Abstract
Background:
Many different genetic and clinical factors have been identified as causes or contributors to atherosclerosis. We present a model of preclinical atherosclerosis based on genetic and clinical data that predicts the presence of coronary artery calcification in healthy Americans of European descent aged 45 to 84 in the Multi-Ethnic Study of Atherosclerosis (MESA).
Methods and Results:
We assessed 712 individuals for the presence or absence of coronary artery calcification, and their genotypes for 2882 single-nucleotide polymorphisms (SNPs). Using these SNPs and relevant clinical data, a Bayesian network that predicts the presence of coronary calcification was constructed. The model contains 13 SNPs (from genes AGTR1, ALOX15, INSR, PRKAB1, IL1R2, ESR2, KCNK1, FBLN5, PPARA, VEGFA, PON1, TDRD6, PLA2G7, and one ancestry informative marker) and 5 clinical variables (sex, age, weight, smoking, and diabetes) and achieves 85% predictive accuracy, as measured by area under the ROC curve (AUC). This is a significant (p < 0.001) improvement upon models using just the SNP data or using just the clinical variables.
Conclusions:
We present an investigation of joint genetic and clinical factors associated with atherosclerosis that shows predictive results for both cases, and enhanced performance for the combination.
Keywords: atherosclerosis, genetics, risk factors
Atherosclerosis is a complex disease with many possible causes, and many adverse clinical outcomes, including heart disease, myocardial infarction, stroke, embolism, thrombosis, and aneurysm. To date, many different genetic1 and clinical factors2 have been identified as causes or contributors to atherosclerosis. We present herein a model of preclinical atherosclerosis based on genetic and clinical data that predicts the presence of coronary artery calcification in healthy Americans of European descent aged 45 to 84.
There has been much progress in identifying genes and single nucleotide polymorphisms (SNPs) associated with atherosclerosis3, or clinical correlates of atherosclerosis (for example, contributing causes like hypertension4 and consequences like myocardial infarction5). There have also been successful predictive models of effects of atherosclerosis, including heart disease6 and thromboembolism7, and these particular models were successful without including genetic information. Additionally, one study considered 30 possible genetic markers8 in a predictive model of atherosclerosis, but concluded that the few SNPs available were unhelpful in predicting atherosclerosis. It seems clear that to achieve a more complete model of atherosclerosis, a multitude of genes and clinical variables must be combined.9
The Multi-Ethnic Study of Atherosclerosis (MESA) recruited a cohort of individuals without clinical evidence for cardiovascular disease (CVD), and then measured coronary artery calcification (CAC), a quantifiable marker of advanced atherosclerosis.10 A large number of candidate genes, 231, related to vascular disease and related phenotypes were selected for genotyping, and extensive clinical and demographic data collected on the subjects. With these combined data, MESA provides an ideal opportunity to investigate both clinical and genetic models of CAC as an indicator of atherosclerosis.
Methods
MESA is a population-based study of 6814 men and women aged 45 to 84 years, free of known CVD at baseline, recruited between 2000 and 2002 from six US communities. The main objective of the study is to determine the characteristics of subclinical cardiovascular disease and its progression. Details of the objectives and design of MESA have been published.10 Institutional review board approval was obtained at all MESA sites and all participants gave their informed consent.
For initial genotyping analyses, a subcohort of 720 subjects was selected from the total MESA cohort of 6814, who both gave informed consent for DNA extraction and genetic sub-study; and had samples in the study DNA laboratory with sufficient DNA. All DNA was of high quality as measured by OD260/OD280, with the mean ratio of 1.77. All DNA was of high molecular weight as determined by gel electrophoresis. For the current study, to minimize the possibility of spurious genetic associations due to population stratification, we analyzed the data from the MESA participants self-identified as having European ancestry. Priority was given to subjects who participated in the MESA 3 additional blood biomarker collection, supplemented by random selection from remaining participant samples to fulfill balanced ethnic group representation and equality by gender. CAC was determined with electron beam or helical CT11. The average Agatston score of two scans was used and the presence of CAC was defined as an Agatston score > 0.
DNA was extracted from peripheral leukocytes isolated from packed cells of anticoagulated blood by use of a commercially available DNA isolation kit (Puregene; Gentra Systems, Minneapolis, MN). The DNA was quantified by determination of absorbance at 260 nm followed by PicoGreen analysis (Molecular Probes, Inc., Eugene, OR). Two vials of DNA were stored per participant at −70 degrees centigrade and subsequently aliquoted for use.
MESA investigators proposed candidate genes for two separate gene marker panels (MESA Candidate Gene Panel 1 and 2), and the genes were priority ranked by contributing investigators. The list of genes included is shown in Supplementary Table 1. For Panel 2, additional weight was given to genes proposed by the MESA Eye ancillary study, a study of retinal microvascular characteristics as predictors of subclinical and clinical cardiovascular diseases. Final priority for both panels was assigned by the MESA Family Study Genetics Committee. SNPs for the chosen genes were selected according to the following criteria. Firstly, SNPs within the proximal and distal 10-kilobase regions 5′ and 3′ to the given candidate gene (NCBI Build 35) were chosen. Next, SNP compatibility with the Illumina GoldenGate technology12,13 as determined by the Assay Design Tool (TechSupport, Illumina, San Diego, CA) was required. Finally, SNPs with minor allele frequency (MAF) greater or equal to 0.05 or a tag (r2 value at least 0.8) for another SNP with MAF > 0.05 as determined by applying the multi-locus or “aggressive” “Tagger” option of Haploview v314,15 using International HapMap project data for CEPH and Yoruban populations (release 19)16 were selected. Due to these competing criteria, a complete set of tagSNPs could not be found for some genes, and additional SNPs were selected from one of the following three sources. 1) LDselect analysis of resequencing information from the Seattle SNPs project if available17,18; 2) Non-synonymous SNPs from dbSNP (release 124)19; 3) SNPs with prior report of association with a phenotype similar or identical to one measured in MESA and proposed by a MESA investigator.
In MESA CG Panel 1, ancestry informative markers (AIMs) were selected from an Illumina proprietary SNP database to maximize the difference in allele frequencies between any pair of ethnic groups: Caucasian- vs African-American; Caucasian- vs Chinese-American; African- vs Chinese-American. For MESA CG Panel 2, additional makers informative for Mexican-American ancestry were selected from published lists20,21.
Genotyping was performed by Illumina Genotyping Services (Illumina Inc., San Diego, CA) using their proprietary GoldenGate assay. The SNPs were typed in two separate panels of 1536 markers, selected to assay multiple phenotype x gene hypotheses. Illumina performed initial quality control in their laboratory to identify samples and SNPs that failed genotyping according to proprietary protocols, and sporadic failed genotypes with gencall quality score <0.25. Of 156 duplicate pairs included in 33 plates of samples typed, Illumina were blinded to 92 pairs. Both unblinded and blinded sample replicate concordance rates were > 99.99%. After removal of failed SNPs and samples, the genotype calling rate was 99.93%, with maximum missing data rate per sample of 2.1%, and maximum missing data per SNP of 4.98%. The cohort genetic data was checked for cryptic sample duplicates and discrepancies in genetically predicted sex (using X markers) versus study database reported sex. Samples with unresolved duplicate and sex discrepancies were removed from the genetic study database.
These criteria resulted in 712 individuals of European descent genotyped at 2882 SNPs. 393 (55.2%) of the cohort was previously determined to have detectable coronary artery calcification at the baseline exam. Missing SNP values in the genotype data were imputed by random assignment according to the marginal frequencies of each SNP across the cohort, in an effort to favor the null hypothesis of no association as much as possible. 593 of the SNPs were removed for having minor allele frequencies lower than 5%. A further 111 autosomal SNPs were rejected because the distribution of their alleles violated Hardy-Weinberg equilibrium (p<0.05)22. These tests resulted in a total of 2177 SNPs being eligible for our model search algorithms.
Additional non-genetic data were available for each individual, and we selected the following for inclusion in our model-building process, on the basis of suspected associations with atherosclerosis or CVD: Age, Sex, Education Level, Income, Smoking Status, Weight, Body Mass Index (BMI), Diabetes, LDL Cholesterol, HDL Cholesterol, Total Cholesterol, Systolic Blood Pressure, Diastolic Blood Pressure, Hypertension, Walking Speed, and Minutes of Exercise per Week. Income and Education are included as potential proxies for other unmeasured environmental factors. In all categories, responses were binned into at most five discrete values using a simple linear binning strategy (different binning strategies did not affect results).
We then sorted all remaining variables by their Bayes factor23, a statistic that assesses the likelihood increase associated with conditioning the outcome (presence of coronary calcification) upon the variable in question. A Bayes factor > 1 (or equivalently, the log of the Bayes factor > 0) for a particular variable indicates that the variable in question is more likely to be probabilistically associated with coronary calcification than to be probabilistically independent of coronary calcification. Bayes factors were computed using conjugate Dirichlet priors as described in Cooper and Herskovitz24. We use the Bayes factors to filter the number of SNPs to a tractable amount: only 50 SNPs had log Bayes factor > 0, and the 17 SNPs with highest Bayes factors were all on the X chromosome. We also note that ten of the clinical variables had log Bayes factor > 0 (age, sex, diabetes, BPS, hypertension, HDL cholesterol, smoking, BMI, total cholesterol, and education). All SNPs and clinical variables with log Bayes factor > 0 are listed in Supplementary Table 2. We used Bayesware Discoverer (www.bayesware.com) to learn a Bayesian network structure on these variables. A Bayesian network is a statistical method with properties that ideally suit it to the task of discovering predictive models of complex diseases25,26. Among these is the ability of the model marginal likelihood to penalize model complexity without need of further adjustment.27 Indeed, Bayesian networks have proved successful in previous prediction problems of complex diseases, including stroke28. We built three separate Bayesian network models: one using the 50 positive-Bayes factor SNPs alone; another using all 16 clinical variables alone; and a third using the combination of the 50 SNPs and 16 clinical variables.
We compared performance of the three different models using the area under the convex hull of the receiver operating characteristic curve (AUC), which measures performance of a classifier at different false positive/false negative thresholds. We then followed the prescription of Lasko et al.29 to compute a p-value comparing the difference of two AUCs, computing the variance of an AUC according to the nonparametric method described by DeLong et al.30
Results
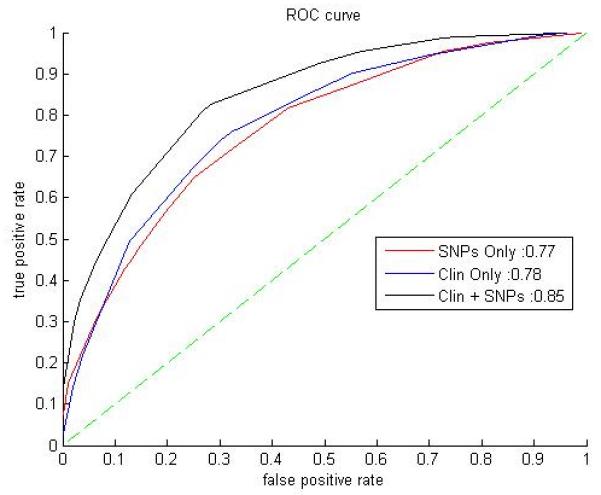
Table 1 shows demographic data for the study participants. We developed a Bayesian network using all available data; the Markov blanket of CAC in the network is depicted in Figure 1. It uses all the variables listed in Table 2 except for SNP rs2380316, on the X-chromosome (variable number 18 in Table 2): five clinical variables and 13 SNPs. This model has an 85% AUC on the entire European MESA cohort for predicting CAC presence (Figure 2).
TABLE 1.
Demographic and coronary artery calcification averages for the 712 genotyped subjects selected from the participants with European ancestry in the MESA cohort.
| MESA Clinical Data mean (SD), n |
All | Men (47%), 332 | Women (53%), 380 |
|---|---|---|---|
| Coronary Artery Calcification | 55%, 393 | 69%, 230 | 43%, 163 |
| Log CAC (In Agatston Score) | 4.4 (1.8), 712 | 4.7 (1.8), 330 | 4.1 (1.8), 380 |
| Age (years) | 61.5 (10.4), 712 | 61.9 (10.2), 332 | 61.1 (10.7), 380 |
| Educationa | 6 (−), 700 | 7 (−), 331 | 5 (−), 369 |
| Incomeb | 11 (−), 698 | 11 (−), 324 | 11 (−), 374 |
| Smoking (current) | 16%, 112 | 16%, 52 | 16%, 60 |
| Smoking (former) | 43%, 304 | 51%, 170 | 35%, 134 |
| Weight (lbs) | 176 (39), 712 | 194 (33), 332 | 159 (36), 380 |
| Body Mass Index (kg/m2) | 28 (5), 712 | 28 (4), 332 | 27 (6), 380 |
| Diabetes (treated) | 4.5%, 32 | 5.7%, 19 | 3.4%, 13 |
| LDL cholesterol (mg/dl) | 115.3 (34.3), 700 | 114.8 (34.8), 325 | 115.6 (33.9), 375 |
| HDL cholesterol (mg/dl) | 52.6 (16.1), 711 | 45.1 (13.7), 331 | 59.1 (15.3), 380 |
| Total Cholesterol (mg/dl) | 195.9 (35.9), 711 | 187.7 (34.6), 331 | 203.1 (35.5), 380 |
| Diastolic Blood Pressure (mmHg) |
70.6 (9.9), 712 | 73.8 (8.7), 332 | 67.8 (10.1), 380 |
| Systolic Blood Pressure (mmHg) |
124.2 (21.1), 712 | 124.0 (18.3), 332 | 124.4 (23.3), 380 |
| Hypertension (JNC VI) | 39%, 712 | 36%, 332 | 42%, 380 |
| Walking Speedc | 2 (−), 709 | 2 (−), 331 | 2 (−), 378 |
| Exercise (MET min/week) | 1690.9 (2404), 710 | 2024.1 (2782), 331 | 1399.7 (1975), 379 |
Mean (standard deviation) or percentage or median (for education, income, and walking speed), followed by N, number of patients with the condition (for smoking, CAC, and diabetes) or with data reported for the attribute in question.
Education completed: 0) no schooling; 1) grades 1-8; 2) grades 9-11; 3) completed high school; 4) some college, no degree; 5) technical school certificate; 6) associate degree; 7) bachelor's degree; 8) graduate, professional school
Yearly family income: 1) < $5000; 2) $5000-7999; 3) $8000-11999; 4) $12000-15999; 5) $16000-19999; 6) $20000-24999; 7) $25000-29999; 8) $30000-34999; 9) $35000-39999; 10) $40000-49999; 11) $50000-74999; 12) $75000-99999; 13) $100000+
Walking Speed: 0) no walking; 1) casual strolling (<2 MPH); 2) average or normal (2-3 MPH); 3) fairly brisk (4-5 MPH); 4) brisk or striding (>5 MPH)
Figure 1.
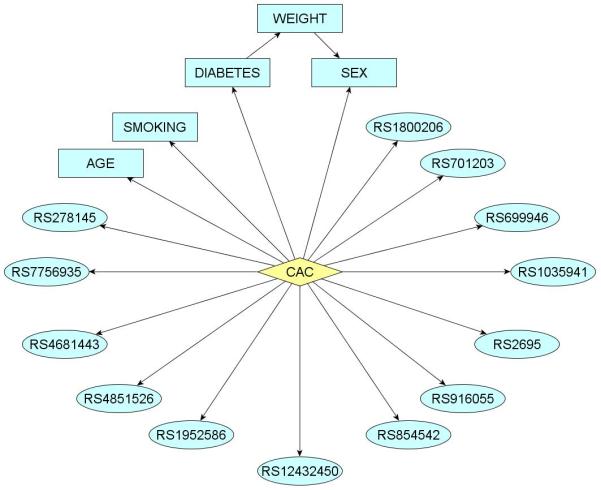
Markov blanket of coronary artery calcification (CAC) in a Bayesian network describing probabilistic dependencies between CAC and SNPs and clinical variables considered. This network achieves 85% AUC on the European MESA cohort.
TABLE 2.
List of all attributes used in our predictive models for coronary artery calcium presence. The individual predictive accuracy of each attribute is reported, along with the accuracy achieved by random guessing. Accuracy is reported as AUC of naïve bayes model involving only the variable in question.
| Variable | ALLELES | GENEa | POSITION/ BAND |
ROLE | Individual Accuracy (AUC)b |
p-value vs. random |
|
|---|---|---|---|---|---|---|---|
| 1 | Age | 69% | < 0.001 | ||||
| 2 | Sex | 65.9% | < 0.001 | ||||
| 3 | Diabetes | 60.2% | 0.015 | ||||
| 4 | Smoking | 60.1% | 0.0017 | ||||
| 5 | Weight | 56.7% | 0.1819 | ||||
| 6 | rs278145 | C/T | PRKAB1 | 12q24.23 | Intron | 58.9% | 0.0162 |
| 7 | rs7756935 | A/C | TDRD6, PLA2G7 |
6p12.3 | Downstream |
58.5% |
0.0043 |
| 8 | rs2695 | A/G | [Selected as an AIM] |
9q21.31 | - |
58.4% |
0.0074 |
| 9 | rs12432450 | C/T | FBLN5 | 14q32.12 | Intron | 58.3% | 0.0275 |
| 10 | rs854542 | A/G | PON1 | 7q21.3 | Downstream | 58.2% | 0.0072 |
| 11 | rs701203 | C/G | KCNK1 | 1q42.2 | Intron | 58.1% | 0.0322 |
| 12 | rs916055 | C/T | ALOX15 | 17p13.2 | 3′UTR | 57.5% | 0.0869 |
| 13 | rs1035941 | C/T | INSR | 19p13.2 | Intron | 57.4% | 0.0778 |
| 14 | rs4851526 | A/G | IL1R2 | 2q11.2 | Intron | 57% | 0.0926 |
| 15 | rs699946 | A/G | VEGFA | 6p21.1 | Promoter | 56.6% | 0.0739 |
| 16 | rs4681443 | A/G | AGTR1 | 3q24 | Intron | 56.5% | 0.0597 |
| 17 | rs1952586 | A/G | ESR2 | 14q23.2 | Intron | 56.1% | 0.3541 |
| 18 | rs2380316 | A/G | WDR44 | Xq24 | Intron | 54.7% | 0.755 |
| Random Guess | 53.8% | - | |||||
| 19 | rs1800206 | C/G | PPARA | 22q13.31 | Coding Exon | 51% | 0.1875 |
Gene symbol key: WDR44 is WD repeat domain 44, AGTR1 is angiotensin II receptor, type 1, ALOX15 is arachidonate 15-lipoxygenase, INSR is insulin receptor, PRKAB1 is protein kinase, AMP-activated, beta 1 non-catalytic subunit, IL1R2 is interleukin 1 receptor, type II, ESR2 is estrogen receptor 2 (ERbeta), KCNK1 is potassium channel K, subfamily 1, FBLN5 is fibulin 5, PPARA is peroxisome proliferative activated receptor alpha, VEGFA is vascular endothelial growth factor A, PON1 is paraoxonase 1, TDRD6 is tudor domain containing 6, and PLA2G7 is phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma).
Predictive accuracy tested by Naïve-Bayes classifier, AUC convex hull reported. Random classifier generated with 1000 realizations, negative classifiers inverted, convex hull AUCs averaged.
Figure 2.
Area under ROC curves (AUCs) for the three Bayesian networks considered. The figure legend indicates the AUCs for each of the models: 85% for SNPs-Clinical, 78% for Clinical-Only, and 77% for SNPs-Only. The dotted line between (0,0) and (1,1) represents the performance of a hypothetical random predictor.
For comparison purposes, we then obtained a Bayesian network using only the genomic data, and the 14 SNPs used in this network are shown in Table 2 (numbers 6-17,19). The model consists of 14 SNPs directly related to the phenotype (CAC); there are no other connections. This model differs from the first by including an extra SNP on the X chromosome, rs2380316, which may act as a proxy for the absent sex attribute. This model achieved an area under the curve of 77% AUC (Figure 2). The combined SNP-clinical model's performance was significantly better than the SNP-only model (p-value < 0.001), using the method of DeLong et al.30
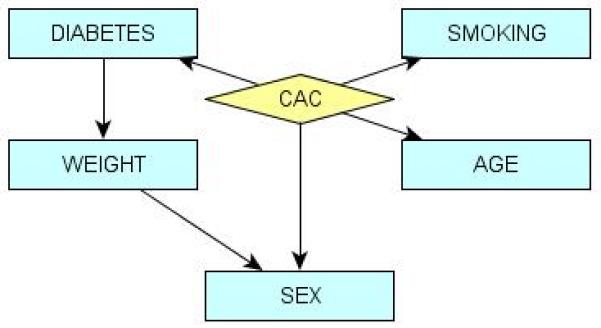
We then compared our results against a Bayesian network constructed from only the clinical data. This model included the features: Sex, Age, Diabetes, Smoking, and Weight (Figure 3). Other attributes did not impact the presence of CAC according to the model. Nevertheless, this model achieved comparable accuracy to the SNP-only model: 78.3% AUC (Figure 2). These were tested for statistical difference from the results of the combined SNP-clinical model and found to be significant (p-value < 0.001, DeLong et al.'s method30), while these results were not significantly different from the performance of the SNP-only model. These observations are summarized in Table 3.
Figure 3.
Markov blanket of coronary artery calcification (CAC) in a Bayesian network describing probabilistic dependencies between CAC and clinical variables considered. This network achieves 78% AUC on the European MESA cohort.
TABLE 3.
Describes the accuracy measurements for the three models developed for predicting coronary artery calcium presence. For each model, we report the Area Under ROC Curve (AUC). The performance of the SNP-Clinical model was found to be a statistically significant improvement over using either category of information separately.
| Model | SNPs Only | Clinical Only | SNP-Clinical |
|---|---|---|---|
| AUC | 77.3% | 78.3% | 85.0% |
| p-value vs. clinical model | .59 | - | < 0.001 |
| p-value vs. SNP model | - | .59 | < 0.001 |
Finally, we attempted to construct models using logistic regression with stepwise selection, of the same types as our Bayesian networks. A logistic regression model constructed from the 50 SNPs and 16 clinical attributes achieved 81.5% AUC (p = .036 vs. 85% AUC of our Bayesian model), using the attributes age, sex, diabetes, smoking, total cholesterol, and six SNPs (numbers 11 and 16 in table 2, an X-linked SNP rs953114, and three SNPs on genes included in the Bayesian model: rs1403543 on AGTR2; rs2498852 at FBLN5; and rs6502804 in ALOX15.) A logistic regression model using only clinical variables performed less well, with an AUC of 78.9% (not significantly different from the Bayesian clinical-only model, p = 0.256), but other researchers have previously obtained a logistic model on the entire European MESA cohort (n = 2619) with AUC of 82%2. Logistic regression limited to using SNPs had predictive accuracy of 66.6% AUC and was significantly worse (p < 1.77 *10−7) than the Bayesian SNP-only model (this regression model contained eight SNPs: two X-linked; two from the combined logistic model, rs2498852 at FBLN5 and rs6502804 on ALOX15; and four listed in table 2, numbers 8, 11, 16, and 17.)
Discussion
Our results show that it is possible to combine genetic data with clinical data and achieve improvements in predictive accuracy. The performance of the combined model (85% AUC) is comparable or better than existing predictors of atherosclerosis. Previous studies have either investigated related but different phenotypes (myocardial infarction, thrombosis, etc.) or had different phenotyping criteria (work by Chen et al.9 checks for a 50% narrowing of any coronary artery – a separate indicator of atherosclerosis, where narrowing of an artery can be due to uncalcified plaques.)
We note that multi-attribute methods are required in order to achieve a well-performing predictive model of atherosclerosis. We tested each individual attribute involved as a single predictor of CAC (summarized in Table 2). Individual attributes were poor predictors of CAC, except for Sex and Age; other attributes only achieved AUCs in the range of 53% to 59%. Table 2 also lists p-values for difference between the performance of a single attribute predictor and a random classifier, and 10 of these are significant at a 0.05-threshold.
The network figure 1 shows that 13 SNPs, located on 12 genes and one AIM, modulate the risk of coronary artery calcification. Note that although ancestry informative markers were included in the genotyping for purposes related to other investigations, it nevertheless turns out that an AIM is included in the model.
Since the SNPs genotyped in the MESA study were chosen as tag SNPs, and these chosen for their candidacy in relation to previous knowledge about the genetics of atherosclerosis and cardiovascular disease, these 12 SNPs represent various connections and relations to atherosclerosis and related physiology. Four of these 12 genes fall into a coherent functional picture related to lipid metabolism, centered on the adipocytokine signaling pathway (PRKAB131, AMPK31, PPARA32, and AMPK, along with INSR, is also related to the insulin signaling pathway33), while the forth is related to HDL- and LDL- bound cholesterols (PON1.34) Five genes represented in our model could be broadly grouped by their relation to vascular inflammation or constriction (AGTR135, VEGFA36), or general inflammation responses (ALOX1537, IL1R238, and PLA2G739). Several genes have previously been associated with atherosclerosis (PON140, ALOX1541, and FBLN5 and ESR242-44). The twelfth gene (KCNK1), a ubiquitous potassium ion channel, and has been linked to heart cells but not vasoconstriction, with differential expression in ventricular and atrial cells.45
Our work here should be considered in light of its scope and limits. First, since our total predictive accuracy is imperfect, we speculate that further genetic investigations will uncover better models. Genome-wide association studies of atherosclerosis, measuring hundreds of thousands of SNPs rather than just thousands, could uncover unknown dependencies between genes in our model, or entirely new genes. Equally, it may be possible that there are further clinical attributes, beyond those collected in MESA or used in our model, which may be of relevance to predicting CAC. Second, our work here is limited to the European cohort of the MESA study, while studies on other ethnic groups remain in preparation. Since we have used the MESA data, our prediction is for the presence or absence of coronary artery calcification, which is only one measure of atherosclerosis. It may be easier to predict atherosclerosis or the onset of cardiovascular disease when using different clinical phenotypes, even different CAC thresholds – CAC raw Agatston score > 400, for example, is frequently considered an indicator of increased CVD risk,46 one we did not consider because the number of MESA patients meeting this threshold is low (n = 83). Further, since atherosclerosis affects such a large proportion of the population, and risk is largely dependent on age and sex, it may be more informative to predict who will get atherosclerosis earlier or later than expected, after more data are available for the onset of atherosclerosis. Last but not least, our models have not been tested on an external replication population. Thus, there may be modifications necessary to our model to achieve robust prediction on different populations; from other studies or from other demographic groups.
This study demonstrates the improved predictive performance of joint consideration of genetic and clinical contributions to a subclinical measure of atherosclerosis. Since the predictive ability of our model is an improvement over the previous best-performing associative tests, this approach has the potential to improve individual-level prediction of CVD risk through the implementation of genetic tests. Further, the model developed in the current study may lead to improved clinical and mechanistic models of atherosclerosis progression.
Clinical Summary.
Pre-symptomatic prediction of late onset diseases is one of the greatest promises of genetic medicine and at the heart of personalized medicine. This study describes a model of preclinical atherosclerosis based on genetic and clinical data. This model predicts the presence of coronary artery calcification with 85% accuracy. We assessed 712 healthy Americans of European descent aged 45 to 84 for the presence of coronary artery calcification, and their genotypes for 2882 single-nucleotide polymorphisms (SNPs), subtle variations in individuals' genomes. We used a methodology known as Bayesian networks, born at the confluence of statistics and Artificial Intelligence, to integrate this genetic data and relevant clinical information into a coherent network model that predicts the presence of coronary calcification. The model contains 13 SNPs and five clinical variables: sex, age, weight, smoking, and diabetes. This model outperforms both the model built from purely genetic data and the model built on clinical information alone. In this way, this study demonstrates the improved predictive performance of joint consideration of genetic and clinical contributions to a subclinical measure of atherosclerosis. Since the predictive ability of our model is an improvement over the previous best-performing associative tests, this approach has the potential to improve individual risk prediction of arteriosclerosis through the development of genetic tests. Further, the model presented in the current study may lead to improved clinical and mechanistic models of atherosclerosis progression and identify novel molecular targets for drug development.
Supplementary Material
Acknowledgements
The authors thank the participants, investigators, and staff of the MESA study for their valuable contributions. We thank reviewers for helpful comments on an earlier version of this manuscript. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org.
Funding Sources: This research was supported in part by NIH grants T32-HL-007427, and U01-HL-065899, R01-HL-071205 and R21-DA-025168 and by contracts N01-HC-95159 through N01-HC-95165 and N01-HC-95169.
Footnotes
Disclosures: none.
References
- 1.Chen Y, Rollins J, Paigen B, Wang X. Genetic and Genomic Insights into the Molecular Baiss of Atherosclerosis. Cell Metabolism. 2007;6:164–179. doi: 10.1016/j.cmet.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bild DE, Detrano R, Peterson D, Guerci A, Liu K, Shahar E, Ouyang P, Jackson S, Saad MF. Ethnic Differences in Coronary Calcification: The Multi-Ethnic Study of Atherosclerosis (MESA) Circulation. 2005;111:1313–1320. doi: 10.1161/01.CIR.0000157730.94423.4B. [DOI] [PubMed] [Google Scholar]
- 3.Benton JL, Ding Z, Tsai MY, Shea S, Rotter JI, Burke GL, Post W. Associates between two common polymorphisms in the ABCA1 gene and subclinical atherosclerosis: Multi-Ethnic Study of Atherosclerosis (MESA) Atherosclerosis. 2007;193:352–360. doi: 10.1016/j.atherosclerosis.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 4.Dries DL, Victor RG, Rame JE, Cooper RS, Wu X, Zhu X, Leonard D, Ho S, Wu Q, Post W, Drazner MH. Corin Gene Minor Allele Defined by 2 MIssense Mutations Is Common in Blacks and Associated With High Blood Pressure and Hypertension. Circulation. 2005;112:2403–2410. doi: 10.1161/CIRCULATIONAHA.105.568881. [DOI] [PubMed] [Google Scholar]
- 5.Ebana Y, Ozaki K, Inoue K, Sato H, Iida A, Lwin H, Saito S, Mizuno H, Takahashi A, Nakamura T, Miyamoto Y, Ikegawa S, Odashiro K, Nobuyoshi M, Kamatani N, Hori M, Isobe M, Nakamura Y, Tanaka T. A functional SNP in ITIH3 is associated with susceptibility to myocardial infarction. Journal of Human Genetics. 2007;52:220–229. doi: 10.1007/s10038-006-0102-5. [DOI] [PubMed] [Google Scholar]
- 6.Wilson PW, D'Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of Coronary Heart Disease Using Risk Factor Categories. Circulation. 1998;97:1837–1847. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 7.Kline JA, Novobilski AJ, Kabrhel C, Richman PB, Courtney MD. Derivation and Validation of a Bayesian Network to Predict Pretest Probability of Venous Thromboembolism. Annals of Emergency Medicine. 2005;45:282–290. doi: 10.1016/j.annemergmed.2004.08.036. [DOI] [PubMed] [Google Scholar]
- 8.Chen Q, Li G, Leong T, Heng C. Predicting Coronary Artery Disease with Medical Profile and Gene Polymorphism Data. Studies in Health Technology and Informatics. 2007;129:1219–1224. [PubMed] [Google Scholar]
- 9.Ghazalpour A, Doss S, Yang X, Aten J, Toomey EM, Van Nas A, Wang S, Drake TA, Lusis AJ. Toward a biological network for atherosclerosis. Journal of Lipid Research. 2004;45:1793–1805. doi: 10.1194/jlr.R400006-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Bild DE, Bluemke DA, Burke GL, Detrano R, Roux AV, Folsom AR, Greenland P, Jacob DR, Kronmal R, Liu K, Nelson JC, O'Leary D, Saad MF, Shea S, Szklo M, Tracy RP. Multi-ethnic study of atherosclerosis: objectives and design. American Journal of Epidemiology. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 11.Carr JJ, Nelson JC, Wong ND, McNitt-Gray M, Arad Y, Jacobs DR, Sidney S, Bild DE, Williams OD, Detrano RC. Calcified Coronary Artery Plaque Measurement with Cardiac CT in Population-based Studies: Standardized Protocol of Multi-Ethnic Study of Atherosclerosis (MESA) and Coronary Artery Risk Development in Young Adults (CARDIA) Study. Radiology. 2005;234:35–43. doi: 10.1148/radiol.2341040439. [DOI] [PubMed] [Google Scholar]
- 12.Gunderson KL, Kruglyak S, Graige MS, Garcia F, Kermani BG, Zhao C, Che D, Dickinson T, Wickham E, Bierle J, Doucet D, Milewski M, Yang R, Siegmund C, Haas J, Zhou L, Oliphant A, Fan J, Barnard S, Chee MS. Decoding randomly ordered DNA arrays. Genome Research. 2004;14:870–877. doi: 10.1101/gr.2255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan JB, Gunderson KL, Bibikova M, Yeakley JM, Chen J, Garcia EW, Lebruska LL, Laurent M, Shen R, Barker D. Illumina universal bead arrays. Methods in Enzymology. 2006;410:57–73. doi: 10.1016/S0076-6879(06)10003-8. [DOI] [PubMed] [Google Scholar]
- 14.de Bakker P. Tagger. 2004 Retrieved from http://www.broad.mit.edu/mpg/tagger.
- 15.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 16.International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 17.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. American Journal of Human Genetics. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.SeattleSNPs. 2007 Retrieved from http://pga.mbt.washington.edu/welcome.html.
- 19.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Geer LY, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Ostell J, Miller V, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Tatusov RL, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Inoformation. Nucleic Acids Research. 2007;35:D5–12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins-Schramm HE, Chima B, Morii T, Wah K, Figueroa Y, Criswell LA, Hanson RL, Knowler WC, Silva G, Belmont JW, Seldin MF. Mexican American ancestry-informative markers: examination of population structure and marker characteristics in European Americans, Mexican Americans, Amerindians and Asians. Human Genetics. 2004;114:263–271. doi: 10.1007/s00439-003-1058-6. [DOI] [PubMed] [Google Scholar]
- 21.Choudhry S, Coyle NE, Tang H, Salari K, Lind D, Clark SL, Tsai H, Naqvi M, Phong A, Ung N, Matallana H, Avila PC, Casal J, Torres A, Nazario S, Castro R, Battle NC, Perez-Stable EJ, Kwok P, Sheppard D, Shriver MD, Rodriguez-Cintron W, Risch N, Ziv E, Burchard EG, Genetics of Asthma in Latino Americans GALA Study Population stratificiation confounds genetic association studies among Latinos. Human Genetics. 2006;118:52–664. doi: 10.1007/s00439-005-0071-3. [DOI] [PubMed] [Google Scholar]
- 22.Wigginton JE, Cutler DJ, Abecasis GR. A Note on Exact Tests of Hardy-Weinberg Equilibrium. American Journal of Human Genetics. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kass RE, Raftery AE. Bayes Factors. J. American Statistical Association. 1995;90:773–795. [Google Scholar]
- 24.Cooper GF, Herskovitz E. A Bayesian method for the induction of probabilistic networks from data. Machine Learning. 1992;9:309–347. [Google Scholar]
- 25.Ramoni MF, Sebastiani P. Bayesian methods. In: Berthold M, Hand DJ, editors. Intelligent data analysis: An introduction. Springer; New York: 2003. pp. 128–166. [Google Scholar]
- 26.Hoh J, Ott J. Mathematical multi-locus approaches to localizing complex human trait genes. Nature Reviews Genetics. 2003;4:701–709. doi: 10.1038/nrg1155. [DOI] [PubMed] [Google Scholar]
- 27.Schwarz GE. Estimating the dimensions of a model. Annals of Statistic. 1978;6:461–464. [Google Scholar]
- 28.Sebastiani P, Ramoni MR, Nolan V, Baldwin CT, Steinberg MH. Genetic dissection and prognostic modeling of overt stroke in sickle cell anemia. Nature Genetics. 2005;37:435–440. doi: 10.1038/ng1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lasko TA, Bhagwat JG, Zou KH, Ohno-Machado L. The use of receiver operating characteristic curves in biomedical informatics. Journal of Biomedical Informatics. 2005;38:404–415. doi: 10.1016/j.jbi.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 30.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the Areas Under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 31.Henin N, Vincent MF, Gruber HE, den Berghe GV. Inhibition of fatty acid and cholesterol synthesis by stimulation of AMP-activated protein kinase. FASEB Journal. 1995;9:541–546. doi: 10.1096/fasebj.9.7.7737463. [DOI] [PubMed] [Google Scholar]
- 32.van Raalte DH, Li M, Pritchard PH, Wasan KM. Peroxisome proliferator-activated receptor (PPAR)-alpha: a pharmacological target with a promising future. Pharmaceutical Research. 2004;21:1531–1538. doi: 10.1023/b:pham.0000041444.06122.8d. [DOI] [PubMed] [Google Scholar]
- 33.Witters LA, Kemp BE. Insulin activation of acetyl-CoA carboxylase accompanied by inhibition of the 5′-AMP-activated protein kinase. Journal Biological Chemistry. 1992;267:2864–2867. [PubMed] [Google Scholar]
- 34.Sumegová K, Blazícek P, Waczulíková I, Zitnanová I, Duracková Z. Activity of paraoxonase 1 (PON1) and its relationship to markers of lipoprotein oxidation in healthy Slovaks. Acta Biochim Pol. 2006;53:783–787. [PubMed] [Google Scholar]
- 35.Liu PY, Christian RC, Ruan M, Miller VM, Fitzpatrick LA. Correlating androgen and estrogen steroid receptor expression with coronary calcification and atherosclerosis in men without known coronary artery disease. J Clin Endorinol Metab. 2005;90:1041–1046. doi: 10.1210/jc.2004-1211. [DOI] [PubMed] [Google Scholar]
- 36.Christian RC, Liu PY, Harrington S, Ruan M, Miller VM, Fitzpatrick LA. Intimal estrogen receptor (ER)beta, but not ERalpha expression, is correlated with coronary calcification and atherosclerosis in pre- and postmenopausal women. J Clin Endocrinol Metab. 2006;91:2713–2720. doi: 10.1210/jc.2005-2672. [DOI] [PubMed] [Google Scholar]
- 37.Brash AR, Boeglin WE, Chang MS. Discovery of a second 15S-lipoxygenase in humans. Proc Natl Academy of Science USA. 1997;94:6148–6152. doi: 10.1073/pnas.94.12.6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rauschmayr T, Groves RW, Kupper TS. Keratinocyte expression of the type 2 interleukin 1 receptor mediates local and specific inhibition of interleukin 1-mediated inflammation. Proc National Academy of Science. 1997;94:5814–5819. doi: 10.1073/pnas.94.11.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stafforini DM, Satoh K, Atkinson DL, Tjoelker LW, Eberhardt C, Yoshida H, Imaizumi T, Takamatsu S, Zimmerman GA, McIntyre TM, Gray PW, Prescott SM. Platelet-activating factor acetylhydrolase deficiency. A missense mutation near the active site of an anti-inflammatory phospholipase. J Clinical Investigation. 1996;97:784–2791. doi: 10.1172/JCI118733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gnasso A, Motti C, Irace C, Gennaro ID, Pujia A, Leto E, Ciamei M, Crivaro A, Bernardini S, Federici G, Cortese C. The Arg allele in position 192 of PON1 is associated with carotid atherosclerosis in subjects with elevated HDLs. Atherosclerosis. 2002;164:289–295. doi: 10.1016/s0021-9150(02)00070-9. [DOI] [PubMed] [Google Scholar]
- 41.McCaskie PA, Beilby JP, Hung J, Chapman CM, McQuillan BM, Powell BL, Thompson PL, Palmer LJ. 15-Lipoxygenase gene variants are associated with carotid plaque but not carotid intima-media thickness. Human Genetics. 2008;123:445–453. doi: 10.1007/s00439-008-0496-6. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura T, Ruiz-Lozano P, Lindner V, Yabe D, Taniwaki M, Furukawa Y, Kobuke K, Tashiro K, Lu Z, Andon NL, Schaub R, Matsumori A, Sasayama S, Chien KR, Honjo T. DANCE, a Novel Secreted RGD Protein Expressed in Developing, Atherosclerotic, and Balloon-injured Arteries. J Biological Chemistry. 1999;274:22476–22483. doi: 10.1074/jbc.274.32.22476. [DOI] [PubMed] [Google Scholar]
- 43.Higuchi S, Ohtsu H, Suzuki H, Shirai H, Frank GD, Eguchi S. Angiotensin II signal transduction through the AT1 receptor: novel insights into mechanisms and pathophysiology. Clin Sci (Lond) 2007;112:417–428. doi: 10.1042/CS20060342. [DOI] [PubMed] [Google Scholar]
- 44.Ribatti D. The crucial role of vascular permeability factor/vascular endothelial growth factor in angiogenesis: a historical review. Br J Haematol. 2005;128:303–309. doi: 10.1111/j.1365-2141.2004.05291.x. [DOI] [PubMed] [Google Scholar]
- 45.Wang Z, Yue L, White M, Pelletier G, Nattel S. Differential distribution of inward rectifier potassium channel transcripts in human atrium versus ventricle. Circulation. 1998;98:2422–2428. doi: 10.1161/01.cir.98.22.2422. [DOI] [PubMed] [Google Scholar]
- 46.Hoffmann U, Brady TJ, Muller J. Use of new imaging techniques to screen for coronary artery disease. Circulation. 2003;108:e50–e53. doi: 10.1161/01.CIR.0000085363.88377.F2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


