Abstract
Serum response factor (SRF) is a transcription factor that binds to a CarG box motif within the serum response element (SRE) of genes that are expressed in response to mitogens. SRF plays essential roles in muscle and nervous system development; however, little is known about the role of SRF during liver growth and function. To examine the function of SRF in the liver, we generated mice in which the Srf gene was specifically disrupted in hepatocytes. The survival of mice lacking hepatic SRF activity was lower than that of control mice; moreover, surviving mutant mice had lower blood glucose and triglyceride levels compared with control mice. In addition, SrfloxP/loxP AlfpCre mice were smaller and had severely depressed levels of serum IGF1. Srf-deficient livers were also smaller than control livers and liver-cell proliferation and viability were compromised. Gene array analysis of SRF depleted livers revealed a reduction in many mRNAs, including those encoding components of the growth hormone/IGF1 pathway, cyclins, several metabolic regulators, and cytochrome p450 enzymes.
Conclusion
SRF is essential for hepatocyte proliferation and survival, liver function, and control of postnatal body growth by regulating hepatocyte gene expression.
Keywords: liver transcription factors, liver development, IGF1, Growth Hormone receptor, liver size
Background
Serum response factor is a transcription factor that regulates immediate-early and muscle gene expression 1. It belongs to the MADS-box family of transcription factors that possess an N-terminal motif capable of binding CArG box elements within target promoters 2,3. Although SRF is widely expressed, its abundance varies between cell types. For example, it is highly expressed in skeletal and cardiac muscle, while expression in the brain, liver, lung, and spleen is relatively lower 4.
SRF plays a key role in activating immediate early genes, including c-fos, egr-1, and jun-B in response to mitogenic signals 3,5–8. Activation of immediate early genes in quiescent cells following the addition of serum is important for cells to exit G0 and reenter the G1 phase of the cell cycle. When anti-SRF antibody was introduced into quiescent rat embryonic fibroblasts, it abolished serum stimulated c-fos induction and suppressed DNA synthesis 9,10. More recently, SRF was found to act downstream of phospho-inositol-3-kinase (PI3K) signaling and to be required for PI3K-induced cell proliferation 11. SRF activity, therefore, appears to be necessary for growth factor-mediated cell cycle progression.
SRF is also essential for mesoderm formation during gastrulation and is required for the function of multiple tissues and organs 12,13. Conditional loss-of-function of SRF in the mouse heart causes defects in cardiac trabeculation, compact layer expansion, and sarcomere assembly, and loss of SRF in cultured cardiac myocytes disrupts the structure of the contractile apparatus 14–17. Deletion of SRF in skeletal muscle also causes severe hypoplasia and mutant mice die during the perinatal period 18. In addition to its roles in normal muscle growth and function, SRF is important in regulating neuronal migration and synaptic plasticity 19,20.
While SRF function has been intensely studied in muscle and the nervous system, little is known about the contribution of SRF to hepatic growth and function. Although relatively low levels of Srf mRNA are expressed in the adult mouse liver 4, a role for SRF during liver regeneration was recently uncovered 21. In these studies, SRF expression was increased in mouse livers following partial hepatectomy and SRF deficient hepatocytes failed to enter the cell cycle or proliferate after surgery. The observed impairment in the regeneration of SRF deficient livers correlated with reduced induction of immediate early genes including JunB, c-fos and pip92. These data seemed provocative and suggested that SRF may not only act in response to liver damage but could contribute toward normal liver function and development. To test this, hepatocyte-specific SRF knockout mice were generated from a strain harboring a conditional null allele of Srf (SrfloxP), which has previously been demonstrated to completely abolish expression of SRF protein in cells that express Cre recombinase 14,20. Analysis of the resulting animals revealed that SRF has a crucial role in regulating hepatocyte proliferation, survival, and normal liver function. As a consequence of loss of hepatic function and liver mass, SRF liver deficient mice display severe defects in body growth.
Results
SRF is expressed throughout hepatogenesis
Srf mRNA was previously identified in the adult and E11.5 mouse liver 4 and has been found to increase in response to partial hepatectomy 21; however, a detailed profile of SRF expression throughout hepatogenesis has not been reported. Using RT-PCR analyses, we identified the presence of Srf mRNA in fetal livers from E10.5, which was the earliest time tested, through to adult (Fig. 1a). This expression profile mirrored that of Hnf4α, an mRNA which is characteristically expressed in hepatocytes 22,23.
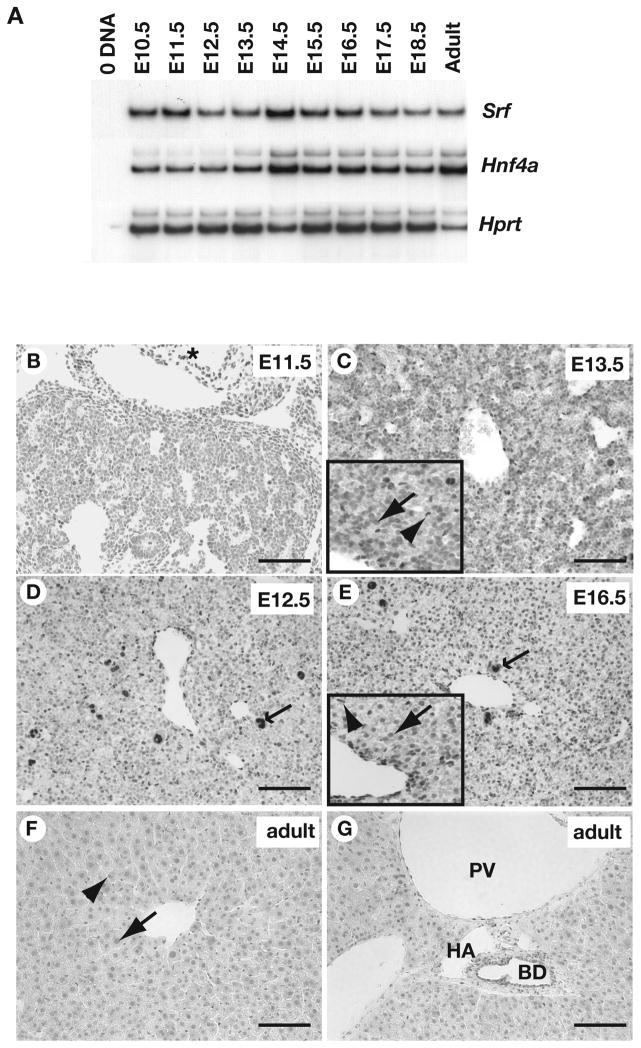
Figure 1. SRF is expressed in multiple cell types in fetal and adult mouse liver.
A). RT-PCR analysis identified Srf, Hnf4a, and Hprt (loading control) mRNAs in livers isolated from mouse embryos at daily intervals between E10.5 and E18.5 as well as in adult livers. B–G) Sections through livers at indicated stages of development in which the presence of SRF was detected in hepatoblasts (arrow), cardiac myocytes (asterisk), sinusoidal endothelial cells (arrowhead), bile ducts (BD), portal vein (PV), hepatic artery (HA), and hematopoietic cells (barbed arrow) by immunohistochemistry. Scale bar = 100μM.
To document the cellular distribution of SRF during liver development, we detected SRF protein in sections of mouse embryos by immunohistochemistry using two different anti-SRF antibodies, which gave indistinguishable results. Between E11.5 and E16.5, the abundance of SRF within the hepatoblasts appeared to be relatively low compared, for example, to cardiac myocytes where SRF levels appeared higher (Fig. 1b, asterisk). By E16.5, SRF staining in hepatoblasts increased and became clearly localized within the nucleus (Fig. 1e), and this distribution was maintained throughout the remainder of development as well as in the adult liver (Fig. 1f–g). In addition to the hepatocytes, SRF was identified in the hepatic endothelial cells and in hematopoietic cells throughout hepatogenesis (Fig. 1b–e). In the adult liver, SRF was present in the endothelial cells of the portal and central veins, the walls of the hepatic artery, as well as in the cholangiocytes of the bile duct (Fig. 1f–g).
Disruption of hepatic SRF results in increased postnatal mortality and impaired body growth
Development of Srf−/− embryos arrests at E6.5–7.0 due to lack of mesoderm formation 12, thereby preventing the use of such embryos to study SRF function in the liver. We, therefore, used the Cre-loxP system to conditionally delete Srf in hepatocytes by breeding SrfloxP/loxP mice (SrfTm1Rmn) 20 with AlfpCre transgenic mice (Tg(Alb1-Cre)1Khk) that express Cre recombinase specifically in the developing hepatocytes starting at E9.5 24. In these mice the Srf promoter and exon 1 are flanked by loxP elements 20. Cre mediated deletion of the intervening sequence results in the absence of a detectable Srf mRNA, SRF DNA binding activity, and SRF protein, and so SrfloxP/loxP mice are believed to harbor a null allele 14,15,20.
To determine whether SrfloxP/loxPAlfpCre mice were viable, we established the genotype of 199 offspring that resulted from SrfloxP/+AlfpCre×SrfloxP/loxP matings. Mice harboring a functional allele of Srf were recovered with normal Mendelian ratios – SrfloxP/loxP (25.1%), SrfloxP/+ (28.6%), and SrfloxP/+AlfpCre (28.1%). In contrast, SrfloxP/loxPAlfpCre mice, contributed only 18.1% of the total offspring suggesting that loss of SRF within hepatocytes leads to increased mortality before weaning, although the penetrance of this phenotype is partial. We also monitored mortality of control and SrfloxP/loxPAlfpCre mice after weaning. Because livers of male and female mice display sexually dimorphic responses to various stresses 25, mice were divided into male and female groups. As shown in Fig. 2a, 10 weeks after birth 96.8% of SrfloxP/+AlfpCre male and 95.2% of SrfloxP/+AlfpCre female mice had survived, which was indistinguishable from wild type mice (not shown). Survival of SrfloxP/loxPAlfpCre female mice, in contrast, dropped to 88.2% and SrfloxP/loxPAlfpCre male mice to 64.3% (Fisher’s exact test, p=0.024). These data demonstrate that loss of SRF function in hepatocytes results in postnatal lethality, which is most predominant in male mice.
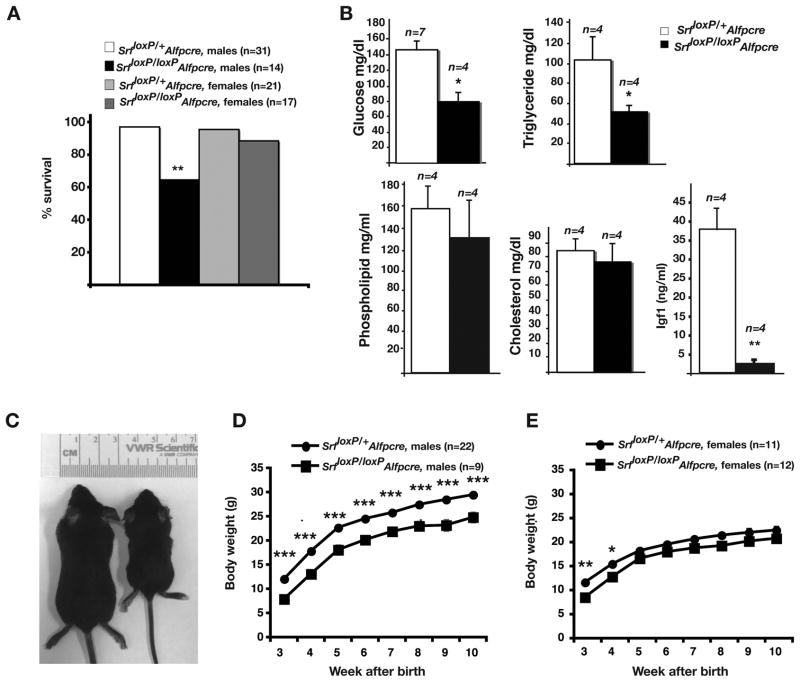
Figure 2. SrfloxP/loxPAlfpCre mice show increased mortality, disrupted serum glucose and triglyceride levels, and diminished body growth.
A) Bar graph showing percent of male and female Srf loxP/+Alfp-Cre and Srf loxP/loxPAlfp-Cre mice surviving until 10-weeks of age. Two-sided Fisher Exact Test, ** p=0.024. B) Bar graphs showing serum levels of glucose, triglycerides, phospholipids, cholesterol and IGF1 in control (Srf loxP/+Alfp-Cre) and mutant (Srf loxP/loxPAlfp-Cre) male mice at 3wks of age; Student’s t-test, * p≤0.01, ** p≤0.001. C) Photograph showing size comparison between control (Srf loxP/+Alfp-Cre; left) and mutant (Srf loxP/loxPAlfp-Cre; right) mice at 3-weeks of age. D, E) Graphs showing difference in body growth over time between control (circles) and mutant (squares) male (D) and female (E) mice. Tukey-Kramer post-hoc multiple comparison test (*p≤0.05, **p≤0.01, *** p≤0.005).
The liver has a central role in controlling nutrient metabolism, and so we considered the possibility that the increase in mortality among SrfloxP/loxPAlfpCre male mice could at least in part reflect a diminished capacity of the liver to control serum nutrient levels. We, therefore, measured the levels of glucose, triglyceride, phospholipid, and cholesterol in 3– week old control and SrfloxP/loxPAlfpCre male mice. As shown in Fig. 2b, although phospholipid and cholesterol levels were not significantly different, blood glucose and triglyceride levels in SrfloxP/loxPAlfpCre mice were significantly lower (Student’s t-test, p ≤ 0.01) than those in control mice. In SrfloxP/loxPAlfpCre mice, blood glucose levels were 54% and triglyceride levels were 49% of control mice. These results show that depletion of SRF in hepatocytes results in deficiencies in carbohydrate and lipid metabolism.
As shown in Fig. 2c, surviving SrfloxP/loxPAlfpCre male mice were considerably smaller than control mice. Although control and SrfloxP/loxPAlfpCre male mice gained weight over time, the weight of SrfloxP/loxPAlfpCre male mice remained significantly and reproducibly lower than those of control mice at any given point, and by 10 weeks surviving SrfloxP/loxPAlfpCre males weighed approximately 85% of controls (Fig. 2d). The weight of SrfloxP/loxPAlfpCre female mice, although less dramatic, was also reproducibly lower over the 10 week time course (Fig. 2e). The difference in weight between control and SrfloxP/loxPAlfpCre female mice was most significant at 3-weeks of age, with mutant mice weighing 73% of control mice, after which time the difference between control and mutant mice narrowed. Circulating IGF1 is a major determinant of body growth 26–28 and so we measured the levels of IGF1 in the serum of SRF-deficient male animals. At 1 week of age male IGF1 levels in SrfloxP/loxPAlfpCre mice were 46% of control levels (p ≤ 0.05; not shown) and by 3 weeks had dropped precipitously to 6% of control levels (p ≤ 0.001) (Fig 2b). Based on these data, we conclude that SRF is required in hepatocytes to maintain an environment that supports normal body growth.
SRF is required for proliferation and survival of hepatocytes
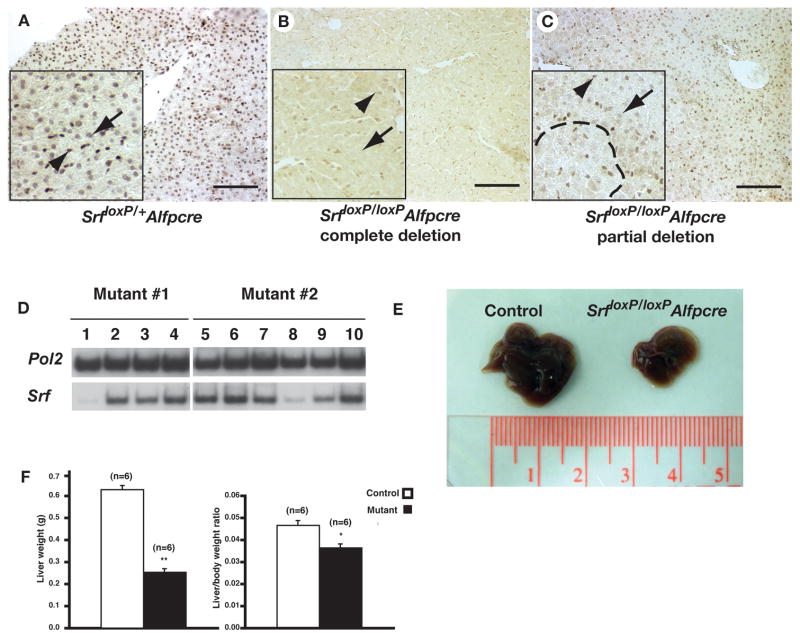
The observation that only a subset of SrfloxP/loxPAlfpCre mice died raised the possibility that there was variation in the efficiency through which Srf was deleted in these animals. We therefore examined expression of SRF protein in SrfloxP/loxPAlfpCre livers by immunohistochemistry (Fig. 3a–c). In contrast to control mice, where SRF was detected throughout the hepatic parenchyma (Fig. 3a), SRF was undetectable in the hepatocytes of a subset (<10%) of SrfloxP/loxPAlfpCre livers (Fig. 3b), regardless of where the section was taken. Such mice looked particularly sickly with standing fur and displaying hunched and lackadaisical behavior suggesting that complete loss of SRF in the liver is not compatible with long term survival and could account for the mortality recorded in Fig. 2a. In contrast to this small subset of sick mice in which SRF was undetectable, the majority (>90%) of the mutant mice displayed variability in the presence of SRF with areas of SRF positive-hepatocytes interspersed with areas of SRF negative hepatocytes (Fig. 3c). Moreover, when small independent regions of individual SrfloxP/loxPAlfpCre livers were isolated and analyzed by RT-PCR to detect Srf mRNA, although the majority of the isolates robustly expressed Srf, regions in which Srf levels were low to undetectable could be recovered (Fig. 3d).
Figure 3. Efficiency of SRF depletion is variable in SrfloxP/loxPAlfpcre livers.
A–C) Immunohistochemistry detecting SRF protein in livers of Srf loxP/+Alfp-Cre (A) and Srf loxP/loxPAlfp-Cre (B, C) mice. Inset shows high-resolution images with hepatocytes indicated by an arrow and endothelial cells by an arrowhead. Dashed line demarcates an area of SRF negative hepatocytes in C. Scale bar = 100μM. D) RT-PCR analysis on multiple liver fragments (numbered) from two Srf loxP/loxPAlfp-Cre mice (mutant #1 and #2) to detect Srf mRNA. Note reduced Srf mRNA levels in samples 1 and 8. Amplification using primers to detect RNA Polymerase II (Pol2) was used to normalize. E) Micrographs comparing the size of typical livers isolated from control (left) and Srf loxP/loxPAlfp-Cre (right) male mice. F) Bar graphs showing the weight of livers (left) and relative body weight to liver ratio (right) in Srf loxP/+Alfp-Cre (white) and Srf loxP/loxPAlfp-Cre (black) mice. Student’s t-test, * p≤0.005, ** p≤0.001.
Although the extent to which SRF was depleted from the livers of mutant mice was often minimal, surviving SrfloxP/loxPAlfpCre mice were consistently smaller. Importantly, the weight of the liver was reduced by 60% in SrfloxP/loxPAlfpCre male mice (p≤0.001) and the liver–to–body weight ratio was significantly lower in mutant male mice (p≤0.005) compared with controls (Figs. 3e–f). These observations seemed at first to contradict our finding that SRF persisted in the livers of the majority of SrfloxP/loxPAlfpCre mice. However, we reasoned that if SRF were necessary for hepatocyte viability then the action of Cre recombinase could result in loss of hepatocytes. This in turn could manifest in smaller livers that were perpetually in a state of regeneration in an attempt to restore normal liver mass. If SRF were also required for hepatocyte proliferation and the action of Cre was less than complete, then an environment would exist that selected for cells that maintained SRF. Such a model is not unprecedented since a similar situation exists for the Alb-uPA transgenic mouse in which hepatocytes that have undergone somatic rearrangement of the hepatotoxic Alb-uPA transgene selectively proliferate and replace the liver mass 29.
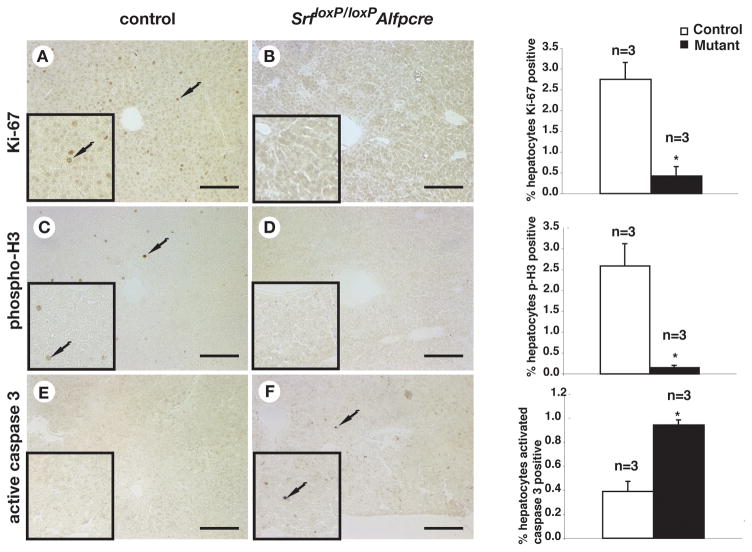
To test this model we first examined whether SRF was required for hepatocyte proliferation. Hepatocytes are generally quiescent in adult mice 30; however, in weanlings hepatocyte proliferation is still relatively high. Cell proliferation was therefore measured in 3-week old control and the rare SrfloxP/loxPAlfpCre mice that lacked detectable SRF expression using immunohistochemistry to identify the cell-cycle markers Ki-67 (G1/S) and phosphohistone-H3 (G2/M), along with the hepatocyte marker HNF4α (not shown). Fig. 4 shows that while proliferating hepatocytes were detectable in control livers, they were exceedingly infrequent in SRF-depleted livers, with depleted livers exhibiting a 16–fold reduction in phosphohistone H3 positive cells (p≤0.01) and a 6–fold reduction in Ki-67 positive cells (p≤0.01). We also measured the incidence of cell death using an antibody to detect activated caspase 3, which revealed a 2.4–fold increase in the number of apoptotic cells (p≤0.01) found in SrfloxP/loxPAlfpCre livers compared with controls (Fig. 4e, f).
Figure 4. SRF is essential for proliferation and viability of hepatocytes.
A–F) Immunohistochemistry detected Ki-67 (A,B), phosphohistone H3 (C,D), and activated caspase 3 (E,F) expressing cells in control (A,C,E) and Srf loxP/loxPAlfp-Cre (B,D,F) livers from three week old male mice. Insets show higher resolution images and adjacent bar graphs show cell counts from three independent control and mutant mouse livers Student’s t-test, *p≤0.05. Arrows indicate stained cells.
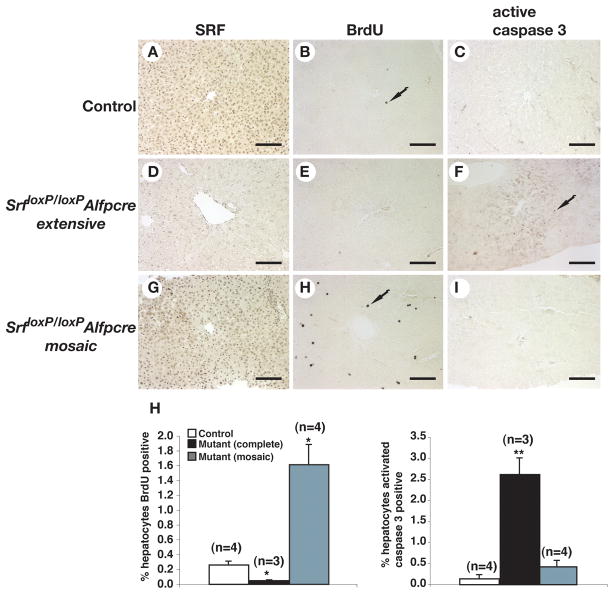
The observation that the absence of SRF results in both reduced proliferation and increased death of hepatocytes in 3–week old mice suggested that, in adult SrfloxP/loxPAlfpCre mice, an environment could exist that would select for proliferating SRF–positive cells. To address this possibility, we compared the extent of proliferation in adult control livers, in SrfloxP/loxPAlfpCre livers that exhibited extensive loss of SRF, as determined by immunohostochemistry for SRF expression in adjacent sections, and in SrfloxP/loxPAlfpCre livers that exhibited a partial loss of SRF, by detecting in vivo BrdU incorporation using immunohistochemistry (Fig. 5). The number of BrdU–positive cells was, as expected, relatively low in adult livers; however, extensive loss of SRF resulted in a 5–fold reduction in proliferating cells (p≤0.05) compared to control livers, which is consistent with the proliferation data obtained from 3 week old mice. When SrfloxP/loxPAlfpCre livers in which deletion of SRF was mosaic were examined, we recorded a significant 6–fold increase in BrdU–positive cells (p≤0.01) compared to control livers. In addition, cell death was examined in the same livers using immunohistochemistry to detect activated caspase 3 (Fig. 5). While the level of apoptotic cell death was relatively low in control hepatocytes and increased only slightly in SrfloxP/loxPAlfpCre mice in which deletion of SRF was mosaic, in SrfloxP/loxPAlfpCre livers harboring extensive loss of SRF, the number of apoptotic cells increased by 19–fold (p≤0.001) compared with control livers. Cumulatively, these analyses demonstrate that SRF is essential for normal hepatocyte proliferation, that the absence of SRF results in loss of hepatocytes by apoptotic cell death, and that the livers of the majority of SrfloxP/loxPAlfpCre mice are in a chronic state of regeneration.
Figure 5. SrfloxP/loxPAlfp-Cre mice are in a perpetual state of liver regeneration.
A–I) Immunohistochemistry detected SRF (A,D,G), incorporated BrdU (B,E,H) and activated caspase-3 (C,F,I) (arrows) in livers of adult control (A,B,C) mice or Srf loxP/loxPAlfp-Cre mice having undergone extensive (D,E,F) or mosaic (G,H,I) deletion of SRF. H) Bar graphs quantifying the number of cells incorporating BrdU or expressing activated caspase 3 in control and experimental animals. Comparison between control and mutant animals; Student’s t-test, *p≤0.01, **p≤0.001.
SRF is required for normal hepatocyte gene expression
Given that SRF is a transcription factor it seemed most likely that the deficiencies observed in SrfloxP/loxPAlfpCre livers reflected a disruption in hepatocyte gene expression. We, therefore, performed oligonucleotide array analyses on liver samples isolated from control and SrfloxP/loxPAlfpCre male mice at 3-weeks of age. Because we had observed mosaicism in depletion of SRF in the mutant mice, SrfloxP/loxPAlfpCre livers were dissected into small fragments and screened by RT-PCR to identify liver fragments with minimal SRF expression (see Fig. 3d). Total RNA isolated from liver fragments with low to undetectable levels of Srf mRNA collected from three independent SrfloxP/loxPAlfpCre mice were used to screen Affymetrix™ GeneChip™ arrays.
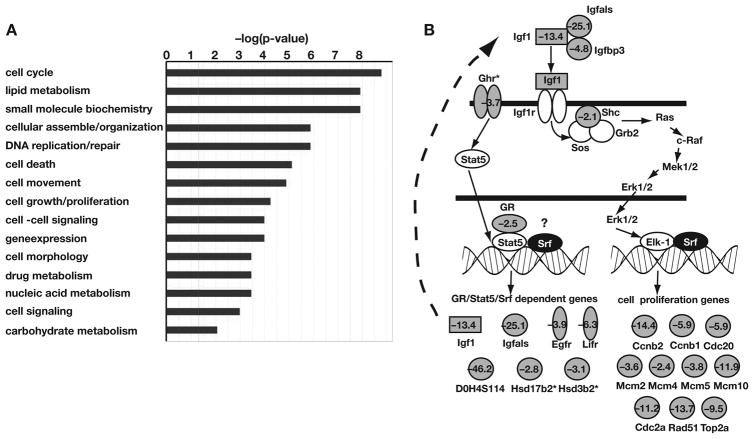
Supplementary tables 1 and 2 list the genes whose average mRNA level in the mutant mice changed by ≥2.0 fold (p≤0.05) compared with the control mice. Using these criteria expression of 294 genes decreased (Table S1) and 109 genes increased (Table S2) in mutant compared to control livers, suggesting that SRF is predominantly an activator of gene expression in hepatocytes. Annotation of these genes by gene ontology implicated SRF in controlling a wide variety of biological processes including cell cycle control, lipid metabolism, DNA replication, and cell death (Fig. 6A), which was consistent with our phenotypic characterization. Analyses of differentially expressed genes using Ingenuity Pathway Analyses™ software, which uses published reports to identify functional relationships between genes, revealed that the expression of cell cycle regulators was particularly impacted by loss of SRF in the liver with changes in expression of 57 mRNAs encoding molecules with a variety of roles in controlling cell proliferation identified. This included disrupted expression of a network of factors whose components center on E2F–mediated cell cycle control and act downstream of Insulin Growth Factor 1 (IGF1) (Fig. 6b). Importantly expression of several mRNAs encoding proteins with integral roles in controlling body growth were substantially reduced, including Growth Hormone Receptor (3.7–fold), Insulin-like Growth Factor 1 (13.4–fold), IGF acid labile subunit (25.1–fold), IGF binding protein 3 (4.8–fold) and the glucocorticoid receptor (2.5–fold). In addition to the disruption of the Growth Hormone/IGF1 pathway and cell cycle regulators, we observed alterations in expression of many genes encoding proteins with other key roles in hepatocyte function, including 38 proteins with roles in lipid metabolism and 10 CypP450 enzymes. While these data support a direct role for SRF in controlling hepatic cell proliferation, they also demonstrate that SRF is an important regulator of hepatocyte function and liver homeostasis.
Figure 6. Loss of SRF disrupts hepatic gene expression.
A) Gene array analyses were performed on liver fragments from control and SrfloxP/loxPAlfpcre livers that had low to undetectable levels of Srf mRNA by RT–PCR. Genes whose expression was significantly (p≤0.05) changed in SRF null livers were categorized based on gene function by Ingenuity Pathway Analysis (IPA). Fischer’s exact test was used to calculate a p-value determining the probability that each function assigned to the data set is due to chance alone. The x-axis lists the top cellular and molecular pathways affected by the loss of SRF in the liver. The y-axis expresses the statistical significance–log(p-value). B) Schematic showing that several genes encoding factors required for IGF1–mediated cell cycle control were negatively affected (grey symbols with fold change) by loss of SRF. (*) denote fold differences that approached significance, but where p>0.05.
Discussion
Our data reveal a crucial role for the transcription factor SRF in regulating liver homeostasis and function; however, the molecular mechanisms underlying the phenotypes associated with loss of hepatic SRF appear to be complex. While we have ascertained that loss of SRF has a significant impact on body growth, whether this represents a direct or indirect effect of loss of SRF is more difficult to definitively establish. Analysis of mRNA in SrfloxP/loxPAlfpCre liver fragments lacking SRF revealed that multiple members of the growth hormone (GH)/IGF1 signaling pathway, which is known to control body growth, were substantially reduced compared to control livers (Fig. 6b). This is provocative because growth hormone receptor–deficient mice 31, liver IGF1 deficient (LID)/acid labile subunit knockout (ALSKO) mice 28, and liver glucocorticoid deficient mice 32 – genes encoding mRNAs whose levels are reduced 3.7, 13.4, 25.1, and 2.5–fold in SRF–deficient hepatocytes, respectively–also exhibit severe defects in post-natal body growth. However, although IGF1 levels are severely depressed in SrfloxP/loxPAlfpCre mice we have shown that, due to selective pressure, the majority of hepatocytes in most SrfloxP/loxPAlfpCre animals continue to express SRF as well as the identified SRF–dependent genes, including IGF1, ALS, and IGFBP3 (not shown). Thus the depletion of serum IGF1 in SrfloxP/loxPAlfpCre mice is likely independent of any regulation of genes involved in IGF signaling by SRF. We, therefore, favor an alternative model to explain the small stature of SrfloxP/loxPAlfpCre mice. In this model the reduction of circulating IGF1 and consequently growth of the mouse is due to the chronic and severe reduction in total liver mass in the mutant animals. Our studies reveal that 90% of SrfloxP/loxPAlfpCre male mice examined appear to exist in a perpetual state of regeneration; however, the size of the livers in these animals is 30% that of normal. Under these circumstances one would predict that the circulating levels of IGF1 and, importantly, its regulatory binding proteins ALS and IGFBP3 would be reduced in mutant mice compared to livers of normal size. Since ALS and IGFBP3 serve to increase IGF1 half-life then a reduction in these proteins, which are primarily synthesized by the liver, should also result in destabilization of both hepatic and peripheral sources of IGF1 resulting in an exacerbated decline in serum IGF1 (Fig. 2b) and growth reduction.
We also demonstrate that SRF is required for expression of several hepatic mRNAs with roles in various aspects of liver physiology. We believe that the observed disruption to hepatic gene expression associated with loss of SRF appears to be cell autonomous because gene array analyses of regions of liver that expressed SRF in SrfloxP/loxPAlfpCre mice were distinct from the changes observed when SRF was absent (not shown). The reduction in expression in some genes is likely to reflect reduced expression of Growth Hormone (GHR) and glucocorticoid (GR) receptors (Fig 6). Growth hormone induces expression of genes through activation of STAT5a/b in concert with GR 32–34. Recent studies in mice lacking either Stat5a/b or GR specifically in hepatocytes have identified a core set of 17 genes whose expression is dependent upon STAT5-GR interactions 35. Of these 17 genes, expression of 7 (Igf1, Igfals, Egfr, Lifr, D0H4S114, Hsd17b2, and Hsd3b2) are also reduced in SrfloxP/loxPAlfpCre mice. Whether this is entirely due to a reduction in GHR and GR or whether SRF also directly controls their expression is yet to be established.
Liver regeneration studies have implicated both autocrine and paracrine signaling mediated by IGF1/GH in controlling hepatocyte proliferation 36,37. IGF1 signals through the Igf1r tyrosine kinase, which activates PI3-kinase and Ras/MEK/ERK (Fig. 6) signal transduction pathways, leading to control of genes involved in cell proliferation and in protecting against apoptotic cell death 38. Elk1, which is a target of Ras/MEK/ERK signaling, has been shown to act in conjunction with SRF to control expression of genes involved in the cell cycle and DNA replication (Fig. 6) 39. This would imply that the block in hepatocyte proliferation observed in SrfloxP/loxPAlfpCre mice could reflect reduced expression of cell cycle genes caused both by loss of IGF1 signaling as well the absence of direct regulation by Elk1/SRF. This model could presumably be addressed by defining the complete repertoire of SRF target genes using genome scale analyses 40.
In summary our data support the proposal that SRF is essential for maintenance of hepatocyte viability and proliferation and that SRF regulates expression of several genes with roles in hepatic function. The fact that livers in SrfloxP/loxPAlfpCre mice exist in a chronic state of regeneration and that stocks of these animals can be maintained with relative ease suggests that these animals could provide a useful model to conduct exogenous cell transplants and facilitate the study of liver progenitor cells. Although our current analyses focused on animals that survived embryonic development, we noted that a subset of SrfloxP/loxPAlfpCre mice, around 30%, died during embryogenesis presumably reflecting a role for SRF in the developing liver. Our results are in contrast with those of Latasa et al who reported that disruption of SRF in hepatocytes resulted in healthy animals born at expected Mendelian ratios 21. The most likely explanation for these differences is that the placement of loxP sites within the Srf gene differs between the strains of mice used in the two studies and therefore could potentially impact the efficiency through which Cre mediates deletion of the Srf gene. The position of loxP sites has been shown recently to quite dramatically influence the efficiency of recombination when Cre activity is analyzed using different reporter mouse strains 41. Regardless, both studies are illuminating with regard to the role of SRF in controlling liver function and the current analyses extend our understanding of the action of SRF on controlling hepatocyte cell proliferation and viability and are consistent with a role for SRF during liver regeneration.
Materials and Methods
Animals
All mice were housed in the MCW Animal Research Center and all procedures obtained IACUC approval. 3-week-old mice, fed ad libitum, were sacrificed to harvest serum and phospholipid, cholesterol, triglyceride, glucose, and IGF1 levels were determined by the Mouse Metabolic Phenotyping Center at the University of Cincinnati.
Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
Reverse Transcriptase PCR was performed as described previously 42 with primers: Hprt AGCGCAAGTTGAATCTGC, AGCGACAATCTACCAGAG); Pol2 CTGATGCGGGTGCTGAGTGAGAAGG, GCGGTTGACCCCATGACGAGTG); Srf (8750F-8995R) AGATCCCTGTCTCTGCAGTTCAGC, GCGTGGCATCCAGGTTCA; Srf (146F-395R) CCATAGGGGCAGGAAAGTGAGG, GGGGTCCGGTTCAGGTT.
Immunohistochemistry
Immunohistochemistry was performed as described previously 42 using the following primary antibodies: anti-HNF4α (Santa Cruz sc-6556, 1:500), anti-SRF (Santa Cruz sc-335, 1:500), anti-SRF (Dr. Misra, 1:2000), anti-Ki-67 (Santa Cruz sc-7846, 1:500), anti-phospho-Histone H3 (Upstate #06–570, 1:1500), anti-activated Caspase-3 (BD Pharmingen 559565, 1:100), anti-BrdU (Accurate Chemical and Scientific Corporation, 1/100). Cell counts using ImageJ software were taken from digital images of 3 sections from each mouse analyzed. To identify cells in S–phase, 50mg/kg body weight of BrdU (Sigma) was introduced into eight-week-old mice by intraperitoneal injection for 2 hours.
Gene expression profiling
Total RNA (15μg) was isolated from three independent control (SrfloxP/+AlfpCre) and mutant (SrfloxP/loxPAlfpCre) 3-week-old mouse liver fragments using RNeasy (Qiagen) and used to prepare biotinylated cRNA following procedures described by Affymetrix. After hybridization to GeneChip Mouse Genome 430 2.0 arrays (Affymetrix), data were collected using GeneChip Operating Software (GCOS) from Affymetrix and dChip v.1.3 software 43 and analyzed using Ingenuity Pathways Analysis (IPA) software.
Supplementary Material
Acknowledgments
Funding for this project was provided by grants from National Institutes of Health (DK66226 and DK55743) and Advancing a Healthier Wisconsin to S.A.D., as well as gifts from the Marcus Family, Sophia Quadracci Memorial Fund, the MCW Digestive Disease center, and Jack and Phoebe Lewis. M.A.B. was the recipient of an NRSA from NIDDK. The authors would like to thank David Ginty and Klaus Kaestner for providing mice and Cheryl Stucky for advice with statistical analysis.
References
- 1.Chai J, Tarnawski AS. Serum response factor: discovery, biochemistry, biological roles and implications for tissue injury healing. J Physiol Pharmacol. 2002;53:147–157. [PubMed] [Google Scholar]
- 2.Sharrocks AD, von Hesler F, Shaw PE. The identification of elements determining the different DNA binding specificities of the MADS box proteins p67SRF and RSRFC4. Nucleic Acids Res. 1993;21:215–221. doi: 10.1093/nar/21.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shore P, Sharrocks AD. The MADS-box family of transcription factors. Eur J Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 4.Belaguli NS, Schildmeyer LA, Schwartz RJ. Organization and myogenic restricted expression of the murine serum response factor gene. A role for autoregulation. J Biol Chem. 1997;272:18222–18231. doi: 10.1074/jbc.272.29.18222. [DOI] [PubMed] [Google Scholar]
- 5.Greenberg ME, Ziff EB. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 6.Hipskind RA, Baccarini M, Nordheim A. Transient activation of RAF-1, MEK, and ERK2 coincides kinetically with ternary complex factor phosphorylation and immediate-early gene promoter activity in vivo. Mol Cell Biol. 1994;14:6219–6231. doi: 10.1128/mcb.14.9.6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodge C, Liao J, Stofega M, et al. Growth hormone stimulates phosphorylation and activation of elk-1 and expression of c-fos, egr-1, and junB through activation of extracellular signal-regulated kinases 1 and 2. J Biol Chem. 1998;273:31327–31336. doi: 10.1074/jbc.273.47.31327. [DOI] [PubMed] [Google Scholar]
- 8.Shaw PE, Schroter H, Nordheim A. The ability of a ternary complex to form over the serum response element correlates with serum inducibility of the human c-fos promoter. Cell. 1989;56:563–572. doi: 10.1016/0092-8674(89)90579-5. [DOI] [PubMed] [Google Scholar]
- 9.Kovary K, Bravo R. The jun and fos protein families are both required for cell cycle progression in fibroblasts. Mol Cell Biol. 1991;11:4466–4472. doi: 10.1128/mcb.11.9.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riabowol KT, Vosatka RJ, Ziff EB, et al. Microinjection of fos-specific antibodies blocks DNA synthesis in fibroblast cells. Mol Cell Biol. 1988;8:1670–1676. doi: 10.1128/mcb.8.4.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poser S, Impey S, Trinh K, et al. SRF-dependent gene expression is required for PI3-kinase-regulated cell proliferation. EMBO J. 2000;19:4955–4966. doi: 10.1093/emboj/19.18.4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arsenian S, Weinhold B, Oelgeschlager M, et al. Serum response factor is essential for mesoderm formation during mouse embryogenesis. Embo J. 1998;17:6289–6299. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parlakian A, Tuil D, Hamard G, et al. Targeted inactivation of serum response factor in the developing heart results in myocardial defects and embryonic lethality. Mol Cell Biol. 2004;24:5281–5289. doi: 10.1128/MCB.24.12.5281-5289.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balza RO, Jr, Misra RP. Role of the serum response factor in regulating contractile apparatus gene expression and sarcomeric integrity in cardiomyocytes. J Biol Chem. 2006;281:6498–6510. doi: 10.1074/jbc.M509487200. [DOI] [PubMed] [Google Scholar]
- 15.Miano JM, Ramanan N, Georger MA, et al. Restricted inactivation of serum response factor to the cardiovascular system. Proc Natl Acad Sci U S A. 2004;101:17132–17137. doi: 10.1073/pnas.0406041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu Z, Yu W, Zhang SX, et al. Conditional mutagenesis of the murine serum response factor gene blocks cardiogenesis and the transcription of downstream gene targets. J Biol Chem. 2005;280:32531–32538. doi: 10.1074/jbc.M501372200. [DOI] [PubMed] [Google Scholar]
- 17.Wiebel FF, Rennekampff V, Vintersten K, et al. Generation of mice carrying conditional knockout alleles for the transcription factor SRF. Genesis. 2002;32:124–126. doi: 10.1002/gene.10049. [DOI] [PubMed] [Google Scholar]
- 18.Li S, Czubryt MP, McAnally J, et al. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc Natl Acad Sci U S A. 2005;102:1082–1087. doi: 10.1073/pnas.0409103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alberti S, Krause SM, Kretz O, et al. Neuronal migration in the murine rostral migratory stream requires serum response factor. Proc Natl Acad Sci U S A. 2005;102:6148–6153. doi: 10.1073/pnas.0501191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramanan N, Shen Y, Sarsfield S, et al. SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat Neurosci. 2005;8:759–767. doi: 10.1038/nn1462. [DOI] [PubMed] [Google Scholar]
- 21.Latasa MU, Couton D, Charvet C, et al. Delayed liver regeneration in mice lacking liver serum response factor. Am J Physiol Gastrointest Liver Physiol. 2007;292:G996–G1001. doi: 10.1152/ajpgi.00493.2006. [DOI] [PubMed] [Google Scholar]
- 22.Duncan SA, Manova K, Chen WS, et al. Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc Natl Acad Sci USA. 1994;91:7598–7602. doi: 10.1073/pnas.91.16.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taraviras S, Monaghan AP, Schutz G, et al. Characterization of the mouse HNF-4 gene and its expression during mouse embryogenesis. Mech Dev. 1994;48:67–79. doi: 10.1016/0925-4773(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 24.Lee CS, Sund NJ, Behr R, et al. Foxa2 is required for the differentiation of pancreatic alpha-cells. Dev Biol. 2005;278:484–495. doi: 10.1016/j.ydbio.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 25.Yokoyama Y, Nimura Y, Nagino M, et al. Current understanding of gender dimorphism in hepatic pathophysiology. J Surg Res. 2005;128:147–156. doi: 10.1016/j.jss.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 26.Clemmons DR. Modifying IGF1 activity: an approach to treat endocrine disorders, atherosclerosis and cancer. Nat Rev Drug Discov. 2007;6:821–833. doi: 10.1038/nrd2359. [DOI] [PubMed] [Google Scholar]
- 27.LeRoith D. Clinical relevance of systemic and local IGF-I: lessons from animal models. Pediatr Endocrinol Rev. 2008;5 (Suppl 2):739–743. [PubMed] [Google Scholar]
- 28.Yakar S, Rosen CJ, Beamer WG, et al. Circulating levels of IGF-1 directly regulate bone growth and density. J Clin Invest. 2002;110:771–781. doi: 10.1172/JCI15463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandgren EP, Palmiter RD, Heckel JL, et al. Complete hepatic regeneration after somatic deletion of an albumin-plasminogen activator transgene. Cell. 1991;66:245–256. doi: 10.1016/0092-8674(91)90615-6. [DOI] [PubMed] [Google Scholar]
- 30.Steer CL. Liver Regeneration. FASEB J. 1995;9:1396–1400. doi: 10.1096/fasebj.9.14.7589980. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Xu BC, Maheshwari HG, et al. A mammalian model for Laron syndrome produced by targeted disruption of the mouse growth hormone receptor/binding protein gene (the Laron mouse) Proc Natl Acad Sci U S A. 1997;94:13215–13220. doi: 10.1073/pnas.94.24.13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tronche F, Opherk C, Moriggl R, et al. Glucocorticoid receptor function in hepatocytes is essential to promote postnatal body growth. Genes Dev. 2004;18:492–497. doi: 10.1101/gad.284704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holloway MG, Cui Y, Laz EV, et al. Loss of sexually dimorphic liver gene expression upon hepatocyte-specific deletion of Stat5a-Stat5b locus. Endocrinology. 2007;148:1977–1986. doi: 10.1210/en.2006-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Udy GB, Towers RP, Snell RG, et al. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci U S A. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engblom D, Kornfeld JW, Schwake L, et al. Direct glucocorticoid receptor-Stat5 interaction in hepatocytes controls body size and maturation-related gene expression. Genes Dev. 2007;21:1157–1162. doi: 10.1101/gad.426007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desbois-Mouthon C, Wendum D, Cadoret A, et al. Hepatocyte proliferation during liver regeneration is impaired in mice with liver-specific IGF-1R knockout. FASEB J. 2006;20:773–775. doi: 10.1096/fj.05-4704fje. [DOI] [PubMed] [Google Scholar]
- 37.Pennisi PA, Kopchick JJ, Thorgeirsson S, et al. Role of growth hormone (GH) in liver regeneration. Endocrinology. 2004;145:4748–4755. doi: 10.1210/en.2004-0655. [DOI] [PubMed] [Google Scholar]
- 38.Dupont J, LeRoith D. Insulin and insulin-like growth factor I receptors: similarities and differences in signal transduction. Horm Res. 2001;55 (Suppl 2):22–26. doi: 10.1159/000063469. [DOI] [PubMed] [Google Scholar]
- 39.Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 40.Sun Q, Chen G, Streb JW, et al. Defining the mammalian CArGome. Genome Res. 2006;16:197–207. doi: 10.1101/gr.4108706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma Q, Zhou B, Pu WT. Reassessment of Isl1 and Nkx2–5 cardiac fate maps using a Gata4-based reporter of Cre activity. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garrison WD, Battle MA, Yang C, et al. Hepatocyte nuclear factor 4alpha is essential for embryonic development of the mouse colon. Gastroenterology. 2006;130:1207–1220. doi: 10.1053/j.gastro.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schadt EE, Li C, Ellis B, et al. Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J Cell Biochem Suppl. 2001;(Suppl 37):120–125. doi: 10.1002/jcb.10073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.