Abstract
Meningiomas are among the most frequent tumors of the brain and spinal cord accounting for 15% to 20% of all central nervous system tumors and frequently associated with neurofibromatosis type 2. In this study, we aimed to unravel molecular meningioma tumorigenesis and discover novel protein biomarkers for diagnostic and/or prognostic purposes and performed in-depth proteomic profiling of meningioma cells compared to human primary arachnoidal cells. We isolated proteins from meningioma cell line SF4433 and human primary arachnoidal cells and analyzed the protein profiles by Gel-nanoLC-MS/MS in conjunction with protein identification and quantification by shotgun nanoLC tandem mass spectrometry and spectral counting. Differential analysis of meningiomas revealed changes in the expression levels of 281 proteins (P < 0.01) associated with various biological functions such as DNA replication, recombination, cell cycle, and apoptosis. Among several interesting proteins, we focused on a subset of the highly significantly up-regulated proteins, the minichromosome maintenance (MCM) family. We performed subsequent validation studies by qRT-PCR in human meningioma tissue samples (WHO grade I: 14 samples, WHO grade II: 7 samples and WHO grade III: 7 samples) compared to arachnoidal tissue controls (from fresh autopsies; 3 samples) and found that MCMs are highly and significantly up-regulated in human meningioma tumor samples compared to arachnoidal tissue controls. We found a significant increase in MCM2 (8 fold) and MCM3 (5 fold), MCM4 (4 fold), MCM5 (4 fold), MCM6 (3 fold), MCM7 (5 fold) expressions in meningiomas. This study suggests that MCM family proteins are up-regulated in meningiomas and can be used as diagnostic markers.
INTRODUCTION
Meningioma is one of the most common central nervous system tumors and accounting for 32.1% of all reported brain tumors.1 They are derived from meningothelial (arachnoid cap) cells. These cells are most common within the arachnoid villi but may be present throughout the craniospinal arachnoid space.2 According to the WHO grading system, these tumors are classified as typical WHO grade I (approximately 91% of meningiomas), atypical WHO grade II (5%), and anaplastic/malignant WHO grade III (4%).2,3 They are most likely to be diagnosed in adults between 40–70 years of age and significantly more common in women than in men with a greater than 2:1 ratio.4 Surgery is the primary and often only choice of treatment for WHO I grade tumors. Complete resection can be achieved in 38–80% of patients, depending on tumor localization. However, despite complete resection, a radiological recurrence or a second primary tumor (SPT) develops in approximately 19% of all cases.6–7 Radiotherapy is used as an additional post-operative treatment in WHO II and III meningiomas,8 and as primary treatment for recurrent or inoperable WHO I meningiomas. Additional radiotherapy could be considered for WHO I tumors if biomarkers were available to identify WHO I meningiomas at risk for recurrence.
As for all cancers, meningioma tumorigenesis is driven by the accumulation of genetic aberrations of which an overview is presented by Riemenschneider et al.8 The deletion in chromosome 22q associated with loss of the neurofibromatosis 2 (NF2) gene has been one of the most common events associated with meningioma tumorigenesis.9,10 The NF2 gene eoncodes a tumor suppressor protein, merlin, also called schwannomin, which is related to ezrin-radixin-moesin (ERM) proteins of the band 4.1 superfamily of membrane-cytoskeletal linkers.11,12 One of our recent studies provided evidence that miRNAs could also contribute to the tumorigenesis of meningiomas; down-regulated microRNA-200a in meningiomas was found to promote tumor growth by reducing E-cadherin and activating the Wnt/β-catenin signaling pathway.13 Several other genetic events such as mutation of TP53, PTEN14 or chromosomal deletions in 1p, 3p, 6q, 10q and 14q15,16 and chromosomal gains in 12q, 15q, 17q and 20q17 have also been pointed.
Apart from genetic aberrations, protein expression changes have been reported. Increased expression of the proliferation marker Ki-67 was described to correlate with a higher risk for recurrence.18,19 Also progesterone receptor expression was found to be inversely related with WHO grade.20–22
Still, knowledge of the biological principles underlying meningioma tumorigenesis is scarce and none of the genetic or protein aberrations has demonstrated sufficient potential to be implemented in routine diagnostics, so as of yet, no markers are available for the prediction of recurrence after resection.
All types of cancer as well as meningioma constitute a major public health problem that presents several challenges to researchers such as identification of biomarkers for improved and early diagnosis, classification of tumors and the definition of targets for more-effective therapeutic precautions.23 Since proteins are the functional output of the genome, responsible for the (tumor) cell’s biology and behavior, and can easily be measured in clinical samples by straight-forward antibody-based detection, they can possibly be very suitable biomarkers. Recently, studies on protein profiling of different types of tumor tissue provided crucial information on the pathogenesis of cancer at the molecular level and additionally supplied multiple protein biomarker candidates for various applications. Proteomics technology enables the simultaneous quantitative investigation of thousands of proteins allowing differential expression profiling of multiple different samples.24
In this study, to unravel molecular meningioma tumorigenesis and discover novel protein biomarkers for diagnostic and/or prognostic purposes, we performed in-depth proteomic profiling of meningioma cells compared to normal human primary arachnoidal cells, the cell origin of meningioma tumors. We identified 281 proteins significantly deregulated in meningiomas (p<0.01). For validation of the differential expression, we focused on a subset of the highly significantly up-regulated proteins, i..e. the minichromosome maintenance (MCM) family. MCM proteins includes six highly conserved DNA binding members, MCM2 through MCM7.25 They are considered to function as licensing components for S-phase of cell cycle.26 The expression of all family members was validated by qRT-PCR in human meningioma tumor samples (n=28) and further western blot demonstrated that MCM proteins are up-regulated in meningiomas compared to arachnoidal controls. All MCM family members exhibited a striking up-regulation as most were undetected in arachnoidal cells and highly expressed in meningioma cells. Interestingly, recent studies have pointed out the role(s) of MCM family members as diagnostic and/or prognostic markers for several malignancies including colon cancer,27 breast and prostate cancers.28,29
MATERIALS AND METHODS
Tumor and Tissue samples
Histopathological primary tumor samples were obtained from the tissue discarded during resection, normal arachnoidal tissues were obtained from autopsies within 5–7 hour of death. Human meningioma samples were collected and de-identified by the Neuro-oncology Tumor Repository, fresh-frozen and stored at −80 °C under IRB protocols approved by Massachusetts General Hospital Committee on Human Research.
Cell lines
Human benign meningioma cell line SF4433, SF4068, and SF3061 were provided by Dr. Anita Lal (University of California, San Francisco) and were immortalized by transduction with expression cassettes for human telomerase and papillomavirus E6/E7.33 Cell lines were cultured at 37 °C and in 5% CO2 in Dulbecco’s Modification of Eagle’s Medium (DMEM) supplemented with 10% FBS (JRH Biosciences, Kansas, USA), 100 U penicillin, and 0.1 mg/ml streptomycin. Primary arachnoidal cells were obtained from Dr. Marianne James (Molecular Neurogenetics Unit, Center for Human Genetic Research, Massachusetts General Hospital, Boston) and cultured in IMEM (Mediatech, Inc. VA, USA) complemented with % 15 FBS (JRH Biosciences, Kansas, USA), 2 mg/ml insulin (Sigma-Aldrich, St. Louis, MO, USA), 100 U penicillin and 0.1 mg/ml streptomycin.
Cell lysis and SDS-PAGE
Meningioma cells, SF4433, and primary arachnoidal cells, AC030, were seeded in 10 cm2 diameter tissue culture plates and cultured for two days until they were full-confluent (4 × 106 cells/plate). Cells were subsequently washed with 1X PBS for two times and tyrpsinized, centrifuged and washed with water for one times to get rid of the excess salts. Cells were scraped in RIPA buffer containing proteinase inhibitor. Each cell line sample was analyzed in duplo. Equal amount of total protein (50 μg) was separated in NuPAGE Novex Bis-Tris Mini Gel (Invitrogen). Gel was stained with Coomassie brilliant blue G-250 (Pierce), washed and each lane was sliced into ten bands using band pattern to guide the slicing. The whole process was performed in keratin-free conditions.
Gel digestion
Before MS analysis, separated proteins were in-gel digested according to the method described by Shevchenko et al.53 Gel lanes corresponding to the different protein samples were sliced into ten bands. The bands were washed/dehydrated three times in 50 mM ABC (ammonium bicarbonate pH 7.9)/50 mM ABC + 50% ACN (acetonitrile). Subsequently, cysteine bonds were reduced with 10 mM dithiotreitol for 1 h at 56 °C and alkylated with 50 mM iodoacetamide for 45 min at RT in the dark. After two subsequent wash/dehydration cycles the bands were dried 10 min in a vacuum centrifuge and incubated overnight with 0.06 μg/μl trypsin at 25 °C. Peptides were extracted once in 1% formic acid and subsequently two times in 50% ACN in 5% formic acid. The volume was reduced to 50 μl in a vacuum centrifuge prior to LC-MS/MS analysis.
NanoLC-MS/MS analysis
Peptides were separated by an Ultimate 3000 nanoLC system (Dionex LC-Packings, Amsterdam, The Netherlands) equipped with a 20 cm × 75 μm ID fused silica column custom packed with 3 μm 120 Å ReproSil Pur C18 aqua (Dr Maisch GMBH, Ammerbuch-Entringen, Germany). After injection, peptides were trapped at 30 μl/min on a 5 mm × 300 μm ID Pepmap C18 cartridge (Dionex LC-Packings, Amsterdam, The Netherlands) at 2% buffer B (buffer A: 0.05% formic acid in MQ; buffer B: 80% ACN + 0.05% formic acid in MQ) and separated at 300 nl/min in a 10–40% buffer B gradient in 60 min. Eluting peptides were ionized at 1.7 kV in a Nanomate Triversa Chip-based nanospray source using a Triversa LC coupler (Advion, Ithaca, NJ). Intact peptide mass spectra and fragmentation spectra were acquired on a LTQ-FT hybrid mass spectrometer (Thermo Fisher, Bremen, Germany). Intact masses were measured at resolution 50.000 in the ICR cell using a target value of 1 × 106 charges. In parallel, following an FT pre-scan, the top 5 peptide signals (charge-states 2+ and higher) were submitted to MS/MS in the linear ion trap (3 amu isolation width, 30 ms activation, 35% normalized activation energy, Q value of 0.25 and a threshold of 5000 counts). Dynamic exclusion was applied with a repeat count of 1 and an exclusion time of 30s.
Database searching, statistics and Ingenuity Pathway Analysis
MS/MS spectra were searched against the human IPI database 3.31(67511 entries) using Sequest (version 27, rev 12), which is part of the BioWorks 3.3 data analysis package (Thermo Fisher, San Jose, CA). MS/MS spectra were searched with a maximum allowed deviation of 10 ppm for the precursor mass and 1 amu for fragment masses. Methionine oxidation and cysteine carboxamidomethylation were allowed as variable modifications, two missed cleavages were allowed and the minimum number of tryptic termini was 1. After database searching the DTA and OUT files were imported into Scaffold 2.01.01 (Proteome software, Portland, OR). Scaffold was used to organize the gel-band data and to validate peptide identifications using the Peptide Prophet algorithm54 only identifications with a probability> 95% were retained. Subsequently, the Protein Prophet algorithm55 was applied and protein identifications with a probability of > 99% with 2 peptides or more in at least one of the samples were retained. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped. For each protein identified, the number of spectra was exported to Excel. The number of spectra per protein per sample was normalized against the total number of measured spectra. The beta-binomial test was performed to identify differentially expressed proteins. The list of differentially expressed proteins, including p-values and fold changes was imported in the online software package Ingenuity (Ingenuity IPA, version 7.6) and pathway and network analysis was performed with only direct relationships.
Quantitative RT-PCR
Quantitative RT-PCR Analysis was performed to determine the mRNA expression levels of MCM proteins. Total RNA was isolated from tissue specimens by using Trizol (Invitrogen) according to the manufacturer instructions and quantified by NanoDrop ND-1000 Spectrophotometer (Thermo Scientific) and stored − 80 °C. Equal amounts (1 μg) of RNA were converted into cDNAs by using Omniscript reverse transcription kit (Qiagen) in accordance with the protocol provided by manufacturer. qRT-PCR was performed in triplicate using ABI PRISM 7000 Sequence Detection system (Applied Biosystems) with the SYBR Green PCR kit from Applied Biosystems and the primers as follows: MCM2: 5′-AGACGAGATAGAGCTGACTG-3′ (F) and 5′-CACCACGTACCTTGTGCTTG-3′ (R);56 MCM3: 5′-CGAGGAAGACCAGGGAATTT-3′ (F) and 5′-AGGCAACCAGCTCCTCAAAG-3′ (R);57 MCM4 (www.genomecenter.ucdavis.edu/.../Human%20E2Fprimerseq.doc): 5′CCACCACCTCCCGTCCTTAA3′ (F) and 5′-AATCACAGCGGCGCTCGTAC-3′ (R); MCM5: 5′CCCATTGGGGTATACACGTC-3′ (F) and 5′ACGGTCATCTTCTCGCATCT-3′ (R:57), MCM6: 5′-ACTAGACAGAAGCGGCTTACTC-3′ (F) and 5′-CTTTTTTCGCTGAACACCGCCAGCT-3′ (R:www.genomecenter.ucdavis.edu/.../Human%20E2Fprimerseq.doc); MCM7: 5′-TCAATTTGTGAGAATGCCAGGCGC-3′ (F) and 5′-CACAGTTACCAACTTCCCCACAGA-3′ (R)58, GAPDH mRNA was used for normalization as described.59
Western Blot
Western blot analysis
Meningioma cells and primary arachnoidal cells were harvested and total protein was separated on a SDS–8% polyacrylamide gel and blotted onto nitrocellulose. The membrane was blocked with 5% nonfat dry milk in TBS-T (TBS containing 0.05% Tween-20) for 2 hrs at 37°C and then rinsed once with TBS-T and washed twice for 15 min and twice for 5 min at room temperature with TBS-T.13 The primary antibodies used were MCM3 (Cell Signaling Technology, #4012) in 1:1000 ratio, and β-actin (Sigma, #A5441) in 1:1000 dilution.
RESULTS
Proteomics protein profiling reveals a set of differential proteins in meningiomas
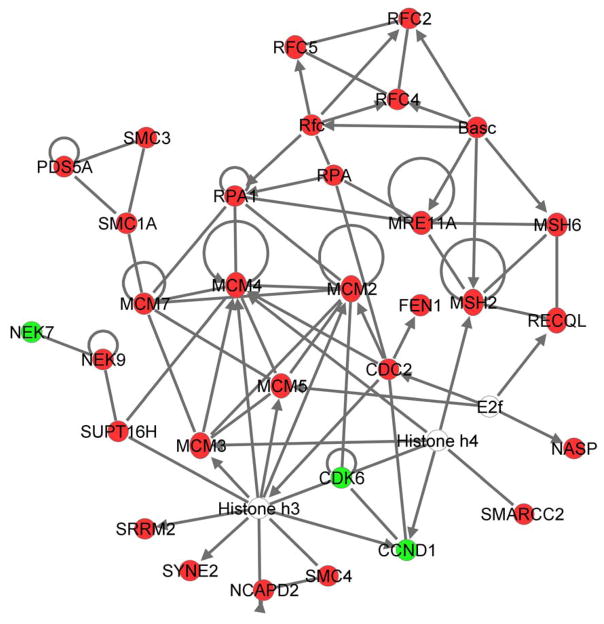
We performed a proteomics study to investigate the protein expression profile in meningioma cells as compared to human primary arachnoidal cells. Based on the findings of others, arachnoidal cells are believed to be the cell of origin of meningioma tumors.30,32 A flow chart of our experimental procedures is shown in Fig. 1. We isolated proteins from meningioma cell line SF443333 and primary arachnoidal cells30 and analyzed, in duplicate, the protein profiles by Gel-nanoLC-MS/MS, i.e. protein separation by 1D SDS-PAGE in conjunction with protein identification and quantification by shotgun nanoLC tandem mass spectrometry and spectral counting. Our approach permitted the simultaneous identification and quantification of over 2,800 proteins; Differential analysis of meningiomas revealed changes in the expression levels of 281 proteins (Supplementary Table 1; P < 0.01) associated with various biological functions such as DNA replication, recombination, cell cycle, and apoptosis. Of these 281 differential proteins 103 were from the cytoplasm, 91 from the nucleus, 37 from the plasma membrane, 19 extracellular and 31 had unknown cellular location. For 133 of these proteins, expression was exclusively detected in either arachnoidal or meningioma cells (on/off regulation). Ninety-seven proteins were found to be detected only in meningioma cells (Table 1), whereas 36 proteins were solely found in arachnoidal cells (Table 2). Within the group of on/off regulated proteins Ingenuity Pathway Analysis (IPA) revealed a significant overrepresentation of proteins involved in cancer (65 out of 133), cellular growth and proliferation (31 out of 133) and DNA replication, recombination and repair (28 out of 133). Network analysis exposed a network of direct relationships between the proteins involved in these cellular processes as depicted in Figure 1. Representative protein families in this network of on/off regulated proteins are the minichromosome maintenance (MCM) family, the replication factor C subunits (RFC proteins) and the structural maintenance of chromosomes (SMC) family (see Figure 2). All three protein families are present in the nucleus and involved in DNA replication. Additionally several other proteins were found to be up-regulated (4–12-fold) such as replication protein A (RPA1) Epiplakin 1(EPPK1), Flap endonuclease 1 (FEN1), BAG family molecular chaperone regulator 3 (BAG3), apoptosis inhibitor isoform 5 (API5), protein tyrosine kinase 7 (PTK7), DNA mismatch repair proteins, MSH2 and MSH6, and double-strand break repair protein MRE11, and DNA repair protein RAD50. Proteins such as AP-1 complex subunit beta-1 (APIB1), G1/S-specific cyclin-D1 (CCND1), cell division protein kinase 6 (CDK6), were found to be detected only in arachnoidal cells (Table 2).
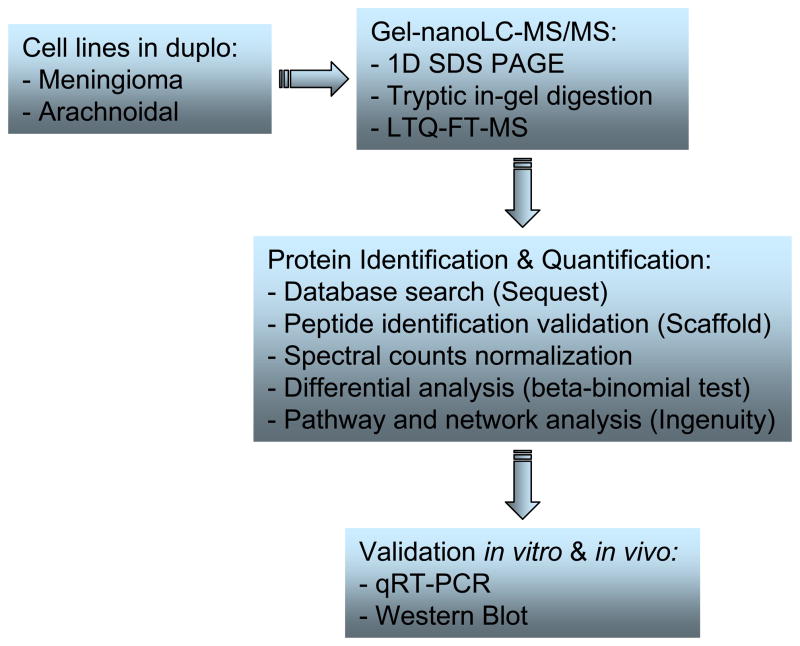
Fig. 1. A schematic presentation of the proteomics workflow applied to the analysis of meningioma and arachnoidal cells.
Proteins from cell lysates were isolated by centrifugation and separated by SDS-PAGE. Separated proteins were digested and peptide extracts are injected to LC column and second separation was performed by chromatography. The samples eluted from the LC column were analyzed by mass spectrometry. The mass spectrum obtained MS/MS are compared with the theoretical spectrums provided by databases. The candidate proteins were validated in vitro and in vivo by qRT-PCR and western blots.
Table 1.
List of proteins exclusively expressed in meningioma cells
| Gene symbol and protein name | Accession number | Normalized spectral counts | P-value | |||
|---|---|---|---|---|---|---|
| Arachnoidal cells in duplo | Meningioma cells in duplo | |||||
| MX1 Interferon-induced GTP-binding protein Mx1 | IPI00167949 | 0 | 0 | 52 | 55 | 0.00005 |
| MCM2 DNA replication licensing factor MCM2 | IPI00184330 | 0 | 0 | 23 | 22 | 0.00017 |
| MCM3 DNA replication licensing factor MCM3 | IPI00013214 | 0 | 0 | 21 | 20 | 0.00019 |
| MCM7 Isoform 1 of DNA replication licensing factor MCM7 | IPI00299904 | 0 | 0 | 21 | 17 | 0.00026 |
| MCM4 DNA replication licensing factor MCM4 | IPI00018349 | 0 | 0 | 19 | 16 | 0.00027 |
| SAMHD1 SAM domain and HD domain-containing protein 1 | IPI00294739 | 0 | 0 | 14 | 15 | 0.00035 |
| MCM5 DNA replication licensing factor MCM5 | IPI00018350 | 0 | 0 | 16 | 13 | 0.00037 |
| RECQL ATP-dependent DNA helicase Q1 | IPI00178431 | 0 | 0 | 11 | 14 | 0.00047 |
| OAS3 2′-5′-oligoadenylate synthetase 3 | IPI00002405 | 0 | 0 | 10 | 11 | 0.00054 |
| SYNE2 Isoform 1 of Nesprin-2 | IPI00239405 | 0 | 0 | 20 | 13 | 0.00055 |
| EPPK1 Epiplakin | IPI00010951 | 0 | 0 | 16 | 11 | 0.00056 |
| CDC2 Hypothetical protein DKFZp686L20222 | IPI00026689 | 0 | 0 | 10 | 10 | 0.00058 |
| HMGB2 High mobility group protein B2 | IPI00219097 | 0 | 0 | 9 | 10 | 0.00064 |
| RPA1 Replication protein A 70 kDa DNA-binding subunit | IPI00020127 | 0 | 0 | 10 | 16 | 0.00074 |
| CLDN11 Claudin-11 | IPI00026053 | 0 | 0 | 9 | 8 | 0.00080 |
| SRRM2 Isoform 1 of Serine/arginine repetitive matrix protein 2 | IPI00782992 | 0 | 0 | 13 | 8 | 0.00096 |
| NASP Isoform 1 of Nuclear autoantigenic sperm protein | IPI00179953 | 0 | 0 | 7 | 8 | 0.00097 |
| SMC4 Isoform 1 of Structural maintenance of chromosomes protein 4 | IPI00411559 | 0 | 0 | 7 | 7 | 0.00107 |
| MCAM Isoform 1 of Cell surface glycoprotein MUC18 precursor | IPI00016334 | 0 | 0 | 6 | 8 | 0.00122 |
| FEN1 Flap endonuclease 1 | IPI00026215 | 0 | 0 | 6 | 8 | 0.00122 |
| RRM1 Ribonucleoside-diphosphate reductase large subunit | IPI00013871 | 0 | 0 | 6 | 9 | 0.00127 |
| FXR1 Isoform 1 of Fragile X mental retardation syndrome-related protein 1 | IPI00016249 | 0 | 0 | 7 | 6 | 0.00130 |
| CRABP2 Cellular retinoic acid-binding protein 2 | IPI00216088 | 0 | 0 | 6 | 6 | 0.00142 |
| RIF1 Isoform 1 of Telomere-associated protein RIF1 | IPI00477805 | 0 | 0 | 5 | 7 | 0.00166 |
| SMARCC2 Isoform 2 of SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 | IPI00150057 | 0 | 0 | 5 | 6 | 0.00173 |
| TMPO Lamina-associated polypeptide 2 isoform alpha | IPI00216230 | 0 | 0 | 8 | 5 | 0.00189 |
| TCEA1 Isoform 1 of Transcription elongation factor A protein 1 | IPI00333215 | 0 | 0 | 5 | 9 | 0.00192 |
| EFTUD2 116 kDa U5 small nuclear ribonucleoprotein component | IPI00003519 | 0 | 0 | 6 | 12 | 0.00195 |
| PGM2 Phosphoglucomutase-2 | IPI00550364 | 0 | 0 | 5 | 5 | 0.00200 |
| RFC4 Replication factor C subunit 4 | IPI00017381 | 0 | 0 | 5 | 5 | 0.00200 |
| VPS13A 351 kDa protein | IPI00251344 | 0 | 0 | 9 | 5 | 0.00217 |
| PDS5A SCC-112 protein | IPI00303063 | 0 | 0 | 4 | 6 | 0.00243 |
| OAS2 Isoform p69 of 2′-5′-oligoadenylate synthetase 2 | IPI00218185 | 0 | 0 | 4 | 7 | 0.00253 |
| GALE UDP-glucose 4-epimerase | IPI00553131 | 0 | 0 | 4 | 7 | 0.00253 |
| NNMT Nicotinamide N-methyltransferase | IPI00027681 | 0 | 0 | 6 | 4 | 0.00257 |
| MRE11A Isoform 1 of Double-strand break repair protein MRE11A | IPI00029159 | 0 | 0 | 5 | 4 | 0.00265 |
| TMEM137;RBM14 Isoform 1 of RNA-binding protein 14 | IPI00013174 | 0 | 0 | 5 | 4 | 0.00265 |
| UBE2C Ubiquitin-conjugating enzyme E2 C | IPI00013002 | 0 | 0 | 5 | 4 | 0.00265 |
| PSMB9 Isoform LMP2.L of Proteasome subunit beta type 9 precursor | IPI00000787 | 0 | 0 | 4 | 8 | 0.00284 |
| TPRKB Isoform 3 of TP53RK-binding protein | IPI00217362 | 0 | 0 | 4 | 4 | 0.00313 |
| NCAPD2 Condensin complex subunit 1 | IPI00299524 | 0 | 0 | 4 | 4 | 0.00313 |
| RCC2 Protein RCC2 | IPI00465044 | 0 | 0 | 4 | 4 | 0.00313 |
| SMCHD1 similar to SMC hinge domain containing 1 | IPI00465022 | 0 | 0 | 8 | 4 | 0.00320 |
| DYSF Dysferlin_v1 | IPI00020210 | 0 | 0 | 4 | 9 | 0.00341 |
| IFIT1 Interferon-induced protein with tetratricopeptide repeats 1 | IPI00018300 | 0 | 0 | 3 | 5 | 0.00399 |
| DNAJB1 DnaJ homolog subfamily B member 1 | IPI00015947 | 0 | 0 | 3 | 5 | 0.00399 |
| TAP1 transporter 1, ATP-binding cassette, sub-family B | IPI00646625 | 0 | 0 | 3 | 5 | 0.00399 |
| GGH Gamma-glutamyl hydrolase precursor | IPI00023728 | 0 | 0 | 3 | 5 | 0.00399 |
| PTK7 PTK7 protein tyrosine kinase 7 isoform c precursor | IPI00168813 | 0 | 0 | 3 | 6 | 0.00413 |
| ANP32E Acidic leucine-rich nuclear phosphoprotein 32 family member E | IPI00165393 | 0 | 0 | 3 | 6 | 0.00413 |
| H1F0 Histone H1.0 | IPI00550239 | 0 | 0 | 3 | 6 | 0.00413 |
| SUPT16H FACT complex subunit SPT16 | IPI00026970 | 0 | 0 | 5 | 3 | 0.00421 |
| SCD Acyl-CoA desaturase | IPI00299468 | 0 | 0 | 5 | 3 | 0.00421 |
| C6orf211 UPF0364 protein C6orf211 | IPI00002270 | 0 | 0 | 5 | 3 | 0.00421 |
| SPC24 Kinetochore protein Spc24 | IPI00168317 | 0 | 0 | 5 | 3 | 0.00421 |
| PPIF Peptidyl-prolyl cis-trans isomerase, mitochondrial precursor | IPI00026519 | 0 | 0 | 3 | 4 | 0.00441 |
| PYGL Glycogen phosphorylase, liver form | IPI00470525 | 0 | 0 | 3 | 4 | 0.00441 |
| C20orf77 Uncharacterized protein C20orf77 | IPI00009659 | 0 | 0 | 3 | 4 | 0.00441 |
| DNMT1 Isoform 1 of DNA (cytosine-5)-methyltransferase 1 | IPI00031519 | 0 | 0 | 4 | 3 | 0.00452 |
| DEK Protein DEK | IPI00020021 | 0 | 0 | 4 | 3 | 0.00452 |
| GNL1 guanine nucleotide binding protein-like 1 | IPI00396387 | 0 | 0 | 4 | 3 | 0.00452 |
| C7orf50 MGC11257 protein | IPI00031651 | 0 | 0 | 4 | 3 | 0.00452 |
| MVK Mevalonate kinase | IPI00010717 | 0 | 0 | 4 | 3 | 0.00452 |
| SMC3 Structural maintenance of chromosomes protein 3 | IPI00219420 | 0 | 0 | 3 | 8 | 0.00576 |
| SMC1A Structural maintenance of chromosomes protein 1A | IPI00291939 | 0 | 0 | 3 | 8 | 0.00576 |
| IFIT3 Interferon-induced protein with tetratricopeptide repeats 3 | IPI00024254 | 0 | 0 | 3 | 3 | 0.00583 |
| FAM129A Niban protein | IPI00328350 | 0 | 0 | 3 | 3 | 0.00583 |
| UBE2E1;UBE2E2 Ubiquitin-conjugating enzyme E2 E1 | IPI00021346 | 0 | 0 | 3 | 3 | 0.00583 |
| FAM128B hypothetical protein LOC80097 | IPI00410094 | 0 | 0 | 3 | 3 | 0.00583 |
| MSH2 DNA mismatch repair protein Msh2 | IPI00017303 | 0 | 0 | 2 | 4 | 0.00800 |
| IGF2BP1 Insulin-like growth factor 2 mRNA-binding protein 1 | IPI00008557 | 0 | 0 | 2 | 4 | 0.00800 |
| RFC2 Isoform 1 of Replication factor C subunit 2 | IPI00017412 | 0 | 0 | 2 | 4 | 0.00800 |
| RFC5 Replication factor C subunit 5 | IPI00031514 | 0 | 0 | 2 | 5 | 0.00806 |
| PLSCR1 Phospholipid scramblase 1 | IPI00005181 | 0 | 0 | 4 | 2 | 0.00837 |
| C10orf35 Uncharacterized protein C10orf35 | IPI00060546 | 0 | 0 | 4 | 2 | 0.00837 |
| SDC1 Syndecan-1 precursor | IPI00002441 | 0 | 0 | 5 | 2 | 0.00872 |
| MSH6 Isoform GTBP-N of DNA mismatch repair protein MSH6 | IPI00384456 | 0 | 0 | 2 | 6 | 0.00930 |
| TK1 Thymidine kinase, cytosolic | IPI00299214 | 0 | 0 | 2 | 6 | 0.00930 |
| DDX58 Isoform 1 of Probable ATP-dependent RNA helicase DDX58 | IPI00654731 | 0 | 0 | 2 | 3 | 0.00974 |
| CBFB core-binding factor, beta subunit isoform 1 | IPI00024871 | 0 | 0 | 2 | 3 | 0.00974 |
| C16orf61 UPF0287 protein C16orf61 | IPI00024619 | 0 | 0 | 2 | 3 | 0.00974 |
| ATG3 Isoform 1 of Autophagy-related protein 3 | IPI00022254 | 0 | 0 | 2 | 3 | 0.00974 |
| NEK9 Serine/threonine-protein kinase Nek9 | IPI00301609 | 0 | 0 | 2 | 3 | 0.00974 |
| SFRS10 Isoform 1 of Splicing factor, arginine/serine-rich 10 | IPI00301503 | 0 | 0 | 2 | 3 | 0.00974 |
| SDHC Succinate dehydrogenase cytochrome b560 subunit, mitochondrial precursor | IPI00016968 | 0 | 0 | 2 | 3 | 0.00974 |
| GNPDA2 Glucosamine-6-phosphate isomerase SB52 | IPI00550894 | 0 | 0 | 2 | 3 | 0.00974 |
| TPD52 Tumor protein D52 | IPI00218323 | 0 | 0 | 2 | 3 | 0.00974 |
| GDA Guanine deaminase | IPI00465184 | 0 | 0 | 2 | 3 | 0.00974 |
| DDX42 Isoform 1 of ATP-dependent RNA helicase DDX42 | IPI00409671 | 0 | 0 | 3 | 2 | 0.00991 |
| FLJ14668 Hypothetical protein FLJ14668 | IPI00303722 | 0 | 0 | 3 | 2 | 0.00991 |
| HSPA4L Heat shock 70 kDa protein 4L | IPI00295485 | 0 | 0 | 3 | 2 | 0.00991 |
| IFIT2 Interferon-induced protein with tetratricopeptide repeats 2 | IPI00018298 | 0 | 0 | 3 | 2 | 0.00991 |
| POLR2J RPB11a protein | IPI00003310 | 0 | 0 | 3 | 2 | 0.00991 |
| CTPS2 CTP synthase 2 | IPI00645702 | 0 | 0 | 3 | 2 | 0.00991 |
| LEPREL1 Prolyl 3-hydroxylase 2 precursor | IPI00217055 | 0 | 0 | 3 | 2 | 0.00991 |
| EXOSC1 3′-5′ exoribonuclease CSL4 homolog | IPI00032823 | 0 | 0 | 3 | 2 | 0.00991 |
| TMPO Isoform Beta of Lamina-associated polypeptide 2, isoforms beta/gamma | IPI00030131 | 0 | 0 | 3 | 2 | 0.00991 |
Table 2.
List of proteins exclusively expressed in arachnoidal cells
| Gene symbol and protein name | Accession number | Normalized spectral counts | P-value | |||
|---|---|---|---|---|---|---|
| Arachnoidal cells in duplo | Meningioma cells in duplo | |||||
| GLS Isoform KGA of Glutaminase kidney isoform, mitochondrial precursor | IPI00289159 | 17 | 18 | 0 | 0 | 0.00025 |
| SDCBP Syntenin isoform 3 | IPI00479018 | 12 | 11 | 0 | 0 | 0.00050 |
| NEK7 Serine/threonine-protein kinase Nek7 | IPI00152658 | 12 | 11 | 0 | 0 | 0.00050 |
| GALNT1 Isoform 1 of Polypeptide N-acetylgalactosaminyltransferase 1 | IPI00025818 | 10 | 13 | 0 | 0 | 0.00060 |
| ITGA2 Integrin alpha-2 precursor | IPI00013744 | 10 | 9 | 0 | 0 | 0.00070 |
| SERPINE1 Plasminogen activator inhibitor 1 precursor | IPI00007118 | 10 | 8 | 0 | 0 | 0.00084 |
| AKR1C3 Aldo-keto reductase family 1 member C3 | IPI00291483 | 7 | 8 | 0 | 0 | 0.00106 |
| HSPB6 Heat-shock protein beta-6 | IPI00022433 | 10 | 7 | 0 | 0 | 0.00110 |
| KCTD12 BTB/POZ domain-containing protein KCTD12 | IPI00060715 | 8 | 6 | 0 | 0 | 0.00136 |
| ATP6V0A1 Isoform 1 of Vacuolar proton translocating ATPase 116 kDa subunit a isoform 1 | IPI00465178 | 6 | 6 | 0 | 0 | 0.00155 |
| QPRT Nicotinate-nucleotide pyrophosphorylase | IPI00300086 | 7 | 5 | 0 | 0 | 0.00186 |
| TM9SF2 Transmembrane 9 superfamily protein member 2 precursor | IPI00018415 | 5 | 6 | 0 | 0 | 0.00192 |
| TMEM173 Transmembrane protein 173 | IPI00257059 | 4 | 6 | 0 | 0 | 0.00274 |
| LOX Protein-lysine 6-oxidase precursor | IPI00002802 | 6 | 4 | 0 | 0 | 0.00275 |
| IKIP IKIP2 | IPI00401791 | 4 | 5 | 0 | 0 | 0.00289 |
| ALDOC Fructose-bisphosphate aldolase C | IPI00418262 | 5 | 4 | 0 | 0 | 0.00290 |
| CRABP1 Cellular retinoic acid-binding protein 1 | IPI00219930 | 5 | 4 | 0 | 0 | 0.00290 |
| TOM1 Target of myb1 | IPI00023191 | 4 | 4 | 0 | 0 | 0.00350 |
| MICA;HLA-A;HLA-A29.1;LOC730410;HLA-B;HLA-C HLA class I histocompatibility antigen, A-23 alpha chain precursor | IPI00472151 | 3 | 5 | 0 | 0 | 0.00455 |
| SLC12A2 Isoform 1 of Solute carrier family 12 member 2 | IPI00022649 | 5 | 3 | 0 | 0 | 0.00458 |
| COL18A1 Isoform 2 of Collagen alpha-1(XVIII) chain precursor | IPI00022822 | 3 | 4 | 0 | 0 | 0.00503 |
| CCND1 G1/S-specific cyclin-D1 | IPI00028098 | 3 | 4 | 0 | 0 | 0.00503 |
| CDK6 Cell division protein kinase 6 | IPI00023529 | 4 | 3 | 0 | 0 | 0.00505 |
| SIL1 Nucleotide exchange factor SIL1 precursor | IPI00296197 | 4 | 3 | 0 | 0 | 0.00505 |
| RND3 Rho-related GTP-binding protein RhoE precursor | IPI00001437 | 4 | 3 | 0 | 0 | 0.00505 |
| ITIH3 Inter-alpha-trypsin inhibitor heavy chain H3 precursor | IPI00028413 | 7 | 3 | 0 | 0 | 0.00547 |
| COMMD7 COMMD7 protein | IPI00641139 | 3 | 3 | 0 | 0 | 0.00673 |
| HCCS Cytochrome c-type heme lyase | IPI00023406 | 3 | 3 | 0 | 0 | 0.00673 |
| KIAA1199 Isoform 1 of Protein KIAA1199 precursor | IPI00376689 | 3 | 3 | 0 | 0 | 0.00673 |
| DNAJC5 Isoform 2 of DnaJ homolog subfamily C member 5 | IPI00023780 | 3 | 3 | 0 | 0 | 0.00673 |
| EHD3 EH domain-containing protein 3 | IPI00021458 | 3 | 3 | 0 | 0 | 0.00673 |
| ETFDH Isoform 1 of Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial precursor | IPI00032875 | 3 | 3 | 0 | 0 | 0.00673 |
| SEC24B Protein transport protein Sec24B | IPI00030851 | 2 | 4 | 0 | 0 | 0.00926 |
| FAH Fumarylacetoacetase | IPI00031708 | 4 | 2 | 0 | 0 | 0.00930 |
| CLU clusterin isoform 1 | IPI00400826 | 5 | 2 | 0 | 0 | 0.00936 |
| RABGGTA Geranylgeranyl transferase type-2 alpha subunit | IPI00022664 | 5 | 2 | 0 | 0 | 0.00936 |
Fig. 2. Most significant network of direct relationship between on/off regulated differential proteins.
Ingenuity Pathway Analysis annotates the differential proteins with biological and cellular functions and then calculates which of these biological and cellular processes are significantly overrepresented within the list of differential proteins. Further, Ingenuity constructs a network of all the proteins involved in the overrepresented biological functions. Figure 2 presents an overview of the proteins exclusively expressed in either meningioma (in red) or arachnoidal (in green) cells and involved in the most significant overrepresented processes namely DNA replication, Recombination, and Repair, Cancer and Cell Cycle; and shows the direct relationships between the proteins as described in literature. Ingenuity constructed a highly significant functional network involved in DNA replication, Recombination, and Repair, Cancer, Cell Cycle of protein exclusively expressed in either meningioma (in red) or arachnoidal (in green) cells.
Validation studies: the possible role(s) of the MCMs in meningioma diagnosis
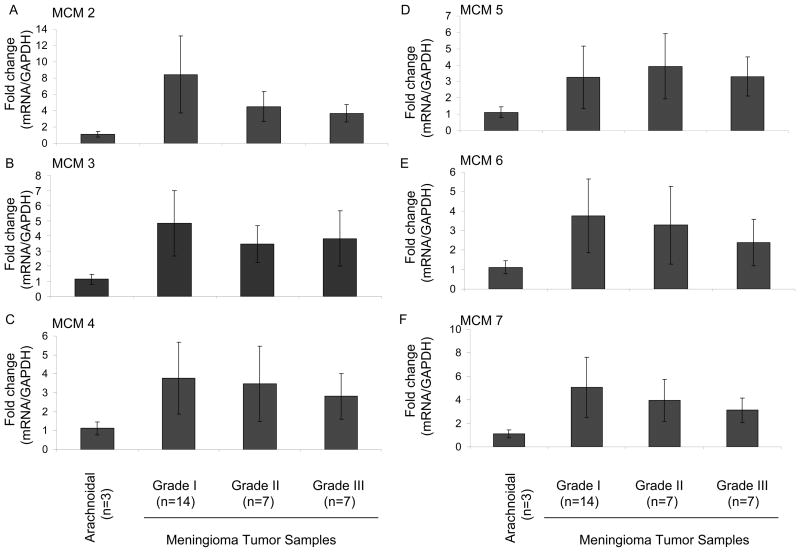
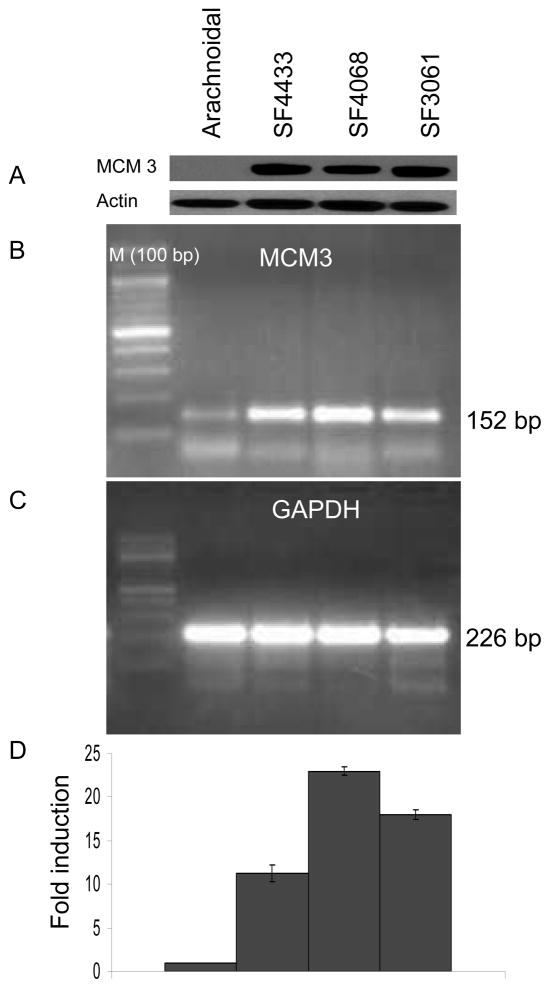
MCM2, MCM3, MCM4, MCM5 and MCM7 were exclusively detected in meningioma cells and MCM6 was found to be over 30-fold upregulated (Table 1). Based on studies emphasizing the functional importance of the MCM family in diagnosis of several malignancies such as MCM2 and MCM5 in colon cancer,27 MCM2 in breast cancer,28 and MCM7 in prostate cancer,29 we decided to follow-up on the differential expression of the MCM family. To validate our proteomics discovery data, we first performed qRT-PCR reactions in meningioma tumor tissue samples WHO grade I, half of which were deleted for the NF2 locus and half were not as determined by comparative genomic hybridization15 (14 samples), WHO grade II (7 samples) and WHO grade III (7 samples) compared to arachnoidal tissue controls (from fresh autopsies; 3 samples). WHO Grade I meningiomas are by far the most common type of meningiomas, representing an initial stage in tumor development, and as such the normal arachnoidal tissue of origin was deemed the best control to look for changes in proteins related to tumorigenesis. As shown in Figure 3, we found a significant increase in MCM2 (8 fold) and MCM3 (5 fold), MCM4 (4 fold), MCM5 (4 fold), MCM6 (3 fold), MCM7 (5 fold) expressions in meningiomas compared to arachnoidal controls. It is possible that MCM family might be also expressed in arachnoidal cells and tissue but their expression levels are below detection limit by MS. However, we did not observe a significant change between meningioma Grade I, II, and III samples. These data suggested that the MCM family proteins are up-regulated in meningiomas and might serve as diagnostic markers. We further evaluated the expression of the MCM family in meningioma cells in western blots. We have chosen MCM3 as a representative member of the family and compared its expression profiles in three different meningioma cell lines, SF4433, SF4068, and SF3061 to primary arachnoidal cells. As shown in Fig. 4, MCM3 proteins were detected in all meningioma cell lines, whereas no expression was found in arachnoidal cells. We have also performed qRT-PCRs for determination of MCM3 expression levels in those cells. As shown in Fig. 4B and D, we found a significant increase in MCM3 expression in all three meningioma cell lines compared to primary arachnoidal cells. Moreover, low level of MCM3 expression was observed in arachnoidal cells compared to SF4433, SF4068 and SF3061 cells.
Fig. 3. Validation of the MCM family proteins in meningiomas by qRT-PCR.
RNAs were isolated from arachnoidal and human meningioma tumor samples and qRT-PCR reactions were performed for MCM2 (A), MCM3 (B), MCM4 (C), MCM5 (D), MCM6 (E), and MCM7 (F) and normalized to GAPDH levels. The expression levels of the MCM transcripts in meningioma tumor samples were compared to human arachnoidal tissues. These experiments were performed in triplicate and the values are expressed as mean ± S.D. P value for every pair-wise comparison in this experiment is below 0.0001 (***p<0.0001).
Fig. 4. MCM3 protein and mRNA expression levels in meningiomas.
MCM3 protein (A) and mRNA levels (B, C, D) of meningioma cell lines, SF4433, SF4068, and SF3061 were compared to primary arachnoidal cells. A: Western blot analysis was performed with antibodies to MCM3 and anti-actin. B, C, and D: In a parallel experiment, total RNA was extracted from human primary arachnoidal cells and all three meningioma cell lines and qRT-PCR reactions were performed for MCM3 and GAPDH mRNAs. The data were normalized to GAPDH mRNA in each sample.
DISCUSSION
To date, to our knowledge there is no report on proteomics based protein profiling in meningiomas compared to arachnoidal tissues, the origin of this tumor. In the present study, we define a meningioma protein signature by Gel-NanoLC-MS/MS profiling of a meningioma cell line. This signature includes 281 differential proteins in meningioma cells as compared with control primary arachnoidal cells. Out of these 281 proteins, 97 were found to be exclusively expressed in meningiomas; whereas 36 proteins were only detected in arachnoidal cells. Because of the striking exclusive expression of all MCM family members, we focused on this family of proteins. We performed subsequent validation studies by qRT-PCR in tissue samples and western blot on the cell lines and found that MCMs are highly and significantly up-regulated in human meningioma tumor samples compared to arachnoidal tissue controls.
MCMs were first discovered in yeast Saccharomyces cerevisiae mutants that had defects in maintaining a simple minichromosome.34 Recent data suggested that these proteins are implicated not only in DNA maintenance but also in many other chromosome processes such as transcription, chromatin remodeling and genome stability.35,36 They are activated by cyclin-dependent kinases, such as Cdc6, Cdt1 and Dbf4/Cdc7 in the early G1 phase of the cell cycle to form the origin complex called the pre-replication complex (pre-RC).37,39 The hexameric MCM component of the pre-RC shows helicase activity that may provide DNA unwinding services during replication.40 Thus, MCM proteins allows the DNA replication machinery to access binding sites on the DNA.40 MCMs are expressed in abundance in all phases of the cell cycle and degraded in quiescence, senescence and differentiation steps thus they can be used as a specific markers of the cell cycle state in tissues.41 This feature of MCM proteins in proliferating cells has led to their potential clinical application as a marker for cancer screening.42 Several studies suggested that increased levels of MCMs can identify not only malignant cells43–48 but also precancerous cells and recurrence of the tumor49–51 indicating that they might also serve as a prognostic tumor marker. Further validation studies need to be performed to resolve whether MCMs might be also prognostic marker for meningiomas.
We found that the combination of 1D SDS-PAGE and shotgun nanoLC-MS/MS was a very valuable approach for proteome analysis, enabling the synchronous identification and quantification of over 2,800 proteins. Besides the MCM family proteins, this analysis has proposed many other proteins that might contribute to meningioma tumorigenesis or be potential biomarkers for diagnosis, prognosis and treatment prediction, such as the RFC proteins, the SMC family proteins and MRE11, MSH6, HDAC2, FEN1, RAD50, and STAT2. To unravel the possible role(s) of these proteins in meningioma tumorigenesis, further investigations are needed.
So far, there has been no biomarker for meningiomas. In this study, we provided evidence that MCMs can serve as diagnostic biomarkers for meningiomas. Further validation is necessary to be able to use the expression of these proteins to predict the change of regrowth after surgery in order to improve medical care in meningioma patients.
Supplementary Material
Acknowledgments
This study was supported by the Children’s Tumor Foundation 2007-01-043 (O.S.) and NINDS NS24279 (O.S.). We thank Ms. Silvina A. Fratantoni for assistance with the SDS-PAGE and in-gel digestion of the samples and Marianne F. James for human primary arachnoidal cells (Massachusetts General Hospital), and Dr. Xandra O. Breakefield for providing laboratory facilities for this work. The VUmc-Cancer Center Amsterdam is acknowledged for financial support for the proteomics infrastructure, TVP and CRJ.
References
- 1.Lamszus K. Meningioma pathology, genetics, and biology. J Neuropathol Exp Neurol. 2004;63(4):275–86. doi: 10.1093/jnen/63.4.275. [DOI] [PubMed] [Google Scholar]
- 2.Ragel BT, Couldwell WT, Gillespie DL, Wendland MM, Whang K, Jensen RL. A comparison of the cell lines used in meningioma research. Surg Neurol. 2008;70(3):295–307. doi: 10.1016/j.surneu.2007.06.031. discussion 307. [DOI] [PubMed] [Google Scholar]
- 3.Ragel B, Jensen RL. New approaches for the treatment of refractory meningiomas. Cancer Control. 2003;10(2):148–58. doi: 10.1177/107327480301000206. [DOI] [PubMed] [Google Scholar]
- 4.Whittle IR, Smith C, Navoo P, Collie D. Meningiomas. Lancet. 2004;363(9420):1535–43. doi: 10.1016/S0140-6736(04)16153-9. [DOI] [PubMed] [Google Scholar]
- 5.Mirimanoff RO, Dosoretz DE, Linggood RM, Ojemann RG, Martuza RL. Meningioma: analysis of recurrence and progression following neurosurgical resection. J Neurosurg. 1985;62(1):18–24. doi: 10.3171/jns.1985.62.1.0018. [DOI] [PubMed] [Google Scholar]
- 6.Stafford SL, Perry A, Suman VJ, Meyer FB, Scheithauer BW, Lohse CM, Shaw EG. Primarily resected meningiomas: outcome and prognostic factors in 581 Mayo Clinic patients, 1978 through 1988. Mayo Clin Proc. 1998;73(10):936–42. doi: 10.4065/73.10.936. [DOI] [PubMed] [Google Scholar]
- 7.Aghi MK, Carter BS, Cosgrove GR, Ojemann RG, Amin-Hanjani S, Martuza RL, Curry WT, Jr, Barker FG. 2nd Long-term recurrence rates of atypical meningiomas after gross total resection with or without postoperative adjuvant radiation. Neurosurgery. 2009;64(1):56–60. doi: 10.1227/01.NEU.0000330399.55586.63. discussion 60. [DOI] [PubMed] [Google Scholar]
- 8.Riemenschneider MJ, Perry A, Reifenberger G. Histological classification and molecular genetics of meningiomas. Lancet Neurol. 2006;5(12):1045–54. doi: 10.1016/S1474-4422(06)70625-1. [DOI] [PubMed] [Google Scholar]
- 9.Menon AG, Gusella JF, Seizinger BR. Progress towards the isolation and characterization of the genes causing neurofibromatosis. Cancer Surv. 1990;9(4):689–702. [PubMed] [Google Scholar]
- 10.Fontaine B, Rouleau GA, Seizinger BR, Menon AG, Jewell AF, Martuza RL, Gusella JF. Molecular genetics of neurofibromatosis 2 and related tumors (acoustic neuroma and meningioma) Ann N Y Acad Sci. 1991;615:338–43. doi: 10.1111/j.1749-6632.1991.tb37776.x. [DOI] [PubMed] [Google Scholar]
- 11.Trofatter JA, MacCollin MM, Rutter JL, Murrell JR, Duyao MP, Parry DM, Eldridge R, Kley N, Menon AG, Pulaski K, et al. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell. 1993;72(5):791–800. doi: 10.1016/0092-8674(93)90406-g. [DOI] [PubMed] [Google Scholar]
- 12.Rouleau GA, Merel P, Lutchman M, Sanson M, Zucman J, Marineau C, Hoang-Xuan K, Demczuk S, Desmaze C, Plougastel B, et al. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature. 1993;363(6429):515–21. doi: 10.1038/363515a0. [DOI] [PubMed] [Google Scholar]
- 13.Saydam O, Shen Y, Wurdinger T, Senol O, Boke E, James MF, Tannous BA, Stemmer-Rachamimov AO, Yi M, Stephens RM, Fraefel C, Gusella JF, Krichevsky AM, Breakefield XO. Down-regulated microRNA-200a in Meningiomas Promotes Tumor Growth by Reducing E-cadherin and Activating the Wnt/{beta} catenin Signaling Pathway. Mol Cell Biol. doi: 10.1128/MCB.00332-09. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joachim T, Ram Z, Rappaport ZH, Simon M, Schramm J, Wiestler OD, von Deimling A. Comparative analysis of the NF2, TP53, PTEN, KRAS, NRAS and HRAS genes in sporadic and radiation-induced human meningiomas. Int J Cancer. 2001;94(2):218–21. doi: 10.1002/ijc.1467. [DOI] [PubMed] [Google Scholar]
- 15.Leone PE, Bello MJ, de Campos JM, Vaquero J, Sarasa JL, Pestana A, Rey JA. NF2 gene mutations and allelic status of 1p, 14q and 22q in sporadic meningiomas. Oncogene. 1999;18(13):2231–9. doi: 10.1038/sj.onc.1202531. [DOI] [PubMed] [Google Scholar]
- 16.Simon M, von Deimling A, Larson JJ, Wellenreuther R, Kaskel P, Waha A, Warnick RE, Tew JM, Jr, Menon AG. Allelic losses on chromosomes 14, 10, and 1 in atypical and malignant meningiomas: a genetic model of meningioma progression. Cancer Res. 1995;55(20):4696–701. [PubMed] [Google Scholar]
- 17.Weber RG, Bostrom J, Wolter M, Baudis M, Collins VP, Reifenberger G, Lichter P. Analysis of genomic alterations in benign, atypical, and anaplastic meningiomas: toward a genetic model of meningioma progression. Proc Natl Acad Sci U S A. 1997;94(26):14719–24. doi: 10.1073/pnas.94.26.14719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakasu S, Li DH, Okabe H, Nakajima M, Matsuda M. Significance of MIB-1 staining indices in meningiomas: comparison of two counting methods. Am J Surg Pathol. 2001;25(4):472–8. doi: 10.1097/00000478-200104000-00006. [DOI] [PubMed] [Google Scholar]
- 19.Perry A, Stafford SL, Scheithauer BW, Suman VJ, Lohse CM. The prognostic significance of MIB-1, p53, and DNA flow cytometry in completely resected primary meningiomas. Cancer. 1998;82(11):2262–9. [PubMed] [Google Scholar]
- 20.Carroll RS, Glowacka D, Dashner K, Black PM. Progesterone receptor expression in meningiomas. Cancer Res. 1993;53(6):1312–6. [PubMed] [Google Scholar]
- 21.Hsu DW, Louis DN, Efird JT, Hedley-Whyte ET. Use of MIB-1 (Ki-67) immunoreactivity in differentiating grade II and grade III gliomas. J Neuropathol Exp Neurol. 1997;56(8):857–65. doi: 10.1097/00005072-199708000-00003. [DOI] [PubMed] [Google Scholar]
- 22.Perry A, Cai DX, Scheithauer BW, Swanson PE, Lohse CM, Newsham IF, Weaver A, Gutmann DH. Merlin, DAL-1, and progesterone receptor expression in clinicopathologic subsets of meningioma: a correlative immunohistochemical study of 175 cases. J Neuropathol Exp Neurol. 2000;59(10):872–9. doi: 10.1093/jnen/59.10.872. [DOI] [PubMed] [Google Scholar]
- 23.Simpson RJ, Dorow DS. Cancer proteomics: from signaling networks to tumor markers. Trends Biotechnol. 2001;19(10 Suppl):S40–8. doi: 10.1016/S0167-7799(01)01801-7. [DOI] [PubMed] [Google Scholar]
- 24.Okamoto H, Li J, Vortmeyer AO, Jaffe H, Lee YS, Glasker S, Sohn TS, Zeng W, Ikejiri B, Proescholdt MA, Mayer C, Weil RJ, Oldfield EH, Zhuang Z. Comparative proteomic profiles of meningioma subtypes. Cancer Res. 2006;66(20):10199–204. doi: 10.1158/0008-5472.CAN-06-0955. [DOI] [PubMed] [Google Scholar]
- 25.Johnson EM, Kinoshita Y, Daniel DC. A new member of the MCM protein family encoded by the human MCM8 gene, located contrapodal to GCD10 at chromosome band 20p12.3-13. Nucleic Acids Res. 2003;31(11):2915–25. doi: 10.1093/nar/gkg395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kearsey SE, Maiorano D, Holmes EC, Todorov IT. The role of MCM proteins in the cell cycle control of genome duplication. Bioessays. 1996;18(3):183–90. doi: 10.1002/bies.950180305. [DOI] [PubMed] [Google Scholar]
- 27.Giaginis C, Georgiadou M, Dimakopoulou K, Tsourouflis G, Gatzidou E, Kouraklis G, Theocharis S. Clinical significance of MCM2 and MCM5 expression in colon cancer: association with clinicopathological parameters and tumor proliferative capacity. Dig Dis Sci. 2009;54(2):282–91. doi: 10.1007/s10620-008-0305-z. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez MA, Pinder SE, Callagy G, Vowler SL, Morris LS, Bird K, Bell JA, Laskey RA, Coleman N. Minichromosome maintenance protein 2 is a strong independent prognostic marker in breast cancer. J Clin Oncol. 2003;21(23):4306–13. doi: 10.1200/JCO.2003.04.121. [DOI] [PubMed] [Google Scholar]
- 29.Padmanabhan V, Callas P, Philips G, Trainer TD, Beatty BG. DNA replication regulation protein Mcm7 as a marker of proliferation in prostate cancer. J Clin Pathol. 2004;57(10):1057–62. doi: 10.1136/jcp.2004.016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.James MF, Lelke JM, Maccollin M, Plotkin SR, Stemmer-Rachamimov AO, Ramesh V, Gusella JF. Modeling NF2 with human arachnoidal and meningioma cell culture systems: NF2 silencing reflects the benign character of tumor growth. Neurobiol Dis. 2008;29(2):278–92. doi: 10.1016/j.nbd.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalamarides M, Niwa-Kawakita M, Leblois H, Abramowski V, Perricaudet M, Janin A, Thomas G, Gutmann DH, Giovannini M. Nf2 gene inactivation in arachnoidal cells is rate-limiting for meningioma development in the mouse. Genes Dev. 2002;16(9):1060–5. doi: 10.1101/gad.226302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleihues P, Burger PC, Scheithauer BW. The new WHO classification of brain tumours. Brain Pathol. 1993;3(3):255–68. doi: 10.1111/j.1750-3639.1993.tb00752.x. [DOI] [PubMed] [Google Scholar]
- 33.Cuevas IC, Slocum AL, Jun P, Costello JF, Bollen AW, Riggins GJ, McDermott MW, Lal A. Meningioma transcript profiles reveal deregulated Notch signaling pathway. Cancer Res. 2005;65(12):5070–5. doi: 10.1158/0008-5472.CAN-05-0240. [DOI] [PubMed] [Google Scholar]
- 34.Tye BK. MCM proteins in DNA replication. Annu Rev Biochem. 1999;68:649–86. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 35.Murakami Y, Ito Y. Transcription factors in DNA replication. Front Biosci. 1999;4:D824–33. doi: 10.2741/murakami. [DOI] [PubMed] [Google Scholar]
- 36.Melendy T, Li R. Chromatin remodeling and initiation of DNA replication. Front Biosci. 2001;6:D1048–53. doi: 10.2741/melendy. [DOI] [PubMed] [Google Scholar]
- 37.Thommes P, Fett R, Schray B, Burkhart R, Barnes M, Kennedy C, Brown NC, Knippers R. Properties of the nuclear P1 protein, a mammalian homologue of the yeast Mcm3 replication protein. Nucleic Acids Res. 1992;20(5):1069–74. doi: 10.1093/nar/20.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kearsey SE, Labib K. MCM proteins: evolution, properties, and role in DNA replication. Biochim Biophys Acta. 1998;1398(2):113–36. doi: 10.1016/s0167-4781(98)00033-5. [DOI] [PubMed] [Google Scholar]
- 39.Ritzi M, Knippers R. Initiation of genome replication: assembly and disassembly of replication-competent chromatin. Gene. 2000;245(1):13–20. doi: 10.1016/s0378-1119(00)00020-2. [DOI] [PubMed] [Google Scholar]
- 40.Laskey RA, Madine MA. A rotary pumping model for helicase function of MCM proteins at a distance from replication forks. EMBO Rep. 2003;4(1):26–30. doi: 10.1038/sj.embor.embor706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoeber K, Tlsty TD, Happerfield L, Thomas GA, Romanov S, Bobrow L, Williams ED, Williams GH. DNA replication licensing and human cell proliferation. J Cell Sci. 2001;114(Pt 11):2027–41. doi: 10.1242/jcs.114.11.2027. [DOI] [PubMed] [Google Scholar]
- 42.Tachibana KE, Gonzalez MA, Coleman N. Cell-cycle-dependent regulation of DNA replication and its relevance to cancer pathology. J Pathol. 2005;205(2):123–9. doi: 10.1002/path.1708. [DOI] [PubMed] [Google Scholar]
- 43.Freeman A, Morris LS, Mills AD, Stoeber K, Laskey RA, Williams GH, Coleman N. Minichromosome maintenance proteins as biological markers of dysplasia and malignancy. Clin Cancer Res. 1999;5(8):2121–32. [PubMed] [Google Scholar]
- 44.Going JJ, Keith WN, Neilson L, Stoeber K, Stuart RC, Williams GH. Aberrant expression of minichromosome maintenance proteins 2 and 5, and Ki-67 in dysplastic squamous oesophageal epithelium and Barrett’s mucosa. Gut. 2002;50(3):373–7. doi: 10.1136/gut.50.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishimi Y, Okayasu I, Kato C, Kwon HJ, Kimura H, Yamada K, Song SY. Enhanced expression of Mcm proteins in cancer cells derived from uterine cervix. Eur J Biochem. 2003;270(6):1089–101. doi: 10.1046/j.1432-1033.2003.03440.x. [DOI] [PubMed] [Google Scholar]
- 46.Meng MV, Grossfeld GD, Williams GH, Dilworth S, Stoeber K, Mulley TW, Weinberg V, Carroll PR, Tlsty TD. Minichromosome maintenance protein 2 expression in prostate: characterization and association with outcome after therapy for cancer. Clin Cancer Res. 2001;7(9):2712–8. [PubMed] [Google Scholar]
- 47.Ramnath N, Hernandez FJ, Tan DF, Huberman JA, Natarajan N, Beck AF, Hyland A, Todorov IT, Brooks JS, Bepler G. MCM2 is an independent predictor of survival in patients with non-small-cell lung cancer. J Clin Oncol. 2001;19(22):4259–66. doi: 10.1200/JCO.2001.19.22.4259. [DOI] [PubMed] [Google Scholar]
- 48.Rodins K, Cheale M, Coleman N, Fox SB. Minichromosome maintenance protein 2 expression in normal kidney and renal cell carcinomas: relationship to tumor dormancy and potential clinical utility. Clin Cancer Res. 2002;8(4):1075–81. [PubMed] [Google Scholar]
- 49.Alison MR, Hunt T, Forbes SJ. Minichromosome maintenance (MCM) proteins may be pre-cancer markers. Gut. 2002;50(3):290–1. doi: 10.1136/gut.50.3.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hunt DP, Freeman A, Morris LS, Burnet NG, Bird K, Davies TW, Laskey RA, Coleman N. Early recurrence of benign meningioma correlates with expression of mini-chromosome maintenance-2 protein. Br J Neurosurg. 2002;16(1):10–5. doi: 10.1080/02688690120114174. [DOI] [PubMed] [Google Scholar]
- 51.Tan DF, Huberman JA, Hyland A, Loewen GM, Brooks JS, Beck AF, Todorov IT, Bepler G. MCM2--a promising marker for premalignant lesions of the lung: a cohort study. BMC Cancer. 2001;1:6. doi: 10.1186/1471-2407-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baia GS, Slocum AL, Hyer JD, Misra A, Sehati N, VandenBerg SR, Feuerstein BG, Deen DF, McDermott MW, Lal A. A genetic strategy to overcome the senescence of primary meningioma cell cultures. J Neurooncol. 2006;78(2):113–21. doi: 10.1007/s11060-005-9076-y. [DOI] [PubMed] [Google Scholar]
- 53.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68(5):850–8. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 54.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 55.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 56.Ishimi Y, Okayasu I, Kato C, Kwon HJ, Kimura H, Yamada K, Song SY. Enhanced expression of Mcm proteins in cancer cells derived from uterine cervix. Eur J Biochem. 2003;270(6):1089–101. doi: 10.1046/j.1432-1033.2003.03440.x. [DOI] [PubMed] [Google Scholar]
- 57.Snyder M, He W, Zhang JJ. The DNA replication factor MCM5 is essential for Stat1-mediated transcriptional activation. Proc Natl Acad Sci U S A. 2005;102(41):14539–44. doi: 10.1073/pnas.0507479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi YK, Yu YP, Zhu ZH, Han YC, Ren B, Nelson JB, Luo JH. MCM7 interacts with androgen receptor. Am J Pathol. 2008;173(6):1758–67. doi: 10.2353/ajpath.2008.080363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.