Abstract
Retinal degeneration resulting in the loss of photoreceptors is the leading cause of blindness. Several therapeutic protocols are under consideration for treatment of this disease. Tissue replacement is one such strategy currently being explored. However, availability of tissues for transplant poses a major obstacle. Another strategy with great potential is the use of adult stem cells, which could be expanded in culture and then utilized to engineer retinal tissue. In this study, we have explored a spontaneously immortalized human retinal progenitor cell line for its potential in retinal engineering using rotary cultures to generate three-dimensional (3D) structures. Retinal progenitors cultured alone or cocultured with retinal pigment epithelial cells form aggregates. The aggregate size increases between days 1 and 10. The cells grown as a 3D culture rotary system, which promotes cell–cell interaction, retain a spectrum of differentiation capability. Photoreceptor differentiation in these cultures is confirmed by significant upregulation of rhodopsin and AaNat, an enzyme implicated in melatonin synthesis (immunohistochemistry and Western blot analysis). Photoreceptor induction and differentiation is further attested to by the upregulation of rod transcription factor Nrl, Nr2e3, expression of interstitial retinal binding protein, and rhodopsin kinase by reverse transcription–polymerase chain reaction. Differentiation toward other cell lineages is confirmed by the expression of tyrosine hydroxylase in amacrine cells, thy 1.1 expression in ganglion cells and calbindin, and GNB3 expression in cone cells. The capability of retinal progenitors to give rise to several retinal cell types when grown as aggregated cells in rotary culture offers hope that progenitor stem cells under appropriate culture conditions will be valuable to engineer retinal constructs, which could be further tested for their transplant potential. The fidelity with which this multipotential cell line retains its capacity to differentiate into multiple cell types holds great promise for the use of tissue-specific adult stem cells for therapy.
Introduction
Retinal degeneration with the loss of photoreceptors is the leading cause of blindness.1–4 Several therapeutic protocols are under consideration for the treatment of the disease.5–7 Tissue replacement/drug delivery is one such strategy, but most transplanted tissues (cells, fetal retinal sheets, etc.) fail to integrate into the host retina.8–10 Additionally, availability and the use of donor tissues will raise further problems, both moral and ethical. One might also speculate that the lack of integration of donor tissue could be due to the architecture of cells and/or tissues transplanted and loss of possible architecture in the host tissue.11,12 Even if tissue replacement were to work, the availability of donor tissues, however, will remain a major impediment in the achievement of this strategy. It may be necessary to explore cell populations (stem cells), which may have the potential to be expanded in vitro and assembled into tissue of choice. When transplanted, these tissues will have the ability to integrate into degenerating structures and/or at least rescue the degenerating retina by secreting neurotrophic factors.
The ability to maintain and expand cultures of undifferentiated progenitors from postmortem fetuses has been expounded by others.13 These authors report that human retinal progenitors give rise to retinal cell types depending on the gestational age of the fetuses.13 Lamba et al.14 reported that human embryonic stem cells can, under appropriate culture conditions, be coaxed to develop into retinal neurons similar to adult retinal stem cells. Although replacement of damaged tissue by fetal tissues and postmortem donor tissue13–15 holds promise, availability and moral ethical issues will pose problems and constraints. As such, the availability of adult stem cells that can be expanded in culture and grown as three-dimensional (3D) constructs seems a pragmatic choice. The rationale for use of adult stem cells for replacement of damaged tissue is strengthened by the identification of neural stem cells that could serve as a potential source for replacement.16–18 It has been reported that when transplanted in retina, neural stem cells are able to differentiate into both neurons and glia.16–18 Neural stem cells, though capable of producing photoreceptors, are often immature and fail to express rhodopsin.19 As such, retinal progenitors will be more suitable and have been identified in rodent retina.20 Increasingly, data confirm that multipotential, multilineage progenitors exist even in brain and retinal tissues of humans. Further, adult stem cells have been identified in retina that are ideal for tissue transplant and/or engineering retina.20–22 Muller-glia and retinal pigment epithelial cells (RPE) are also being explored for their potential in tissue replacement.23,24 Tissue engineering as a field is gaining momentum in the treatment of a variety of diseases (liver, pancreas, skin, bone, etc.).25,26 The success achieved by Atala with construction of urinary bladder and other tissues currently in clinical use is phenomenal and raises hope for construction of other complex tissues in vitro.27 The question that still remains is, if tissues can be generated from tissue-specific stem cells. Great progress has been made with 3D cultures of freshly, dissociated tissues. Can the same be achieved with embryonic stem cells, fetal progenitor cells, adult stem cells, and/or cell lines?
In the past, we have reported that multipotential retinal cell lines (spontaneously immortalized and SV-40T immortalized) can grow in the NASA-developed horizontally rotating culture vessel (bioreactor) and form 3D retina-like structures. Various retinal cell types can be identified in these constructs morphologically and biochemically, with some photoreceptors expressing rudimentary outer segments.28,29 The model system, though very useful, had some problems. Cells had to be maintained on cytodex beads that were unsuitable for transplant. Because no suitable biodegradable substrates have been identified to grow these cells in the bioreactor, we have explored other options. In this study, we have explored the substrate-free, 3D rotary culture initially reported by Moscona30 in the sixties and seventies and subsequently reassessed and improved by others.31,32 Sophisticated models of “Retinospheroids, stratospheroids” culture methodology have illuminated not only the role of cell–cell interaction and recognition in retinal development, but also the role played by RPE.31
In this study, retinal progenitors (nontransformed cell line) were grown alone or as a coculture with RPE cell line (D407) as aggregate culture for days 1–10. These aggregates showed a higher degree of maturation reflected by upregulation of several proteins expressed in mature retinal neurons, including photoreceptors. The emergence of photoreceptors from progenitors was also further confirmed by the expression of Nrl, rhodopsin kinase, and interstitial retinal binding protein by reverse transcription–polymerase chain reaction (RT-PCR). Additionally, maturation toward photoreceptor phenotype was substantiated by the expression of D2, D3, and D4 receptors localized to photoreceptors by immunocytochemistry and cone β transducin GNB3 expressed in cones. Further, the presence of ganglion cells and amacrine cells in cell aggregates was confirmed by the expression of thy 1.1 and tyrosine hydroxylase, respectively (Western blot and immunohistochemistry). The presence of cone photoreceptors was confirmed by the expression of calbindin protein and transducin GNB3, and the presence of bipolar neurons was confirmed by the expression of protein kinase C-α (PKCα) (immunohistochemistry and Western blot analysis).
A higher degree of differentiation and histotypic reaggregation is usually demonstrated in primary cultures of dissociated retinal fetal and adult cells when grown as 3D cultures. What is unique in this study is that we report a cell line that has been in culture for years, is closer to the in vivo developmental state of developing retina, and is still capable of such reproductive fidelity when grown as a 3D model system. This provides hope that stem cells, when grown in 3D constructs, might prove a very efficient source of tissue repair.
Materials and Methods
Cell culture
Retinal progenitor cells (cell line)33,34 in passage 45–47 were grown alone at a density of 2 × 106 cells/5 mL medium in a 25 mL conical flask on a rotary shaker at rpm 90 at 37°C or cocultured with RPE cell line D407 at passage 67 (1 × 105 cells/5 mL medium in 25 mL flask). Fresh medium was added every 24 h. The cells were maintained routinely in Dulbecco's modified Eagle's medium–nutrient mixture F-12 (Ham) 1:1 (Gibco, Bethesda, MD) supplemented with 10% fetal bovine serum (Hyclone Laboratories, Logan, UT), 10% serum plus supplement (JRH Biosciences, Lenexa, KS), 2 mM L-glutamine, 0.075% sodium bicarbonate, penicillin 100 units/mL, and streptomycin 100 μg/mL (Gibco). Samples were collected on days 1, 3, 5, 7, and 10 for scanning immunophenotyping, RT-PCR, and Western blot analysis, and processed by our previously published protocols.27,28 D407 cell line is a generous gift from Dr. Richard Hunt, Department of Microbiology, University of South Carolina, Medical School, Columbia, SC.
Immunophenotyping of cells grown as rotary cultures
Cells grown in rotary cultures were either characterized as aggregates or as dissociated cells after mild trypsinization. Both aggregates and dissociated cells were allowed to attach on polylysine-coated coverslips. Immunophenotyping was performed 24 h postattachment. Cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min at room temperature and permeabilized with 0.2% triton × 100 at 37°C for 10 min. Nonspecific binding was blocked by treatment with 10% goat serum overnight at 4°C or at 37°C for 1 h. Controls were included in each determination by the omission of primary and secondary antibodies. Cells were reacted with primary antibody at room temperature for 1 h and secondary antibody for 1 h at room temperature. Secondary antibodies used were goat anti-rabbit Alexa fluor 488 dilution (1:500) color green, and goat anti-rabbit Alexa fluor 594 dilution (1:500) color red (Molecular Probes, Eugene, Oregon). Nuclear staining is DAP1 blue. All proteins are green except neurofilament protein, which is identified by the color red. Superimposed blue (DAP1) over red neurofilament proteins looks purplish in color. Primary antibodies PKCα, D4 receptor, NF-L, NF-M, calbindin, tyrosine hydroxylase, and thy 1.1 were procured from Santa Cruz (Santa Cruz, CA) and were used at a dilution of 1:50 and 1:200 for immunohistochemistry, and 1:200 and 1:2000 for Western blot analysis. Antibodies AaNat, rhodopsin 1D4, Nr2e3, GNB3, and D2D3 were procured from Chemicon (Temecula, CA) and used at a dilution of 1:200 for immunohistochemistry and at 1:1000 for Western blot analysis. NSE was procured from Zymed (San Francisco, CA) and used at a 1:500 dilution, and actin antibody from Sigma-Aldric Corp. (St. Louis, MO) was used at a 1:500 dilution.
Western blot analysis
Whole-cell extracts were prepared from the rotating cultured cells. Samples of cell aggregates, retinal progenitors, and cocultures of retinal progenitors with D407 were collected from the rotating culture at specific times. The cell aggregates were washed thrice with PBS and were lysed with lysis buffer, which consisted of 20 mM Tris, pH 7.5, 150 mM sodium chloride (NaCl), 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM ethylene glycol tetraacetic acid (EGTA), 1% Triton X-100, 0.2 mM 4(2 Aminoethyl) benzene sulfonyl fluoride hydrochloride (AEBSF), 0.5 mM Benzamidine, 2 μg/mL Aprotinin, 0.5 mM Leupeptin, and 5 mg/mL pepstatin A. Cell debris and detergent insoluble material were removed by centrifugation at 15,000 rpm for 15 min at 4°C. The protein content of the whole-cell extract was determined using the BCA protein assay reagent kit (Pierce, Rockford, IL) according to manufacturer's instructions. Whole-cell extract containing 50 μg proteins was separated by electrophoresis on 10% sodium dodecyl sulphate polyacrymide gels. After the separation, the proteins were electrophoretically transferred to a polyvinylidene difluoride membrane (Immobilon R; Millipore, Bedford, MA). The nonspecific binding sites were blocked by immersing the membrane in 5% (W/V) nonfat dry milk powder in tris-buffered saline-tween-20 (TBS-T) (25 mM Tris-HC1, pH 6.5, 150 mM NaCl, and 0.05% Tween-20) for 2 h at room temperature. The antibody for neuron-specific enolase was obtained from Zymed, and all other primary and secondary antibodies were purchased from Chemicon. Expression of specific proteins was determined by binding to specific primary antibodies (dilution is as indicated), PKCα (1:500), NSE (1:500), TH (1:100), Calbindin (1:500), and Dopamine receptor D4 antibodies (1:300). Detection was done with enhanced chemiluminescence (Amersham, Arlington Heights, IL) using secondary antibodies conjugated with peroxidase and exposed to X-ray film. The level of expression was determined using densitometer and Scion Image analysis software (Frederick, MD).
RT-PCR analysis
Aggregates of progenitors and a coculture of progenitors with D407 were harvested from the rotating cultures at specified times. The aggregates were washed thrice with PBS. Total cellular RNA was isolated by adding Trizol reagent (Gibco Industries Inc., Langley, OK) directly to the cells or aggregates according to manufacturer's recommendations. The yield of total RNA obtained was determined spectrophotometrically at A260/280 nm. All reagents were purchased from Perkin Elmer (Waltham, MA). The sequence of primer pairs was obtained from the GenBank database (see Table 1). Two μg of total RNA from each sample was reverse transcripted using random hexamer primer according to the manufacturer's specifications. The amplification reactions were carried out in a total volume of 100 μL with 5 units of Taq DNA polymerase. Typically, after denaturation of 5 min at 94°C, samples were subject to 35 cycles of denaturation (30 s at 94°C), annealing (60 s at 55°C), and elongation (60 s at 72°C), followed by a final extension at 72°C for 5 min. For determination of quality and PCR condition, β-actin was used as a control. Ten μL of PCR products was obtained on indicated cycles, and reactions were stopped with an equal amount of 2× sample buffer. All RT-PCR determinations were routinely controlled by conducting PCR, by omitting the reverse transcription and by including a PCR negative control in which all the components were included but no cDNA template was added. RT-PCR reaction products were separated by electrophoresis on a 2% agarose gel, and the ethidium bromide–stained bands were visualized by ultra violet transillumination, and quantified. The levels of expression were determined by densitometry. All the images were saved in TIFF format. The target PCR products were analyzed by determining the intensity of bands by Scion Image, which is a PC version of the image analysis software initially developed by the National Institutes of Health (NIH).
Table 1.
Primers for Reverse Transcription-Polymerase Chain Reaction
| Primers | Sequence (5′–3′) | Size (bp) |
|---|---|---|
| NRL | Forward: ggg ggc tgg gga ggc att gggf | 347 |
| Reverse: agc tgg gcg gcc agg cgg g | ||
| Rhodopsin kinase | Forward: ccc tcc gcg aca gcc tca gcc | 371 |
| Reverse: tca gga agt aca ggc tgc cc | ||
| IRBP | Forward: cag gaa gcc atc cag cag gcc | 380 |
| Reverse: gcc tcc tgt gca gtg ccg g | ||
| mGluR6 | Forward: gcc gct gaa gaa gga gca g | 211 |
| Reverse: gga gac gga gct ggc cga ggc g | ||
| β-Actin | Forward: agc tta tgg atg atg ata tcg c | 591 |
| Reverse: ctt aat gtc acg cac gat ttc c |
Results
Description of the nontransformed human progenitor cell line
We have previously established and characterized a cell line from spontaneously aborted human fetus (first trimester) by SV-40T transfection.33 While we were establishing primary cultures for another cell line, one clone (208) became spontaneously immortalized and was subsequently cloned and subcloned establishing a cell line.34 We have been able to passage these cells up to the 65th passage. This cell line is very similar to our SV-40T immortalized retinal cell line KGLDMSM. The cell line grows in a contact inhibited manner with a flat layer of precursors and cells with neuronal phenotype (arrows) overlaying the flat layer of cells (Fig. 1). We have looked for the expression of SV-40T protein in this nontransformed cell line to rule out inadvertent exposure. The results were negative (data not included).
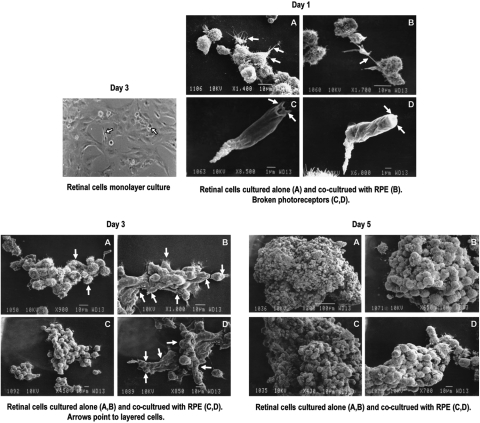
FIG. 1.
Morphological analysis of cells grown as monolayer or in rotary culture by phase and scanning electron micrography. Note the flat layer of cells in monolayer culture overlayed by neurons (arrows). The increasing size of the aggregates in cultures is evident between days 1 and 7 (scanning electron microscopy). No significant differences were noted in aggregates generated by coculture of progenitors with RPE. Some degree of layering is seen [Day 3 (B) and (D), arrows]. In Day 1 (C) and (D) (arrows), note broken photoreceptors.
Morphological analysis of rotary aggregate cultures
To determine the potential of retinal progenitors to differentiate as aggregate culture and into various retinal cells, retinal progenitors (2 × 106 in 25 mL flasks) were either cultured alone or cocultured with the RPE cell line D407 (1 × 105/25 mL flask). Samples were analyzed on days 1, 3, 5, 7, and 10, by both phase microscopy and scanning electron microscopy. Fresh medium was added every day. Aggregates began to form within the first few hours of culturing. Initial aggregate size varied between 30 and 100 cells (Fig. 1; day 1, A and B). By day 3, cell aggregates were increasing in size due to cell–cell aggregation and proliferation. Between days 5 and 10, cell aggregates were too large to identify individual cells (Fig. 1; day 5, A–D). Occasionally, cells fell off from these aggregates and resembled rod, cone photoreceptors with broken outer segments (Fig. 1; day 1, C and D, arrows). Layering of cells occurred between days 3 and 10. The bottom-most cells in the column resembled ganglion, and the outer-most cells resembled photoreceptor phenotype (Fig. 1; day 3, B and D, arrows).
Characterization of rotary aggregate culture immunophenoyping and Western blot analysis
To more precisely determine if the progenitor cells will differentiate into various retinal cells in aggregated rotary culture, immunohistochemical analysis and Western blotting were performed. Cell aggregates were analyzed on specific days (1, 3, 5, 7, and 10) by immunophenotyping (day 10; data not included). Characterization was done on aggregates allowed to attach, as well as on cells dispersed with mild trypsinization and cells replated on polylysine-coated slides. Immunolabeling was performed 24 h postattachment, and Western blot analysis was performed on aggregates on respective days (Fig. 5).
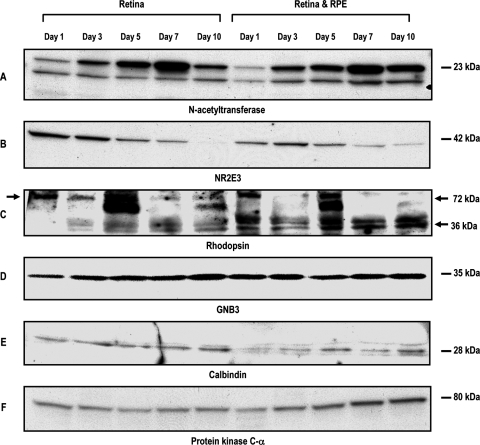
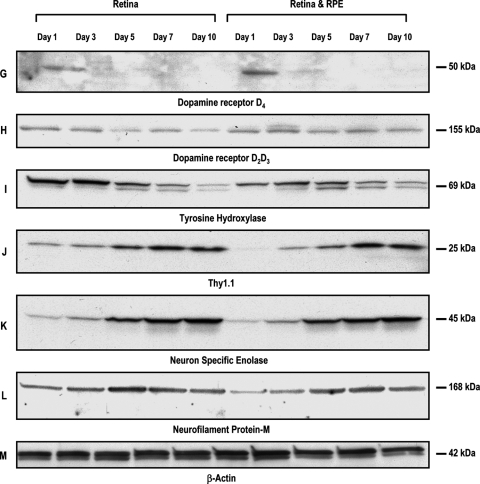
FIG. 5.
Identification of proteins expressed in retinal progenitors cultured alone or as a coculture with RPE by Western blot analysis. Proteins expressed are identified by name and letter. This figure represents the composite of various retinal proteins expressed in rotary cultured cells. Note in this figure, an 8–10-fold increase in AaNat expression between days 1 and 7, both in progenitors cultured alone or as a coculture with RPE (A). Rod transcription factor Nr2e3 (B) is highest on day 1 and declines subsequently with increasing days in culture. Rod photoreceptor differentiation is confirmed by expression of rhodopsin monomers and dimers (36 and 72 kDa) (C) and transcription factor. There was a significant increase in GNB3, a protein expressed in cones (D), but not in calbindin (E). Protein kinase Cα (F), a protein expressed in rod and cone bipolars, shows a slight increase with increasing days in culture in retinal and RPE cocultures, but not in retinal cells cultured alone. Tyrosine hydroxylase (I), a protein expressed in amacrine cells, shows the highest level on day 2 in retinal progenitors cultured alone. There is, however, no change in expression of TH in cocultured cells, Thy 1.1 (J), NSE (K), neurofilament protein (L). The proteins expressed in ganglion cells show an increase in expression with increasing age in culture. Although the D4 level (G) shows a decline in Western blot analysis, immunohistochemical analysis reflects an increased intensity of label in cells expressing D4. Some of the other proteins represented in this Western blot were not significantly altered. Control protein β-actin levels remained relatively unchanged.
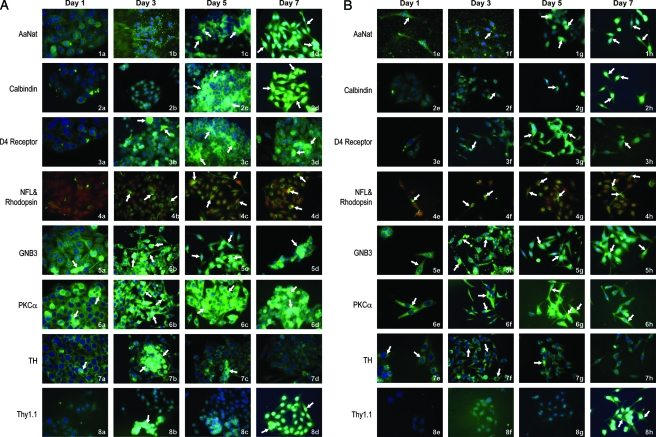
The identification of cells generated in rotary culture by immunophenotyping
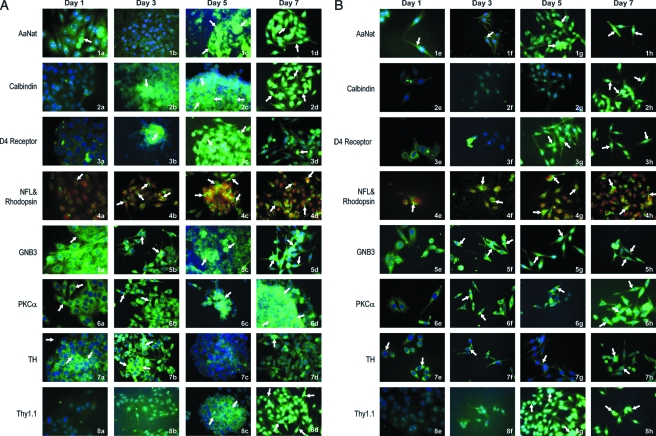
Figure 2 represents the composite of cells grown as aggregates and immunophenotyped as aggregates (A) or dispersed cells (B) on days 1–7. Both aggregates and dispersed cells were immunophenotyped 24 h postsampling and allowed to attach on polylysine-coated coverslips for 24 h. The antibodies used for identification of the specific retinal cell types are those listed in Materials and Methods along with the source and dilutions used. Figure 2A represents aggregates on specific days, and 2B a sample of aggregates dispersed and allowed to reattach. The primary antibodies in the composite are listed by number and name. Samples on increasing days are labeled, for example, AaNat expression (1a–1h). Secondary antibodies used are goat anti-mouse Alexa 488 (green; Molecular Probes) and goat anti-rabbit Alexa 594 (red, Molecular Probes); the blue stain in the nuclei is DAP1.
FIG. 2.
Immunohistochemical analysis to identify retinal neurons generated in rotary culture. This figure represents the composite of immunohistochemical analysis performed on aggregates and dispersed cells on specific days. In this composite, (A) represents aggregates and (B) represents dispersed cells. Both aggregates and dispersed cells were allowed to attach for 24 h, and immunohistochemical analysis was performed 24 h postsampling. The secondary antibody used is goat anti-mouse Alexa 488 (color green) in all determinations with the exception of neurofilament protein that is goat anti-rabbit, Alexa fluor 594 (color red). Nuclei are stained with DAP1. Note the cells labeled for AaNat: (A) (1a–1d) aggregates and (B) (1e–1h) dispersed cells showing increased intensity of label between days 1 and 7 (arrows). Similarly, calbindin (2a–2h) and D4 receptor (3a–3h) show increased expression with increasing culture age. Specifically in dispersed cells, the label is in the cells with bipolar phenotype representing possible photoreceptors (3g and 3h, arrows). In cells labeled for rhodopsin (green) and neurofilament (red) (4a–4h), an increased polarized distribution of rhodopsin with increasing days in culture (4c, 4d, 4g, and 4h, arrows). Note the increased expression of cone transducin GNB3 in cells with bipolar phenotype (5d, 5g, and 5h, arrows). Also note increased PKCα expression between days 1 and 7, specifically 6f–6h (arrows). In the composite, 7a–7h depicts expression of tyrosine hydroxylase in possible amacrine cells (arrows). Note in 8a–8h the increased expression of thy 1.1 in possible ganglion cells with increasing days in culture (8c, 8d, 8g, and 8h).
Tables 2 and 3 represent the summary of data for intensity of label for various retinal proteins (antigens) with specific antibodies. Notice the overall increase in the expression of specific marker proteins with increasing days in culture. This possibly reflects the increased maturation profile of retinal neurons generated from progenitors. In Figure 2A and B we see an increased expression of AaNat both in aggregates and dispersed cells with increasing days in culture. In Figure 2A and B (1a–1h), cells labeled for AaNat with bipolar phenotype are possibly photoreceptors (1d and 1h, arrows). This is further confirmed by the slightly increased expression of calbindin with increasing days in culture (2a–2h); some layering is also seen in aggregates in 2c. Receptor D4 is not expressed in undifferentiated precursors on day 1 (3a); however, with increasing days in culture there is expression in polarized cells (3d and 3h, arrows), which are possibly photoreceptors (3d, 3g, and 3h). Photoreceptor phenotype of cells generated in rotary cultures is represented by Figure 2 (4a–4h). The cells are double-labeled rhodopsin antibody 1D4 (the secondary antibody is Alexa 488 green). The secondary antibody for the identification of neurofilament protein is goat anti-rabbit Alexa 594 red. Note in Figure 2 (4b) progenitor cells expressing a low level of rhodopsin in undifferentiated rounded progenitors (arrows). Note the continuously increased expression and polarized distribution of rhodopsin in cells with bipolar photoreceptor phenotype (arrows) with increasing days in culture, possibly representing mature photoreceptors (4c, 4d, 4g, and 4h, arrows). Figure 2 (5a–5h) depicts cells labeled for cone transducin GNB3; note the bipolar phenotype of cells (5f–5h). Note PKCα expression represented in Figure 2 (6a–6h); the distribution is mostly cytoplasmic. The polarized bipolar-like cells (6f–6h; days 3, 5, and 7) are labeled for PKCα, a protein expressed in rod and cone bipolars.
Table 2.
Expression of Retinal Proteins in Progenitors in Rotary Cultures
| |
Day 1 |
Day 3 |
Day 5 |
Day 7 |
||||
|---|---|---|---|---|---|---|---|---|
| Marker proteins (antigens) | a | b | a | b | a | b | a | b |
| AaNat | + | − | − | − | +++ | +++ | +++ | + |
| Calbindin | − | − | + | − | ++ | − | +++ | ++ |
| D4 Receptor | − | − | + | − | +++ | ++ | + | + |
| GNB3 | ++ | + | + | ++ | ++ | + | +++ | + |
| NFL & Rhodopsin | − | − | + | + | ++ | ++ | +++ | ++ |
| PKCα | + | − | ++ | + | + | + | +++ | +++ |
| TH | + | + | +++ | + | − | − | + | + |
| Thy 1.1 | − | − | + | − | + | ++++ | +++ | ++ |
The data presented are from four separate experiments employed 300–500 immunophenotyped cells scored by two different investigators in a blind study. +, faint expression of antigens; ++, medium labeling; +++, intense labeling for antigen; −, no expression of antigens. Data represent intensity of label. a, Aggregates; b, dispersed cells.
Table 3.
Expression of Retinal Proteins in Retinal Progenitors Cocultured with RPE in Rotary Cultures
| |
Day 1 |
Day 3 |
Day 5 |
Day 7 |
||||
|---|---|---|---|---|---|---|---|---|
| Marker proteins (antigens) | a | b | a | b | a | b | a | b |
| AaNat | − | − | + | − | ++ | + | +++ | + |
| Calbindin | − | − | − | − | ++ | − | +++ | + |
| D4 Receptor | − | − | + | − | ++ | ++ | + | + |
| GNB3 | + | − | +++ | ++ | ++ | ++ | + | ++ |
| NFL & Rhodopsin | − | − | − | − | ++ | ++ | ++ | ++ |
| PKCα | + | + | ++ | ++ | +++ | +++ | ++ | ++ |
| TH | − | + | ++ | + | + | − | − | − |
| Thy 1.1 | − | − | + | − | + | − | +++ | ++ |
The data presented are from four separate experiments employed 300–500 immunophenotyped cells scored by two different investigators in a blind study. +, Faint expression of antigens; ++, medium labeling; +++, intense labeling for antigens; −, no expression of antigens. Data represent intensity of label. a, Aggregates; b, dispersed cells.
RPE, retinal pigment epithelial cells.
An increased intensity of label for protein tyrosine hydroxylase expressed in amacrine cells was noted on days 1 and 3 in aggregate culture. Similarly, the cells positive for thy 1.1 (ganglion cells) show an increased expression on days 5 and 7 (Fig. 2A, B).
Immunophenotyping of retinal neurons generated when retinal progenitors are cocultured with RPE
To test if coculturing retinal progenitors with RPE will alter retinal differentiation, retinal progenitors cocultured with RPE were analyzed by immunophenotyping and Western blot analysis between days 1 and 10. No clear differences were noted in the pattern of protein expression in retinal progenitors cultured alone or cocultured with RPE (Fig. 3A, samples analyzed as aggregates; Fig. 3B, samples analyzed as dispersed cells). Note the increased intensity of label for most antibodies tested for identification of retinal cells, both for aggregates and dispersed cells (Fig. 3A, B; Table 3). Figure 3 is the composite of samples processed between days 1 and 7 as aggregates (A) and dispersed cells (B) on specific days. All samples were processed 24 h postsampling and reattached on polylysine-coated coverslips. Specific antibodies are both named and numbered; for example, Figure 3 (1a–1h) represents expression of AaNat on days 1–7. Figure 3 (2c and 2d) depicts an increase in the intensity of label for calbindin expression (arrows); however, no significant changes were noted by Western blot analysis. Note similarly cells labeled for D4 receptor expression (3b–3d and 3h). Figure 3A and B (4a–4h) represents cells labeled for rhodopsin and neurofilament protein. Note the polarized distribution of rhodopsin (greenish yellow; Fig. 3A, B [4c, 4d, 4g, and 4h, arrows]). Also, there is an increased expression of GNB3 between days 1 and 7 (5b, 5c, 5g, and 5h). Similarly, note for PKCα (6a–6h) increased intensity of label (6b, 6c, and 6f–6h, arrows). The slight increase in intensity of label in Figure 3 (7b and 7f) and (8b, 8d, and 8h) reflects increased expression of tyrosine hydroxylase and thy 1.1, respectively. There is a very close correspondence between immunohistochemical and Western blot analysis for proteins, AaNAT, rhodopsin, GNB3, tyrosine hydroxylase, and Thy 1.1. However, we do not see the same for PKCα, calbindin, and D4 receptor, possibly due to the sensitivity of antibodies for specific analyses.
FIG. 3.
Immunohistochemical analysis in retinal progenitors cocultured with RPE in rotary culture. (A) Aggregates analyzed on various days, and (B) dispersed cells. Immunohistochemical studies were performed 24 h postsampling. The secondary antibody is goat anti-mouse Alexa 488 (green) in the identification of all retinal neuronal proteins except neurofilament protein, which is goat anti-rabbit Alexa fluor 594 (red), and the blue stain in the nuclei is DAP 1. There is increased expression in most retinal proteins with increasing days in culture. Note the increased expression of rhodopsin in cell aggregates and dispersed cells with increasing age (arrows) (4a–4h) for days 1–7. A similar increase is noted for GNB3 (5a–5h). PKCα expression in bipolar cells is mostly cytoplasmic (6a, 6c, and 6f–6h, arrows). Tyrosine hydroxylase expression in cells is pointed out in 7a–7h. Note the increased intensity of label green in 8a–8h with increased age (arrows identifying possible ganglion cells, 8h). Color images available online at www.liebertonline.com/ten.
High-magnification composites to depict differentiation of specific retinal cell types
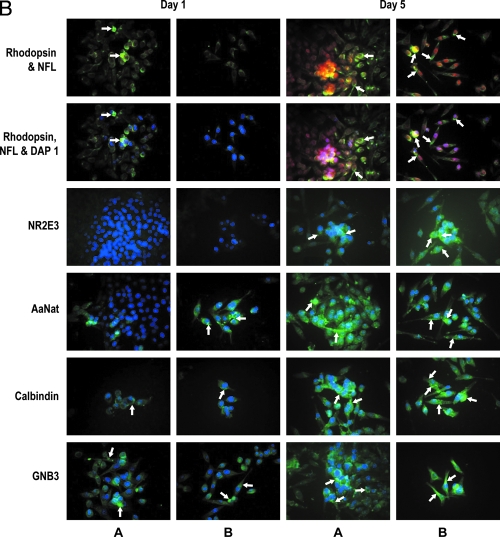
Figure 4A is a high-magnification photomicrograph of specific parts of different experiments. Figure 4A (4a1) represents increased expression of rhodopsin (green) between days 1 and 5. Note that on day 1, the undifferentiated progenitors are beginning to express rhodopsin (arrowheads) in a somewhat polarized manner. By day 5, rhodopsin is expressed in very polarized photoreceptors (arrows) in both aggregates and dispersed cells. The red stain in the nuclei represents neurofilament protein. The same pattern is noted (arrows) for both aggregates and dispersed cells. Note the TH expression (days 1, 5, and 7, in amacrine cell punctuate label) in (4a2) and thy 1.1 expression on days 1, 5, and 7 (ganglion cells) (4a3). Bipolar cells expressing PKCα are shown in 4a4 on days 1 and 5. Note cytoplasmic expression in elongated bipolar-like cells (4a4 arrows) and a photoreceptor cell expressing D4 receptor in 4a5 (day 1, arrowhead).
FIG. 4.
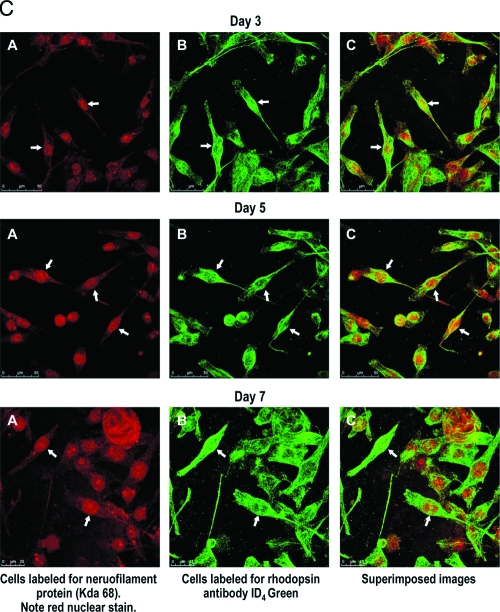
(A) High-magnification composite to depict differentiation of specific retinal cell types. Immunohistochemical analysis. Portions of different experiments enlarged to show distribution of labeled cells. Note in 4a1, large cell aggregates and dispersed cells labeled for rhodopsin (photoreceptors) neurofilament protein (neurons including photoreceptors). The diffused label for rhodopsin (day 1, arrowheads) becomes increasingly polarized and intense by day 5 (arrows), reflecting photoreceptor differentiation. Also, the increase in the expression of tyrosine hydroxylase (4a2, arrows) possible amacrine cells is noted. Punctuate label of individual cells and aggregates labeled for thy 1.1; arrows point to possible ganglion cells (4a3). Note the bipolar phenotype of cells expressing PKCα (4a4). Note cells labeled for D4 receptor (4a5); also visible is one photoreceptor-like cell expressing D4 receptor (arrowhead). (B) Photoreceptor differentiation in rotary culture. Note photoreceptor differentiation with increasing days in culture (A, aggregates; B, dispersed cells). Note the increased expression of rhodopsin, transcription factor Nr2e3, AaNat, calbindin, and GNB3 on days 1 and 5 (arrows). (C) Confocal analysis to confirm photoreceptor differentiation. Letter A indicates cells labeled for neurofilament protein 68 kDa (red); letter B indicates cells labeled for rhodopsin (green); letter C indicates superimposed images. Note the increased intensity of label for neurofilament protein (kDa 68) and rhodopsin (green) between days 3 and 7. Color images available online at www.liebertonline.com/ten.
Photoreceptor differentiation in rotary cell culture
Figure 4B shows increased photoreceptor differentiation confirmed by upregulation of photoreceptor-specific proteins (rhodopsin, Nr2e3, AaNat, calbindin, and GNB3 between days 1 and 5). Note the dramatic increase in green label (arrows) on day 5 rotary cultured cells analyzed as aggregates (A) or dispersed cells (B).
Confocal microscopy to confirm photoreceptor differentiation
Figure 4C represents photoreceptor cells labeled for rhodopsin and neurofilament protein analyzed by confocal microscopy, for days 3, 5, and 7. Note the increased expression of photoreceptor-specific proteins between days 3, 5, and 7 (arrows). Figure 4A shows cells labeled for neurofilament protein kDa 68, Figure 4B shows cells labeled for rhodopsin, and Figure 4C shows superimposed images.
Western blot analysis
Figure 5 is a composite representing proteins expressed in rotary cultures of retinal progenitors cultured alone or as a coculture with RPE. Proteins are represented by name and letters. AaNat is a protein expressed mostly in photoreceptors, and possibly cells in INL upregulated 8–10-fold in aggregate culture (A). The same was reflected in samples assessed by immunophenotyping. Transcription factor Nr2e3 is expressed at a slightly higher level on days 1 and 3 and declines thereafter (B). Rhodopsin levels are increased significantly between days 1 and 5, both in retinal progenitors cultured alone or cocultured with RPE. Both rhodopsin monomers 36 kDa and dimers 72 kDa were expressed (C). Cone transducin GNB3 almost doubled between days 1 and 10 (D). Western blot analysis reflected that calbindin levels remained relatively unchanged in the 10-day culture period by Western blot analysis (E); however, a slight increase in intensity was noted at cellular level by immunohistochemical analysis. D4 levels dropped after the day 1 culture by Western blot analysis (G), but for the cells expressing D4, the labeling intensity by immunohistochemical analysis had increased considerably. Receptor D2D3 levels were unchanged in the 10-day culture period (H).
Similarly tyrosine hydroxylase (TH) levels expressed in amacrine cells dropped between days 1 and 7 in cells processed by Western blot analysis (I). Immunohistochemical data point to increased intensity in labeling of cells expressing TH, possibly pointing to increased levels in cells expressing TH but possibly no increase in the number of cells expressing TH. Enhanced ganglion cell differentiation in aggregate cultures was confirmed by upregulation of thy 1.1, NSE, and NFm between days 1 and 10, by both immunophenotyping and Western blot analysis (J–L). Bipolar cell differentiation was confirmed by the expression of PKCα (F). Transcription factor Nr2e3 and β transducin GNB3 confirm rod and cone photoreceptor differentiation in this system (B and D).
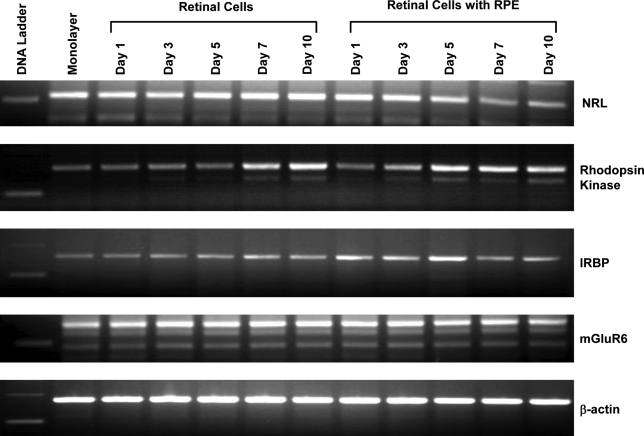
RT-PCR analysis to confirm photoreceptor and bipolar cell differentiation in rotary cultures
Figure 6 confirms photoreceptor and bipolar differentiation in this system. Photoreceptor cell commitment transcription factors, Nrl (RT-PCR) and Nr2e3 (immunohistochemistry), were expressed in these cultures. Nrl, a transcription factor implicated in rod cell commitment, shows two to three times upregulation in rotary cultures by RT-PCR. Rhodopsin kinase expressed in rod and cone photoreceptors was doubled, and interstitial retinal binding protein was slightly upregulated. The presence of mGluR6 expressed in (bipolar) cells was also confirmed by RT-PCR with similar levels in progenitors cultured alone or cocultured with RPE. Coculturing retinal progenitors with RPE did not show any significant change in the expression of various messages. Nrl expression actually declined in cocultured cells on days 7 and 10. Rhodopsin kinase levels were similar in retinal progenitors cultured alone or cocultured with RPE.
FIG. 6.
RT-PCR analysis to confirm maturation of progenitors in rotary culture, specifically photoreceptors. Note the increased expression of rod transcription factor and Nrl expression in retinal progenitors cultured alone between days 1 and 10. In retinal progenitors cocultured with RPE, a slight decline in Nrl expression was noted between days 1 and 10. Rhodopsin kinase, a protein expressed in rod and cone photoreceptors, shows an increase with increasing days in rotary cultures, in retinal progenitors cultured alone or as cocultures with RPE. A slight increase in rod bipolar cells marked mGluR6 was also noted. No significant change in interstitial retinal binding protein (IRBP), another retinal-specific protein was noted. β-Actin used as a control showed no significant change.
Discussion
Retinal degenerative diseases, such as retinitis pigmentosa and age-related macular degeneration, remain the leading cause of blindness in humans. Currently available therapeutic measures, although reflecting a major advance, have had limited efficacy.35–39 Novel strategies under investigation for treatment of retinal diseases currently include pharmacological intervention, transplantation of donor cells, and transplantation of electronic chips to stimulate the brain.40,41 Animal models of retinal degeneration have provided some valuable insights into the survival and integration of transplanted tissues.42,43
Tissue engineering provides an alternate strategy for the treatment of diseases, specifically retinal degeneration, where implanted tissues have restored vision in animal models to a degree.
In this study, an established cell line was tested for its potential in the generation of 3D retina-like structures. Current research on the 3D culture system is more vital and promising than ever.44–47 The utility of 3D systems is underscored by their potential use in drug delivery, study of metastasis, and tissue engineering for therapeutic purposes. The advantage of the 3D cell culture system is its defined geometry that makes it possible to directly relate structure to function and that approximates an in vivo state more closely by allowing spatial resolution. Combining 3D approaches with molecular analysis has clearly demonstrated that in comparison to monolayer cultures, 3D systems have many advantages. Spheroid cultures developed by Layer et al.32 point to the role of RPE in retinospheroids and stratospheroids generation when cocultured with retinal cells, as well as the role that is played by Muller cells. Cells grown in 3D constructs express receptors and extracellular matrices differently. The development of artificial organs has been attempted for liver, pancreas, bone, and other tissues by 3D growth. Aggregate cultures have been generated from dissociated primary retinal tissues from both fetal and adult retina.48–50 The ability to maintain and expand primitive undifferentiated human retinal progenitors in cultures will be valuable in the study of human retinal genes, and possibly could be used for treatment of degenerative retinal diseases.
We have been successful in generating progenitor cell lines with and without gene manipulation (transfection). We have previously explored the differentiation potential of these cells by manipulating external cues that triggered differentiation into various retinal cell types. We have also previously reported that 3D structures can be generated using the multilineage cell lines. These progenitors when grown attached to cytodex beads in the NASA-developed rotating culture vessel system and formed large 3D constructs.50,51 We realized that until well-defined biocompatible substrates could be generated, we will be unable to test these transplants in an animal model.
As such, we have revisited the 3D rotary culture technology developed by Moscona30 half a century ago and redefined by others, and report that a cell line that has been in culture for years can be grown in rotary aggregate cultures to generate many retinal cell types. These cells retain a spectrum of differentiation capability and express upregulation of specific retinal markers such as tyrosine hydroxylase and dopamine receptors, and demonstrate significant upregulation of AaNat, an enzyme implicated melatonin synthesis expressed in photoreceptors. In addition, RT-PCR data confirm the expression of photoreceptor marker rhodopsin kinase. The expression of transcription factor Nrl interstitial retinal binding protein, and Nr2e3 in this system is confirmed by immunohistochemistry and Western blot analysis. This finding is exciting in that neural retina leucine protein zipper (Nrl), a basic motif leucine zipper transcription factor that is preferentially expressed in rod photoreceptors, is expressed in this system, and it acts synergistically with crx to regulate rhodopsin transcription. Missense mutation in human Nrl have been associated with autosomal dominant retinitis pigmentosa. The photoreceptors in Nrl−/− retina have cone phenotype.52,53 It is believed that Nrl transcript expression is upstream of Nr2e3 in photoreceptor development. Nr2e3 is also expressed in the outer nuclear layer of human retina and possibly has a role in determining photoreceptor phenotype during human retinogenesis.54 We are aware of a recent report that suggests that Nr2e3 may also be expressed in RPE and Muller cells. The consensus is that Nrl acts as a “molecular switch” during rod cell development by specifically modulating rod-specific genes while simultaneously inhibiting the S cone pathway through the activation of Nr2e3. It has been postulated that Nrl−/− mouse have photoreceptors that are cone–rod intermediates.55–57 The expression of rhodopsin kinase (rods and cones), calbindin, GNB3 (cones), and transcription factors Nrl and Nr2e3 confirms that progenitors grown as a rotary culture mature into rod and cone photoreceptors.
Tyrosine hydroxylase expression (amacrine cells) peaks on days 1–3. The D2D3 receptors remain unchanged. The D4 receptor peaks on day 1, while thy 1.1 and NSE expressed in ganglion cells are upregulated twofold to threefold between days 1 and 7. Levels of PKCα expression (rod, cone bipolars, and blue cone) show a slight increase when retinal progenitors are cocultured with RPE. Calbindin does not show significant changes during 1–7 days by Western blot analysis.
High-level expression of enzyme AaNat in these cultures warrants discussion. It is well established that vertebrate retina contains autonomous circadian clocks.58 Melatonin and dopamine are mutually inhibitory neuromodulators, respectively, mediating effects of dark and light necessary for retinal rhythmicity and circadian clocks. AaNat is a key enzyme in serotonin-melatonin synthesis. Dopamine plays an important role in regulation of AaNat acting via D2/D4-like receptors present in photoreceptors.59 In mammalian retina, AaNat mRNA has been localized to the photoreceptor layer.59–61 There are also data to point out that circadian rhythmicity can be generated independent of photoreceptors. Plasticity in the retinal circadian system is further suggested by upregulation of AaNat transcripts in the inner retina of RCS rats with photoreceptor degeneration.62 Thus, melatonin regulation in part is controlled by photoreceptors other than those known for vision. These photoreceptors appear to comprise a subset of melopsin-positive retinal ganglion cells that project directly to SCN.63–66 Similarly, light-induced circadian phase shifts have been demonstrated in humans with complete vision loss.67 Increased AaNat expression seen in our rotary cultures could be due to photoreceptor maturation such as rods and cones or melopsin-containing photoreceptors in INL, but there is equal possibility that it reflects general maturation of retinal tissue and is not directly related to photoreceptor maturation.68,69 Studies in developing rodents indicate that MT1 and MT2 receptor levels and D2/D4 levels peak at different developmental stages, suggesting that besides its role as an oscillator, melatonin could be implicated in developmental regulations and morphogenesis.68 However, in our studies, expression of several photoreceptor-specific markers (rhodopsin kinase, NRL, and GNB3) along with AaNat upregulation possibly points to photoreceptor maturation in our system.
In this paper, we offer proof of the concept that progenitor cells in aggregated rotary cultures reproduce the genetic program of multipotential retinal progenitors with fidelity. This could be extremely valuable in refining the conditions for engineering retinal tissue that could be subsequently extended to retinal stem cells from ciliary epithelium in the eye.
In this study, we report for the first time that an established cell line that has been cultured for greater than 12 years reproduces with fidelity in 3D rotary aggregate cultures, the developmental program of the multipotential progenitors. It is remarkable that progenitor cell lines retain the potential for differentiation in rotating culture. The rotary culture system provides a higher degree of differentiation attested by the presence of several retinal differentiation–specific proteins, primarily photoreceptors, but also ganglion cells, bipolar cells, and cone photoreceptors. The next question is, are the retinal layers orientated properly and will the function be restored when these constructs are tested as transplants?
Acknowledgments
The authors thank Ms. Renarder Pressley for excellent secretarial assistance, Mr. Patrick Abramson for graphics, Mr. L. Brako for outstanding scanning electron micrography, Mr. Andrew Shaw for confocal microscopy, and Ms. Suzanne Alexander for editorial help (Morehouse School of Medicine, Atlanta, GA). The authors are profoundly grateful to Dr. Richard Hunt (Department of Microbiology, University of South Carolina Medical School, Columbia, SC) for sharing RPE cell line D407. The authors thank Dr. R. Kumar (Morehouse School of Medicine) for RT-PCR analysis. The authors also thank Dr. M. Smith, Dr. M. Thierry-Palmer, and Dr. Harris-Hooker (Morehouse School of Medicine) for their moral support. Profound thanks to Dr. J. Duke (University of Texas, Health Sciences Center, Houston, TX) for her valuable scientific advice. The authors acknowledge input of Dr. G. Tosini (Neuroscience Institute, Morehouse School of Medicine) and Dr. M. Iuvone (Department of Pharmacology, Emory University, Atlanta, GA) for their scientific advice. The authors also acknowledge Darlene P. Kelly, Information Specialist (Multi-Media Center, Morehouse School of Medicine), for her help. The research was supported by NASA Grant NCC 9-112 (K.D.) and NIH NCRR RCMI Core Grant 12RR03034. Special thanks to Drs. R. Bellamkonda and R. Nerem, Georgia Institute of Technology (EEC-9731643 NSF).
Disclosure Statement
No competing financial interests exist.
References
- 1.Berson E.L. Retinitis pigmentosa: the Friedenwald Lecture. Invest Ophthalmol Vis Sci. 1993;34:1659. [PubMed] [Google Scholar]
- 2.Gregory C.Y. Bird A.C. Cell loss in retinal dystrophies by apoptosis-death by informed consent! Br J Ophthalmol. 1995;79:186. doi: 10.1136/bjo.79.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heckenlively J.R. Bouchman J. Friedman J. Diagnosis and classification of retinitis pigmentosa. In: Heckenlively J.R., editor. Retinitis Pigmentosa. Philadelphia: J.B. Lippincott; 1988. p. 21. [Google Scholar]
- 4.Phelan J.K. Bok D. A brief review of retinitis pigmentosa and the identified retinitis pigmentosa genes. Mol Vis. 2000;6:116. [PubMed] [Google Scholar]
- 5.Das T. del Cerro M. Jalali S. Rao V.S. Gullapalli V.K. Little C. Loreto D.A. Sharma S. Sreedharan A. del Cerro C. Rao G.N. The transplantation of human fetal neuroretinal cells in advanced retinitis pigmentosa patients: results of a long-term safety study. Exp Neurol. 1999;157:58. doi: 10.1006/exnr.1998.6992. [DOI] [PubMed] [Google Scholar]
- 6.Humayun M.S. de Juan E., Jr. Weiland J.D. Dagnelie G. Katona S. Greenberg R. Suzuki S. Pattern electrical stimulation of human retina. Vision Res. 1999;39:2569. doi: 10.1016/s0042-6989(99)00052-8. [DOI] [PubMed] [Google Scholar]
- 7.Algvere P.V. Berglin L. Gouras P. Sheng Y. Transplantation of fetal retinal pigment epithelium in age-related macular degeneration with subfoveal neovascularization. Graefes Arch Clin Exp Ophthalmol. 1994;232:707. doi: 10.1007/BF00184273. [DOI] [PubMed] [Google Scholar]
- 8.Litchfield T.M. Whiteley S.J. Lund R.D. Transplantation of retinal pigment epithelial, photoreceptor and other cells as treatment for retinal degeneration. Exp Eye Res. 1997;64:655. doi: 10.1006/exer.1996.0253. [DOI] [PubMed] [Google Scholar]
- 9.Radtke N.D. Aramant R.B. Seiler M. Petry H.M. Preliminary report: indications of improved visual function following retina sheet transplantation in retinitis pigmentosa patients. Am J Ophthalmol. 1999;128:384. doi: 10.1016/s0002-9394(99)00250-0. [DOI] [PubMed] [Google Scholar]
- 10.Sieving P.A. Caruso R.C. Tao W. Coleman H.R. Thompson D.J. Fullmer K.R. Bush R.A. Ciliary neurotrophic factor (CNTF) for human retinal degeneration: phase I trial of CNTF delivered by encapsulated cell intraocular implants. Proc Natl Acad Sci USA. 2006;103:3896. doi: 10.1073/pnas.0600236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milam A.H. Li Z.Y. Fariss R.N. Histopathology of the human retina in retinitis pigmentosa. Prog Retin Eye Res. 1998;17:175. doi: 10.1016/s1350-9462(97)00012-8. [DOI] [PubMed] [Google Scholar]
- 12.Humayun M.S. Prince M. de Juan E., Jr. Barron Y. Moskowitz M. Klock I.B. Milam A.H. Morphometric analysis of the extramacular retina from postmortem eyes with retinitis pigmentosa. Invest Ophthalmol Vis Sci. 1999;40:143. [PubMed] [Google Scholar]
- 13.Gamm D.M. Nelson A.D. Svendsen C.N. Human retinal progenitor cells grown as neurospheres demonstrate time-dependent changes in neuronal and glial cell fate potential. Ann NY Acad Sci. 2005;1049:107. doi: 10.1196/annals.1334.011. [DOI] [PubMed] [Google Scholar]
- 14.Lamba D.A. Karl M.O. Ware C.B. Reh T.A. Efficient generation of retinal progenitor cells from human embryonic stem cells. Proc Natl Acad Sci USA. 2006;103:12769. doi: 10.1073/pnas.0601990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelley M.W. Turner J.K. Reh T.A. Regulation of proliferation and photoreceptor differentiation in fetal human retinal cell cultures. Invest Ophthalmol Vis Sci. 1995;36:1280. [PubMed] [Google Scholar]
- 16.Akita J. Takahashi M. Hojo M. Nishida A. Haruta M. Honda Y. Neuronal differentiation of adult rat hippocampus-derived neural stem cells transplanted into embryonic rat explanted retinas with retinoic acid pretreatment. Brain Res. 2002;954:286. doi: 10.1016/s0006-8993(02)03356-5. [DOI] [PubMed] [Google Scholar]
- 17.Yang P. Seiler M.J. Aramant R.B. Whittemore S.R. Differential lineage restriction of rat retinal progenitor cells in vitro and in vivo. J Neurosci Res. 2002;69:466. doi: 10.1002/jnr.10320. [DOI] [PubMed] [Google Scholar]
- 18.Chacko D.M. Das A.V. Zhao X. James J. Bhattacharya S. Ahmad I. Transplantation of ocular stem cells: the role of injury in incorporation and differentiation of grafted cells in the retina. Vision Res. 2003;43:937. doi: 10.1016/s0042-6989(02)00688-0. [DOI] [PubMed] [Google Scholar]
- 19.Young M.J. Ray J. Whiteley S.J. Klassen H. Gage F.H. Neuronal differentiation and morphological integration of hippocampal progenitor cells transplanted to the retina of immature and mature dystrophic rats. Mol Cell Neurosci. 2000;16:197. doi: 10.1006/mcne.2000.0869. [DOI] [PubMed] [Google Scholar]
- 20.Ahmad I. Tang L. Pham H. Identification of neural progenitors in the adult mammalian eye. Biochem Biophys Res Commun. 2000;270:517. doi: 10.1006/bbrc.2000.2473. [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharya S. Jackson J.D. Das A.V. Thoreson W.B. Kuszynski C. James J. Joshi S. Ahmad I. Direct identification and enrichment of retinal stem cells/progenitors by Hoechst dye efflux assay. Invest Ophalmol Vis Sci. 2003;44:2764. doi: 10.1167/iovs.02-0899. [DOI] [PubMed] [Google Scholar]
- 22.Klassen H. Ziaeian B. Kirov I.I. Young M.J. Schwartz P.H. Isolation of retinal progenitor cells from post-mortem human tissue and comparison with autologous brain progenitors. J Neurosci Res. 2004;77:334. doi: 10.1002/jnr.20183. [DOI] [PubMed] [Google Scholar]
- 23.Tropepe V. Coles B.L. Chiasson B.J. Horsford D.J. Elia A.J. McInnes R.R. van der Kooy D. Retinal stem cells in the adult mammalian eye. Science. 2000;287:2032. doi: 10.1126/science.287.5460.2032. [DOI] [PubMed] [Google Scholar]
- 24.Fischer A.J. Reh T.A. Muller glia are a potential source of neural regeneration in the postnatal chicken retina. Nat Neurosci. 2001;4:247. doi: 10.1038/85090. [DOI] [PubMed] [Google Scholar]
- 25.Bonassar L.J. Vacanti C.A. Tissue engineering: the first decade and beyond. J Cell Biochem. 1998;Suppl. 30–31:297. [PubMed] [Google Scholar]
- 26.Levenberg S. Huang N.F. Lavik E. Rogers A.B. Itskovitz-Eldor J. Langer R. Differentiation of human embryonic stem cells on three-dimensional polymer scaffolds. Proc Natl Acad Sci USA. 2003;100:12741. doi: 10.1073/pnas.1735463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atala A. Tissue engineering and regenerative medicine: concepts for clinical application. Rejuvenation Res. 2004;7:15. doi: 10.1089/154916804323105053. [DOI] [PubMed] [Google Scholar]
- 28.Dutt K. Harris-Hooker S. Ellerson D. Layne D. Kumar R. Hunt R. Generation of 3D retina-like structures from a human retinal cell line in a NASA bioreactor. Cell Transplant. 2003;12:717. doi: 10.3727/000000003108747334. [DOI] [PubMed] [Google Scholar]
- 29.Kumar R. Dutt K. Enhanced neurotrophin synthesis and molecular differentiation in non-transformed human retinal progenitor cells cultured in a rotating bioreactor. Tissue Eng. 2006;12:141. doi: 10.1089/ten.2006.12.141. [DOI] [PubMed] [Google Scholar]
- 30.Moscona A.A. Cell suspension from organ rudiments of chick embryos. Exp Cell Res. 1952;3:535. [Google Scholar]
- 31.Rothermel A. Willbold E. Degrip W.J. Layer P.G. Pigmented epithelium induces complete retinal reconstitution from dispersed embryonic chick retinae in reaggregation culture. Proc Biol Sci. 1997;264:1293. doi: 10.1098/rspb.1997.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Layer P.G. Rothermel A. Willbold E. Inductive effects of the retinal pigmented epithelium (RPE) on histogenesis of the avian retina as revealed by retinospheroid technology. Semin Cell Dev Biol. 1998;9:257. doi: 10.1006/scdb.1998.0234. [DOI] [PubMed] [Google Scholar]
- 33.Dutt K. Scott M. Wang M. Semple E. Sharma G.P. Srinivasan A. Establishment of a human retinal cell line by transfection of SV40 T antigen gene with potential to undergo neuronal differentiation. DNA Cell Biol. 1994;13:909. doi: 10.1089/dna.1994.13.909. [DOI] [PubMed] [Google Scholar]
- 34.Ezeonu I. Wang M. Kumar R. Dutt K. Density-dependent differentiation in nontransformed human retinal progenitor cells in response to basic fibroblast growth factor- and transforming growth factor-alpha. DNA Cell Biol. 2003;22:607. doi: 10.1089/104454903770238085. [DOI] [PubMed] [Google Scholar]
- 35.Bok D. Retinal transplantation and gene therapy. Present realities and future possibilities. Invest Ophthalmol Vis Sci. 1993;34:473. [PubMed] [Google Scholar]
- 36.Kaplan H.J. Tezel T.H. Berger A.S. Wolf M.L. Del Priore L.V. Human photoreceptor transplantation in retinitis pigmentosa. A safety study. Arch Ophthalmol. 1997;115:1168. doi: 10.1001/archopht.1997.01100160338012. [DOI] [PubMed] [Google Scholar]
- 37.Ehinger B. Bergstrom A. Seiler M. Aramant R.B. Zucker C.L. Gustavii B. Adolph A.R. Ultrastructure of human retinal cell transplants with long survival times in rats. Exp Eye Res. 1991;53:447. doi: 10.1016/0014-4835(91)90162-8. [DOI] [PubMed] [Google Scholar]
- 38.LaVail M.M. Yasumura D. Matthes M.T. Drenser K.A. Flannery J.G. Lewin A.S. Hauswirth W.W. Ribozyme rescue of photoreceptor cells in P23H transgenic rats: long-term survival and late-stage therapy. Proc Natl Acad Sci USA. 2000;97:11488. doi: 10.1073/pnas.210319397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sahel J.A. Mohand-Said S. Leveillard T. Hicks D. Picaud S. Dreyfus H. Rod-cone interdependence: implications for therapy of photoreceptor cell diseases. Prog Brain Res. 2001;131:649. doi: 10.1016/s0079-6123(01)31051-8. [DOI] [PubMed] [Google Scholar]
- 40.Otani A. Dorrell M.I. Kinder K. Moreno S.K. Nusinowitz S. Banin E. Heckenlively J. Friedlander M. Rescue of retinal degeneration by intravitreally injected adult bone marrow-derived lineage-negative hematopoietic stem cells. J Clin Invest. 2004;114:765. doi: 10.1172/JCI21686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pardue M.T. Phillips M.J. Yin H. Sippy B.D. Webb-Wood S. Chow A.Y. Ball S.L. Neuroprotective effect of subretinal implants in the RCS rat. Invest Ophthalmol Vis Sci. 2005;46:674. doi: 10.1167/iovs.04-0515. [DOI] [PubMed] [Google Scholar]
- 42.Bennett J. Tanabe T. Sun D. Zeng Y. Kjeldbye H. Gouras P. Maguire A.M. Photoreceptor cell rescue in retinal degeneration (rd) mice by in vivo gene therapy. Nat Med. 1996;2:649. doi: 10.1038/nm0696-649. [DOI] [PubMed] [Google Scholar]
- 43.Lem J. Flannery J.G. Li T. Applebury M.L. Farber D.B. Simon M.I. Retinal degeneration is rescued in transgenic rd mice by expression of the cGMP phosphodiesterase beta subunit. Proc Natl Acad Sci USA. 1992;89:4422. doi: 10.1073/pnas.89.10.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mueller-Klieser W. Three-dimensional cell cultures: from molecular mechanisms to clinical applications. Am J Physiol. 1997;273:C1109. doi: 10.1152/ajpcell.1997.273.4.C1109. [DOI] [PubMed] [Google Scholar]
- 45.Polak J. Hench L. Gene therapy progress and prospects: in tissue engineering. Gene Ther. 2005;12:1725. doi: 10.1038/sj.gt.3302651. [DOI] [PubMed] [Google Scholar]
- 46.Lu L. Yaszemski M.J. Mikos A.G. Retinal pigment epithelium engineering using synthetic biodegradable polymers. Biomaterials. 2001;22:3345. doi: 10.1016/s0142-9612(01)00172-7. [DOI] [PubMed] [Google Scholar]
- 47.Young M.J. Borras T. Walter M. Ritch R. Tissue bioengineering: potential applications to glaucoma. Arch Ophthalmol. 2005;123:1725. doi: 10.1001/archopht.123.12.1725. [DOI] [PubMed] [Google Scholar]
- 48.Engelhardt M. Wachs F.P. Couillard-Despres S. Aigner L. The neurogenic competence of progenitors from the postnatal rat retina in vitro. Exp Eye Res. 2004;78:1025. doi: 10.1016/j.exer.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Rothermel A. Biedermann T. Weigel W. Kurz R. Ruffer M. Layer P.G. Robitzki A.A. Artificial design of three-dimensional retina-like tissue from dissociated cells of the mammalian retina by rotation-mediated cell aggregation. Tissue Eng. 2005;11:1749. doi: 10.1089/ten.2005.11.1749. [DOI] [PubMed] [Google Scholar]
- 50.Andressen C. Briese V. Ulfig N. Progenitor cells from human embryonic retina—proliferation and preferential differentiation into ganglion cells. Neuroembyology. 2003;2:123. [Google Scholar]
- 51.Dutt K. Sanford G. Harris-Hooker S. Brako L. Kumar R. Sroufe A. Melhado C. Three-dimensional model of angiogenesis: coculture of human retinal cells with bovine aortic endothelial cells in the NASA bioreactor. Tissue Eng. 2003;9:893. doi: 10.1089/107632703322495547. [DOI] [PubMed] [Google Scholar]
- 52.Mears A.J. Kondo M. Swain P.K. Takada Y. Bush R.A. Saunders T.L. Sieving P.A. Swaroop A. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 53.Haider N.B. Jacobson S.G. Cideciyan A.V. Swiderski R. Streb L.M. Searby C. Beck G. Hockey R. Hanna D.B. Gorman S. Duhl D. Carmi R. Bennett J. Weleber R.G. Fishman G.A. Wright A.F. Stone E.M. Sheffield V.C. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet. 2000;24:127. doi: 10.1038/72777. [DOI] [PubMed] [Google Scholar]
- 54.Corbo J.C. Cepko C.L. A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet. 2005;1(ell) doi: 10.1371/journal.pgen.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haider N.B. Naggert J.K. Nishina P.M. Excess cone cell proliferation due to lack of a functional NR2E3 causes retinal dysplasia and degeneration in rd7/rd7 mice. Hum Mol Genet. 2001;10:1619. doi: 10.1093/hmg/10.16.1619. [DOI] [PubMed] [Google Scholar]
- 56.Milam A.H. Rose L. Cideciyan A.V. Barakat M.R. Tang W.X. Gupta N. Aleman T.S. Wright A.F. Stone E.M. Sheffield V.C. Jacobson S.G. The nuclear receptor NR2E3 plays a role in human retinal photoreceptor differentiation and degeneration. Proc Natl Acad Sci USA. 2002;99:473. doi: 10.1073/pnas.022533099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen J. Rattner A. Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci. 2005;25:118. doi: 10.1523/JNEUROSCI.3571-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Besharse J.C. Iuvone P.M. Pierce M.E. Regulation of rhythmic photoreceptor metabolism: a role for post-receptoral neurons. Prog Retin Res. 1988;7:21. [Google Scholar]
- 59.Nir I. Haque R. Iuvone P.M. Diurnal metabolism of dopamine in the mouse retina. Brain Res. 2000;870:118. doi: 10.1016/s0006-8993(00)02409-4. [DOI] [PubMed] [Google Scholar]
- 60.Tosini G. Chaurasia S.S. Michael Iuvone P. Regulation of arylalkylamine N-acetyltransferase (AANAT) in the retina. Chronobiol Int. 2006;23:381. doi: 10.1080/07420520500482066. [DOI] [PubMed] [Google Scholar]
- 61.Iuvone P.M. Tosini G. Pozdeyev N. Haque R. Klein D.C. Chaurasia S.S. Circadian clocks, clock networks, arylakylamine N-acetyltransferase, and melatonin in the retina. Prog Retin Eye Res. 2005;24:433. doi: 10.1016/j.preteyeres.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 62.Sakamoto K. Liu C. Tosini G. Circadian rhythms in the retina of rats with photoreceptor degeneration. J Neurochem. 2004;90:1019. doi: 10.1111/j.1471-4159.2004.02571.x. [DOI] [PubMed] [Google Scholar]
- 63.Bellingham J. Foster R.G. Opsins and mammalian photoentrainment. Cell Tissue Res. 2002;309:57. doi: 10.1007/s00441-002-0573-4. [DOI] [PubMed] [Google Scholar]
- 64.Brainard G.C. Rollag M.D. Hanifin J.P. Photic regulation of melatonin in humans: ocular and neural signal transduction. J Biol Rhythms. 1997;12:537. doi: 10.1177/074873049701200608. [DOI] [PubMed] [Google Scholar]
- 65.Berson D.M. Dunn F.A. Takao M. Phototransduction by retinal ganglion cells that set the circadian clock. Science. 2002;295:1070. doi: 10.1126/science.1067262. [DOI] [PubMed] [Google Scholar]
- 66.Reppert S.M. Godson C. Mahle C.D. Weaver D.R. Slaugenhaupt S.A. Gusella J.F. Molecular characterization of a second melatonin receptor expressed in human retina and brain: the (Mel1b) melatonin receptor. Proc Natl Acad Sci USA. 1995;92:8734. doi: 10.1073/pnas.92.19.8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Czeisler C.A. Shanahan T.L. Klerman E.B. Martens H. Brotman D.J. Emens J.S. Klein T. Rizzo J.F., 3rd. Suppression of melatonin secretion in some blind patients by exposure to bright light. N Engl J Med. 1995;332:6. doi: 10.1056/NEJM199501053320102. [DOI] [PubMed] [Google Scholar]
- 68.Scher J. Wankiewicz E. Brown G.M. Fujieda H. MT(1) melatonin receptor in the human retina: expression and localization. Invest Ophthalmol Vis Sci. 2002;43:889. [PubMed] [Google Scholar]
- 69.Weaver D.R. The roles of melatonin in development. Adv Exp Med Biol. 1999;460:199. doi: 10.1007/0-306-46814-x_22. [DOI] [PubMed] [Google Scholar]