Abstract
Porphyromonas gingivalis, a causative agent of periodontitis, has at least two types of thin, single-stranded fimbriae, termed FimA and Mfa1 (according to the names of major subunits), which can be discriminated by filament length and by the size of their major fimbrilin subunits. FimA fimbriae are long filaments that are easily detached from cells, whereas Mfa1 fimbriae are short filaments that are tightly bound to cells. However, a P. gingivalis ATCC 33277-derived mutant deficient in mfa2, a gene downstream of mfa1, produced long filaments (10 times longer than those of the parent), easily detached from the cell surface, similar to FimA fimbriae. Longer Mfa1 fimbriae contributed to stronger autoaggregation of bacterial cells. Complementation of the mutant with the wild-type mfa2 allele in trans restored the parental phenotype. Mfa2 is present in the outer membrane of P. gingivalis, but does not co-purify with the Mfa1 fimbriae. However, co-immunoprecipitation demonstrated that Mfa2 and Mfa1 are associated with each other in whole P. gingivalis cells. Furthermore, immunogold microscopy, including double labelling, confirmed that Mfa2 was located on the cell surface and likely associated with Mfa1 fimbriae. Mfa2 may therefore play a role as an anchor for the Mfa1 fimbriae and also as a regulator of Mfa1 filament length. Two additional downstream genes (pgn0289 and pgn0290) are co-transcribed with mfa1 (pgn0287) and mfa2 (pgn0288), and proteins derived from pgn0289, pgn0290 and pgn0291 appear to be accessory fimbrial components.
INTRODUCTION
Porphyromonas gingivalis is a Gram-negative, black-pigmented, obligate anaerobe that has been implicated in adult periodontitis (Lamont & Jenkinson, 1998; Socransky & Haffajee, 2005), which is a major cause of tooth loss in the adult population. Periodontitis and infection with this organism are also thought to be associated with several systemic diseases, including diabetes, preterm birth, coronary heart disease and atherosclerosis (Dasanayake et al., 2003; Gibson et al., 2006). Fimbriae are one of the major colonization factors of P. gingivalis, and contribute to the formation of mixed-species biofilms on oral surfaces (Jenkinson & Lamont, 2005; Kolenbrander et al., 2002; Yoshimura et al., 2009). Fimbriae also play a major role in adhesion to and invasion of gingival epithelial cells by P. gingivalis (Andrian et al., 2006; Lamont & Jenkinson, 1998).
P. gingivalis has at least two types of fimbriae, namely FimA and Mfa1 fimbriae, following the names of their major subunit proteins, whose gene loci are separated from each other on the chromosome (Hamada et al., 2002; Ogawa et al., 1994; Yoshimura et al., 1984, 1989). Well-characterized strains such as ATCC 33277 and 381 have been shown to express both fimbriae types (Hayashi et al., 2000; Naito et al., 2008; Park et al., 2005; Yoshimura et al., 1989). However, neither fimbriae type is produced in the sequenced strain W83 (Nelson et al., 2003; Suzuki et al., 1988; Yoshimura et al., 1989). There are several reports describing a third type of fimbria or an outer membrane protein of 53 kDa in 381 (Arai et al., 2000; Hongyo et al., 1997, 1998). This protein seems to have a strong homology with Mfa1 fimbriae, although the extent to which this is distributed among P. gingivalis strains remains to be established (Murakami et al., 2002; Yoshimura et al., 1989). The Mfa1 fimbriae have been less characterized than the FimA fimbriae, partly because the former appear to be much shorter filaments than the latter, and are difficult to purify (Hamada et al., 1996; Ogawa et al., 1995). In later studies, Mfa1 fimbriae were purified from lysed cells of a fimA mutant and found indeed to be short filaments of uniform length (average 103 nm) (Park et al., 2005). Very little is known about the function(s) of Mfa1 fimbriae, although recent reports show that Mfa1 fimbriae are involved in coadhesion with Streptococcus gordonii (Chung et al., 2000; Lamont et al., 2002; Park et al., 2005), autoaggregation and colonization (Lin et al., 2006), and, like FimA fimbriae, they can stimulate potent inflammatory responses (Hajishengallis et al., 2002; Hiramine et al., 2003; Takahashi et al., 2006).
During the course of isolation and purification of Mfa1 and FimA fimbriae from various strains, we found that an mfa1 mutant complemented by the introduction of the wild-type mfa1 gene in trans (cSMF1) (Park et al., 2005) had an aberrant phenotype and produced Mfa1 fimbriae as long filaments, loosely attached to and therefore easily shed from the cells, similar to the FimA fimbriae of strains ATCC 33277 or 381. Based on these observations and our previous finding that mfa1 and the adjacent downstream gene pg0179 (equivalent to pgn0288 in 33277) (Naito et al., 2008) are co-transcribed (Chung et al., 2000), we hypothesized that cSMF1 has a defect in the downstream gene, and that this defect causes the aberrant Mfa1 fimbrial phenotype, presumably due to a polar effect of the insertional mutation in cSMF1 (Lamont et al., 2002), which is complemented with mfa1 only (Park et al., 2005). Here, we report the characterization of the pgn0288 gene (hereafter designated mfa2) downstream of mfa1 and its protein product in ATCC 33277.
METHODS
Bacterial strains and growth conditions.
All P. gingivalis and Escherichia coli strains used in this study are listed in Table 1. P. gingivalis ATCC 33277 (33277) and its derivatives were grown on blood agar plates [Brucella HK agar base (Kyokuto) supplemented with 5 % (w/v) laked rabbit blood, 2.5 μg haemin ml−1, 5.0 μg menadione ml−1 and 0.01 % (w/v) DTT] at 37 °C under anaerobic conditions [10 % (v/v) CO2, 10 % (v/v) H2 and 80 % (v/v) N2]. Cells were cultivated in sTSB liquid medium [trypticase soy broth supplemented with 0.25 % (w/v) yeast extract, 2.5 μg haemin ml−1, 5.0 μg menadione ml−1 and 0.01 % (w/v) DTT] or general anaerobic GAM broth (Nissui) at 37 °C for 30–48 h under anaerobic conditions. When necessary, chloramphenicol (Cm; 4 μg ml−1), erythromycin (Em; 10 μg ml−1) or tetracycline (Tc; 2 μg ml−1) was added to the medium. E. coli strains were grown in Luria–Bertani medium supplemented, when necessary, with ampicillin (100 μg ml−1), kanamycin (50 μg ml−1) or erythromycin (200 μg ml−1).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics* | Source or reference |
|---|---|---|
| P. gingivalis strains | ||
| ATCC 33277 | Type strain, producing both FimA and Mfa1 fimbriae, Gmr | ATCC |
| SMF1 | Derivative of ATCC 33277 with an insertional inactivation of the mfa1 gene, Emr | Lamont et al. (2002); Park et al. (2005) |
| JI-1 | Derivative of ATCC 33277 with fimA deletion by cat, Cmr | This study |
| JI-2 | Derivative of ATCC 33277 with an insertional inactivation of the mfa2 gene, Emr | This study |
| JI-3 | JI-2 containing pTCBex-mfa2, a complemented strain, Emr, Tcr | This study |
| JI-12 | Derivative of JI-2 with fimA deletion by cat, Emr, Cmr | This study |
| JI-4 | JI-12 containing pTCBex-mfa2, a complemented strain, Emr, Cmr, Tcr | This study |
| E. coli strains | ||
| DH5 α | F−φ80lacZ ΔM15 Δ(lacZYA-argF) U169 deoR recA1endA1hsdR17( |
Takara |
| BL21 | F−ompT hsdSB ( |
GE Healthcare Bio-Science |
| S17-1 | RecA−, ΩRP4-Tc : : Mu-Kn : : T7), Tpr | Gardner et al. (1996) |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) F80lacZ ΔM15 ΔlacC74 recA1 araD139 Δ(ara-leu) 7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| Plasmids | ||
| pCR-TOPO | Cloning vector, Kmr | Invitrogen |
| pCRJI-2 | pCR-TOPO derivative carrying a 1.6 kb fragment containing mfa2 | This study |
| pVA2198 | Plasmid used for a drug cassette, ermF-ermAM, Emr | Fletcher et al. (1997) |
| pTCBex | Derivative of pT-COW, an expression vector, containing the promoter region of fimR, Apr in E. coli, Tcr in P. gingivalis | Nagano et al. (2007) |
| pTCBex-mfa2 | pTCBex derivative carrying the coding region of mfa2, Apr in E. coli, Tcr in P. gingivalis | This study |
| pGEX-6P | GST gene fusion and expression vector, Apr | GE Healthcare Bio-Science |
*ATCC, American Type Culture Collection; Gmr, gentamicin resistance; cat, chloramphenicol acetyltransferase; Cmr, chloramphenicol resistance; Emr, erythromycin resistance; Tcr, tetracycline resistance; Tpr, trimethoprim resistance; Kmr, kanamycin resistance; Apr, ampicillin resistance.
DNA manipulations.
Standard techniques were used for purification and manipulation of DNA (Nagano et al., 2005; Nishiyama et al., 2007). Restriction endonucleases, DNA ligase and related enzymes were purchased from Takara Bio or New England Biolabs. The Zero Blunt TOPO PCR cloning kit was from Invitrogen. The oligonucleotides used for PCR or RT-PCR were synthesized by Sigma Genosys. DNA sequencing was carried out using the ABI PRISM version 1.1 kit and ABI PRISM 3100-Avant Genetic Analyzer (Applied Biosystems). Constructs for generating mutants or recombinant protein production were sequenced to rule out unintended base changes.
Construction of P. gingivalis mutant and complementation strains.
The mfa2 gene (pgn0288) downstream of mfa1 was amplified as an approximately 1.6 kb fragment from the 33277 chromosome by PCR using primers AGU-101F and AGU-101R (see Table 2 for primer sequences), designed based on the sequence of corresponding gene pg0179 in the P. gingivalis W83 genome (Nelson et al., 2003). The amplified fragment was cloned into the plasmid vector pCR-TOPO with the Zero Blunt TOPO PCR cloning kit to construct pCRJI-2 (Table 1). This was used for sequencing mfa2 and for construction of the mfa2 mutant.
Table 2.
Primers for PCR and RT-PCR used in this study
| Primer | Sequence (5′ to 3′)* | Relevant information |
|---|---|---|
| AGU-101F | GGGAAGATCATAATACAAATGAGG | For mutation of mfa2 |
| AGU-101R | ACTCGCTTTCCCCTCTTTCT | |
| AGU-102BF | TTTTAAAACAGGATCCATGAACAAACGGAAGCATAT | For complementation of mfa2 |
| AGU-102HR | CTTTTTTCTCAAGCTTTTAAAGTTCTATTTCGTAAC | |
| AGU-103BF | GTGGATCCCCTCGAGGAGTATATGTCAA | For production of recombinant Mfa2 |
| AGU-103SR | TCGTCGACTTAAAGTTCTATTTCGTAAC | |
| mfa1F_RT | GTTCTGTTTTGGCTACCATTACG | For RT-PCR of mfa1 |
| mfa1R_RT | TTGATGCTCTTGATGTGGATATG | |
| mfa2F_RT | AATGTGTTTGAGGATGTCCAGTT | For RT-PCR of mfa2 |
| mfa2R_RT | CATTGATACAGCGCAAGTTGATA | |
| 0289F_RT | ATATCGTGGAAGGAGTGGAAAAT | For RT-PCR of pgn0289 |
| 0289R_RT | ATAAACCTCCAGATAGGCAGAGC | |
| 0290F_RT | TACGCTATGAAAGGAATCAAGGA | For RT-PCR of pgn0290 |
| 0290R_RT | GTAACAATCCGTAGTGCCGTTAC | |
| 0291F_RT | TAACAGTACCTGTCGATGTGGTG | For RT-PCR of pgn0291 |
| 0291R_RT | TCTCGTCTGTAGCGGTTAGTTTC |
*Underlined sequences in AGU-102BF, AGU-102HR, AGU-103BF and AGU-103SR indicate BamHI, HindIII, BamHI and SalI sites, respectively.
A fimA mutant (JI-1) was constructed by the PCR-based overlap extension method as described previously (Nagano et al., 2005). An mfa2 mutant (JI-2) that had an ermF–ermAM cassette (Fletcher et al., 1997) insertion at the NaeI site (50 bp) in the mfa2 coding region cloned in pCRJI-2 was constructed as described previously (Hasegawa et al., 2003; Hongo et al., 1999). A fimA and mfa2 double mutant (JI-12) was also constructed by the same techniques. The complemented strains JI-3 and JI-4 were constructed by the introduction of an expression vector, pTCBex-mfa2, a pT-COW derivative (Gardner et al., 1996). In brief, the entire coding region of mfa2 in 33277 was amplified by PCR using AGU-102BF and AGU-102HR with BamHI and HindIII tags, respectively, and the resulting fragment was then cloned downstream of the fimR promoter region in pTCBex (Nagano et al., 2007), digested with the appropriate enzymes. The resulting vector pTCBex-mfa2 was transferred into P. gingivalis JI-2 or JI-12 via conjugation from E. coli S17-1. P. gingivalis with pTCBex (empty vector) was used as a negative control where appropriate, although these data are not shown.
Production of recombinant protein.
To construct a plasmid expressing recombinant Mfa2, the region of mfa2 encoding the putative mature product (nucleotides 121–975) without the N-terminal leader sequences was amplified by PCR using AGU-103BF and AGU-103SR. The resulting fragment was cloned into expression vector pGEX-6P (GE Healthcare Bio-science), yielding plasmid pGEX-′Mfa2 encoding glutathione S-transferase (GST)-fused recombinant Mfa2 (GST-r′Mfa2). The plasmid was introduced into E. coli strain BL21 for overproduction and purification of GST-r′Mfa2.
Preparation of cellular fractions.
P. gingivalis strains were anaerobically cultured for 24–48 h, and cells were harvested by centrifugation, washed once, and disrupted by chemical reagents (BugBuster, Novagen) for immunoprecipitation, sonication (Nagano et al., 2005) or in a French pressure cell (Park et al., 2005). Undisrupted cells were removed by centrifugation at 1000 g for 10 min. The supernatant was used as a whole-cell lysate. The envelope was collected as a pellet by centrifugation of whole-cell lysate at 100 000 g for 60 min at 4 °C and washed once as described previously (Nagano et al., 2005). The outer and inner membranes were separated by the differential extraction method (Murakami et al., 2002).
Purification and preparation of fimbriae, proteins and antibody.
Mfa1 fimbriae were purified from JI-1 (ΔfimA) as described previously (Park et al., 2005), except for the absence of detergent. FimA fimbriae and Mfa1 fimbriae were purified from washing solutions of SMF1 and JI-12 (ΔfimA and Δmfa2), respectively (Yoshimura et al., 1984). Purification of GST-r′Mfa2 was carried out with a GST affinity column (Glutathione Sepharose 4B, GE Healthcare Bio-science), following the manufacturer's instructions. Purity was examined by SDS-PAGE and Coomassie brilliant blue R-250 (CBB) staining. Protein concentration was measured by using the Micro BCA Protein Assay kit (Pierce Biotechnology). Polyclonal antibody against Mfa1 and GST-r′Mfa2 was raised in rabbits as described previously (Sakakibara et al., 2007; Yoshimura et al., 1989). Another specific antiserum against Mfa1 for double labelling immunogold microscopy was raised in chickens. Briefly, purified Mfa1 protein obtained by dissection of bands in SDS-PAGE gels using pure Mfa1 fimbriae was mixed with Freund's complete adjuvant, and the mixture was injected into chickens subcutaneously four times at 2-week intervals.
SDS-PAGE and Western immunoblotting.
SDS-PAGE and Western immunoblotting were carried out as described previously (Murakami et al., 2002; Nagano et al., 2005). Proteins were separated with a 12 % (w/v) SDS-PAGE gel and visualized by staining with CBB. For Western immunoblotting, proteins were transferred onto a nitrocellulose membrane (Hybond ECL nitrocellulose membrane, GE Healthcare Bio-science) and detected by using antibody raised against Mfa1 (Yoshimura et al., 1989) or Mfa2 and peroxidase-conjugated goat anti-rabbit IgG (MP Biomedicals). Signals were visualized with 0.01 % (w/v) 4-chloro-1-naphthol in 20 mM Tris/HCl (pH 7.5) containing 0.5 M NaCl supplemented with hydrogen peroxide (Nagano et al., 2005). To increase sensitivity for detection, ECL Plus Western immunoblotting detection reagents (GE Healthcare Bio-science) were used for visualization according to the manufacturer's instructions.
Detection of Mfa1 fimbrilin, minor components and Mfa2 protein.
Mfa1 in whole culture, cells and culture supernatant was detected after precipitation with cold TCA, as described previously (Hongo et al., 1999), except for the use of 2 M Tris to neutralize samples. Minor components of Mfa1 fimbriae and Mfa2 were detected by CBB dye staining and Western immunoblotting after SDS-PAGE.
Protein analysis by MS.
The minor components of Mfa1 fimbriae were identified by MALDI-TOF MS (Masuda et al., 2006). CBB-stained protein bands were excised and digested with trypsin. The peptides were extracted and concentrated, and analysed using a 4800 MALDI TOF/TOF analyser (Applied Biosystems). The identities of the proteins were deduced from MS peaks via the peptide mass fingerprinting methods in Mascot (http://www.matrixscience.com/). The proteins were identified according to the significance criteria of the search program (P<0.05).
Electron microscopy.
P. gingivalis cells and fimbriae were examined with a transmission electron microscope (Carl-Zeiss LEO LIBRA120 or JEM-1200EX, JEOL). An aliquot (10 μl) of bacterial culture or the purified fimbriae was placed on a high-resolution carbon substrate (Ohkenshoji). Then samples were negatively stained with 1 % (w/v) ammonium molybdate for 2 min. For immunogold staining, bacterial cells were fixed with 4 % (w/v) paraformaldehyde in 0.1 M PBS, pH 7.4, at 37 °C for 1 h. The specimens were dehydrated, immersed in L-R White (London Resin), and embedded in gelatin capsules. The resin was polymerized at 55 °C for 24 h. Ultra-thin sections were cut and collected on 400-mesh nickel grids (Ohkenshoji). The sections were incubated with 1 % (w/v) BSA in PBS (BSA/PBS) for 30 min at room temperature, then incubated with anti-rMfa2 rabbit antiserum overnight at 4 °C, followed by washing with PBS. The sections were then incubated in anti-rabbit IgG labelled with 20 nm gold (EY Laboratories), diluted in BSA/PBS (1 : 50), followed by incubation for 1 h and washing with PBS. Chicken antiserum against Mfa1 and goat anti-chicken Ig conjugated with 6 nm gold particles (Abcam) were used for double-labelling experiments. Then they were fixed again with 2 % glutaraldehyde, and stained with uranyl acetate and lead citrate. The samples were observed under a JEM-1210 electron microscope (JEOL).
Autoaggregation assay.
Cells were grown as described above, harvested by centrifugation at 8000 g for 10 min, gently washed once with PBS (pH 6.0), and suspended in the same buffer. The OD600 value of the cell suspension was measured and adjusted by dilution with the buffer to 1.0. Aliquots (2 ml) in test tubes (13 mm diameter) were shaken at 37 °C at a speed of 130 strokes per minute. At various time points, OD600 values of the suspension were measured with a spectrophotometer (CO8000 cell density meter).
Total RNA isolation and RT-PCR.
Total RNA was isolated from P. gingivalis 33277 by the RiboPure-Bacteria kit (Ambion). The extracted RNA was treated with RNase-free DNase I (Ambion). Superscript III (Invitrogen) was used to generate cDNA from RNA (1 μg) templates with a random primer, as described by the manufacturer. The resulting cDNA was used as a template for following standard PCR (Takara Ex Taq). The primers used for standard PCR (Table 2) were specific for the mfa1, mfa2, pgn0289, pgn0290 or pgn0291 genes. Controls without reverse transcriptase were included in all experiments.
Immunoprecipitation assay.
Immunoprecipitation assays were performed to determine the interaction between Mfa1 and Mfa2 by using the ProFound co-immunoprecipitation kit based on a direct immobilization of an antibody to aldehyde-activated, beaded agarose (Pierce Biotechnology). Briefly, a 33277 cell pellet was lysed with BugBuster (Novagen) containing proteinase inhibitors. Anti-Mfa2, anti-Mfa1 or anti-OmpA (Pgm6/7) antiserum (Nagano et al., 2005) was immobilized on the agarose gel beads, washed, and incubated overnight with whole-cell lysate at 4 °C. Beads were then washed and resuspended in SDS-PAGE loading buffer. Samples were separated by SDS-PAGE, and Western immunoblotting was performed with anti-Mfa2 or anti-Mfa1 antiserum.
RESULTS
Genes downstream of mfa1 in ATCC 33277
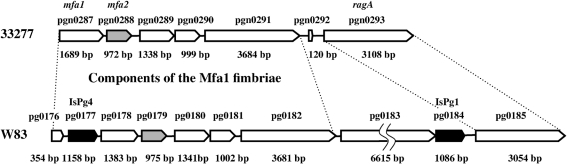
In this study we focused on the mfa1 gene, encoding the major subunit (fimbrilin) of Mfa1 fimbriae, and genes downstream of mfa1. The complete genome sequences of 33277 and strain W83 are now available (Naito et al., 2008; Nelson et al., 2003). Since mfa1 in W83 is interrupted by an insertion sequence, to study Mfa1 fimbriae, we have used 33277 expressing both FimA fimbriae (Yoshimura et al., 1984) and Mfa1 fimbriae (Park et al., 2005), which present morphologically as long and short filaments, respectively. Sequencing of mfa2 (corresponding to pgn0288 in 33277 or pg0179 in W83) was carried out in 33277 [our sequence data for mfa2 (pgn0288) was matched completely to mfa2 in the 33277 genome database], and the sequence of a further downstream region from mfa2 was taken from the 33277 genome database. At least five genes (mfa1 to pgn0292) prior to ragA with the same transcriptional direction were found at this locus in 33277, and thus represent a possible gene cluster for Mfa1 fimbriae (Fig. 1). In contrast, W83 appears to have nine ORFs prior to ragA, including IsPg4 (pg0177) disrupting mfa1, pg0183 and IsPg1 (pg0184). Among these, pg0183 is not present in 33277 (Hall et al., 2005). mfa2 has a high homology (98 % at the DNA level) between 33277 and W83. blastp analysis showed that Mfa2 has some homologies to corresponding regions of ORFs immediately downstream of fimA [comprising pgn0181 (29 % identity and 49 % similarity at the N-terminal region in 33277) and pg2133 (26 % identity and 43 % similarity at the N-terminal region in W83)]. Mfa2 showed homology to the hypothetical protein BT1062 of Bacteroides thetaiotaomicron VPI-5482 (E value 8×10−12) and the hypothetical protein BF2211 in Bacteroides fragilis YCH46 (E value 6×10−11) in the global value, but the function of Mfa2 could not be predicted.
Fig. 1.
Mfa1 fimbriae-associated genes in P. gingivalis ATCC 33277. Mfa1 fimbrial genes in 33277 are shown in parallel with those in W83. The mfa1 gene has been previously sequenced in ATCC 33277 and 381 and deposited as a 67 kDa fimbrilin and a cell surface protein (AB016284 and D28770), respectively (Hamada et al., 2002; Ogawa et al., 1994), although the corresponding gene of W83 is split into pg0176 and pg0178 by insertion of pg0177 (ISPg4, transposase) (Park et al., 2005). W83 produces neither FimA nor Mfa1 fimbriae, presumably due to disruption of fimS (Hayashi et al., 2000) and mfa1, respectively. The mfa2 gene (annotated as pgn0288 in 33277 or pg0179 in W83, indicated by the grey bars with pointed ends) in 33277 was sequenced in this study, and the 33277 genome database was utilized for the sequence of genes downstream of mfa2. The gene products of pgn0289, pgn0290 and pg0291 were identified as minor components of pure Mfa1 fimbriae in JI-1 (ΔfimA) by MALDI-TOF MS, as described in the text (and also shown in Fig. 3, Table 3).
Characterization of a mutant deficient in mfa2 and its complementation strain
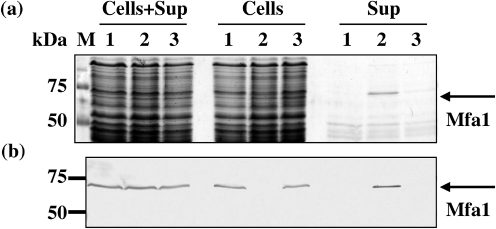
A knockout mutant of mfa2 and its complemented strain were constructed. The mutant JI-2 had mfa2 disrupted with an antibiotic resistance cassette, and the complemented strain JI-3 carried an expression vector pTCBex-mfa2 in JI-2 (Nagano et al., 2007). Furthermore, to perform the morphological study of Mfa1 fimbriae, we constructed another mfa2 mutant, JI-12 (ΔfimA and Δmfa2), and its mfa2-complemented strain JI-4 (see Methods). The presence and location of Mfa1 fimbriae in the mfa2 mutant JI-2 strain were investigated. In the absence of Mfa2, retention of Mfa1 fimbriae on the cells was diminished, and Mfa1 fimbriae appeared mostly in the cell supernatant (Fig. 2a, b, lane 2 in Sup). The complemented strain JI-3 demonstrated a phenotype similar to the parent, with Mfa1 fimbriae firmly bound to the cell surface and absent from the supernatant (Fig. 2a, b, lane 3 in Sup). Total expression levels of Mfa1 fimbriae among the parental, JI-2 and JI-3 strains were consistent (Fig. 2b, lanes 1–3 in Cells+Sup), as determined by densitometric analysis of the Mfa1 protein bands present in the cell and supernatant fractions (data not shown). Furthermore, numerous long filamentous structures and some vesicles were observed in the culture supernatant from JI-12 (ΔfimA and Δmfa2) by electron microscopy (data not shown).
Fig. 2.
Unusual distribution of filaments of the Mfa1 fimbriae in the mfa2 mutant. (a) Partial protein profile of the parental 33277 (lane 1), mfa2 mutant JI-2 (lane 2) and complemented mutant strain JI-3 (lane 3). Whole cultures (Cells+Sup), cells and culture supernatants (Sup) were treated with cold TCA in order to recover all proteins and to destroy intrinsic, strong proteolytic activities of this organism (Nishiyama et al., 2007). The TCA precipitates were neutralized and separated by SDS-PAGE, followed by CBB staining. (b) Western immunoblotting of panel (a) using anti-Mfa1 serum. A band in lane 2 in Sup of (a), indicated by an arrow, was identified as Mfa1 by antiserum [lane 2 in Sup of (b)].
Analysis of minor/accessory proteins in Mfa1 fimbriae and localization of Mfa2
The Mfa1 fimbriae could be purified from whole-cell lysate of 33277 or its fimA-defective mutant JI-1 by complete cell disruption using a French pressure cell, as described in Methods. They were not washed from the cell surface by the mild washing method that removes FimA fimbriae (Yoshimura et al., 1984).
A major band (74 kDa) and at least nine minor protein bands were detected in Mfa1 fimbriae purified from JI-1 (ΔfimA), when a large amount of the fimbriae was applied (Fig. 3, lane Mfa1). The major and minor protein bands were cut from gels and analysed using MALDI-TOF MS. All the proteins were identified through searching the NCBI database for peptide mass fingerprinting as follows. Bands 1 and 2 (approximately 150 and 130 kDa, respectively) were gene products of pgn0291 in 33277 that have a conserved amino acid sequence of von Willebrand factor A in an N-terminal region, bands 3–8 including the major band (74–50 kDa) were all matched to a gene product of pgn0287 (annotated as the 67 kDa major outer membrane protein that is an earlier name for Mfa1) (Ogawa et al., 1994), band 9 (40 kDa) was a putative lipoprotein (corresponding to pgn0289), and band 10 (30 kDa) was antigen PG49 (corresponding to pgn0290), interestingly indicating that all the minor bands were gene products either of mfa1 or its downstream genes pgn0289, pgn0290 and pgn0291, as shown in Table 3. Therefore, Mfa1 fimbriae appear to have at least three minor components, associated with the major protein, expressed from the downstream three genes of mfa2. Because the association between the major and minor proteins seems to be stable during purification, the minor proteins identified seem to be inherent to Mfa1 fimbriae. Hereafter, the term ‘accessory’ rather than ‘minor’ proteins will be used. The gene product of mfa2, deduced to be a 37 kDa protein from the nucleotide sequence, was not detected in the purified Mfa1 fimbrial preparation (Fig. 3, lane Mfa1).
Fig. 3.
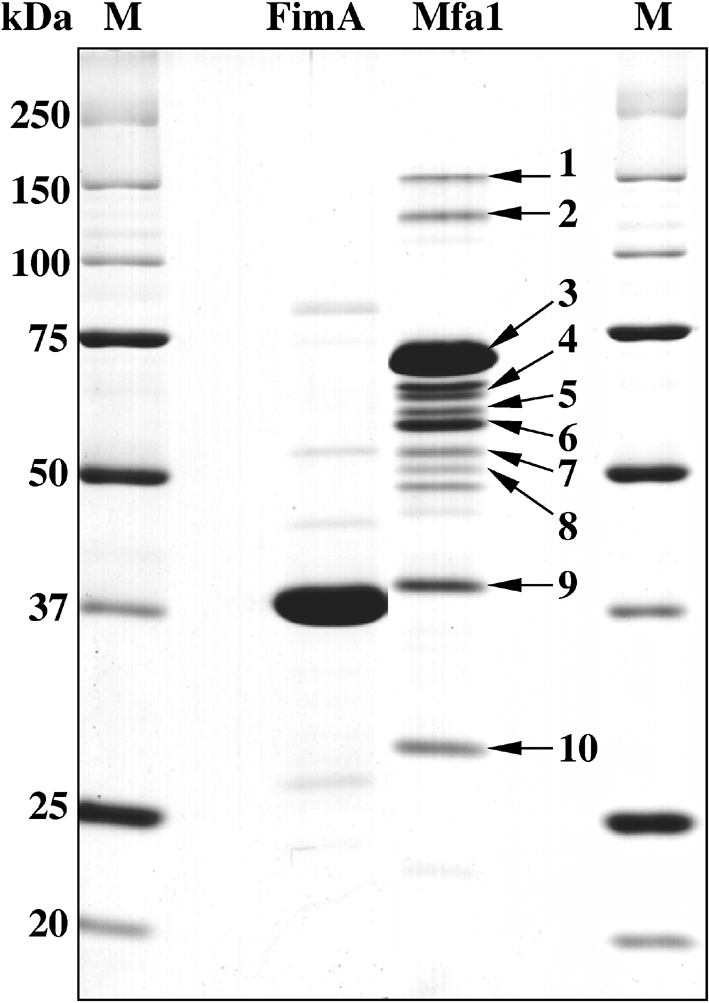
Identification of the minor components of the Mfa1 fimbriae. A typical SDS-PAGE pattern of highly purified P. gingivalis Mfa1 fimbriae is shown together with that of FimA fimbriae. Both FimA (lane FimA) and Mfa1 fimbriae (lane Mfa1) were purified without using any detergent from the mutants SMF1 (Δmfa1) and JI-1 (ΔfimA), respectively. Protein bands are numbered from 1 to 10 together with arrows to identify them by MALDI-TOF MS (see also Table 3).
Table 3.
Identification of proteins by MALDI-TOF MS peptide mass fingerprinting
| Protein number* | Probability† | Description (corresponding CDS‡ number in P. gingivalis ATCC 33277 genome database) (Naito et al., 2008) | Mr (kDa) | Accession number§ |
|---|---|---|---|---|
| 1 | 111 | von Willebrand factor A domain protein (pgn0291) | 134.65 | 34540039 |
| 2 | 90 | |||
| 3 | 71 | Pg-II fimbriae a (pgn0287)|| | 60.79 | 22255314 |
| 4 | 51 | |||
| 5 | 101 | |||
| 6 | 53 | |||
| 7 | 93 | |||
| 8 | 77 | |||
| 9 | 88 | Lipoprotein, putative (pgn0289) | 50.01 | 34540037 |
| 10 | 81 | Immunoreactive 32 kDa antigen PG49 (pgn0290) | 37.14 | 34540038 |
*Numbers refer to designations of protein bands in Fig. 3.
†Probabilities were based on the molecular weight search (MOWSE) scoring algorithm.
‡Protein-coding sequence.
§Nonredundant protein database at the NCBI (http://www.ncbi.nlm.nih.gov/).
||Pg-II fimbriae a is the gene product of Pg-II fim a (designation from Ogawa et al., 1995) from P. gingivalis strain BH18/10 and is another name for the Mfa1 protein, both of which were originally called minor fimbriae (Hamada et al., 1996).
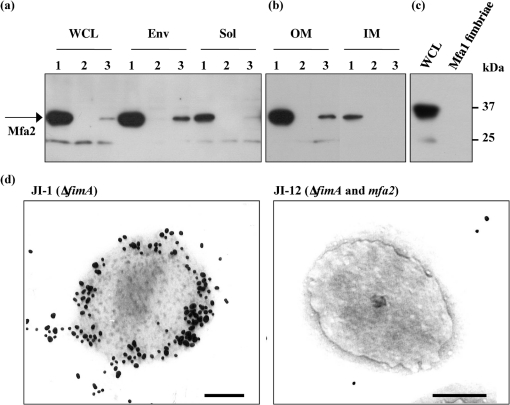
The localization of Mfa2 was examined using various cell fractions of the wild-type, its mutant (Δmfa2) and the complemented strain (JI-3). As shown in Fig. 4(a), mature Mfa2 was detected as the 35 kDa protein in the whole-cell lysate and predominantly in the envelope fraction from the wild-type (Fig. 4a, lane 1 in WCL and Env), but not the mutant (Fig. 4a, lane 2 in WCL and Env). Much less Mfa2 was detected in the soluble fraction of 33277 (Fig. 4a, lane 1 in Sol). This is consistent with the observation that Mfa1 fimbriae, purified from the soluble fraction, did not carry Mfa2, as mentioned above. To demonstrate exact localization of Mfa2, the envelope fraction was then separated into the outer membrane and inner membrane fraction. As shown in Fig. 4(b), Mfa2 was detected mainly in the outer membrane fraction and was only slightly present in the inner membrane. The same was true of the mfa2-complemented strain JI-3, although much less Mfa2 appeared to be expressed. However, Mfa2 in JI-3 was strictly localized in the envelope and outer membrane fraction, strongly indicating that Mfa2 was originally present in the outer membrane. To confirm that the gene product of mfa2 was absent in Mfa1 fimbriae, we carried out Western blotting with Mfa2 antiserum. As shown in Fig. 4(c), lane Mfa1 fimbriae, Mfa2 was not detected in Mfa1 fimbriae, although Mfa2 was clearly detected as a 35 kDa protein in whole-cell lysate (Fig. 4c, lane WCL). In addition, the localization of Mfa2 in the cell was demonstrated by immunogold electron microscopy (Fig. 4d, left panel). The cell surface of JI-1 was stained with gold particles. In contrast, JI-12 was only sparsely labelled (Fig. 4d, right panel). These data indicate that Mfa2 is localized on the cell surface, likely at the base of the Mfa1 fimbriae in the outer membrane.
Fig. 4.
Localization of Mfa2. (a) Localization of Mfa2 in the P. gingivalis cell. P. gingivalis whole-cell lysate (WCL), prepared by physical lysis, was fractionated into envelope (Env) and soluble (Sol) fractions, and Mfa2 was detected by Western blotting as described above. Lanes: 1, parental strain 33277; 2, JI-2; 3, JI-3. The band immunoreactive to anti-Mfa2 antiserum, which has a molecular mass of 24 kDa, appeared to cross-react with the antibody, but is unrelated to Mfa2 as it was present in the mfa2 mutant. (b) Localization of Mfa2 in the outer membrane fraction. The envelope fractions were separated into outer membrane (OM) and inner membrane (IM) fractions, and Western blotting using Mfa2 antiserum was performed. The lanes are the same as for (a). (c) Absence of Mfa2 in the purified Mfa1 fimbriae. Mfa2 was detected by Western blotting. Lane WCL, whole-cell lysate of JI-1 (1 μg protein); lane Mfa1 fimbriae, the purified Mfa1 fimbriae from JI-1 (5 μg protein). (d) Immunogold electron microscopy. Ultra-thin sections of JI-1 (ΔfimA) and JI-12 (ΔfimA and Δmfa2) cells were incubated with anti-Mfa2 serum, followed by labelling with 20 nm colloidal gold-labelled goat anti-rabbit serum. Bars, 200 nm.
Mfa1 and Mfa2 are associated with each other in cells
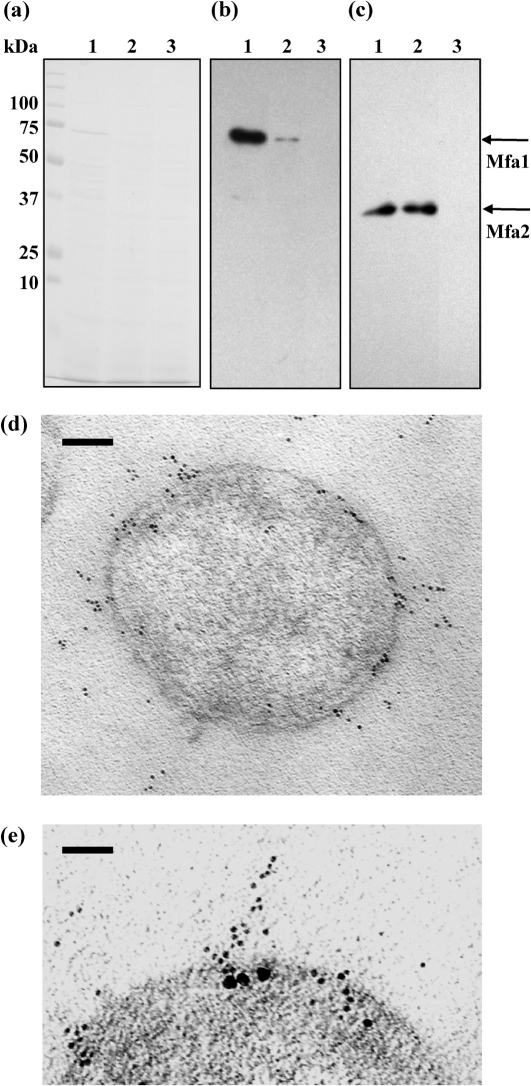
To determine whether Mfa2 interacts with Mfa1 in P. gingivalis cells, a whole-cell lysate, prepared by chemical lysis, was analysed by an immunoprecipitation assay. The whole-cell lysate from 33277 was immunoprecipitated by the addition of anti-Mfa1 or anti-Mfa2 serum, and the precipitate was separated by SDS-PAGE, followed by Western blotting with anti-Mfa2 or anti-Mfa1 serum. As shown in Fig. 5(a), proteins in immunoprecipitates with either antiserum were almost indistinguishable on SDS-PAGE gels, except for a 74 kDa protein band in lane 1, which appeared to be Mfa1, based on the results of subsequent Western blotting. Mfa2 was precipitated together with Mfa1 by the addition of anti-Mfa1 serum (Fig. 5c, lane 1). Also, Mfa1 was co-precipitated together with Mfa2 by the addition of anti-Mfa2 serum (Fig. 5b, lane 2). However, neither Mfa1 nor Mfa2 was precipitated by the addition of either pre-immune serum (data not shown) or anti-OmpA serum, which is specific to major outer membrane proteins (Pgm6/7) in this organism (lanes 3 in Fig. 5b, c), indicating that the co-precipitation of Mfa1 and Mfa2 with each antiserum is specific.
Fig. 5.
Physical association and co-localization between Mfa1 and Mfa2. Mfa1 or Mfa2 was immunoprecipitated from whole-cell lysate of 33277 with anti-Mfa1 or anti-Mfa2 serum. The precipitates were separated by SDS-PAGE (a), followed by Western blotting using either anti-Mfa1 (b) or anti-Mfa2 serum (c). Lanes: 1, 33277 precipitate with anti-Mfa1; 2, 33277 precipitate with anti-Mfa2; 3, 33277 precipitate with anti-OmpA (Pgm6/7) as a negative control. The positions of Mfa1, Mfa2 and molecular mass markers are indicated. (d) Single-labelled, immunogold electron microscopy. Ultra-thin sections of JI-1 (ΔfimA) were reacted with chicken anti-Mfa1, followed by 6 nm gold-labelled anti-chicken Ig. Bar, 100 nm. (e) Double-labelling immunogold microscopy. The sections were first reacted with chicken anti-Mfa1 and rabbit anti-Mfa2 serum, followed by incubation with 6 nm gold-labelled anti-chicken Ig and 20 nm gold-labelled anti-rabbit IgG. Bar, 50 nm.
In order to confirm this association, immunogold microscopy, including double-labelling experiments, was performed using the two antisera. To do this, we raised specific antiserum to Mfa1 in chickens. Using this chicken anti-Mfa1 serum and 6 nm gold-labelled anti-chicken Ig, we first examined Mfa1 fimbriae of thin-sectioned JI-1 cells and then the association of Mfa1 fimbriae and Mfa2 using the two antisera, followed by incubation with 6 nm gold-labelled anti-chicken Ig and 20 nm gold-labelled anti-rabbit IgG, respectively. Many 6 nm gold particles (for Mfa1) were deposited around the cell (Fig. 5d) and appeared to be along filaments, and two types of gold particles were clearly detected around the cell in Fig. 5(e); apparently, several 20 nm gold particles (for Mfa2) were localized on the cell surface, as shown in Fig. 4(d), and were closely associated with an array of 6 nm gold particles representing Mfa1 fimbriae. These results are consistent with the idea that Mfa2 plays an anchoring role for the Mfa1 fimbriae in the outer membrane.
Electron micrographs of P. gingivalis cells and longer Mfa1 fimbriae purified from JI-12
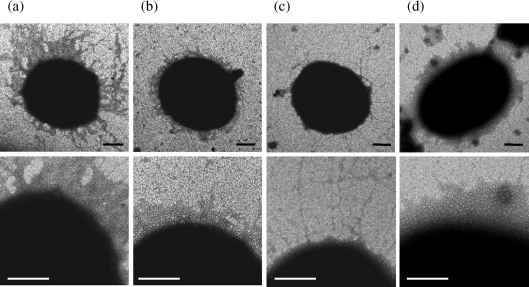
Cells from 33277, JI-1 (ΔfimA) and JI-12 (ΔfimA and Δmfa2) were negatively stained and examined by electron microscopy. Long filaments, characteristic of FimA fimbriae (Yoshimura et al., 1984), were mainly seen on the surface of 33277 cells, as shown in Fig. 6(a). Only short filaments characteristic of Mfa1 fimbriae were detected in JI-1 (Fig. 6b), as shown elsewhere (Hamada et al., 1996), and confirmed with Mfa1 antibodies (Park et al., 2005). Interestingly, the double mutant JI-12 extruded long filaments from the cell surface (Fig. 6c). Furthermore, as mentioned above, mutant Mfa1 fimbriae tend to be easily released from the cell surface into the supernatant (Fig. 2a, b). Since our previous paper reported that the length of Mfa1 fimbriae purified from KDP98, another fimA-deficient mutant, is about 103 nm (Park et al., 2005), they became remarkably longer in the mutant. The aberrant phenotype of Mfa1 fimbriae in JI-12 was restored to normal in an mfa2-complemented strain (JI-4) carrying pTCBex-mfa2, suggesting that the loss of mfa2 was responsible for the aberrancy (Fig. 6d).
Fig. 6.
Electron micrographs of the cell and cell surface of the parental and mutant strains. A part of the cell surface of each strain is enlarged in the lower panel. (a) ATCC 33277, (b) JI-1 (ΔfimA), (c) JI-12 (ΔfimA and Δmfa2), (d) complemented strain JI-4. All strains were negatively stained with 1 % (w/v) ammonium molybdate. Bars, 100 nm.
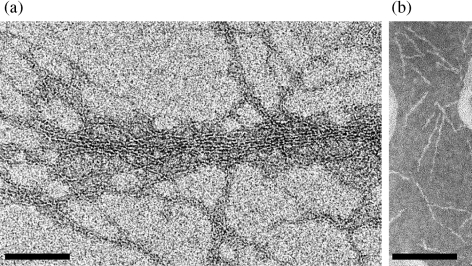
Longer Mfa1 fimbriae were easily purified from JI-12 cells by the mild washing method for FimA fimbriae, and long filaments similar to those partially purified from the culture supernatant (described above and data not shown) were again observed. Purified Mfa1 fimbriae from JI-12 indeed showed much longer filaments than those of parent Mfa1 fimbriae under electron microscopy (Fig. 7). The majority of fibres traversed throughout whole fields (Fig. 7a), reminiscent of long FimA fimbriae in 381 (Yoshimura et al., 1984). On the other hand, the majority of fibres of the purified fimbriae from JI-1 (ΔfimA) were approximately 100 nm long (Fig. 7b), as seen in our previous observation (Park et al., 2005). The length of Mfa1 fimbriae from JI-12 was estimated to be 1 μm or more (approximately 10 times longer than those of the parent), based on examination of five independent micrographs.
Fig. 7.
Effect of mfa2 on the Mfa1 fimbriae length. (a) Electron micrograph of the purified Mfa1 fimbriae from JI-12 (ΔfimA and Δmfa2) negatively stained with 1 % (w/v) ammonium molybdate. The samples were observed under a Carl-Zeiss LEO LIBRA120 electron microscope. Bar, 100 nm. (b) Electron micrograph of the purified Mfa1 fimbriae from JI-1 (ΔfimA). The sample was negatively stained with 1 % (w/v) ammonium molybdate and observed under a JEM-1200EX (JEOL). Bar, 100 nm.
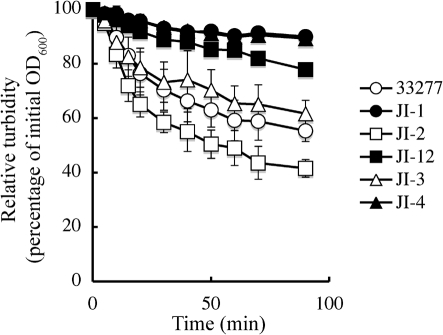
Autoaggregation activities of the mfa2 mutants
Since Mfa1 fimbriae are reported to be involved in P. gingivalis autoaggregation (Lin et al., 2006), autoaggregation activities in various strains were examined. As we previously reported (Nishiyama et al., 2007), loss of FimA fimbriae was confirmed to result in much less autoaggregation (Fig. 8, compare open circles with filled circles), suggesting a reduced contribution of normal Mfa1 fimbriae to autoaggregation in this assay. Indeed, the mutant (JI-12) having longer Mfa1 fimbriae, but no FimA fimbriae, did not show a strong autoaggregation (Fig. 8, filled squares). However, the mfa2 mutant, producing longer Mfa1 and having original long FimA fimbriae, resulted in a stronger autoaggregation than the wild-type (Fig. 8, compare open squares with open circles), implying a positive effect of long Mfa1 fimbriae. The mfa2-complemented strain JI-3 (equivalent to the wild-type) gave a similar autoaggregation pattern to the wild-type (compare open triangles with open circles). Another mfa2-complemented strain, JI-4 (equivalent to JI-1), was indistinguishable from JI-1 in the autoaggregation pattern (compare filled triangles with filled circles), suggesting that no matter how long they are, Mfa1 fimbriae alone contribute marginally to autoaggregation.
Fig. 8.
Time-course of autoaggregation of the wild-type and various mutants. The OD600 value of each cell suspension was measured at the indicated time points. Relative turbidities, defined as the relative OD600 value (as a percentage) normalized to the initial value of each suspension, are plotted against incubation time. All assays were performed in triplicate and the means±sd are shown. ○, Wild-type strain ATCC 33277; •, JI-1 (ΔfimA); □, JI-2 (Δmfa2); ▪, JI-12 (ΔfimA and Δmfa2); ▵, JI-3 (JI-2 containing pTCBex-mfa2, complemented strain); ▴, JI-4 (JI-12 containing pTCBex-mfa2, complemented strain).
Transcriptional analysis of the mfa1 and downstream genes
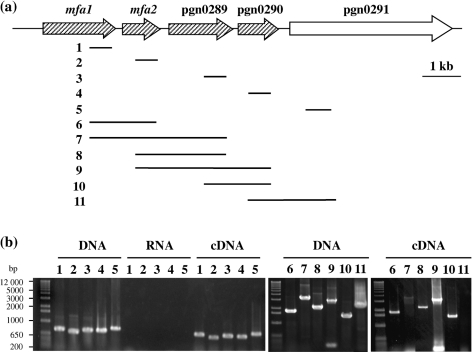
Mfa1 and related minor proteins are encoded in a chromosomal region containing five genes that have the same transcriptional direction (Fig. 1 and Fig. 9a). We extended our earlier analysis of mfa1 and mfa2 co-transcription (Chung et al., 2000) by RT-PCR throughout the mfa1–pgn0291 cluster in 33277 using the primers listed in Table 2. Amplification with intragenic primer pairs for the genes mfa1–pgn0291 demonstrated that each of the genes was expressed (Fig. 9b, lanes 1–5 under cDNA). The same primers were used in pairs that spanned each intergenic region. Results in Fig. 9(b), lanes 6–10 under cDNA, show that mRNAs were detected that spanned mfa1 and mfa2, mfa1 and pgn0289, mfa2 and pgn0289, mfa2 and pgn0290, and pgn0289 and pgn0290. The mRNA that spanned mfa1 and pgn0289 was detected, although the detected band was weak (Fig. 9b, lane 7 under cDNA). This is presumably due to its being the longest target, which may be close to the PCR detection limit. However, P. gingivalis mRNA that spans pgn0290 and pgn0291 was not detected, although mRNA of pgn0291 itself was detected (Fig. 9b, lane 11 under cDNA). These data indicate that the mfa1 operon can be transcribed as a four-gene polycistronic message encompassing mfa1–pgn0290.
Fig. 9.
Transcriptional analysis of the genes for Mfa1 fimbrial components. (a) The predicted PCR fragments from mfa1 to pgn0291 are indicated as thin lines at the right side of the numbers (1–11). The possible operon is shaded. Gene annotation corresponds to the 33277 genome database. (b) RT-PCR of 33277 mRNAs from mfa1 to pgn0291. RT-PCR amplification of intragenic and intergenic segments within the mfa1 gene cluster was carried out with primer pairs listed in Table 2. Results labelled as DNA indicate that PCR was carried out using chromosomal DNA as a template, showing that each primer set was appropriate, and those labelled as RNA indicate that RT-PCR was carried out without reverse transcriptase as negative controls, showing that the RNA preparation was pure, without contamination of DNA. Therefore, results labelled as cDNA indicate the presence or absence of mRNA. The lane numbers correspond to those in panel (a). Lanes: 1, primers for mfa1; 2, primers for mfa2; 3, primers for pgn0289; 4, primers for pgn0290; 5, primers for pgn0291; 6, primers for a region spanning mfa1 and mfa2; 7, primers for a region spanning mfa1 and pgn0289; 8, primers for a region spanning mfa2 and pgn0289; 9, primers for a region spanning mfa2 and pgn0290; 10, primers for a region spanning pgn0289 and pgn0290; 11, primers for a region spanning pgn0290 and pgn0291.
DISCUSSION
P. gingivalis ATCC 33277 possesses two types of morphologically distinct long (FimA) and short (Mfa1) fimbriae (Naito et al., 2008), although both are thin, single-stranded filaments with a diameter of 5–6.5 nm (Park et al., 2005; Yoshimura et al., 1984). In order to investigate the role of mfa2, downstream of mfa1, experiments were conducted in a fimA-knockout background to prevent any misidentification of fimbriae of aberrant length. Indeed, double mutant JI-12 (ΔfimA and Δmfa2) clearly formed unusual longer filaments, easily shed from the cell surface, and strains JI-3 and JI-4 complemented with the wild-type mfa2 allele recovered the phenotype of the parent. Collectively, these results indicate that mfa2 plays a role in the normal formation of Mfa1 filaments of short, uniform size (Park et al., 2005) that are tightly bound to the cell surface. Longer Mfa1 fimbriae emerged on the cell surface of JI-2 (Δmfa2) and induced enhanced autoaggregation activity, to some extent presumably in collaboration with long FimA fimbriae, although Mfa1 fimbriae themselves did not contribute much to autoaggregation in the absence of FimA fimbriae [Fig. 8, see JI-1 (ΔfimA) and JI-12 (ΔfimA and Δmfa2)].
Antibodies to rMfa2 showed that mature Mfa2 resided in the outer membrane, consistent with the concept that Mfa2 plays a role as an anchor in order to tightly connect filaments to the outer membrane following polymerization of fimbrilins into filaments. Thus, Mfa2 may play roles as an anchor and as a regulator of their length. We also showed that purified Mfa1 fimbriae have at least three accessory proteins, as do FimA fimbriae (Nishiyama et al., 2007; Yoshimura et al., 1993), and that Mfa2 is not present as an accessory component (Figs 3 and 4c, Table 3). However, Mfa2 was found to associate with Mfa1 fimbrilin in cells and was likely present on the base of the Mfa1 fimbrial structure (Fig. 5), supporting an anchor function for Mfa2. Whole-cell lysates for immunoprecipitation were prepared by a chemical lysis method using a commercial solution including a detergent mix, different from that for purification of Mfa1 fimbriae. For purification of Mfa1 fimbriae, bacterial cells were physically disrupted with a French pressure cell (Park et al., 2005). We assume that harsh, mechanical disruption of bacterial cells, such as by French press, tends to leave Mfa2 in the outer membrane and to release filaments of Mfa1 fimbriae into solution, which may reflect the structure and function of Mfa2 in the membrane.
By RT-PCR, mfa1 and mfa2 (equivalent to pgn0288) were confirmed to be co-transcribed, and the genes pgn0289, pgn0290 and pgn0291, encoding the three accessory components detected in this study, were shown to be indeed transcribed in cells. In addition, mfa1–pgn0290 appeared to be co-transcribed. We also examined expression of three downstream genes from mfa2 in JI-2 (Δmfa2), because of the possibility that the mutation in mfa2 could result in a polar effect on these genes. RT-PCR products of the expected sizes for pgn0289, pg0290 and pgn0291 were detected from JI-2. Moreover, longer Mfa1 fimbriae purified from JI-12 (ΔfimA and Δmfa2) also contained three accessory proteins as their gene products (data not shown). Taken together, the three genes pgn0289, pg0290 and pgn0291, which were transcribed as a large mRNA spanning mfa1 to pgn0290 (Fig. 9), seem to be at least in part independent of upstream mfa2 and partially expressed, although further work is needed to confirm this.
A similarity between FimA and Mfa1 fimbriae in P. gingivalis is notable with regard to the possession of three minor protein components, and the presence of similar gene orders and clusters consisting of five genes. However, FimA and Mfa1 have different biological activities and may thus be operational in differing environmental situations. The gene corresponding to mfa2 in the fimA cluster has been shown to be ORF1 (equivalent to pgn0181 in 33277 or pg2133 in W83), located between fimA and fimC in 33277 and 381 (Naito et al., 2008; Nishikawa et al., 2004; Nishiyama et al., 2007), and its gene product and function have yet to be identified. However, the mfa2-corresponding gene (designated fimB) appears to be truncated by a point mutation, creating a stop codon at the middle portion in both strains (Naito et al., 2008; Nishiyama et al., 2007; Watanabe et al., 1996), but not in W83 (Nelson et al., 2003) or OMZ 314 (type II in fimA) (Kato et al., 2007). Therefore, ORF1 in 33277 and 381 encodes a putative 15 kDa protein (Nishiyama et al., 2007), but the corresponding genes (pg2133 for W83) in W83 and OMZ 314 encode a putative 34 kDa protein (Kato et al., 2007; Nelson et al., 2003), a size equivalent to Mfa2.
We assume that both strain 33277 and strain 381 carry FimA fimbriae (both fimA genotype I), easily shed from the cell surface, with unusually long filaments due to the mutation in the fimB gene, equivalent to mfa2 in Mfa1 fimbriae. Evidence for this is as follows. (1) The two strains are believed to be siblings. (2) We have not thus far encountered any P. gingivalis strains carrying FimA fimbriae that could be shed from the cell surface by the mild, selective washing method, except for the two strains 33277 and 381 (Yoshimura et al., 1984). (3) FimA fimbriae in several strains, OMZ314, HW24D1 (both genotype II), MPWIb-01 (type Ib, a close variant of genotype I in fimA, kindly provided by M. Miura, Kyushu University) and ESO101 (genotype I) (Kato et al., 2007; Miura et al., 2005; Nakagawa et al., 2002), were not selectively solubilized from the cell surface by the washing method, which is a reliable procedure for isolation from 33277 and 381 (our unpublished data). (4) In all these strains, most of the FimA and Mfa1 fimbriae can be released into the soluble fraction by French pressure disruption (Murakami et al., 2002). (5) It is also pertinent to mention that complete deletion of ORF1 (pgn0181) does not produce any apparent differences in FimA fimbriae in 33277 (Nishiyama et al., 2007). Taken together, these data suggest that mfa2 and fimB play a similar role in the Mfa1 and FimA fimbriae of P. gingivalis, and that strains 33277 and 381 may be natural mutants in FimA fimbriae. However, this hypothesis needs to be examined further.
Fimbriae, pili or curli are adhesive hair-like organelles that project from the cell surface of a wide variety of Gram-negative as well as Gram-positive bacteria (Barnhart & Chapman, 2006; Capitani et al., 2006; Pizarro-Cerda & Cossart, 2006). The morphogenesis or biogenesis of some of these, especially in E. coli, has been extensively studied (Kuehn et al., 1994; Piatek et al., 2005; Wu & Fives-Taylor, 2001), and several systems such as the chaperone/usher pathway (Piatek et al., 2005), type II secretion (Hansen & Forest, 2006) and secretion/nucleation (Barnhart & Chapman, 2006) have been proposed for their generation. PapH, similar to Mfa2, with roles in anchoring fimbriae to cells and in modulating their length, has been reported in E. coli Pap pili (Baga et al., 1987; Verger et al., 2006). papH is located adjacent to and downstream of papA, which encodes the major pilin subunit, a similar situation to the configuration of mfa1 and mfa2. The corresponding gene fimI, downstream of fimA in E. coli type I pili, has not yet been assigned a function (Sauer et al., 2004; Schilling et al., 2001). Mfa1 and FimA fimbriae in P. gingivalis are structurally, genetically and biosynthetically different from those in E. coli (Nishiyama et al., 2007; Yoshimura et al., 1984, 1993). Therefore, elucidation of the structure and function of Mfa2 and ORF1 could bring new insights in fimbrial research that extend beyond the paradigm of E. coli.
P. gingivalis, a periodontopathogen, proteomically most similar to B. thetaiotaomicron and B. fragilis, also a member of the Cytophaga–Flavobacteria–Bacteroides group (Nelson et al., 2003), appears to use a novel fimbrial biogenesis system via a lipoprotein precursor and using Arg-gingipain (RGP), a major arginine-specific proteinase, as the maturation enzyme (Shoji et al., 2004). Our findings on Mfa2 function do not necessarily conflict with the lipoprotein precursor biogenesis as proposed previously (Shoji et al., 2004). Lipoprotein precursors of fimbrilin could be processed by RGP in the outer membrane, followed by assembly into filaments with the involvement of Mfa2.
In conclusion, our study has highlighted that Mfa2, the product of mfa2 downstream of the major subunit gene mfa1, may be involved as an anchor of filaments as well as a regulator in the filament length of Mfa1 fimbriae in P. gingivalis.
Acknowledgments
We thank Kunihiko Kobayashi (College of Life and Health Sciences, Chubu University) and Yuka Mitamura-Kondo (Electron Microscopy Laboratory, Aichi-Gakuin University) for their helpful, technical advice. We also thank the Institute for Genomic Research (TIGR) for making the P. gingivalis W83 genomic sequence freely available to the public. This work was supported by Grants-in-Aid for Scientific Research (20890248 to Y. H., 15591958 and 20592165 to Y. M., 19592139 to K. N. and 15591957 to F. Y.) from the Japan Society for the Promotion of Science (JSPS) and the AGU High-Tech Research Center Project and Strategic Research AGU-Platform Formation from The Ministry of Education, Culture, Sports, Science and Technology, Japan, the Furukawa foundation to Y. H., and DE12505 from the National Institute of Dental and Craniofacial Research (NIDCR) (to R. J. L.).
Abbreviations
CBB, Coomassie brilliant blue R-250
GST, glutathione S-transferase
Footnotes
The GenBank/EMBL/DDBJ accession number for the mfa2 sequence of P. gingivalis ATCC 33277 is AB360435.
References
- Andrian, E., Grenier, D. & Rouabhia, M. (2006). Porphyromonas gingivalis-epithelial cell interactions in periodontitis. J Dent Res 85, 392–403. [DOI] [PubMed] [Google Scholar]
- Arai, M., Hamada, N. & Umemoto, T. (2000). Purification and characterization of a novel secondary fimbrial protein from Porphyromonas gingivalis strain 381. FEMS Microbiol Lett 193, 75–81. [DOI] [PubMed] [Google Scholar]
- Baga, M., Norgren, M. & Normark, S. (1987). Biogenesis of E. coli Pap pili: papH, a minor pilin subunit involved in cell anchoring and length modulation. Cell 49, 241–251. [DOI] [PubMed] [Google Scholar]
- Barnhart, M. M. & Chapman, M. R. (2006). Curli biogenesis and function. Annu Rev Microbiol 60, 131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capitani, G., Eidam, O., Glockshuber, R. & Grutter, M. G. (2006). Structural and functional insights into the assembly of type 1 pili from Escherichia coli. Microbes Infect 8, 2284–2290. [DOI] [PubMed] [Google Scholar]
- Chung, W. O., Demuth, D. R. & Lamont, R. J. (2000). Identification of a Porphyromonas gingivalis receptor for the Streptococcus gordonii SspB protein. Infect Immun 68, 6758–6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasanayake, A. P., Russell, S., Boyd, D., Madianos, P. N., Forster, T. & Hill, E. (2003). Preterm low birth weight and periodontal disease among African Americans. Dent Clin North Am 47, 115–125. [DOI] [PubMed] [Google Scholar]
- Fletcher, H. M., Morgan, R. M. & Macrina, F. L. (1997). Nucleotide sequence of the Porphyromonas gingivalis W83 recA homolog and construction of a recA-deficient mutant. Infect Immun 65, 4592–4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, R. G., Russell, J. B., Wilson, D. B., Wang, G. R. & Shoemaker, N. B. (1996). Use of a modified Bacteroides–Prevotella shuttle vector to transfer a reconstructed β-1,4-d-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B14. Appl Environ Microbiol 62, 196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, F. C., III, Yumoto, H., Takahashi, Y., Chou, H. H. & Genco, C. A. (2006). Innate immune signaling and Porphyromonas gingivalis-accelerated atherosclerosis. J Dent Res 85, 106–121. [DOI] [PubMed] [Google Scholar]
- Hajishengallis, G., Martin, M., Sojar, H. T., Sharma, A., Schifferle, R. E., DeNardin, E., Russell, M. W. & Genco, R. J. (2002). Dependence of bacterial protein adhesins on Toll-like receptors for proinflammatory cytokine induction. Clin Diagn Lab Immunol 9, 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, L. M., Fawell, S. C., Shi, X., Faray-Kele, M. C., Aduse-Opoku, J., Whiley, R. A. & Curtis, M. A. (2005). Sequence diversity and antigenic variation at the rag locus of Porphyromonas gingivalis. Infect Immun 73, 4253–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada, N., Sojar, H. T., Cho, M. I. & Genco, R. J. (1996). Isolation and characterization of a minor fimbria from Porphyromonas gingivalis. Infect Immun 64, 4788–4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada, N., Watanabe, K., Arai, M., Hiramine, H. & Umemoto, T. (2002). Cytokine production induced by a 67-kDa fimbrial protein from Porphyromonas gingivalis. Oral Microbiol Immunol 17, 197–200. [DOI] [PubMed] [Google Scholar]
- Hansen, J. K. & Forest, K. T. (2006). Type IV pilin structures: insights on shared architecture, fiber assembly, receptor binding and type II secretion. J Mol Microbiol Biotechnol 11, 192–207. [DOI] [PubMed] [Google Scholar]
- Hasegawa, Y., Nishiyama, S., Nishikawa, K., Kadowaki, T., Yamamoto, K., Noguchi, T. & Yoshimura, F. (2003). A novel type of two-component regulatory system affecting gingipains in Porphyromonas gingivalis. Microbiol Immunol 47, 849–858. [DOI] [PubMed] [Google Scholar]
- Hayashi, J., Nishikawa, K., Hirano, R., Noguchi, T. & Yoshimura, F. (2000). Identification of a two-component signal transduction system involved in fimbriation of Porphyromonas gingivalis. Microbiol Immunol 44, 279–282. [DOI] [PubMed] [Google Scholar]
- Hiramine, H., Watanabe, K., Hamada, N. & Umemoto, T. (2003). Porphyromonas gingivalis 67-kDa fimbriae induced cytokine production and osteoclast differentiation utilizing TLR2. FEMS Microbiol Lett 229, 49–55. [DOI] [PubMed] [Google Scholar]
- Hongo, H., Osano, E., Ozeki, M., Onoe, T., Watanabe, K., Honda, O., Tani, H., Nakamura, H. & Yoshimura, F. (1999). Characterization of an outer membrane protein gene, pgmA, and its gene product from Porphyromonas gingivalis. Microbiol Immunol 43, 937–946. [DOI] [PubMed] [Google Scholar]
- Hongyo, H., Kurihara, H., Kokeguchi, S., Miyamoto, M., Maeda, H., Hayakawa, M., Abiko, Y., Takashiba, S. & Murayama, Y. (1997). Molecular cloning and characterization of the gene encoding 53 kD outer membrane protein of Porphyromonas gingivalis. Microbios 92, 47–57. [PubMed] [Google Scholar]
- Hongyo, H., Kokeguchi, S., Kurihara, H., Miyamoto, M., Maeda, H., Takashiba, S. & Murayama, Y. (1998). Comparative study of two outer membrane protein genes from Porphyromonas gingivalis. Microbios 95, 91–100. [PubMed] [Google Scholar]
- Jenkinson, H. F. & Lamont, R. J. (2005). Oral microbial communities in sickness and in health. Trends Microbiol 13, 589–595. [DOI] [PubMed] [Google Scholar]
- Kato, T., Kawai, S., Nakano, K., Inaba, H., Kuboniwa, M., Nakagawa, I., Tsuda, K., Omori, H., Ooshima, T. & other authors (2007). Virulence of Porphyromonas gingivalis is altered by substitution of fimbria gene with different genotype. Cell Microbiol 9, 753–765. [DOI] [PubMed] [Google Scholar]
- Kolenbrander, P. E., Andersen, R. N., Blehert, D. S., Egland, P. G., Foster, J. S. & Palmer, R. J., Jr (2002). Communication among oral bacteria. Microbiol Mol Biol Rev 66, 486–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn, M. J., Jacob-Dubuisson, F., Dodson, K., Slonim, L., Striker, R. & Hultgren, S. J. (1994). Genetic, biochemical, and structural studies of biogenesis of adhesive pili in bacteria. Methods Enzymol 236, 282–306. [DOI] [PubMed] [Google Scholar]
- Lamont, R. J. & Jenkinson, H. F. (1998). Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev 62, 1244–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont, R. J., El-Sabaeny, A., Park, Y., Cook, G. S., Costerton, J. W. & Demuth, D. R. (2002). Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148, 1627–1636. [DOI] [PubMed] [Google Scholar]
- Lin, X., Wu, J. & Xie, H. (2006). Porphyromonas gingivalis minor fimbriae are required for cell–cell interactions. Infect Immun 74, 6011–6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda, T., Murakami, Y., Noguchi, T. & Yoshimura, F. (2006). Effects of various growth conditions in a chemostat on expression of virulence factors in Porphyromonas gingivalis. Appl Environ Microbiol 72, 3458–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, M., Hamachi, T., Fujise, O. & Maeda, K. (2005). The prevalence and pathogenic differences of Porphyromonas gingivalis fimA genotypes in patients with aggressive periodontitis. J Periodontal Res 40, 147–152. [DOI] [PubMed] [Google Scholar]
- Murakami, Y., Imai, M., Nakamura, H. & Yoshimura, F. (2002). Separation of the outer membrane and identification of major outer membrane proteins from Porphyromonas gingivalis. Eur J Oral Sci 110, 157–162. [DOI] [PubMed] [Google Scholar]
- Nagano, K., Read, E. K., Murakami, Y., Masuda, T., Noguchi, T. & Yoshimura, F. (2005). Trimeric structure of major outer membrane proteins homologous to OmpA in Porphyromonas gingivalis. J Bacteriol 187, 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano, K., Murakami, Y., Nishikawa, K., Sakakibara, J., Shimozato, K. & Yoshimura, F. (2007). Characterization of RagA and RagB in Porphyromonas gingivalis: study using gene-deletion mutants. J Med Microbiol 56, 1536–1548. [DOI] [PubMed] [Google Scholar]
- Naito, M., Hirakawa, H., Yamashita, A., Ohara, N., Shoji, M., Yukitake, H., Nakayama, K., Toh, H., Yoshimura, F. & other authors (2008). Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res 15, 215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, I., Amano, A., Ohara-Nemoto, Y., Endoh, N., Morisaki, I., Kimura, S., Kawabata, S. & Hamada, S. (2002). Identification of a new variant of fimA gene of Porphyromonas gingivalis and its distribution in adults and disabled populations with periodontitis. J Periodontal Res 37, 425–432. [DOI] [PubMed] [Google Scholar]
- Nelson, K. E., Fleischmann, R. D., DeBoy, R. T., Paulsen, I. T., Fouts, D. E., Eisen, J. A., Daugherty, S. C., Dodson, R. J., Durkin, A. S. & other authors (2003). Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol 185, 5591–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa, K., Yoshimura, F. & Duncan, M. J. (2004). A regulation cascade controls expression of Porphyromonas gingivalis fimbriae via the FimR response regulator. Mol Microbiol 54, 546–560. [DOI] [PubMed] [Google Scholar]
- Nishiyama, S., Murakami, Y., Nagata, H., Shizukuishi, S., Kawagishi, I. & Yoshimura, F. (2007). Involvement of minor components associated with the FimA fimbriae of Porphyromonas gingivalis in adhesive functions. Microbiology 153, 1916–1925. [DOI] [PubMed] [Google Scholar]
- Ogawa, T., Mori, H., Yasuda, K. & Hasegawa, M. (1994). Molecular cloning and characterization of the genes encoding the immunoreactive major cell-surface proteins of Porphyromonas gingivalis. FEMS Microbiol Lett 120, 23–30. [DOI] [PubMed] [Google Scholar]
- Ogawa, T., Yasuda, K., Yamada, K., Mori, H., Ochiai, K. & Hasegawa, M. (1995). Immunochemical characterisation and epitope mapping of a novel fimbrial protein (Pg-II fimbria) of Porphyromonas gingivalis. FEMS Immunol Med Microbiol 11, 247–255. [DOI] [PubMed] [Google Scholar]
- Park, Y., Simionato, M. R., Sekiya, K., Murakami, Y., James, D., Chen, W., Hackett, M., Yoshimura, F., Demuth, D. R. & Lamont, R. J. (2005). Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect Immun 73, 3983–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatek, R., Zalewska, B., Bury, K. & Kur, J. (2005). The chaperone–usher pathway of bacterial adhesin biogenesis – from molecular mechanism to strategies of anti-bacterial prevention and modern vaccine design. Acta Biochim Pol 52, 639–646. [PubMed] [Google Scholar]
- Pizarro-Cerda, J. & Cossart, P. (2006). Bacterial adhesion and entry into host cells. Cell 124, 715–727. [DOI] [PubMed] [Google Scholar]
- Sakakibara, J., Nagano, K., Murakami, Y., Higuchi, N., Nakamura, H., Shimozato, K. & Yoshimura, F. (2007). Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiology 153, 866–876. [DOI] [PubMed] [Google Scholar]
- Sauer, F. G., Remaut, H., Hultgren, S. J. & Waksman, G. (2004). Fiber assembly by the chaperone–usher pathway. Biochim Biophys Acta 1694, 259–267. [DOI] [PubMed] [Google Scholar]
- Schilling, J. D., Mulvey, M. A. & Hultgren, S. J. (2001). Structure and function of Escherichia coli type 1 pili: new insight into the pathogenesis of urinary tract infections. J Infect Dis 183 (Suppl. 1), S36–S40. [DOI] [PubMed] [Google Scholar]
- Shoji, M., Naito, M., Yukitake, H., Sato, K., Sakai, E., Ohara, N. & Nakayama, K. (2004). The major structural components of two cell surface filaments of Porphyromonas gingivalis are matured through lipoprotein precursors. Mol Microbiol 52, 1513–1525. [DOI] [PubMed] [Google Scholar]
- Socransky, S. S. & Haffajee, A. D. (2005). Periodontal microbial ecology. Periodontol 2000 38, 135–187. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., Yoshimura, F., Takahashi, K., Tani, H. & Suzuki, T. (1988). Detection of fimbriae and fimbrial antigens on the oral anaerobe Bacteroides gingivalis by negative staining and serological methods. J Gen Microbiol 134, 2713–2720. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., Davey, M., Yumoto, H., Gibson, F. C., III & Genco, C. A. (2006). Fimbria-dependent activation of pro-inflammatory molecules in Porphyromonas gingivalis infected human aortic endothelial cells. Cell Microbiol 8, 738–757. [DOI] [PubMed] [Google Scholar]
- Verger, D., Miller, E., Remaut, H., Waksman, G. & Hultgren, S. (2006). Molecular mechanism of P pilus termination in uropathogenic Escherichia coli. EMBO Rep 7, 1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, K., Onoe, T., Ozeki, M., Shimizu, Y., Sakayori, T., Nakamura, H. & Yoshimura, F. (1996). Sequence and product analyses of the four genes downstream from the fimbrilin gene (fimA) of the oral anaerobe Porphyromonas gingivalis. Microbiol Immunol 40, 725–734. [DOI] [PubMed] [Google Scholar]
- Wu, H. & Fives-Taylor, P. M. (2001). Molecular strategies for fimbrial expression and assembly. Crit Rev Oral Biol Med 12, 101–115. [DOI] [PubMed] [Google Scholar]
- Yoshimura, F., Takahashi, K., Nodasaka, Y. & Suzuki, T. (1984). Purification and characterization of a novel type of fimbriae from the oral anaerobe Bacteroides gingivalis. J Bacteriol 160, 949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, F., Watanabe, K., Takasawa, T., Kawanami, M. & Kato, H. (1989). Purification and properties of a 75-kilodalton major protein, an immunodominant surface antigen, from the oral anaerobe Bacteroides gingivalis. Infect Immun 57, 3646–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, F., Takahashi, Y., Hibi, E., Takasawa, T., Kato, H. & Dickinson, D. P. (1993). Proteins with molecular masses of 50 and 80 kilodaltons encoded by genes downstream from the fimbrilin gene (fimA) are components associated with fimbriae in the oral anaerobe Porphyromonas gingivalis. Infect Immun 61, 5181–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, F., Murakami, Y., Nishikawa, K., Hasegawa, Y. & Kawaminami, S. (2009). Surface components of Porphyromonas gingivalis. J Periodontal Res 44, 1–12. [DOI] [PubMed] [Google Scholar]