Abstract
How cell type-specific differences in chromatin conformation are achieved, and their contribution to gene expression are incompletely understood. Here we identify a cryptic upstream orchestrator of interferon-γ (Ifng) transcription, which is embedded within the human IL26 gene, compromised of a single CTCF-binding site and retained in all mammals, even surviving near-complete deletion of IL26 in rodents. CTCF and cohesins occupy this element in vivo in a cell-type non-specific manner. This element is approximated with two other sites located within the first intron and downstream of Ifng, where CTCF, cohesins and T-bet bind in a Th1-specific manner. These interactions, approximation of other elements within the locus to each other and to Ifng, and robust Ifng expression are dependent on CTCF and T-bet. The results demonstrate that cooperation between architectural (CTCF) and transcriptional enhancing (T-bet) factors and the elements to which they bind is required for proper Th1-specific expression of Ifng.
Naïve CD4+ T cells can differentiate into one of several helper cell lineages, including Th1 and Th2 cells, whose distinct functions allow them to play non-redundant roles in host defense (Zhu and Paul, 2008). The functional specialization of T helper cells results from differences in the abundance of lineage-specific transcription factors and from T helper cell subset-specific epigenetic modifications that collaborate to initiate and to maintain appropriate programs of gene expression (Lee et al., 2006; Wilson et al., 2009; Zhu and Paul, 2008). These epigenetic modifications include DNA methylation, post-translational histone modifications, nucleosome position and density and higher order structural organization of the genome. The latter mechanism has received increasing attention of late, following the recognition that the three-dimensional structure of DNA can be altered in a cell type-specific manner through chromatin looping to bring distal regulatory elements in proximity to one another and the promoters of their target genes, or vice versa, to facilitate proper expression (Apostolou and Thanos, 2008; Decker, 2008; Fraser and Bickmore, 2007; Lee et al., 2006). In the immune system, such structural changes may be important for properly ordered T cell receptor and immunoglobulin gene rearrangement and for Th2 cytokine gene expression (Cai et al., 2006; Lee et al., 2006; Skok et al., 2007; Spilianakis and Flavell, 2004)
The Th2 cytokine locus contains the Il4, Il5, and Il13 genes and the constitutively expressed Rad50 gene. The linear relationships between these genes are conserved in mammals, and in part for this reason, the Th2 locus has provided an important model for identifying distal cis-regulatory elements and elucidating the mechanisms by which they coordinately regulate nearby clustered genes (Ansel et al., 2006). Coordinate regulation of these genes may be achieved in part through chromatin looping, which can bring the Il4, Il5, and Il13 gene promoters into proximity to each other and to the Th2 cytokine locus control region (LCR) to promote their expression (Cai et al., 2006; Spilianakis and Flavell, 2004).
By contrast to the Th2 cytokine locus where co-expressed cytokine genes are clustered together, the nearest upstream neighbors of IFNG, which encodes the signature Th1 cytokine IFN-γ, are IL22 and IL26. These genes encode cytokines expressed mainly by Th17 cells rather than Th1 cells (McGeachy and Cua, 2008), and the nearest downstream coding gene is ~500 kb away. The insertion of repetitive elements and other complex structural rearrangements beginning ~70 kb upstream of mouse Ifng deleted IL26 in a common rodent ancestor (Schoenborn et al., 2007; She et al., 2008). This observation initially led us and others to search for cis-regulatory elements proximal to these structural alterations, resulting in the identification of several enhancers located within an ~120 kb region surrounding mouse Ifng (Hatton et al., 2006; Lee et al., 2004; Schoenborn et al., 2007; Shnyreva et al., 2004). However, there is increasing evidence that only a fraction of the cis-regulatory elements responsible for differences in appearance, behavior, function and disease risk are identified by such focused experimental approaches and by searches for regions of extended conservation of non-coding sequences (CNSs) as compared to unbiased approaches (Donnelly, 2008; Wray, 2007).
Here, using next-generation sequencing to map cis-regulatory elements (denoted by the presence of DNaseI hypersensitive sites) in an unbiased, high-resolution and comprehensive manner, we identify a cryptic IFNG regulatory element embedded within the neighboring IL26 gene. This element consists of a single binding site for the insulator factor CTCF (CCCTC-binding factor). This site is present in all mammals and has been selectively retained in rodents despite the lack of surrounding sequence conservation and near total deletion of IL26 from their genomes (Schoenborn et al., 2007; She et al., 2008). This element was occupied in vivo by CTCF and cohesins, as were two other elements located within and downstream of Ifng. These three CTCF binding sites and multiple interposed enhancers were approximated to each other and the Ifng promoter through Th1-specific chromatin looping. This looping and robust Ifng expression were dependent on normal levels of CTCF, and robust CTCF binding and CTCF-dependent chromatin looping were in turn dependent on the Th1 lineage-specifying transcription factor T-bet (Szabo et al., 2000).
RESULTS
Cell-specific chromatin accessibility and in vivo CTCF occupancy at the human IFNG locus
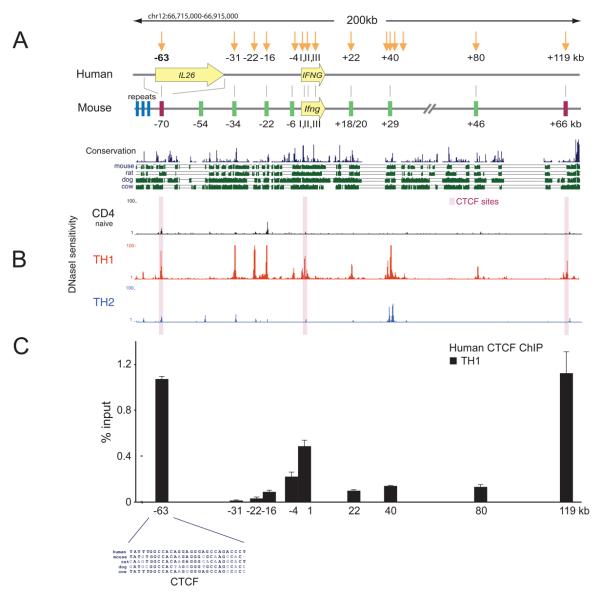
In rodents, the insertion of repetitive elements and other structural rearrangements in a common ancestor (Schoenborn et al., 2007; She et al., 2008) resulted in deletion of IL26, which in humans and other mammalian species is located ~40-70kb upstream of IFNG (Figure1A). These alterations make the murine locus difficult to analyze. Thus, to determine whether Th1-specific cis-regulatory elements might be present in addition to those previously identified in the mouse Ifng locus (Hatton et al., 2006; Lee et al., 2004; Schoenborn et al., 2007; Shnyreva et al., 2004), we performed an unbiased search in the human IFNG locus, which lacks the upstream structural rearrangements and repetitive elements that make the mouse locus difficult to analyze. This was done by applying to human naïve, Th1, and Th2 CD4+ T cells an approach we recently developed (digital DNaseI) for genome-wide high-resolution mapping of DNaseI hypersensitive sites (DHSs) (Hesselberth et al., 2009), which mark cis-regulatory regions of all types (Stalder et al., 1980; Wu, 1980). This revealed Th1-restricted DHSs at the promoter and within introns 1 and 3 of IFNG, which are orthologous to previously-described mouse Ifng DHSs I, II and III (Agarwal and Rao, 1998). Th1 cells also exhibited DHSs at −31kb, −22kb, −16kb, −4kb, +22kb, +40kb and +80 kb relative to the transcriptional start site of IFNG (Figure 1B); only two of these sites were similarly evident in the other cell types — the DHS at −16kb in naïve CD4+ T cells; 2 of the 3 DHSs clustered at +40kb in Th1 cells were also detected in Th2 cells. With the exception of DHS-22, each of these sites corresponds to regions of non-coding conservation, DHSs and peaks of T cell subset-specific chromatin modifications found at orthologous locations in the mouse Ifng locus (Figure 1A) (Hatton et al., 2006; Lee et al., 2004; Schoenborn et al., 2007; Shnyreva et al., 2004).
Figure 1. Cell-specific chromatin accessibility and CTCF occupancy in the IFNG locus.
(A) Schematic depicting the location of the human IL26 and IFNG genes and DNaseI hypersensitive sites found in human Th1 cells (downward arrows, shown with distance from the IFNG 5′ end) in relation to the orthologous gene and regulatory elements in the mouse Ifng locus reported previously (Schoenborn et al., 2007). Nearly all of the mouse IL26 gene has been deleted as a result of complex structural rearrangements, which include a set of tandem repeats shown just upstream of the mouse −70kb region where the CTCF-binding site orthologous to the human −63kb CTCF-binding site located in an intron of IL26 is present. Shown below is PhastCons conservation score and evolutionary conservation relative to mouse, rat, dog, and cow. (B) Chromatin accessibility shown as the density of mapped DNaseI cleavage sites (in a 150bp sliding window) from human naïve, Th1 and Th2 CD4+ T cells. DHSs correspond with peaks in the density profiles. (C) CTCF ChIP analysis in human Th1 cells. Results are the mean ± SD of 3 experiments. (D) Zoomed view of conserved CTCF binding motifs within DHSs located in human at −63 kb and mouse at −70kb relative to IFNG.
Inspection of the digital DNaseI profiles also disclosed a DHS located ~63kb upstream of human IFNG within the major intron of IL26, which was strong in Th1 cells and detectable though very weak in naïve and Th2 CD4+ T cells (Figure 1B). This element and another DHS that was located 119kb downstream of human IFNG, and detectable only in Th1 cells, did not coincide with previously identified mouse Ifng regulatory elements. Comparative genomic analysis of DHS-63 revealed that the core ~20 nucleotides of this element were shared among mammals (Figure 1D) and had survived the rodent-specific structural alterations that removed nearly all of IL26. This observation suggested that this previously cryptic element was functionally significant.
Consistent with this possibility, these ~20 nucleotides correspond precisely with a consensus binding site for CTCF (Figure 1D), a constitutive regulatory factor known to participate in chromatin looping and chromatin domain formation (Bushey et al., 2008; Cuddapah et al., 2009; Majumder et al., 2008; Phillips and Corces, 2009; Splinter et al., 2006). Remarkably, these nucleotides were precisely and selectively retained in the mouse within an ancestral remnant of IL26 located 70kb upstream of Ifng. CTCF binding motifs were also present within the DHS at +119 and within the Th1-specific DHS located at +1kb in intron 1 (Supplementary Figures 1 and 2), but not within the other DHSs. To determine if CTCF occupied these sites in human Th1 cells, we performed CTCF ChIP, which demonstrated CTCF occupancy at DHSs −63kb, +1kb and +119kb but not at the other DHSs (Figure 1C).
CTCF occupies orthologous sequences at the mouse Ifng locus and colocalizes with cohesins
To determine if CTCF could bind to orthologous sequences in mouse cells, we first used electrophoretic mobility shift assays, which demonstrated binding of recombinant murine CTCF to these specific nucleotide sequences (Supplementary Figure 1). Binding was blocked by cold competitor oligonucleotides but not by oligonucleotides in which the central bases of the CTCF consensus sequence were mutated.
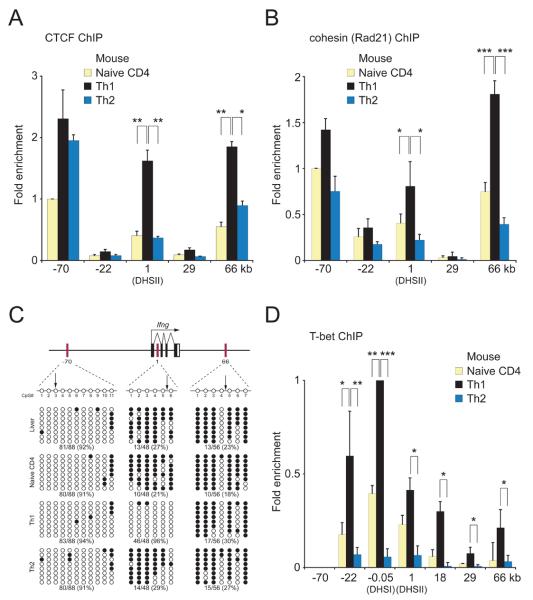
We next performed ChIP in murine naïve, Th1 and Th2 CD4+ T cells. CTCF occupancy was detected at the mouse −70kb region (orthologous to human DHS-63kb) in naïve CD4+ T cells; CTCF occupancy increased modestly and to a similar degree in Th1 and Th2 cells (Figure 2A). By contrast, CTCF occupancy at +1kb (located in the first intron of Ifng and corresponding to DHSII) and at +66kb (the mouse ortholog of human DHS+119kb) was somewhat above background in naïve CD4+ T cells, and was clearly and selectively increased in Th1 but not Th2 cells. CTCF occupancy was not detected at the −22kb and +29kb enhancers.
Figure 2. CTCF, cohesin, T-bet binding and DNA methylation state of CTCF-binding sites in the mouse Ifng locus.
(A) CTCF ChIP analysis in mouse naive, Th1 and Th2 CD4+ T cells. Results are the mean ± SD of 3 experiments; results are shown relative to the binding of CTCF at −70kb in naive T cells, which represented 0.9±0.3 % of input and was assigned a value of 1. P values (calculated by Student's t test) ≤0.05 are shown. (B) Rad21 (cohesin) ChIP analysis in mouse naive, Th1 and Th2 CD4+ T cells. Results are the mean ± SD of 2 experiments for naive T cells and 3 experiments for Th1 and Th2 cells; results are shown relative to the binding of cohesin at −70kb in naive T cells, which represented 2.6±0.3 % of input and was assigned a value of 1. (C) CD4+ T cell subset-specific CpG methylation at the CTCF-binding elements as determined by bisulfite sequencing. Arrows mark the positions of predicted CTCF-binding sites. Hepatocytes were used as a control. Rows, sequences of cloned alleles; filled circles, methylated CpG; open circles, unmethylated CpG. Below plots, the fraction and percentage of unmethylated CpGs. (D) T-bet ChIP analysis in mouse naive, Th1 and Th2 CD4+ T cells. Results are the mean ± SD of 2 experiments for naive T cells and 4 experiments for Th1 and Th2 cells; results are shown relative to the binding of T-bet at the Ifng promoter (DHSI) in naive T cells, which represented 4.9±0.4 % of input and was assigned a value of 1. ***, p<0.005; **p<0.01; *p<0.05
CTCF has been shown recently to colocalize to a large degree with cohesins and perhaps to recruit cohesins to specific locations in the genome (Parelho et al., 2008; Wendt et al., 2008). Consistent with these findings, when we performed ChIP using antibodies to the Rad21 component of the cohesin complex, Rad21 was found to colocalize with CTCF at the Ifng locus (Figure 2B).
CTCF occupancy at the +1kb and +66kb regions parallels T-bet occupancy not DNA demethylation
We next explored possible mechanisms for differences in CTCF binding between Th1 and Th2 cells. CTCF is known to preferentially associate with unmethylated CpG dinucleotides (Kanduri et al., 2000; Ling et al., 2006), but we found no consistent relationship between CTCF occupancy and CpG methylation at the −70kb or +66kb regions, since nearly all CpGs at −70kb were unmethylated and nearly all at +66kb were methylated, regardless of cell type. However, CpGs at +1kb were unmethylated in a Th1-specific fashion (Figure 2C, Supplementary Figure 2), as previously reported for this region (Lee et al., 2006; Wilson et al., 2009).
Although CTCF occupancy was not clearly correlated with the degree of DNA demethylation, a relationship was observed between T-bet and Th1-specific CTCF occupancy. The +1kb and +66 kb regions, which were occupied by CTCF and cohesins in a Th1-specific manner (Figure 2A,B), were also occupied by T-bet in Th1 cells (Figure 2D). Conversely, T-bet was not bound at the −70kb region, where CTCF and cohesin occupancy was not Th1-specific. Nor was T-bet occupancy sufficient for CTCF binding at the locus, because in Th1 cells (and to a limited degree in naïve cells) T-bet but not CTCF occupied the Ifng promoter (DHSI) and the −22kb and +18kb enhancers (Figure 2A, D), which lack CTCF consensus sequences (not shown).
The CTCF-binding sites can function as insulators
CTCF has been reported to be involved in regulatory domain boundary functions (Phillips and Corces, 2009), therefore we tested these three CTCF-binding sites for enhancer blocking and chromatin barrier activity (Bushey et al., 2008; Gaszner and Felsenfeld, 2006; Schoenborn et al., 2007). These assays revealed that the −70kb CTCF-binding element exhibited both barrier and enhancer-blocking functions comparable to the c-MYC insulator and superior to the +1kb and +66kb regions (Supplementary Figure 3). These activities were CTCF-specific, because they were abolished following mutation of the CTCF motifs. We also tested all of the aforementioned elements for enhancer or silencer activity in vitro, with negative results (Supplementary Figure 4).
Th1-specific chromatin looping in the Ifng locus
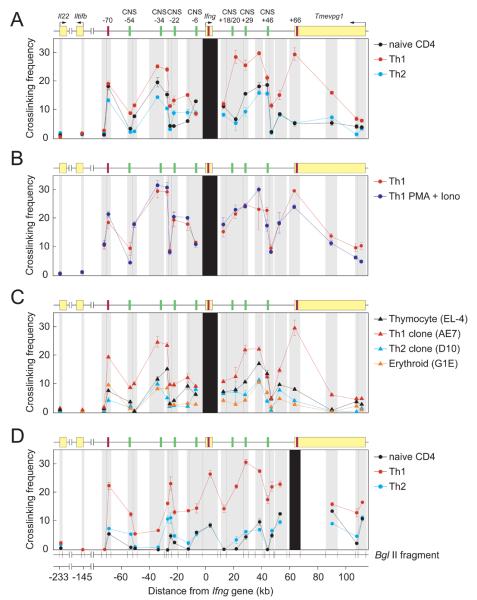
CTCF has been proposed to exert its functional effect on transcription of nearby genes through a mechanism involving chromatin looping (Bushey et al., 2008; Gaszner and Felsenfeld, 2006; Majumder et al., 2008; Phillips and Corces, 2009; Splinter et al., 2006; Williams and Flavell, 2008). We therefore used the chromosome conformation capture (3C) assay (Dekker, 2006; Tolhuis et al., 2002) to interrogate interactions between Ifng and 350 kb of surrounding sequences, including the CTCF-binding elements at −70kb and +66 kb, other known regulatory elements and control regions (Supplementary Figure 5). This analysis revealed strong interaction of the −70kb and +66 kb sites with Ifng in Th1 cells (Figure 3A). Two additional regions were also approximated to Ifng: the −40 to −27 kb fragment, encompassing the conserved −34kb enhancer, and the broad region containing the conserved +18/20kb and +29kb enhancers. Approximation of these distal regulatory elements to Ifng and to each other was similar in Th1 cells before and after activation with PMA plus ionomycin (Figure 3B), as was Ctcf and Tbx21 (encoding T-bet) mRNA abundance, whereas expression of Ifng increased >25-fold with activation (data not shown). Thus, Th1-specific chromatin looping and Ifng locus architecture was established during cellular differentiation and was not modified further by activation-induced Ifng expression.
Figure 3. Three-dimensional conformation of the mouse Ifng locus.
Relative cross-linking frequencies between a fixed anchor fragment bearing the Ifng gene (A-C) or the +66 CTCF site (D) and other BglII fragments using primary mouse naïve, Th1 and Th2 CD4+ T cells (A,B,D), primary mouse Th1 cells with or without restimulation with PMA + ionomycin for 6 hr (B), or Th1 (AE7), Th2 (D10G4.1), T cell progenitor (EL-4 thymocyte) or erythroid progenitor (G1E) cell lines (C). Black shading represents the position of the anchor fragment, and the locations and widths of gray shading indicate the positions and sizes of the BglII fragments whose cross-linking frequency to the anchor fragments was assessed. The location of the mouse Ifng and Il22 genes and their orientation are shown in the cartoon above. Iltifb, a degenerate pseudogene derived from an inverted duplication of Il22 and located between Il22 and Ifng, and the annotated non-coding transcript Tmevpg1 located downstream of Ifng, are also shown (Schoenborn et al., 2007). Data (error bars, s.d.) are representative of three independent experiments. Each signal was normalized to control templates and to interactions within the Gapdh locus.
These interactions were reduced in Th2 cells and intermediate between Th1 and Th2 cells in naïve CD4+ T cells (Figure 3A), paralleling differences in CTCF occupancy at +1kb and +66kb (Figure 2A). These interactions were also substantially greater in the AE7 Th1 cell line than in D10 Th2, T cell progenitor (EL-4 thymocyte) or erythroid progenitor (G1E) cell lines (Gregory et al., 1999) (Figure 3C). These findings were reproduced when the 3C assays were done with the +66 (Figure 3D) or −70 kb (Supplementary Figure 6) CTCF-binding elements rather than Ifng as the reference point, with the exception that the −34kb enhancer interacted less with these CTCF-binding elements than with Ifng.
shRNA-mediated knockdown of CTCF impairs the 3-dimensional conformation of the Ifng locus and Ifng expression
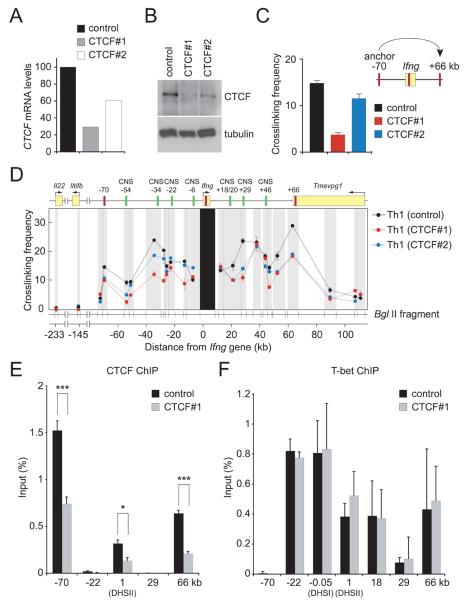
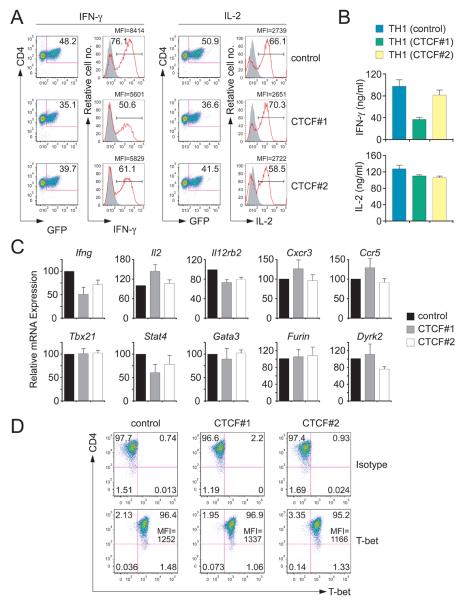
We next sought to determine whether CTCF is required for the formation of this Th1-specific Ifng locus conformation and for robust Ifng expression by Th1 cells. Transduction of Th1 cells with retroviruses expressing CTCF shRNAs (Supplementary Figure 7) led to marked (CTCF#1) to moderate (CTCF#2) reductions in CTCF mRNA and protein compared to cells transduced with the control retrovirus (Figure 4A,B). These reductions were directly paralleled by reduced interactions between the −70 and +66kb CTCF-binding elements, by reduced interactions between these elements, Ifng and the enhancers interposed between them (Figure 4C,D) and by reduced occupancy by CTCF at the −70kb, +1kb and +66 kb elements (Figure 4E). Conversely, CTCF knockdown had no effect on T-bet occupancy (Figure 4F).
Figure 4. shRNA-mediated knockdown of CTCF impairs CTCF binding at the Ifng locus and the 3-dimensional conformation of the locus.
Naive CD4+ T cells were cultured under Th1-polarizing conditions and transduced on day 1 with a bicistronic retroviral vector expressing GFP and CTCF#1, CTCF#2 or control (scrambled CTCF#1 sequence) shRNAs. GFP+ cells were purified by flow cytometric cell sorting on day 6 and CTCF mRNA and protein abundance were determined (A) by real-time RT-PCR normalized to β-actin and expressed as percentage of the control and (B) by Western blotting with tubulin serving as a loading control. (C) Relative cross-linking frequencies between the −70kb and +66 kb CTCF elements and (D) between Ifng as the fixed anchor fragment and other BglII fragments for Th1 cells transduced with CTCF#1, CTCF#2, or control retroviruses are shown by the red, blue, and black lines, respectively. Data (error bars, s.d.) are representative of 4 independent experiments. CTCF (E) and T-bet (F) occupancy at the Ifng locus in Th1 cells transduced with control or CTCF#1 shRNAs; only p values (calculated by Student's t test) ≤0.05 are shown. Data (error bars, s.d.) are representative of 4 independent experiments. ***p<0.005; *p<0.05.
The reduced interactions between regulatory elements observed in Th1 cells transduced with CTCF shRNAs compared to the control shRNA were paralleled by a reduction in the percentage of cells producing IFN-γ, the amounts produced per cell (as indicated by the mean fluorescence intensity, MFI) the amounts secreted into culture supernatants and Ifng mRNA abundance (Figure 5A-C). In contrast to IFN-γ, there was little or no effect of CTCF knockdown on the production of IL-2 (Figure 5A-C) or TNF (data not shown) or in the fraction of Th1 cells containing T-bet or amounts of T-bet they contained (Figure 5D). Tbx21 mRNA, mRNAs encoding the Th1 genes Cxcr3, Ccr5, Il12rb2 and Furin; mRNAs of the nearest upstream (Mdm1, Il22) and downstream (Dyrk2) gene neighbors of Ifng on mouse chromomome 10, and Gata3 mRNA were comparably abundant in control and CTCF shRNA-transduced cells (Figure 5C). Although Stat4 expression was reduced in CTCF shRNA-transduced cells, this is unlikely to be an important cause of reduced Ifng expression because expression of Furin, which is strongly Stat4-dependent (Pesu et al., 2006; Thieu et al., 2008), was not reduced by CTCF knockdown.
Figure 5. shRNA-mediated knockdown of CTCF impairs Ifng expression.
(A) Intracellular staining for the indicated cytokines in Th1 cells transduced with the control shRNA or CTCF shRNAs #1 or #2. The percentage of IFN-γ+ and IL-2+ cells among GFP+ Th1 cells and the mean fluorescence intensity (MFI) for cytokine containing cells are shown 6 hr after restimulation with PMA + ionomycin. (B) Concentrations (mean ± SD) of the indicated cytokines in culture supernatants measured by ELISA. (C) Expression of mRNA (mean ± SD) relative to cells transduced with the control shRNA for the indicated genes. (D) The percentage of Th1 cells containing T-bet protein and the amount as indicated by MFI are shown along with cells stained with an isotype control antibody.
CTCF-binding, Th1-specific Ifng locus conformation and Ifng expression are abrogated in the absence of T-bet
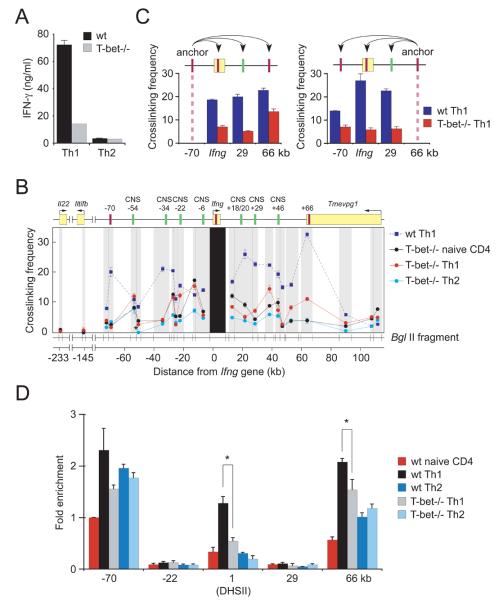
A major consequence of the CTCF-driven interactions is Th1-specific juxtaposition of the conserved enhancers at −34kb, +18/20kb, and +29kb with Ifng (Figure 3). In Th1 cells, CTCF and T-bet together occupy the +1kb and+66 kb regions (Figure 2); the −34kb, −22kb and +18/20 kb enhancers and the Ifng promoter are also occupied by T-bet (Chang and Aune, 2005; Hatton et al., 2006; Shnyreva et al., 2004). These observations suggested that T-bet might potentiate Ifng expression in part by promoting chromatin looping and CTCF binding. To test this possibility, we studied interactions in T-bet-deficient T cells, in which STAT4- and TCR-dependent activation of Ifng is intact but T-bet-dependent facilitation is absent. Consistent with previous reports (Szabo et al., 2000; Thieu et al., 2008), T-bet-deficient Th1 cells produced 4-5-fold less IFN-γ than wildtype cells (Figure 6A). This reduction was paralleled not only by reduced interactions of these enhancers with Ifng, but also by reduced interactions of the −70kb and +66kb CTCF-binding elements with Ifng and with one another (Figure 6 B,C). As a result, and in clear contrast to wildtype cells (Figure 3), the conformations of the Ifng locus in T-bet−/− Th1, Th2 and naïve CD4+ T cells did not differ greatly (Figure 6B). Consistent with these effects on chromatin looping, T-bet was required for the Th1-specific binding of CTCF at the +1kb and +66kb elements but not at the −70kb element (Figure 6D), where CTCF occupancy was not Th1-specific. These results suggest that T-bet regulates locus conformation, at least in part, through control of CTCF-binding and CTCF-dependent chromatin looping.
Figure 6. Impaired Ifng expression and three-dimensional organization of the Ifng locus in T-bet-deficient Th1 cells.
(A) Concentrations of IFN-γ (mean ± SD) in culture supernatants of wildtype (WT) and Tbx21-/- CD4+ T cells cultured in Th1 or Th2 conditions for 6 days. (B) Relative cross-linking frequencies between Ifng as the fixed anchor fragment and other BglII fragments and (C) between the −70kb CTCF or +66 CTCF element and the indicated BglII fragments for WT Th1 CD4+ T cells (dashed dark blue lines) and Tbx21-/- naive, Th1, and Th2 CD4+ T cells (black, red, and light blue lines, respectively). Results are presented as in Figure 3. (D) CTCF ChIP in naïve, Th1 and Th2 CD4+ T cells from WT or Tbx21-/- mice. Results are the mean ± SD of 2 experiments; results are shown relative to wildtype naïve T cells at −70kb, which represented 1.05±0.17% of input; only p values (calculated by Student's t test) ≤0.05 are shown. *p<0.05.
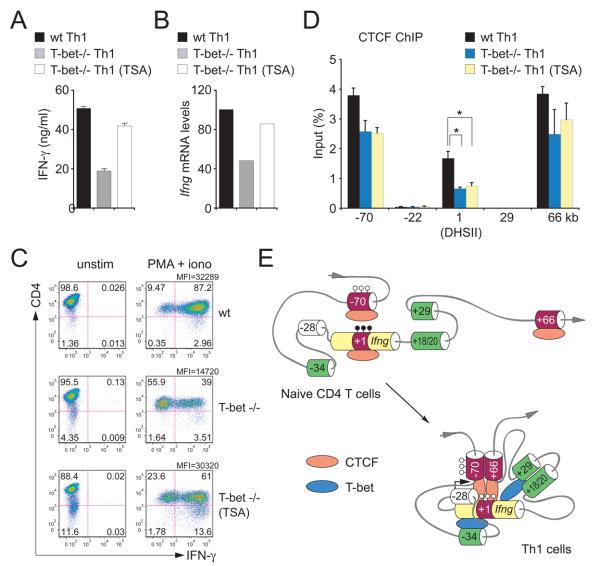
How might T-bet promote the binding of CTCF to the intronic and +66kb elements? We first considered the possibility that T-bet might interact with CTCF. To address this possibility, we performed immunoprecipitation followed by Western blotting with T-bet and CTCF antibodies. In primary Th1 cells, we readily detected T-bet but not CTCF in T-bet immunoprecipitates and readily detected CTCF but not T-bet in CTCF immunoprecipitates (data not shown). Similar results were obtained in EL-4 cells (which express endogenous CTCF but not endogenous T-bet) in which epitope-tagged T-bet was over-expressed (data not shown). Next we considered the possibility that chromatin modifications induced by T-bet (Miller et al., 2008; Wilson et al., 2009) might promote CTCF occupancy at the Ifng locus. To address this possibility, we differentiated T-bet-deficient CD4 T cells in Th1 conditions in the presence or absence of the histone deacetylase inhibitor trichostatin A (TSA), and compared IFN-γ production in these cells to wildtype Th1 cells. IFN-γ secretion and Ifng mRNA expression were augmented by TSA and were comparable to wildtype Th1 cells (Figure7A-C). However, TSA treatment did not result in increased occupancy of CTCF at the Ifng locus (Figure 7D). These results indicate that neither direct interaction between T-bet and CTCF nor T-bet-induced histone acetylation account for T-bet-dependent enhancement of CTCF occupancy.
Figure 7. In T-bet-deficient Th1 cells, the histone deacetylase inhibitor trichostatiin A (TSA) restores IFN-γ production and 3-dimensional locus conformation but not CTCF occupancy at the Ifnglocus.
(A) Concentrations of IFN-γ in culture supernatants, (B) Ifng mRNA expression, (C) intracellular IFN-γ and (D) CTCF ChIP for wildtype and T-bet-/- CD4+ T cells differentiated in Th1 conditions for 5 days with or without TSA. Data (error bars, s.d.) are representative of 4 independent experiments. *, p<0.05. (E) Proposed model for the three-dimensional conformation of the Ifng locus. In naïve CD4+ T cells, CTCF is bound primarily at −70kb and T-bet is not bound. Upon Th1 differentiation, T-bet binds to the Ifng promoter and to the CNS-34, CNS+18/20 and CNS+29 enhancers, and CTCF binds strongly at +1kb, in intron 1 of Ifng, and at +66kb in addition to −70kb. This binding contributes to and is required for the juxtaposition of each of these distal regulatory elements to Ifng and its promoter. In this active locus conformation, the CNS-34 enhancer is in close proximity to Ifng but more distant from the +1 and +66 CTCF-binding elements. Filled and open lollipops represent methylated and unmethylated CpGs, respectively.
DISCUSSION
Using a newly developed approach to map DHSs and starting with human cells in which the structural rearrangements that complicate analysis of the mouse Ifng locus are not present, we identified a DHS that represents a previously cryptic upstream locus orchestrator. This element consists of a single CTCF binding motif and is conserved in all mammals, even in rodents in which it is present within a sea of non-conserved sequences, which masked earlier detection by conventional computational searches for CNSs. This element was occupied by CTCF in situ.
This upstream CTCF-binding element was co-occupied by cohesins, had insulator activity and cooperated with two other CTCF/cohesin-binding elements (one located within the first intron of Ifng and coinciding with DHSII, and another newly identified by the presence of a DHS at +119kb in the human and located at +66kb in the mouse) to approximate interposed enhancers to the Ifng promoter in a Th1-specific and CTCF-dependent manner. In doing so, the upstream and downstream CTCF-binding elements demarcate a Th1 domain surrounding Ifng and its enhancers in which intervening non-regulatory sequences are looped out (Figure 7E). This looping segregates Ifng from the upstream Th17 cytokine genes encoding IL-22 and (in humans) IL-26 and from other sequences downstream. These findings are consistent with the emerging evidence that insulators interact with each other to create a physical, spatial and functional boundary that separates cis-regulatory elements and chromatin domains of one locus from its surroundings (Bushey et al., 2008; Gaszner and Felsenfeld, 2006; Hou et al., 2008). Within this domain, the +1kb intronic CTCF-binding element might in principle act as a barrier to transcription. However, while CTCF can block enhancer-promoter interactions when bound between these elements, CTCF can be dynamically evicted from transcribed regions by elongating pol II complexes (Bushey et al., 2008; Lefevre et al., 2008).
We found that the Ifng locus is largely unstructured in naïve CD4+ T cells, and became more compact through chromatin looping in Th1 cells and more linear in Th2 cells. By contrast, the Th2 cytokine locus assumes a pre-poised conformation in naïve T cells and NK cells, and this conformation is maintained in resting Th1 and Th2 cells (Spilianakis and Flavell, 2004). CTCF was recently shown to bind to 4 sites in the Th2 cytokine locus both in Th1 and Th2 cells (Ribeiro de Almeida et al., 2009), but whether it contributes to the pre-poised conformation of the locus observed in T and NK cells (Spilianakis and Flavell, 2004) was not explored. Th2-specific conformational changes occur when Th2 cells are activated. Activation induces expression of the architectural factor SATB1 (special AT-rich sequence binding protein 1), which binds to CNS1, CNS2 and multiple other sites in the Th2 cytokine locus to drive the formation of additional chromatin loops and more intimate interactions of the Il4, Il5 and Il13 promoters with each other and a number of other cis-regulatory elements (Cai et al., 2006). These interactions are lost and Th2 cytokine expression is compromised when SATB1 abundance is reduced (Cai et al., 2006). Thus, SATB1 appears to play a Th2-specific architectural role at the Th2 cytokine locus analogous to the Th1-specific effects of CTCF at the Ifng locus. However, the strategy is somewhat different — CTCF demarcates boundaries and chaperones interposed enhancers to a single, central target Ifng, whereas SATB1 binds at multiple sites in the Th2 cytokine locus to promote locus contraction in response to TCR-driven activation of Th2 cells. What might be the raison d'etre for this strategic difference? The Th2 locus may have the luxury of deferring its conformational changes until cells are activated because no lineage-forbidden cytokine genes are nearby that might be inappropriately activated if not constitutively constrained. By contrast, it may be important for Th1 cells to segregate Ifng from nearby Th17 cytokines or repressive chromatin even when at rest. These differences notwithstanding, the findings at the Ifng and Th2 cytokine loci together suggest that the cell type-specific differences in three-dimensional locus conformation promoted by CTCF and SATB1, respectively, facilitate high-level cytokine gene expression, though additional mechanisms may also be involved.
Our findings are consistent with and illuminate the mechanisms for findings reported recently while our work was being completed. Ribiero de Almeida et al (2009) observed an ~50% reduction in IFN-γ producing cells when CD4 T cells from CTCF-conditional knockout mice were cultured in Th1 conditions. However, CTCF occupancy at the Ifng locus and other possible mechanisms by which CTCF might facilitate IFN-γ production were not assessed, and the possibility that the difference in IFN-γ production was secondary to the intra-thymic developmental bottleneck these T cells experienced could not be excluded. Hadjur et al (2009) demonstrated that cohesins bind with CTCF at the IFNG locus in human Th1 cells, and that cohesins help to promote chromatin looping and IFN-γ production, but did not determine whether CTCF promoted IFN-γ production nor explore the basis for the Th1-specificity of CTCF and cohesin binding or actions. Our results demonstrate that CTCF acts to establish a Th1-specific Ifng locus architecture and to promote Ifng expression as naïve CD4 T cells differentiate into Th1 effectors, and show that these actions of CTCF and its occupancy at the Ifng locus are T-bet-dependent.
CTCF is a ubiquitous insulator and architectural factor. The vast majority of sites occupied by CTCF throughout the genome are common to all cell types, suggesting that it plays a general role in genome organization (Cuddapah et al., 2009; Kim et al., 2007; Phillips and Corces, 2009; Xie et al., 2007). Nonetheless, CTCF promoted the formation of an Ifng locus conformation conducive to robust Ifng expression in a Th1-specific manner. Cell context-dependent differences in CTCF-mediated chromatin looping and insulator function have been reported at the β-globin, H19/Igf2 and MHC class II loci (Ling et al., 2006; Majumder et al., 2008; Splinter et al., 2006). The context-dependent effects of CTCF may result from post-translational modifications affecting its ability to interact with other factors, differences in the abundance of such factors or epigenetic alterations that regulate its binding.
At the Ifng locus, the ability of CTCF to promote a three-dimensional locus architecture conducive to efficient Ifng expression correlated closely with the extent of its binding. In naïve CD4 T cells, CTCF was bound primarily at the upstream −70kb element, and the Th1 domain surrounding Ifng was not yet established (Figure 7F). Upon Th1 differentiation, T-bet abundance and occupancy at the Ifng promoter and at the +1kb and +66kb sites and at other sites within the locus increased and led to increased occupancy of the +1kb and +66kb sites by CTCF and cohesins. How T-bet promotes CTCF occupancy of these sites remains uncertain, though it does not appear to do so by direct interaction with CTCF or by augmenting histone acetylation at this locus. And while our results strongly suggest that CTCF promotes Th1-specific Ifng locus architecture and expression is mediated by binding to these three elements, mutation of these elements will be required to demonstrate this directly.
The data nonetheless show that a key and previously unappreciated Th1-specific regulatory effect of T-bet at the Ifng locus is to condition the binding of CTCF and thereby convert this ubiquitous architectural factor into a Th1-specific regulator of chromatin looping at the Ifng locus. This looping in turn helps to drive the juxtaposition of T-bet-binding enhancers and the flanking CTCF-binding elements to Ifng and to promote Ifng expression. These results also illuminate one mechanism giving rise to the small fraction of CTCF-binding sites in the genome that are cell lineage-specific (Cuddapah et al., 2009; Kim et al., 2007; Xie et al., 2007), and demonstrate a presumably widespread mechanism of cooperation between orchestrating (CTCF) and lineage-specific transcriptional enhancer (T-bet) factors in cell-specific gene regulation.
EXPERIMENTAL PROCEDURES
Mice
C57BL/6 and T-bet−/− mice from the Jackson Laboratory and P25 TCR transgenic mice (Tamura et al., 2004) were housed in specific pathogen-free conditions in the University of Washington Animal Facility. All experiments were approved by the Institutional Animal Care and Use Committee of the University of Washington.
Cell culture
Naive (CD44lo) CD4+ T cells were purified from P25 TCR transgenic mice by flow cytometric cell sorting and either analyzed directly or stimulated with 30 μg/ml of cognate peptide-25 (United Biochemical Research) in the presence of antigen presenting cells (CD4, CD8 and NK1.1-depleted B6 splenocytes), expanded under Th1 or Th2 conditions for 5 days and then analyzed for cytokine and transcription factor expression as described (Schoenborn et al., 2007). Naïve CD4+ T-bet−/− T cells were expanded in a similar manner with the exception that they were stimulated with CD3/CD28 T cell expander beads (Dynal Biotech). Where indicated, T-bet −/− T cells were expanded in Th1 conditions for 5 days in the presence of trichostatin A (12nM). For intracellular analysis of T-bet abundance, cells were stained for 30 min with anti-T-bet (4B10; Santa Cruz) or mouse IgG1 isotype (P3; eBioscience) in Hank's balanced-salt solution containig 1% BSA, 10 mM HEPES, and 0.5% saponin. EL-4 (Schoenborn et al., 2007), AE7 and D10.G4.1(Zheng and Flavell, 1997) and G1E-ER (Gregory et al., 1999) cells were maintained as described. Human naïve (CD45RA+) CD4+ T cells were isolated using an Automacs (Miltenyi), stimulated with human CD3+CD28 T cell expander beads in Aim V medium (Invitrogen) containing 2% human type AB serum in Th1 (10 ng/ml huIL-12, 5 μg/ml anti-IL4) or Th2 (10ng/ml huIL-4, 10 μg/ml anti-IFN-γ) conditions then expanded for 6-7 days in medium plus IL-2. Studies with human cells were approved by the University of Washington Human Subjects Division Institutional Review Board.
Digital DNase I sample preparation, library construction and sequencing
Cells were washed with PBS, pelleted and resuspended in ice-cold Buffer A (15mM Tris-Cl, pH 8.0, 15mM NaCl, 60mM KCl, 1mM EDTA, pH 8.0, 0.5mM EGTA, pH 8.0, 0.5mM spermidine). Nuclei were isolated by adding 2x lysis buffer (Buffer A containing 0.2% IGEPAL), mixing by inversion and incubating on ice for 8 minutes. Nuclei were then pelleted, resuspended and washed with Buffer A. DNaseI digestions (40-80U/mL) were carried out for 3 minutes at 37°C, then stop buffer (50mM Tris-Cl, pH 8.0, 100mM NaCl, 0.10% SDS, 100mM EDTA, pH 8.0, 10ug/mL RNAse A) was added followed by incubation at 55°C for 15 minutes, addition of proteinase K overnight incubation at 55°C. DNA was purified by gentle phenol chloroform extraction, then double-cut fragments of 100-500bp were isolated by sucrose gradient centrifugation as described (Sabo et al., 2006) with minor modification. Digital DNaseI libraries were constructed according to Illumina's protocol by end repair of 50 ng of purified DNA, 3′ adenine addition, column purification, adapter ligation, and 16 cycles of amplification. Amplified libraries were purified, quantified and sequenced by the University of Washington, High-Throughput Genomics Unit using an Illumina Genome Analyzer to produce 18-25 million uniquely mapping 27-mer reads per cell type (Hesselberth et al., 2009). The 5′ end of each uniquely mapping read from a digital DNaseI library corresponds to the DNaseI cleavage site. The density of DNaseI cleavage sites in a 150bp sliding (step 20bp) window was computed across the entire genome for each cell type and formatted for display as a track in the UCSC browser. Additionally, the number of DNaseI cleavages per nucleotide was computed and formatted for display as a UCSC track.
ChIP, DNA methylation, and boundary element assays
ChIP was performed with rabbit anti-CTCF (07-729; Millipore), anti-Rad21 (ab992; Abcam) or anti-T-bet (sc-21003; Santa Cruz) and quantified on an ABI PRISM 7300 system as described (Schoenborn et al., 2007). CpG methylation in the Ifng locus was quantified by sequencing of genomic DNA after bisulfite modification and PCR amplification.(Schoenborn et al., 2007). Insulator assays were performed using linearized constructs, as described (Schoenborn et al., 2007); significance was determined with a two-tailed, unpaired Student's t-test using Prism 4.0 software.
3C analysis
The 3C assay was done as described(Dekker, 2006; Tolhuis et al., 2002) with some modifications. 107 cells in 10 ml of RPMI 1640 + 10% FBS were crosslinked with 2% formaldehyde for 10 min, after which the reaction was quenched by addition of glycine (final concentration 0.125 M). Cells were lysed using ice-cold lysis buffer (10 mM Tris, pH 8.0, 10 mM NaCl, 0.2% NP-40) containing complete protease inhibitor cocktail (Roche) for 45 min. Nuclei were resuspended in 0.5 ml of restriction enzyme buffer containing 0.3% SDS then incubated for 1 h at 37°C on a rotator. Triton X-100 was added (final concentration 1.8%), then nuclei were incubated for 1 h to sequester the SDS. Crosslinked DNA was digested overnight with 400 U BglII, SDS was added (final concentration 1.3%), and the restriction enzyme was inactivated. The reaction was diluted with 8 ml of ligation buffer (50 mM Tris, pH 8.0, 10 mM MgCl2, 10 mM DTT, 1 mM ATP, and 1 mg/ml bovine serum albumin), and Triton X-100 was added to 1% followed by incubation for 1 h on a rotator. DNA fragments were ligated with 4,000 U of T4 ligase for 4 h at 16°C followed by 30 min at RT. Crosslinks were reversed by incubation with proteinase K overnight at 65°C. The samples were further incubated for 30 min at 37°C with RNase, and the DNA was purified by phenol extraction and ethanol precipitation. Ligation products were quantified in triplicate by quantitative TaqMan real-time PCR as described (Hagege et al., 2007). To correct for differences in ligation and PCR efficiency between different templates, we used a control template containing all possible ligation products. Equimolar amounts of three BAC clones spanning the mouse Ifng locus (RP23-401E11, RP23-325C14, and RP23-55O21) and a BAC spanning the mouse Gapdh locus (RP23-410F11) were mixed, then digested and ligated as described above; this product was used as the DNA reference standard. Ligation frequencies between the analyzed pairs were normalized to those detected between two restriction fragments in the Gapdh locus. Primers and probes for these assays and all other assays are listed in Supplementary Methods.
Expression of the zinc finger domains of CTCF and EMSA
Mouse CTCF cDNA was PCR-amplified and cloned into the pET15b. The His-tagged CTCF zinc finger domains were expressed in BL21 E. coli, purified by Ni-NTA resin (Qiagen), and concentrated by Microcon (Millipore). EMSA probes were generated as described(Sekimata and Homma, 2004). Binding was done in 10 μl of binding buffer (10 mM HEPES, pH 7.9, 1 mM dithiothreitol, 5 mM MgCl2, 0.5 mM ZnCl2, 60 mM KCl, 0.05% Nonidet P-40, 200 ng poly(dI·dC), 10% glycerol, and 50 μg/ml BSA) for 30 min at RT with the labeled probe and 2 μg of purified His-tagged CTCF. Unlabeled double-stranded oligonucleotides were added as competitors at 200-fold molar excess. Binding reactions were resolved by PAGE followed by autoradiography.
Co-immunoprecipitation assays
Whole-cell extracts were prepared from primary Th1 cells or from EL-4 cells that had been transfected with V5 epitope-tagged T-bet expression constructs as described (Miller et al., 2008) and incubated with antibodies to CTCF (07-729, Millipore), T-bet (4B10; Santa Cruz Biotechnologies) or to the V5 epitope (R960-25; Invitrogen). The immunocomplexes were then incubated with protein G beads for 1-2 h, washed and used for Western analysis.
CTCF shRNA retrovirus production and transduction
The target sequences for mouse CTCF shRNA (CTCF#1, CTCF#2, and control – containing scrambled CTCF#1 sequences) were designed according to protocol(Olson et al., 2006) and are listed in Supplementary Table 3. These oligonucleotides were used as templates to perform PCR with primers mir30-f and mir30-r, and the products were cloned into MSCV/LTR-U6miR30-PIG. (Dickins et al., 2005) Retroviruses were produced by transfecting Phoenix-Eco packaging cells and used to transduce T cells(Shnyreva et al., 2004). After 5 d of retroviral transduction, GFP+ cells were isolated by flow cytometric cell sorting and analyzed. RNA was isolated with the RNeasy kit (Qiagen), cDNA was synthesized with SuperScript II reverse transcriptase (Invitrogen) and mouse CTCF mRNA expression was assessed by quantitative real-time PCR using primers CTCF-f and CTCF-r and normalized to β-actin using primers actin-f and actin-r (Supplementary Table 4) (Shnyreva et al., 2004). For Western blotting, 20 μg of cell lysates were resolved on 7.5% SDS-PAGE gels, transferred to nitrocellulose membranes, which were incubated with a 1/500 dilution of mouse monoclonal anti-CTCF or anti-α-tubulin, HRP-conjugated anti-mouse antibody, then developed with ECL. Other cells were restimulated with 1 μM ionomycin (Sigma) and 25 ng/ml PMA (Sigma) for 6 h for analysis of cytokine production by ELISA or flow cytometric detection of intracellular cytokine production.
qRT-PCR
Presynthesized TaqMan Gene Expression assays (Applied Biosystems) were used for amplification of mRNA transcripts of Tbx21 (Mm0045096_m1), Gata3 (Mm00484683_m1), Stat4 (Mm00448890_m1), Il2 (Mm99999222_m1), Cxcr3 (Mm00438259_m1), Ccr5 (Mm01216171_m1), Dyrk2 (Mm01165529_m1), Furin (Mm00440646_m1), Il12rb2 (Mm00434200_m1), IL22 (Mm00444241_m1), Mdm1 (Mm00487650_m1). Target gene value was calculated relative to eukaryotic 18S rRNA (4319413E; Applied Biosystems) expression.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Stuart Orkin and Gerd Blobel for providing G1E cells, Matthias Merkenschlager for useful discussions, Ashutosh Chaudhry for the retroviral construct, and Alicia Brasfield for animal care. This work was supported in part by funding from NIH grants R01-AI071282 (C.B.W), R01-HD18184 (C.B.W.), N01-AI40069 (C.B.W. and J.A.S), U54-HG004592 (J.A.S.), R01-GM71923 (J.A.S.), R01-GM71852 (J.A.S.), R01-AI061061 (A.S.W) and the American Cancer Society (A.S.W) and by NIH predoctoral training grant GMT3207270 (S.A.M).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Apostolou E, Thanos D. Linking differential chromatin loops to transcriptional decisions. Mol Cell. 2008;29:154–156. doi: 10.1016/j.molcel.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Bushey AM, Dorman ER, Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell. 2008;32:1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Chang S, Aune TM. Histone hyperacetylated domains across the Ifng gene region in natural killer cells and T cells. Proc Natl Acad Sci U S A. 2005;102:17095–17100. doi: 10.1073/pnas.0502129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker J. Gene regulation in the third dimension. Science. 2008;319:1793–1794. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Gregory T, Yu C, Ma A, Orkin SH, Blobel GA, Weiss MJ. GATA-1 and erythropoietin cooperate to promote erythroid cell survival by regulating bcl-xL expression. Blood. 1999;94:87–96. [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. 2009 doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- Hatton RD, Harrington LE, Luther RJ, Wakefield T, Janowski KM, Oliver JR, Lallone RL, Murphy KM, Weaver CT. A distal conserved sequence element controls Ifng gene expression by T cell and NK cells. Immunity. 2006 doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Zhao H, Tanimoto K, Dean A. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc Natl Acad Sci U S A. 2008;105:20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DU, Avni O, Chen L, Rao A. A distal enhancer in the interferon-gamma (IFN-gamma) locus revealed by genome sequence comparison. J Biol Chem. 2004;279:4802–4810. doi: 10.1074/jbc.M307904200. [DOI] [PubMed] [Google Scholar]
- Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Lefevre P, Witham J, Lacroix CE, Cockerill PN, Bonifer C. The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol Cell. 2008;32:129–139. doi: 10.1016/j.molcel.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Majumder P, Gomez JA, Chadwick BP, Boss JM. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J Exp Med. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeachy MJ, Cua DJ. Th17 cell differentiation: the long and winding road. Immunity. 2008;28:445–453. doi: 10.1016/j.immuni.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated, but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activatin of developmental gene expression. Genes Dev. 2008;22:2980–2983. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson A, Sheth N, Lee JS, Hannon G, Sachidanandam R. RNAi Codex: a portal/database for short-hairpin RNA (shRNA) gene-silencing constructs. Nucleic Acids Res. 2006;34:D153–157. doi: 10.1093/nar/gkj051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Pesu M, Muul L, Kanno Y, O'Shea JJ. Proprotein convertase furin is preferentially expressed in T helper 1 cells and regulates interferon gamma. Blood. 2006;108:983–985. doi: 10.1182/blood-2005-09-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro de Almeida C, Heath H, Krpic S, Dingjan GM, van Hamburg JP, Bergen I, van de Nobelen S, Sleutels F, Grosveld F, Galjart N, Hendriks RW. Critical role for the transcription regulator CCCTC-binding factor in the control of Th2 cytokine expression. J Immunol. 2009;182:999–1010. doi: 10.4049/jimmunol.182.2.999. [DOI] [PubMed] [Google Scholar]
- Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimata M, Homma Y. Sequence-specific transcriptional repression by an MBD2-interacting zinc finger protein MIZF. Nucleic Acids Res. 2004;32:590–597. doi: 10.1093/nar/gkh249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, Cheng Z, Zollner S, Church DM, Eichler EE. Mouse segmental duplication and copy number variation. Nat Genet. 2008;40:909–914. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnyreva M, Weaver WM, Blanchette M, Taylor SL, Tompa M, Fitzpatrick DR, Wilson CB. Evolutionarily conserved sequence elements that positively regulate IFN-gamma expression in T cells. Proc Natl Acad Sci U S A. 2004;101:12622–12627. doi: 10.1073/pnas.0400849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skok JA, Gisler R, Novatchkova M, Farmer D, de Laat W, Busslinger M. Reversible contraction by looping of the Tcra and Tcrb loci in rearranging thymocytes. Nat Immunol. 2007;8:378–387. doi: 10.1038/ni1448. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder J, Larsen A, Engel JD, Dolan M, Groudine M, Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980;20:451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- Tamura T, Ariga H, Kinashi T, Uehara S, Kikuchi T, Nakada M, Tokunaga T, Xu W, Kariyone A, Saito T, et al. The role of antigenic peptide in CD4+ T helper phenotype development in a T cell receptor transgenic model. Int Immunol. 2004;16:1691–1699. doi: 10.1093/intimm/dxh170. [DOI] [PubMed] [Google Scholar]
- Thieu VT, Yu Q, Chang HC, Yeh N, Nguyen ET, Sehra S, Kaplan MH. Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination. Immunity. 2008;29:679–690. doi: 10.1016/j.immuni.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Williams A, Flavell RA. The role of CTCF in regulating nuclear organization. J Exp Med. 2008;205:747–750. doi: 10.1084/jem.20080066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci U S A. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W-P, Flavell R. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–598. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.