Abstract
Higher-order multi-protein complexes such as RNA polymerase II (Pol II) complexes with transcription initiation factors are often not amenable to X-ray structure determination. Here, we show that protein cross-linking coupled to mass spectrometry (MS) has now sufficiently advanced as a tool to extend the Pol II structure to a 15-subunit, 670 kDa complex of Pol II with the initiation factor TFIIF at peptide resolution. The N-terminal regions of TFIIF subunits Tfg1 and Tfg2 form a dimerization domain that binds the Pol II lobe on the Rpb2 side of the active centre cleft near downstream DNA. The C-terminal winged helix (WH) domains of Tfg1 and Tfg2 are mobile, but the Tfg2 WH domain can reside at the Pol II protrusion near the predicted path of upstream DNA in the initiation complex. The linkers between the dimerization domain and the WH domains in Tfg1 and Tfg2 are located to the jaws and protrusion, respectively. The results suggest how TFIIF suppresses non-specific DNA binding and how it helps to recruit promoter DNA and to set the transcription start site. This work establishes cross-linking/MS as an integrated structure analysis tool for large multi-protein complexes.
Keywords: higher-order protein complex, integrated structure analysis, mass spectrometry, multi-dimensional structure and dynamics of biological macromolecules, transcription and its regulation
Introduction
Protein crystallography has been the primary source of structural insights into multi-protein complexes for decades. However, only homogenous, stoichiometric, stable, and rigid complexes that are available in sufficient amounts generally form crystals of sufficient quality for X-ray analysis. Therefore, core complexes are often resolved by crystallography whereas the position of additional, peripheral factors remains elusive.
Cross-link analysis can provide positional information on flexible, transient, and modular higher-order multi-protein complexes, by mapping regions of spatial proximity. Cross-linking and mass spectrometry (MS) have first been used for the analysis of a multi-protein complex a decade ago (Rappsilber et al, 2000). After long development (Sinz, 2006) cross-link sites are now identified by database searches in a similar way to protein modification sites (Maiolica et al, 2007; Rinner et al, 2008). This revealed the organization of the 180 kDa Ndc80 complex, the largest complex analysed to date, at peptide resolution (Maiolica et al, 2007) and guided the X-ray analysis of the complex (Ciferri et al, 2008). Here, we show that cross-linking can be used to study even larger complexes in synergy with established structural biology techniques.
We applied our approach to a major unresolved question in molecular biology, the structure of the RNA polymerase II (Pol II) transcription initiation complex. Transcription initiation at eukaryotic protein-coding genes requires Pol II and the basal transcription factors (TFs) IIB, -D, -E, -F, and -H (Reinberg et al, 1998). Although the crystal structure of the 12-subunit Pol II is known (Armache et al, 2005), structural information on the complex of Pol II with initiation factors remains limited (Kostrewa et al, 2009).
Here, we investigate the structure of Pol II in complex with TFIIF. TFIIF was first identified based on its tight interaction with Pol II (Sopta et al, 1985). In the yeast Saccharomyces cerevisiae, about half of Pol II is bound by TFIIF (Rani et al, 2004). Yeast TFIIF comprises the essential subunits Tfg1 and Tfg2, and the non-essential subunit Tfg3 (Henry et al, 1994). Human TFIIF consists of homologues to Tfg1 (Rap74) and Tfg2 (Rap30), but lacks a Tfg3 homologue (Henry et al, 1994). Rap74 comprises an N-terminal region that binds Rap30 (Wang and Burton, 1995), a charged, central region, and a C-terminal domain that binds the phosphatase Fcp1(Chambers et al, 1995; Kobor et al, 2000). Rap30 comprises an N-terminal region that binds Rap74 (Yonaha et al, 1993), a central region that binds Pol II (Sopta et al, 1989; McCracken and Greenblatt, 1991), and a C-terminal domain (Garrett et al, 1992; Tan et al, 1994b). The N-terminal regions of Rap74 and Rap30 form a dimerization domain with a triple-barrel fold (Gaiser et al, 2000), whereas the C-terminal domains of Rap74 and Rap30 form winged helix (WH) domains (Groft et al, 1998; Kamada et al, 2001).
TFIIF is required for initiation at TATA-containing and TATA-less promoters (Burton et al, 1988), reduces the affinity of Pol II to DNA (Garrett et al, 1992), and prevents interaction of Pol II with non-specific DNA (Killeen and Greenblatt, 1992). TFIIF is required for stable pre-initiation complex formation (Tan et al, 1995), and for normal start site selection (Pinto et al, 1994; Ghazy et al, 2004; Freire-Picos et al, 2005). In initially transcribing complexes, TFIIF stimulates phosphodiester bond formation and stabilizes a short DNA–RNA hybrid (Funk et al, 2002; Khaperskyy et al, 2008). During elongation in vitro, TFIIF reduces the time of Pol II pausing (Flores et al, 1989; Price et al, 1989; Bengal et al, 1991; Chafin et al, 1991; Tan et al, 1994b) and suppresses backtracking and RNA cleavage induced by TFIIS (Elmendorf et al, 2001; Zhang et al, 2003).
To understand the multiple TFIIF functions, and the architecture of the Pol II initiation complex, detailed structural knowledge of the Pol II–TFIIF complex is required. Electron microscopy (EM) of complexes of Pol II with endogenous TFIIF and recombinant Tfg2 at 18 Å resolution suggested that Tfg2 extends along the polymerase cleft and Tfg1 binds around the Rpb4/7 subcomplex and the clamp on the Rpb1 side of the cleft (Chung et al, 2003). Site-specific radical-generating probing however placed TFIIF on the other side of the cleft near Rpb2 (Chen et al, 2007).
Here, we used protein cross-linking coupled to MS to first analyse the free, 12-subunit Pol II, a complex of 513 kDa. Agreement of the data with the crystal structure shows for the first time that the method can be applied to such large complexes. This establishes cross-linking coupled to MS as a tool for the structural analysis of large multi-protein complexes. We then apply the approach to the Pol II–TFIIF complex that was purified as a stoichiometrically homogeneous complex from yeast cells using a new protocol. This complex comprises 15 polypeptides and has a total molecular weight of 670 kDa. The resulting detailed map of cross-links between Pol II and TFIIF, together with previous crystallographic data and molecular modelling, unravels the architecture of the Pol II–TFIIF complex and provides insights into the function of TFIIF during transcription.
Results
MS cross-link analysis of Pol II
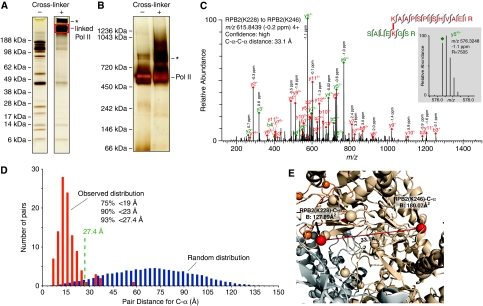
To test whether we could extend our cross-link analysis to large multi-protein complexes, we analysed the 12-subunit 513 kDa Pol II, for which a crystal structure is available (PDB 1WCM) (Armache et al, 2005). Pol II was obtained as described (Sydow et al, 2009). 30 μg of Pol II was subjected to cross-linking with the label-free cross-linker Bis (sulphosuccinimidyl) suberate (BS3, Thermo Fisher Scientific) (see Materials and methods). BS3 reacts with primary amines in lysine side chains and protein N-termini. The amines must be <11.4 Å apart, the maximal length of the BS3 spacer. Adding 16 Å to this, two times the length of a lysine side chain (6–6.5 Å) including an estimated coordinate error for mobile surface residues (1.5 Å), defines the maximal C-α distance of linkable lysine residues, 27.4 Å, when comparing our cross-link data with the available crystallographic data. We used a charge-based enrichment strategy for cross-linked peptides (Maiolica et al, 2007; Rinner et al, 2008) and high-resolution MS for peptide and fragment detection (see Materials and methods). We identified 146 linkage pairs in 429 mass spectra matching to cross-linked peptides (Supplementary Tables 1 and 2). In our subsequent analysis we focussed on those 106 linkage pairs that had both linked residues present in the Pol II structure.
Our cross-link data reflected accurately the structural features of Pol II. The observed cross-links were significantly different from a random selection of all possible pairs in the structure (P-value of 3 × 10−87) (Figure 1D). The C-α distances of 99 pairs fell below 27.4 Å, 95 (90%) fell below 23 Å, 79 (75%) fell below 19 Å. Only five of our high-confidence links (see Materials and methods) and two of low confidence cannot be explained with the crystal structure. This is apparently because of cross-linking being conducted in solution, allowing internal movement of protein regions that are fixed at certain positions in the crystal structure because of crystal lattice restraints. Indeed, six of these seven links involve residues with high B-factors in or proximal to the mobile clamp domain of Pol II (Figure 1E; Supplementary Figure S2). Thus, only a single lower confidence link appears to be false. We conclude that our cross-link analysis returns accurate distance constrains in the context of a large, multi-protein complex, with an experimentally determined error rate in the order of 1%, with 1 of the 106 observed cross-links being false.
Figure 1.
MS-coupled cross-linking analysis of Pol II. (A) SDS–PAGE analysis and (B) native gel electrophoresis of Pol II and BS3 cross-linked Pol II. Cross-linked Pol II was excised from the SDS–PAGE gel and analysed (red box). A higher-order linkage product (asterisk) was excluded, most likely corresponding to a Pol II dimer, also observed on the native gel in the absence and increased in the presence of cross-linker (asterisk). (C) High-resolution fragmentation spectrum of a cross-linked peptide. The linkage site Rpb2 K228–Rpb2 K246 was observed in the cross-linked peptide SALEK(xl)GSR/K(xl)AAPSPISHVAEIR (m/z 615.8439, 4+). Extensive ion series for both peptides are observed in the high-resolution fragmentation spectrum and provide high confidence in the match. (D) C-α distance distribution for experimentally observed lys–lys pairs (red bars) and a random probability distribution (blue bars) within Pol II. The approximate cross-link limit for BS3 of 27.4 Å is indicated by a dashed line. Observed links falling below this limit are in agreement with the X-ray structure of Pol II (PDB 1WCM); observed links exceeding this limit are potentially in conflict with the known structure. (E) Zoom into 1WCM showing Rpb2 K228 and Rpb2 K246 (red sphere). The link spans 33.1 Å and is thus 5.7 Å longer than the maximal distance the cross-linker plus side chains of lysine can bridge (27.4 Å). The crystallographic B-factor is 128 Å2 for Rpb2 K228 and 180 Å2 for Rpb2 K246, indicating both residues as likely mobile. Both residues are in loop regions.
Preparation and characterization of the Pol II–TFIIF complex
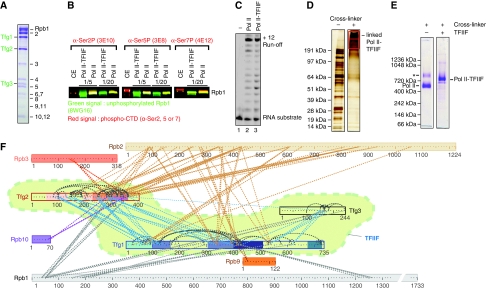
To subject the even larger, scarce, and fragile Pol II–TFIIF complex to cross-linking, we established a new protocol for its large-scale preparation. Extensive attempts to obtain S. cerevisiae TFIIF after co-expression of its subunits in Escherichia coli were unsuccessful. Previous purification of endogenous Pol II–TFIIF complex resulted in low yields and partially degraded Tfg1 (Chung et al, 2003). We therefore prepared a yeast strain that over-expresses the three TFIIF subunits and contains a tandem affinity purification (TAP) tag on Tfg2 (see Materials and methods). We could obtain up to 2 mg of pure Pol II–TFIIF complex after TAP and size exclusion chromatography.
Pol II–TFIIF complex preparations contained the 12 Pol II subunits and three TFIIF subunits in apparently stoichiometric amounts (Figure 2A). Pol II was not phosphorylated, as judged from western blotting with antibodies specific against phosphorylation at the C-terminal repeat domain (CTD) residues Ser2, Ser5, or Ser7 (Figure 2B). In an RNA extension assay (see Materials and methods and (Brueckner et al, 2007), the Pol II–TFIIF complex was as active as free Pol II (Figure 2C). Thus, the new protocol provided previously unavailable amounts of pure, homogeneous, and catalytically active yeast Pol II–TFIIF complex.
Figure 2.
Preparation and MS-coupled cross-linking analysis of the complete Pol II–TFIIF complex. (A) SDS–PAGE analysis of pure Pol II–TFIIF complex. Protein identity was confirmed by MS (not shown). (B) Western blot analysis of the phosphorylation state of the Pol II CTD residues Ser2, Ser5, and Ser7; 10 μl of 5- or 20-fold dilutions of pure Pol II and Pol II–TFIIF complexes at 1 mg/ml were subjected to 6% SDS–PAGE. After blotting on a nitrocellulose membrane (GE Healthcare), dual labelling was performed with antibodies that recognize unphosphorylated CTD (8WG16, green) and antibodies 3E10, 3EB, and 4E12 (Chapman et al, 2007), specific for phopshorylated CTD serines 2, 5, and 7 (Ser2P, Ser5P, and Ser7P, respectively). Yeast crude extract (CE) was used as control. (C) RNA extension assay of Pol II and Pol II–TFIIF in vitro (see Materials and methods). (D) SDS–PAGE analysis of Pol II–TFIIF complex and BS3 cross-linked Pol II–TFIIF complex. Cross-linked Pol II–TFIIF complex was excised from the SDS–PAGE gel in two bands and analysed (red box). (E) Native gel electrophoresis of BS3 cross-linked Pol II–TFIIF complex with BS3 cross-linked Pol II complex for comparison. The native gel shows absence of a dimer complex for BS3 cross-linked Pol II–TFIIF complex. (F) Cross-link map for TFIIF in complex with Pol II. Observed links from TFIIF to Pol II (dashed lines) are colour coded by the respective Pol II subunit. Links between TFIIF subunits (blue) and within TFIIF subunits (grey). For colour coding of domains in TFIIF see Figure 3A.
Cross-link analysis of Pol II–TFIIF complex
We next cross-linked and analysed the Pol II–TFIIF complex (Figure 2D–F), comprising 15 subunits with a total molecular weight of 670 kDa. Using 200 μg of purified complex allowed for elaborate fractionation and more comprehensive analysis. We identified by MS 402 linkage sites of which 220 fell within TFIIF and 182 between Pol II and TFIIF (Supplementary Tables 3 and 4). Data covering residue pairs within Pol II were again obtained but not included in the analysis. The quality of the MS data allowed confident assignment of 224 linkage sites and revealed a further 178 sites with lower confidence. There was no confidence bias between intra- and inter-protein links. Of the 220 linkage sites within TFIIF 149 were within proteins and 71 between proteins. In total, 253 inter-protein and 149 intra-protein links were identified. In comparison, the previous study on the Ndc80 complex had identified 13 inter-protein and 12 intra-protein links (Maiolica et al, 2007). This advancement in number of detected linkage pairs apparently results from improved MS equipment including high-resolution fragmentation spectra, an additional fractionation step, and the larger size of the analysed complex, providing more possible linkage sites. The cross-link data obtained here for Pol II and Pol II–TFIIF complex are the largest collection to date and will provide a valuable resource to understand method-specific aspects such as the fragmentation behaviour of cross-linked peptides.
Yeast TFIIF domain structure
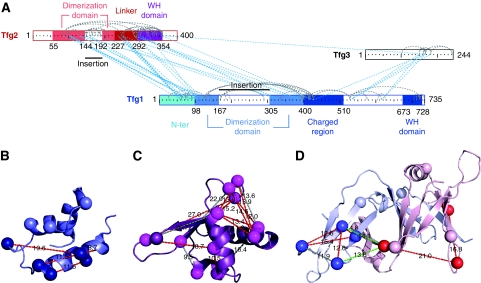
To build a model of the Pol II–TFIIF complex, we first modelled domains of yeast TFIIF based on the three known domain structures for human TFIIF (Figure 3). We first obtained sequence alignments between the human and yeast sequences using HHPred (Soding et al, 2005) (Supplementary Figure S3). We then modelled the yeast domains with program MODELLER. The sequence conservation for the two WH domains was high, making modelling straightforward. The Tfg1 WH domain spans residues 673–728, whereas the Tfg2 WH domain spans residues 292–354 (Figure 3). For the Tfg1–Tfg2 dimerization domain, modelling was hampered because of low sequence conservation and uncertainty with respect to the N-terminal border of the domain in Tfg1 (Chen et al, 2007). The modelling suggested that the dimerization domain encompasses Tfg1 residues 98–400 with a non-conserved insertion at 167–305, and Tfg2 residues 55–227, with a non-conserved insertion at 144–192. Following the N-terminal dimerization domain, Tfg1 contains a ‘charged region' (residues 400–510).
Figure 3.
TFIIF domain architecture. (A) Schematic representation of TFIIF subunits and domains. Links between TFIIF subunits (blue) and within TFIIF subunits (grey). (B, C, D) Cross-links confirm domain modelling of yeast sequences into the human crystal structures for (B) the Tfg1 WH domain, (C) the Tfg2 WH domain, and (D) the dimerization domain of Tfg1 (blue) and Tfg2 (red). Lysine residues (sphere for c-α atom) and observed links (dashed lines, red for high confidence, grey for low confidence, green for inter-protein Tfg1–Tfg2) with distance found in the respective homology model.
The domain homology models, and the proposed domain structure of yeast TFIIF subunits, could be validated with the set of distance restraints within and between TFIIF subunits that were obtained as part of the cross-link analysis of the Pol II–TFIIF complex. The homology models for the three yeast TFIIF domains agree with the distance restraints provided by cross-links (Figure 3B–D). We observed eight cross-links within the dimerization domain, four within the Tfg1 WH domain, and 15 within the Tfg2 WH domain. In the domain models, the cross-linked residue pairs are all within the permitted distance of 27.5 Å for C-α atoms. Taken together, the cross-linking and modelling reveal that the two large yeast TFIIF subunits form three structured domains in the Pol II-bound state. The N-terminal regions of subunits Tfg1 and Tfg2 form a dimerization domain, whereas WH domains are present in the C-terminal regions of the subunits.
Location of TFIIF on Pol II
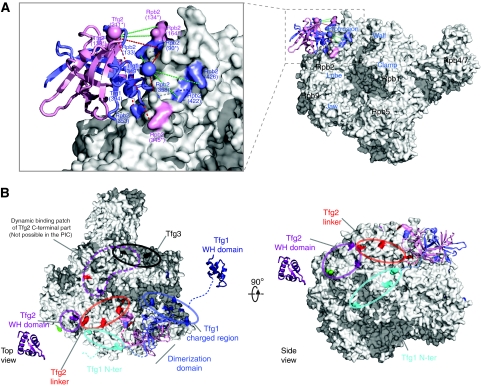
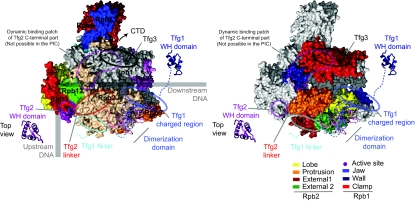
The cross-linking with the Pol II–TFIIF complex revealed an extensive network of proximities between TFIIF subunits Tfg1 and Tfg2 and the second largest Pol II subunit Rpb2, whereas only few cross-links were observed, which involve other subunits (Figure 2). TFIIF is positioned mostly on the Rpb2 side of the Pol II cleft (Figure 4; Supplementary Figure S4). The TFIIF dimerization domain is located at the Pol II lobe. The dimerization domain was placed manually on the Pol II surface, using only high-confidence cross-links and only cross-linking residues that were located either within the dimerization domain and Pol II structure or not more than 10 residues away in sequence. Enough spatial restraints were available to position and orient the dimerization domain (Figure 4A; Supplementary Figure S5 and Supplementary Table 5). One high-confidence cross-link is not satisfied by this position of the dimerization domain (Tfg1 394 to Rpb2 228). Satisfying this constraint moves the domain along its axis into the cleft, also agreeing with some previously reported evidence (Chen et al, 2007) (Supplementary Figure S6). In this position, sufficient space remains for the DNA to pass below the domain. Satisfying this constraint, however, conflicts a number of constraints at the other end of the domain. The data hence support two overlapping positions for the dimerization domain and indicate residual mobility for this domain. This could indicate an open and closed form for the cleft with the TFIIF dimerization domain acting as a lid, open to allow entry of the DNA and closed during initial transcription.
Figure 4.
Architecture of the Pol II–TFIIF complex. (A) The TFIIF dimerization domain has been positioned on the Pol II surface based on a series of cross-links between Pol II and the dimerization domain. Cross-link sites on the Pol II surface (slate and pink, matching the colour code of the dimerization domain), cross-link sites in TFIIF (sphere for C-α atom), cross-links used for positioning the dimerization domain (red dashed line) and for validation (green dashed line). For linkage sites that are absent from the Pol II structure or the model of the Tfg1–Tfg2 dimerization domain the nearest residue that is present is highlighted and labelled together with an asterisk (compare with Supplementary Table 3 and Supplementary Table 5). (B) Location of high-confidence cross-linking sites on Pol II surface coloured according to cross-linked TFIIF domains (represented in Figure 3). The dimerization domain has been placed on the Pol II surface; the location of other TFIIF regions is indicated. Two views are used, the top view and the side view, related by a 90° rotation around the horizontal axis. For linkage sites that are absent from the Pol II structure or the model of the Tfg1–Tfg2 dimerization domain the nearest residue that is present is highlighted (compare with Supplementary Table 3 and Supplementary Figures S4 and S5).
TFIIF regions extending from the dimerization domain locate to neighbouring surfaces on Pol II. The Tfg1 region N-terminal to the dimerization domain cross-links to external 1 domain of Rpb2. The Tfg1 charged region C-terminal to the dimerization domain cross-links around the Rpb1 jaw at the downstream end of the cleft (Figure 5).
Figure 5.
Architecture of the Pol II initiation complex. Pol II is represented in top view. The path of the DNA in a closed promoter initiation complex is indicated as a thick grey line (Kostrewa et al, 2009). Pol II subunits (left) and domains (right) are highlighted in canonical colours. The position of TFIIF regions is indicated. The point of attachment of the linker to the CTD of Rpb1 is depicted as an arrow.
The Tfg1 WH domain is apparently highly mobile, as no cross-links to Pol II were obtained. Our data on the Tfg2 WH domain and the dimerization domain show that cross-linking can capture dynamic structures. However, there is currently no data that establishes a limit beyond which interactions are too dynamic to be captured by cross-linking using N-hydroxysuccinimide esters, such as BS3 used in our study. The existence of an upper limit has been shown at least for formaldehyde cross-linking (Schmiedeberg et al, 2009). Our data within the Tfg1 WH domain show that we are principally able to cross-link this domain and detect such cross-links. Absence of data linking this domain to the rest of the complex indicates therefore that cross-linking requires a minimal amount of interaction that is not present in this case. The domain being held in close proximity to the complex through the Tfg1 linker region alone is insufficient for detectable cross-linking.
The Tfg2 linker C-terminal to the dimerization domain extends along the Rpb2 protrusion over the side of Pol II. This path leads to cross-links of the Tfg2 WH domain with the protrusion on the upstream face of Pol II (Figure 5). In the free Pol II–TFIIF complex, the Tfg2 WH domain is apparently not restricted to this location, as cross-links to the Pol II wall and clamp were also obtained. These interactions on wall and clamp are likely dynamic as the same sites also cross-link to the Tfg2 linker and C-terminal region (Figure 4; Supplementary Figure S7) and emphasize the ability of cross-linking to capture such transient interactions. Additional density at this location was observed in the previous EM structure (Chung et al, 2003). However, this alternative location of the Tfg2 WH domain cannot be adopted in an initiation complex, as it overlaps with the path of the DNA (Chen and Hahn, 2004; Kostrewa et al, 2009). Tfg2 WH binding at the protrusion is likely stabilized on binding of DNA and/or initiation factors. Subunit Tfg3 is apparently mobile, as only a few cross-links to the Pol II clamp were observed (Figure 4; Supplementary Table 3 and Supplementary Figure S4), consistent with previous cross-links of clamp residues to a small, not identified protein in PICs (Chen et al, 2007).
Discussion
Architecture of the Pol II–TFIIF complex
Knowing the three dimensional structure of the transcription initiation complex is of fundamental importance for our understanding of how gene promoters are recognized and used to start transcription. To arrive at the initiation complex structure, the location of the initiation factor modules on the Pol II surface must be determined. Here, we show for the first time that cross-linking and MS can be used to analyse spatial proximities within a large, 15-subunit 670 kDa multi-protein complex. This goes beyond the previously analysed four-subunit 176 kDa Ndc80 complex (Maiolica et al, 2007) and proves the value of this technology for the analysis of large multi-protein complexes.
Our work determines the three-dimensional architecture of the Pol II–TFIIF complex by use of a developing technology, cross-linking/MS, that shows the locations of the different parts of yeast TFIIF on the Pol II surface and reveals dynamic aspects of this interaction. The N-terminal Tfg1–Tfg2 dimerization domain anchors TFIIF on the Pol II lobe near the location of downstream DNA in initiation and elongation complexes (Chen and Hahn, 2004; Kettenberger et al, 2004). The C-terminal WH domains of Tfg1 and Tfg2 are mobile, but the Tfg2 WH domain can reside at the Pol II protrusion near upstream DNA in the initiation complex (Chen and Hahn, 2004). The linker between the dimerization domain and the Tfg2 WH domain runs along the protrusion, whereas the charged region connecting the dimerization domain to the Tfg1 WH domain resides at the Rpb1 jaw.
Our results are consistent with previously reported cross-links between linker-containing amino acids in the TFIIF dimerization domain and the Pol II lobe (Chen et al, 2007), but not with most of the densities observed in the previous electron microscopic analysis of the Pol II–TFIIF complex (Chung et al, 2003). Our results significantly extend previous TFIIF location analysis as they suggest the orientation of the dimerization domain and the location of the other TFIIF regions. Modelling with the use of the previously reported initiation complex model (Chen and Hahn, 2004) shows that our data are broadly consistent with the reported cross-links of human TFIIF subunits Rpb30 and Rap74 to promoter DNA positions −44 to −12 and −19 to −8, respectively, upstream of the transcription start site (Kim et al, 2000).
TFIIF function during transcription
Our results help to understand the mechanisms that TFIIF uses to accomplish its multiple functions during transcription. TFIIF has been implicated in the suppression of non-specific DNA binding to Pol II (Conaway and Conaway, 1990; Killeen and Greenblatt, 1992), in stable recruitment of promoter DNA to Pol II (Flores et al, 1991; Tan et al, 1995), in setting the transcription start site (Pinto et al, 1994; Ghazy et al, 2004; Freire-Picos et al, 2005), and in the stimulation of early RNA elongation and the suppression of abortive transcription (Tan et al, 1994a; Yan et al, 1999).
Non-specific DNA binding to Pol II likely occurs through association of DNA with the downstream cleft, because it is the only extensively positively charged surface on Pol II except for the hybrid site, which however binds A-form nucleic acids rather than B-DNA (Cramer et al, 2001). DNA association with the cleft may be suppressed by either stabilizing a closed state of the clamp, or by transient occupancy of part of the cleft with a TFIIF domain. The cluster of cross-links between the downstream cleft and the charged region in Tfg1 (Figures 3 and 4) suggest that TFIIF may prevent non-specific DNA binding by placing an unstructured, predominantly negatively charged protein region in the cleft that repels DNA. Consistently, the bacterial initiation factor σ70 contains a negatively charged region (region 1.1) that also resides in the downstream cleft (Murakami et al, 2002).
The function of TFIIF in promoter DNA recruitment may result at least in part from interactions of the WH domain in Tfg2 with promoter DNA upstream of the transcription start site. This domain has been implicated in DNA binding (Tan et al, 1994b; Kamada et al, 2001). Indeed, upstream DNA in an initiation complex would pass near the location of the Tfg2 WH domain on the protrusion (Figure 5) (Chen and Hahn, 2004). Some mobility of the WH domain may be required to allow for flexibility in DNA interactions to accommodate different promoters. The Tfg2 linker binds to the protrusion and apparently positions the Tfg2 WH domain in an initiation complex, explaining why the human Tfg2 homolog Rap30 is sufficient to recruit Pol II into an initiation complex (Flores et al, 1991). A resulting stabilization of the protrusion domain may at least in part underlie the ability of TFIIF to stimulate early elongation and to suppress abortive transcription, because the base of the protrusion domain is intimately connected with the domain binding the DNA–RNA hybrid.
Our results show that Tfg3 cross-links to the clamp of Pol II, which is not far from the CTD linker of Pol II revealed recently in S. pombe Pol II structure (Spahr et al, 2009). Furthermore, interactions between Tfg1 WH and the CTD phosphatase Fcp1 were reported (Chambers et al, 1995; Kobor et al, 2000), suggesting a possible close proximity between the CTD of Pol II, Tfg1 WH, and Tfg3. The absence of any cross-link data involving the CTD is consistent with this entire region of the TFIIF–Pol II complex, including CTD, Tfg1 WH, and Tfg3, being highly mobile.
The function of TFIIF in setting the transcription start site likely results from a role in stabilizing an open promoter complex during scanning for an initiator sequence in the DNA template strand. Some mutations in TFIIF, which shift the start site, are located within the dimerization domain and destabilize this domain and reduce its binding to Pol II (Chen et al, 2007). Mutation of yeast Tfg1 residue E346 in the dimerization domain (D95 in human Rap74) has a defect in start site selection (Ghazy et al, 2004; Khaperskyy et al, 2008). Mutation of the adjacent residue G363 suppresses defects in start site selection caused by mutations in TFIIB or Rpb1 (Freire-Picos et al, 2005). Some mutations in Pol II, which shift the start site, are located in the lobe (Trinh et al, 2006), and may also decrease the binding of the TFIIF dimerization domain. Loss of the Pol II subunit Rpb9 also leads to start site shifts (Ziegler et al, 2003), likely because Rpb9 buttresses the Rpb2 lobe (Figure 5). Indeed, Rpb9 deletion decreases TFIIF binding affinity (Ziegler et al, 2003).
Our results show that the Tfg1 charged region binds to the Pol II cleft. This is consistent with a role of this domain in stimulating elongation (Kephart et al, 1994), and explains why mutations in the charged region (human residues L155, I176, or M177) cause defects in stimulating formation of the first phosphodiester bond during initiation (Ren et al, 1999). This region likely resides at the base of the lobe and in the downstream cleft and may influence the conformation or dynamics of the mobile trigger loop at the floor of the polymerase cleft that together with the bridge helix constitutes the ratchet required for RNA synthesis and translocation.
Cross-linking and structural biology of large assemblies
Protein cross-linking coupled to MS has allowed us to extend the Pol II structure to a 15-subunit, 670 kDa complex of Pol II with the initiation factor TFIIF at peptide resolution. We have shown the ability of cross-linking in conjunction with MS to capture interactions in a fragile complex of such size and thus expand our structural understanding from a stable complex core to the more elusive periphery. Furthermore, dynamic aspects of the Pol II–TFIIF complex have been captured. The absence of data placing the internally well-covered Tfg1 WH domain on the large Pol II surface indicates that cross-linking requires a minimum of specific interactions and structure. Our analysis reveals that cross-linking/MS has now reached a level of maturity that will see it integrate seamlessly with the established toolbox of integrated structural biology to increase our structural and mechanistic insight into large multi-protein complexes.
Materials and methods
Preparation of Pol II
Endogenous complete Pol II was purified as described earlier (Sydow et al, 2009) except that the final gel filtration step was performed in presence of buffer B (10 mM HEPES pH 8.0, 200 mM potassium acetate, 1 mM EDTA, 1 mM DTT, 10% glycerol). Fractions that contained pure and stoichiometric Pol II were concentrated to 0.7 mg/ml and flash-frozen in liquid nitrogen in buffer B containing 10% glycerol.
Preparation of Pol II–TFIIF complex
A yeast over-expression cassette containing the S. cerevisiae ADH1 promotor and terminator sequences was subcloned into E. coli–yeast shuttle integrative vectors YIplac128, YIplac204, and YIplac211 (Gietz and Sugino, 1988). These vectors contain markers (LEU2, TRP1, and URA3) that complement specific auxotrophic mutations in yeast strain DSY5 (Dualsystems Biotech) and allow selection of transformants containing the corresponding plasmids. Genes coding for yeast Tfg1, Tfg2, and Tfg3 were amplified from yeast genomic DNA and subcloned into plasmids YIplac128, YIplac204, and YIplac211, respectively. A TAP tag was added at the C-terminus of Tfg2. The YIplac204 plasmid was linearized with EcoRV within the TRP1 gene, and used to transform the DSY5 strain. The resulting strain was recovered on YPD selective plate lacking tryptophan. From a single clone, a culture was grown and the corresponding cell pellet was transformed with the YIplac211 plasmid, linearized with StuI restriction enzyme within the URA3 gene. The resulting strain was recovered on a YPD selective plate lacking uracil and transformed with the YIplac128 plasmid, linearized with EcoRV within the LEU2 gene. The resulting strain DSY5-Int3 was recovered on YPD selective plate lacking leucine and stored at −80°C.
The strain DSY5-Int3, which contained the genes for the three TFIIF subunits each under the control of the ADH1 promotor, was grown overnight in a 200 l fermenter at 30°C. Cells were collected at OD600=3–4 and lysed by bead beating (BioSpec) in buffer A (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 10 mM β-mercaptoethanol, 1 mM PMSF, 1 mM benzamidine, 200 μM pepstatin, 60 μM leupeptin). After filtration, the lysate was cleared by centrifugation (60 min, 8000 g), and ultracentrifugation (90 min, 30 000 g). The supernatant was incubated overnight at 4°C with IgG beads pre-equilibrated with buffer A. The protein was eluted by TEV cleavage and purified by size exclusion chromatography (Superose 6, GE Healthcare) in buffer B (10 mM HEPES pH 8.0, 200 mM potassium acetate, 1 mM EDTA, 1 mM DTT, 10% glycerol). Fractions that contained pure and stoichiometric Pol II–TFIIF complex were concentrated to 0.8 mg/ml and flash-frozen in liquid nitrogen in buffer B containing 10% glycerol.
RNA extension assays
An amount of 4 pmol of complete Pol II or Pol II–TFIIF complex was incubated for 30 min at 20°C with 2 pmol of a pre-annealed with minimal nucleic acid scaffold (template DNA, 3′-GCTCAGCCTGGTCCG-5′; non-template DNA, 5′-CACACAGTCAG-3′; 6-carboxyfluoresceine (FAM) 5′ end-labelled RNA, 5′-UGCAUAAAGACCAGGC-3′). The complexes were incubated in the presence of 1 mM NTPs at 28°C for 20 min in transcription buffer (5 mM HEPES pH 7.3, 40 mM ammonium sulphate, 10 μM ZnCl2, 5 mM DTT). For gel electrophoresis, reactions were stopped by addition of an equal volume of 2 × loading buffer (8 M urea, 2 × TBE) and incubation for 5 min at 95°C. The FAM-labelled RNA extension products were separated by denaturing gel electrophoresis (0.5 pmol RNA per lane, 0.4 mm 15–20% polyacrylamide gels containing 8 M urea, 50–55°C) and visualized with a Typhoon 9400 phosphoimager (GE Healthcare).
Protein cross-linking
The mixing ratio of BS3 to complex was determined for Pol II using 2.5 μg aliquots and using a protein-to-cross-linker molar ratio of 1:200, 1:600, 1:1800, 1:5400, and 1:16 200, respectively (Supplementary Figure S1). As the best condition we chose the ratio that was sufficient to convert most of the individual Pol II subunits into a high molecular weight band as judged by SDS–PAGE.
The purified Pol II complex (50 μl containing 35 μg) was mixed with 150 μg BS3 (Thermo Fisher Scientific) dissolved in 70 μl cross-link buffer (10 mM HEPES pH 8.0, 200 mM potassium acetate) and incubated on ice for 2 h. The reaction was stopped by adding 1 μl of 2.5 M ammonium bicarbonate for 45 min on ice. The reaction mix was separated on a NuPAGE 4–12% Bis–Tris gel using MES running buffer and Coomassie blue stain.
The purified TFIIF–Pol II complex (250 μl containing 200 μg) was mixed with 1 mg BS3 (Thermo Fisher Scientific) dissolved in 470 μl cross-link buffer (10 mM HEPES pH 8.0, 200 mM potassium acetate) and incubated on ice for 2 h. The reaction was stopped by adding 50 μl of 2.5 M ammonium bicarbonate for 45 min on ice. The reaction mix was separated on a NuPAGE 4–12% Bis–Tris gel using MES running buffer and Coomassie blue stain.
Sample preparation for MS analysis
Bands from the SDS–PAGE gel corresponding to cross-linked complexes were excised and the proteins reduced/alkylated and digested using trypsin following standard protocols. In addition, for Pol II–TFIIF the area between the cross-linked complex and above the Rpb1 subunit was excised and analysed. The MS raw data for this fraction were combined with the main band and both were treated as one from thereon. Pol II cross-linked peptides were fractionated using SCX-StageTips (Ishihama et al, 2006) following the published protocol for linear peptides (Rappsilber et al, 2007) and desalted using StageTips (Rappsilber et al, 2003) before MS analysis. TFIIF–Pol II cross-linked peptides were desalted using StageTips and fractionated using strong cation exchange chromatography (200 × 2.1 mm Poly SULFOETHYLA column; Poly LC, Columbia, MD, USA) as described (Chen and Rappsilber, manuscript in preparation). Briefly, peptides were separated using solvent A (5 mM KH2PO4, 10% acetonitrile, pH 3.0), solvent B (solvent A with 1 M KCl), a flow rate of 200 μl/min, and a gradient consisting of 5 min at 100% solvent A followed by 20 min transition to 60% solvent B with a curve gradient (curve 8 equation, CHROMELEON software v.6.80; Dionex), 1 min at 60% solvent B. Fractions were collected every 1 min. Only fractions 14–26 were retained and desalted using StageTips for subsequent LC–MS/MS analysis.
Mass spectrometry
Peptides were loaded directly onto the analytical column, packed with C18 material (ReproSil-Pur C18-AQ 3 μm; Dr Maisch GmbH, Ammerbuch-Entringen, Germany) using a self-assembled particle frit into the spray emitter (Ishihama et al, 2002), at a flow rate of 0.7 μl/min. A linear gradient going from 5% acetonitrile in 0.5% acetic acid to 23% acetonitrile in 0.5% acetic acid in 90 min eluted the peptides at 0.3 μl/min into an LTQ-Orbitrap classic. Peptides were analysed using a high/high strategy, detecting them at high resolution in the Orbitrap, and analysing their fragments also in the Orbitrap. FTMS spectra were recorded at 100 000 resolution. The three highest intensity peaks with a charge state of three or higher were selected in each cycle for iontrap fragmentation and Orbitrap detection of the fragments at 7500 resolution. Dynamic exclusion was set to 90 s and repeat count was 1. This resulted in a cycle time of up to 5 s and an average cycle time of 3 s.
Database searching
The MS raw files were processed into peak lists using MaxQuant (Cox and Mann, 2008) at default parameters except for ‘top MS/MS peaks per 100 Da' being set to 200. Searches were conducted against a database containing the sequences of the 12 Pol II subunits and the three TFIIF subunits from S. cerevisiae using in-house Xi program. Search parameters were MS accuracy, 6 ppm; MS/MS accuracy, 20 ppm; enzyme, trypsin; specificity, fully tryptic; allowed number of missed cleavages, four; fixed modifications, carbamidometylation on cysteine; variable modifications, oxidation on methionine and BS3 mono-link reacted with water or ammonia on lysine and protein N-termini. No linkage specificity of BS3 was assumed at the point of search. However, all identified peptides contained either a lysine residue or a protein N-terminus at the most likely linkage position as determined by observed fragments. As decoy search, 30 Da were subtracted from the mass of BS3. Post search filters of 3 ppm for the recalibrated precursor masses and 6 ppm for the recalibrated fragment masses were applied. The candidate sites as returned by automated matching of fragmentation spectra to cross-linked peptides were manually validated using in-house Xaminatrix program and sorted into high and low confidence. High confidence was attributed to a match of a cross-linked peptide to a spectrum when both peptides had at least four uniquely observed fragments and all major peaks of the spectrum were accounted for. Low confidence meant that one peptide was matching essentially all observed fragments and the second peptide had up to three observed fragments. All matches had to be highest ranking and unambiguous in the target and decoy search.
Supplementary Material
Supplementary Data
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Review Process File
Acknowledgments
We thank Lau Sennels, Flavia de Lima Alves (University of Edinburgh), Dietmar Martin, Kerstin Maier, Heidi Feldmann, Gerrit Daubner (Gene Center Munich), Martin Heidemann (Helmholtz Zentrum München), and Marian Kalocay (Max-Planck Institute for Biochemistry, Martinsried) for help. We thank Steve Hahn for communicating results before publication. TK was supported by a Boehringer Ingelheim Fonds fellowship. JCBW was supported by the Centre for Systems Biology, a Centre for Integrative Systems Biology (CISB) funded by BBSRC and EPSRC, reference BB/D019621/1. PC was supported by the Deutsche Forschungsgemeinschaft, the Sonderforschungsbereich SFB646, the SFB TR5, the EU research grant network 3D Repertoire, LMUexcellent, the Nanosystems Initiative Munich, the Fonds der chemischen Industrie, and the Jung-Stiftung. JR was supported by the Marie Curie Excellence Grant MS-MODIB and the Wellcome Trust, reference 84229/Z/07/Z. JR is a Wellcome Trust Senior Research Fellow.
Footnotes
The authors declare that they have no conflict of interest.
References
- Armache KJ, Mitterweger S, Meinhart A, Cramer P (2005) Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J Biol Chem 280: 7131–7134 [DOI] [PubMed] [Google Scholar]
- Bengal E, Flores O, Krauskopf A, Reinberg D, Aloni Y (1991) Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol Cell Biol 11: 1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueckner F, Hennecke U, Carell T, Cramer P (2007) CPD damage recognition by transcribing RNA polymerase II. Science 315: 859–862 [DOI] [PubMed] [Google Scholar]
- Burton ZF, Killeen M, Sopta M, Ortolan LG, Greenblatt J (1988) RAP30/74: a general initiation factor that binds to RNA polymerase II. Mol Cell Biol 8: 1602–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chafin DR, Claussen TJ, Price DH (1991) Identification and purification of a yeast protein that affects elongation by RNA polymerase II. J Biol Chem 266: 9256–9262 [PubMed] [Google Scholar]
- Chambers RS, Wang BQ, Burton ZF, Dahmus ME (1995) The activity of COOH-terminal domain phosphatase is regulated by a docking site on RNA polymerase II and the by theneral transcription factors IIF and IIB. J Biol Chem 270: 1–8 [DOI] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D (2007) Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318: 1780–1782 [DOI] [PubMed] [Google Scholar]
- Chen HT, Hahn S (2004) Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell 119: 169–180 [DOI] [PubMed] [Google Scholar]
- Chen H-T, Warfield L, Hahn S (2007) The positions of TFIIF and TFIIE in the RNA polymerase II transcription initiation complex. Nat Struct Mol Biol 8: 696–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WH, Craighead JL, Chang WH, Ezeokonkwo C, Bareket-Samish A, Kornberg RD, Asturias FJ (2003) RNA polymerase II/TFIIF structure and conserved organization of the initiation complex. Mol Cell 12: 1003–1013 [DOI] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, Salek M, Rappsilber J, Moores CA, Salmon ED, Musacchio A (2008) Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway JW, Conaway RC (1990) An RNA polymerase II transcription factor shares functional properties with Escherichia coli sigma 70. Science 248: 1550–1553 [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Kornberg RD (2001) Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292: 1863–1876 [DOI] [PubMed] [Google Scholar]
- Elmendorf BJ, Shilatifard A, Yan Q, Conaway JW, Conaway RC (2001) Transcription factors TFIIF, ELL, and Elongin negatively regulate SII-induced nascent transcript cleavage by non-arrested RNA polymerase II elongation intermediates. J Biol Chem 276: 23109–23114 [DOI] [PubMed] [Google Scholar]
- Flores O, Lu H, Killeen M, Greenblatt J, Burton ZF, Reinberg D (1991) The small subunit of transcription factor IIF recruits RNA polymerase II into the preinitiation complex. Proc Natl Acad Sci USA 88: 9999–10003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores O, Maldonado E, Reinberg D (1989) Factors involved in specific transcription by mammalian RNA polymerase II. Factors IIE and IIF independently interact with RNA polymerase II. J Biol Chem 264: 8913–8921 [PubMed] [Google Scholar]
- Freire-Picos MA, Krishnamurthy S, Sun ZW, Hampsey M (2005) Evidence that the Tfg1/Tfg2 dimer interface of TFIIF lies near the active center of the RNA polymerase II initiation complex. Nucleic Acids Res 33: 5045–5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk JD, Nedialkov YA, Xu D, Burton ZF (2002) A key role for the alpha 1 helix of human RAP74 in the initiation and elongation of RNA chains. J Biol Chem 277: 46998–47003 [DOI] [PubMed] [Google Scholar]
- Gaiser F, Tan S, Richmond TJ (2000) Novel dimerization fold of RAP30/RAP74 in human TFIIF at 1.7 A resolution. J Mol Biol 302: 1119–1127 [DOI] [PubMed] [Google Scholar]
- Garrett KP, Serizawa H, Hanley JP, Bradsher JN, Tsuboi A, Arai N, Yokota T, Arai K, Conaway RC, Conaway JW (1992) The carboxyl terminus of RAP30 is similar in sequence to region 4 of bacterial sigma factors and is required for function. J Biol Chem 267: 23942–23949 [PubMed] [Google Scholar]
- Ghazy MA, Brodie SA, Ammerman ML, Ziegler LM, Ponticelli AS (2004) Amino acid substitutions in yeast TFIIF confer upstream shifts in transcription initiation and altered interaction with RNA polymerase II. Mol Cell Biol 24: 10975–10985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534 [DOI] [PubMed] [Google Scholar]
- Groft CM, Uljon SN, Wang R, Werner MH (1998) Structural homology between the Rap30 DNA-binding domain and linker histone H5: implications for preinitiation complex assembly. Proc Natl Acad Sci USA 95: 9117–9122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry NL, Campbell AM, Feaver WJ, Poon D, Weil PA, Kornberg RD (1994) TFIIF-TAF-RNA polymerase II connection. Genes Dev 8: 2868–2878 [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Rappsilber J, Andersen JS, Mann M (2002) Microcolumns with self-assembled particle frits for proteomics. J Chromatogr A 979: 233–239 [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Rappsilber J, Mann M (2006) Modular stop and go extraction tips with stacked disks for parallel and multidimensional Peptide fractionation in proteomics. J Proteome Res 5: 988–994 [DOI] [PubMed] [Google Scholar]
- Kamada K, De Angelis J, Roeder RG, Burley SK (2001) Crystal structure of the C-terminal domain of the RAP74 subunit of human transcription factoar IIF. Proc Natl Acad Sci USA 98: 3115–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kephart DD, Wang BQ, Burton ZF, Prices DH (1994) Functional analysis of Drosophila factor 5 (TFIIF), a general transcription factor. J Biol Chem 269: 13536–13543 [PubMed] [Google Scholar]
- Kettenberger H, Armache K-J, Cramer P (2004) Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell 16: 955–965 [DOI] [PubMed] [Google Scholar]
- Khaperskyy DA, Ammerman ML, Majovski RC, Ponticelli AS (2008) Functions of Saccharomyces cerevisiae TFIIF during transcription start site utilization. Mol Cell Biol 28: 3757–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killeen MT, Greenblatt JF (1992) The general transcription factor RAP30 binds to RNA polymerase II and prevents it from binding nonspecifically to DNA. Mol Cell Biol 12: 30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Ebright RH, Reinberg D (2000) Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 288: 1418–1422 [DOI] [PubMed] [Google Scholar]
- Kobor MS, Simon LD, Omichinski J, Zhong G, Archambault J, Greenblatt J (2000) A motif shared by TFIIF and TFIIB mediates their interaction with the RNA polymerase II carboxy-terminal domain phosphatase Fcp1p in Saccharomyces cerevisiae. Mol Cell Biol 20: 7438–7449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P (2009) RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature 462: 323–330 [DOI] [PubMed] [Google Scholar]
- Maiolica A, Cittaro D, Borsotti D, Sennels L, Ciferri C, Tarricone C, Musacchio A, Rappsilber J (2007) Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol Cell Proteomics 6: 2200–2211 [DOI] [PubMed] [Google Scholar]
- McCracken S, Greenblatt J (1991) Related RNA polymerase-binding regions in human RAP30/74 and Escherichia coli σ70. Science 253: 900–902 [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA (2002) Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296: 1285–1290 [DOI] [PubMed] [Google Scholar]
- Pinto I, Wu W-H, Na JG, Hampsey M (1994) Characterization of sua7 mutations defines a domain of TFIIB involved in transcripiton start site selection in yeast. J Biol Chem 269: 30569–30573 [PubMed] [Google Scholar]
- Price DH, Sluder AE, Greenleaf AL (1989) Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol Cell Biol 9: 1465–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani PG, Ranish JA, Hahn S (2004) RNA polymerase II (Pol II)-TFIIF and Pol II-mediator complexes: the major stable Pol II complexes and their activity in transcription initiation and reinitiation. Mol Cell Biol 24: 1709–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75: 663–670 [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2: 1896–1906 [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Siniossoglou S, Hurt EC, Mann M (2000) A generic strategy to analyze the spatial organization of multi-protein complexes by cross-linking and mass spectrometry. Anal Chem 72: 267–275 [DOI] [PubMed] [Google Scholar]
- Reinberg D, Orphanides G, Ebright R, Akoulitchev S, Carcamo J, Cho H, Cortes P, Drapkin R, Flores O, Ha I, Inostroza JA, Kim S, Kim TK, Kumar P, Lagrange T, LeRoy G, Lu H, Ma DM, Maldonado E, Merino A et al. (1998) The RNA polymerase II general transcription factors: past, present, and future. Cold Spring Harb Symp Quant Biol 63: 83–103 [DOI] [PubMed] [Google Scholar]
- Ren DL, Lei L, Burton ZF (1999) A region within the RAP74 subunit of human transcription factor IIF is critical for initiation but dispensable for complex assembly. Mol Cell Biol 19: 7377–7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinner O, Seebacher J, Walzthoeni T, Mueller LN, Beck M, Schmidt A, Mueller M, Aebersold R (2008) Identification of cross-linked peptides from large sequence databases. Nat Methods 5: 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedeberg L, Skene P, Deaton A, Bird A (2009) A temporal threshold for formaldehyde crosslinking and fixation. PLoS One 4: e4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinz A (2006) Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom Rev 25: 663–682 [DOI] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33 (Web Server issue): W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopta M, Burton ZF, Greenblatt J (1989) Structure and associated DNA-helicase activity of a general transcription initiation factor that binds to RNA polymerase II. Nature 341: 410–414 [DOI] [PubMed] [Google Scholar]
- Sopta M, Carthew RW, Greenblatt J (1985) Isolation of three proteins that bind to mammalian RNA polymerase II. J Biol Chem 260: 10353–10360 [PubMed] [Google Scholar]
- Spahr H, Calero G, Bushnell DA, Kornberg RD (2009) Schizosacharomyces pombe RNA polymerase II at 3.6-A resolution. Proc Natl Acad Sci USA 106: 9185–9190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydow JF, Brueckner F, Cheung AC, Damsma GE, Dengl S, Lehmann E, Vassylyev D, Cramer P (2009) Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA. Mol Cell 34: 710–721 [DOI] [PubMed] [Google Scholar]
- Tan S, Aso T, Conaway RC, Conaway JW (1994a) Roles for both the RAP30 and RAP74 subunits of transcription factor IIF in transcription initiation and elongation by RNA polymerase II. J Biol Chem 269: 25684–25691 [PubMed] [Google Scholar]
- Tan S, Conaway RC, Conaway JW (1995) Dissection of transcription factor TFIIF functional domains required for initiation and elongation. Proc Natl Acad Sci USA 92: 6042–6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Garrett KP, Conaway RC, Conaway JW (1994b) Cryptic DNA-binding domain in the C terminus of RNA polymerase II general transcription factor RAP30. Proc Natl Acad Sci USA 91: 9808–9812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh V, Langelier MF, Archambault J, Coulombe B (2006) Structural perspective on mutations affecting the function of multisubunit RNA polymerases. Microbiol Mol Biol Rev 70: 12–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BQ, Burton ZF (1995) Functional domains of human RAP74 including a masked polymerase binding domain. J Biol Chem 270: 27035–27044 [DOI] [PubMed] [Google Scholar]
- Yan Q, Moreland RJ, Conaway JW, Conaway RC (1999) Dual roles for transcription factor IIF in promoter escape by RNA polymerase II. J Biol Chem 274: 35668–35675 [DOI] [PubMed] [Google Scholar]
- Yonaha M, Aso T, Kobayashi Y, Vasavada H, Yasukochi Y, Weissman SM, Kitajima S (1993) Domain structure of a human general transcription initiation factor, TFIIF. Nucleic Acids Res 21: 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Yan H, Burton ZF (2003) Combinatorial control of human RNA polymerase II (RNAP II) pausing and transcript cleavage by transcription factor IIF, hepatitis delta antigen, and stimulatory factor II. J Biol Chem 278: 50101–50111 [DOI] [PubMed] [Google Scholar]
- Ziegler LM, Khaperskyy DA, Ammerman ML, Ponticelli AS (2003) Yeast RNA polymerase II lacking the Rpb9 subunit is impaired for interaction with transcription factor IIF. J Biol Chem 278: 48950–48956 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Review Process File