) Dev Biol. 2006;300:35–48. doi: 10.1016/j.ydbio.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Tu Q, Brown CT, Davidson EH, Oliveri P. Sea urchin Forkhead gene family: phylogeny and embryonic expression. Dev Biol. 2006;300:49–62. doi: 10.1016/j.ydbio.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 9.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 10.Su YH, Li E, Geiss GK, Longabaugh WJ, Kramer A, Davidson EH. A perturbation model of the gene regulatory network for oral and aboral ectoderm specification in the sea urchin embryo. Dev Biol. 2009;329:410–421. doi: 10.1016/j.ydbio.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci U S A. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peter IS, Davidson EH. The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stage. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.10.037. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pehrson JR, Cohen LH. The fate of the small micromeres in sea urchin development. Dev Biol. 1986;113:522–526. doi: 10.1016/0012-1606(86)90188-0. [DOI] [PubMed] [Google Scholar]
- 14.Ransick A, Cameron RA, Davidson EH. Postembryonic segregation of the germ line in sea urchins in relation to indirect development. Proc Natl Acad Sci USA. 1996;93:6759–6763. doi: 10.1073/pnas.93.13.6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliveri P, Carrick DM, Davidson EH. A regulatory gene network that directs micromere specification in the sea urchin embryo. Dev Biol. 2002;246:209–228. doi: 10.1006/dbio.2002.0627. [DOI] [PubMed] [Google Scholar]
- 16.Smith J, Davidson EH. Regulative recovery in the sea urchin embryo and the stabilizing role of fail-safe gene network wiring. Proc Natl Acad Sci USA. 2009;106:18291–18296. doi: 10.1073/pnas.0910007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ransick A, Davidson EH. cis-regulatory processing of Notch signaling input to the sea urchin glial cells missing gene during mesoderm specification. Dev Biol. 2006;297:587–602. doi: 10.1016/j.ydbio.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 18.Sherwood DR, McClay DR. LvNotch signaling mediates secondary mesenchyme specification in the sea urchin embryo. Development. 1999;126:1703–1713. doi: 10.1242/dev.126.8.1703. [DOI] [PubMed] [Google Scholar]
- 19.Sweet HC, Gehring M, Ettensohn CA. LvDelta is a mesoderm-inducing signal in the sea urchin embryo and can endow blastomeres with organizer-like properties. Development. 2002;129:1945–1955. doi: 10.1242/dev.129.8.1945. [DOI] [PubMed] [Google Scholar]
- 20.Ransick A, Davidson EH. A complete second gut induced by transplanted micromeres in the sea urchin embryo. Science. 1993;259:1134–1138. doi: 10.1126/science.8438164. [DOI] [PubMed] [Google Scholar]
- 21.Ransick A, Davidson EH. Micromeres are required for normal vegetal plate specification in sea urchin embryos. Development. 1995;121:3215–3222. doi: 10.1242/dev.121.10.3215. [DOI] [PubMed] [Google Scholar]
- 22.Sethi AJ, Angerer RC, Angerer LM. Gene regulatory network interactions in sea urchin endomesoderm induction. PLoS Biol. 2009;7:e1000029. doi: 10.1371/journal.pbio.1000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wikramanayake AH, Peterson R, Chen J, Huang L, Bince JM, McClay DR, Klein WH. Nuclear beta-catenin-dependent Wnt8 signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis. 2004;39:194–205. doi: 10.1002/gene.20045. [DOI] [PubMed] [Google Scholar]
- 24.Ransick A, Davidson EH. Late specification of Veg1 lineages to endodermal fate in the sea urchin embryo. Dev Biol. 1998;195:38–48. doi: 10.1006/dbio.1997.8814. [DOI] [PubMed] [Google Scholar]
- 25.Davidson EH. Network design principles from the sea urchin embryo. Curr Opin Genetics Dev. 2009;19:1–6. doi: 10.1016/j.gde.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Revilla-i-Domingo R, Minokawa T, Davidson EH. R11: a cis-regulatory node of the sea urchin embryo gene network that controls early expression of SpDelta in micromeres. Dev Biol. 2004;274:438–451. doi: 10.1016/j.ydbio.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 27.Barolo S, Stone T, Bang AG, Posakony JW. Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless. Genes Dev. 2002;16:1964–1976. doi: 10.1101/gad.987402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveri P, Davidson EH. Development. Built to run, not fail. Science. 2007;315:1510–1511. doi: 10.1126/science.1140979. [DOI] [PubMed] [Google Scholar]
- 29.Oliveri P, Davidson EH, McClay DR. Activation of pmar1 controls specification of micromeres in the sea urchin embryo. Dev Biol. 2003;258:32–43. doi: 10.1016/s0012-1606(03)00108-8. [DOI] [PubMed] [Google Scholar]
- 30.Smith J, Kraemer E, Liu H, Theodoris C, Davidson E. A spatially dynamic cohort of regulatory genes in the endomesodermal gene network of the sea urchin embryo. Dev Biol. 2008;313:863–875. doi: 10.1016/j.ydbio.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gurdon JB. A community effect in animal development. Nature. 1988;336:772–774. doi: 10.1038/336772a0. [DOI] [PubMed] [Google Scholar]
- 32.Bolouri H, Davidson EH. The gene regulatory network basis of the “community effect,” and analysis of a sea urchin embryo example. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.06.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minokawa T, Wikramanayake AH, Davidson EH. cis-Regulatory inputs of the wnt8 gene in the sea urchin endomesoderm network. Dev Biol. 2005;288:545–558. doi: 10.1016/j.ydbio.2005.09.047. [DOI] [PubMed] [Google Scholar]
- 34.Nam J, Su YH, Lee PY, Robertson AJ, Coffman JA, Davidson EH. Cis-regulatory control of the nodal gene, initiator of the sea urchin oral ectoderm gene network. Dev Biol. 2007;306:860–869. doi: 10.1016/j.ydbio.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Range R, Lapraz F, Quirin M, Marro S, Besnardeau L, Lepage T. Cis-regulatory analysis of nodal and maternal control of dorsal-ventral axis formation by Univin, a TGF-beta related to Vg1. Development. 2007;134:3649–3664. doi: 10.1242/dev.007799. [DOI] [PubMed] [Google Scholar]
- 36.Range RC, Venuti JM, McClay DR. LvGroucho and nuclear beta-catenin functionally compete for Tcf binding to influence activation of the endomesoderm gene regulatory network in the sea urchin embryo. Dev Biol. 2005;279:252–267. doi: 10.1016/j.ydbio.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 37.Davidson EH. How embryos work: a comparative view of diverse modes of cell fate specification. Development. 1990;108:365–389. doi: 10.1242/dev.108.3.365. [DOI] [PubMed] [Google Scholar]
- 38.Davidson EH. Gene Activity in Early Development. Academic Press; Orlando, FL: 1987. [Google Scholar]
- 39.Smith J, Theodoris C, Davidson EH. A gene regulatory network subcircuit drives a dynamic pattern of gene expression. Science. 2007;318:794–797. doi: 10.1126/science.1146524. [DOI] [PubMed] [Google Scholar]
- 40.Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126:345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- 41.Ransick A, Rast JP, Minokawa T, Calestani C, Davidson EH. New early zygotic regulators expressed in endomesoderm of sea urchin embryos discovered by differential array hybridization. Dev Biol. 2002;246:132–147. doi: 10.1006/dbio.2002.0607. [DOI] [PubMed] [Google Scholar]
- 42.Evans TC, Crittenden SL, Kodoyianni V, Kimble J. Translational control of maternal glp-1 mRNA establishes an asymmetry in the C. elegans embryo. Cell. 1994;77:183–194. doi: 10.1016/0092-8674(94)90311-5. [DOI] [PubMed] [Google Scholar]
- 43.Owraghi M, Broitman-Maduro G, Luu T, Roberson H, Maduro MF. Roles of the Wnt effector POP-1/TCF in the C. elegans endomesoderm specification gene network. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumano G, Nishida H. Ascidian embryonic development: an emerging model system for the study of cell fate specification in chordates. Dev Dyn. 2007;236:1732–1747. doi: 10.1002/dvdy.21108. [DOI] [PubMed] [Google Scholar]
- 45.Smith J, Davidson EH. Gene regulatory network subcircuit controlling a dynamic spatial pattern of signaling in the sea urchin embryo. Proc Natl Acad Sci USA. 2008;105:20089–20094. doi: 10.1073/pnas.0806442105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amore G, Davidson EH. cis-Regulatory control of cyclophilin, a member of the ETS-DRI skeletogenic gene battery in the sea urchin embryo. Dev Biol. 2006;293:555–564. doi: 10.1016/j.ydbio.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 47.Kurokawa D, Kitajima T, Mitsunaga-Nakatsubo K, Amemiya S, Shimada H, Akasaka K. HpEts, an ets-related transcription factor implicated in primary mesenchyme cell differentiation in the sea urchin embryo. Mech Dev. 1999;80:41–52. doi: 10.1016/s0925-4773(98)00192-0. [DOI] [PubMed] [Google Scholar]
- 48.Mahmud AA, Amore G. The surprising complexity of the transcriptional regulation of the spdri gene reveals the existence of new linkages inside sea urchin's PMC and Oral Ectoderm Gene Regulatory Networks. Dev Biol. 2008;322:425–434. doi: 10.1016/j.ydbio.2008.07.036. [DOI] [PubMed] [Google Scholar]
- 49.Otim O, Amore G, Minokawa T, McClay DR, Davidson EH. SpHnf6, a transcription factor that executes multiple functions in sea urchin embryogenesis. Dev Biol. 2004;273:226–243. doi: 10.1016/j.ydbio.2004.05.033. [DOI] [PubMed] [Google Scholar]
- 50.Bolouri H, Davidson EH. Transcriptional regulatory cascades in development: initial rates, not steady state, determine network kinetics. Proc Natl Acad Sci USA. 2003;100:9371–9376. doi: 10.1073/pnas.1533293100. [DOI] [PMC free article] [PubMed] [Google Scholar]