Abstract
Many vertebrate species use ultraviolet (UV) vision for such behaviors as mating, foraging, and communication. UV vision is mediated by UV-sensitive visual pigments, which have the wavelengths of maximal absorption (λmax) at ~360 nm, whereas violet (or blue) vision is mediated by orthologous pigments with λmax values of 390–440 nm. It is widely believed that amino acids in transmembrane (TM) I–III are solely responsible for the spectral tuning of these SWS1 pigments. Recent molecular analyses of SWS1 pigments, however, show that amino acids in TM IV–VII are also involved in the spectral tuning of these pigments through synergistic interactions with those in TM I–III. Comparisons of the tertiary structures of UV and violet pigments reveal that the distance between the counterion E113 in TM III and amino acid sites 87–93 in TM II is narrower for UV pigments than for violet pigments, which may restrict the access of water molecules to the Schiff base pocket and deprotonate the Schiff base nitrogen. Both mutagenesis analyses of E113Q and quantum chemical calculations strongly suggest that unprotonated Schiff base-linked chromophore is responsible for detecting UV light.
Keywords: Visual pigments, Crystal structure, UV vision, Violet vision, Vertebrates
1. Introduction
It was only the early 1990s that vision scientists started to realize that a wide range of vertebrate species use ultraviolet (UV) vision (Jacobs, 1992). Today, many fish, amphibian, reptilian, avian, and mammalian species are known to use UV vision in such behaviors as foraging, mating, and communication (e.g., Yokoyama, 2002). UV vision is mediated by visual pigments that have the wavelength of maximum absorption (λmax) at ~360 nm. These UV pigments and orthologous violet (or blue) pigments with λmax values of 390–440 nm belong to a short wavelength-sensitive type 1 (SWS1) pigment group (Yokoyama and Yokoyama, 1996; Yokoyama, 2000a; Ebrey and Koutalos, 2001). Each of these visual pigments consists of a transmembrane (TM) protein, an opsin, and the 11-cis-retinal chromophore. The protonated Schiff base-linked chromophore in solution absorbs light at 440 nm, whereas the unprotonated Schiff base-linked chromophore in solution has a λmax value of 365 nm (for a review, see Yokoyama, 2002). By interacting with an opsin, however, the chromophore in a visual pigment can detect a wide range of λmax values between 360 and 560 nm, which is known as the spectral tuning of visual pigments (Kochendoerfer et al., 1999).
Over the last few years, substantial progress has been made in identifying amino acids that are responsible for the spectral tuning of SWS1 pigments. In particular, by analyzing the absorption spectra of violet pigments in human, bovine, guinea pig, elephant, wallaby, frog, and avian ancestors (Yokoyama and Shi, 2000; Babu et al., 2001; Shi et al., 2001; Cowing et al., 2002; Fasick et al., 2002; Shi and Yokoyama, 2003; Parry et al., 2004; Yokoyama et al., 2005; Yokoyama and Tada, in press), a total of 11 amino acid sites in transmembrane (TM) helices I–III that are involved in the spectral tuning of SWS1 pigments have been identified. By engineering various ancestral pigments, it was also shown that most SWS1 pigments in vertebrate ancestors, including that in the common ancestor, were UV-sensitive (Shi and Yokoyama, 2003). Therefore, UV pigments in most contemporary species inherited the ancestral phenotype directly, whereas violet pigments have evolved from ancestral UV pigments by accumulating various amino acid replacements at least at the 11 critical sites. The avian lineage is the exception, where the ancestral pigment acquired violet-sensitivity and some descendants regained UV-sensitivity more recently (Shi and Yokoyama, 2003). Mutagenesis analyses of engineered ancestral pigments also show that the restoration of UV pigments in the avian lineage has been accomplished by a single amino acid change, S90C (Shi and Yokoyama, 2003; see also Yokoyama and Shi, 2000; Yokoyama et al., 2000; Wilkie et al., 2000).
In these analyses, one critical observation has received little attention; that is, the magnitude of λmax-shift caused by amino acid changes in a UV pigment and that caused by the reverse mutations in a closely related violet pigment can differ significantly (Shi et al., 2001). The search for the cause of the non-symmetrical λmax-shifts between the frog and its ancestral SWS1 pigments has led us to discover a new aspect of spectral tuning of SWS1 pigments. Namely, the spectral tuning of SWS1 pigments are controlled not only by amino acids in TM I–III but also by strong synergistic interactions within and between amino acids in TM I–III and IV–VII. Furthermore, using the crystal structure of the bovine rhodopsin (Palczewski et al., 2000; Okada et al., 2002), it is now possible to explore the chemical causes of spectral tuning of SWS1 pigments as well. By modeling tertiary structures of SWS1 pigments, we show that there exists a strong correlation between the molecular structures of SWS1 pigments and their λmax values. Both mutagenesis experiments of the counterion E113 and excitation energies of visual pigments evaluated by quantum chemical computations strongly suggest that the unprotonated Schiff base-linked chromophore is the chemical cause of UV-sensitivity.
2. Materials and methods
2.1. Construction of chimeric SWS1 pigments and site-directed mutagenesis
We engineered SWS1 pigments of major vertebrate ancestors by introducing a total of ~70 amino acid changes into several extant pigments, one of which is the ancestral amphibian pigment between African clawed frog (Xenopus laevis) and salamander (Ambystoma trigrinum) (ancestral pigment c; Shi and Yokoyama, 2003). The opsin cDNAs of the frog and ancestral amphibian pigment have been subcloned into pBlue-script vector KS(+). All mutant pigments with single and multiple amino acid changes were generated by using QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), and the primers were designed using the protocol recommended by the manufacture. Various chimeric pigments were constructed by recombining their cDNA fragments of restriction enzyme recognition sites, HindIII, BsmBI, and SphI, which digest the SWS1 opsin genes at codon sites 67, 99, and 152, and separate TM I and II, TM II and III, and TM III and IV, respectively. The amino acid sequences of all mutant pigments were confirmed by Sequitherm Excel II long-read kit (Epicentre Technologies, Madison, WI) with dye labeled M13 forward and reverse primers. Reactions were run on a LI-COR 4200LD automated DNA sequencer (LI-COR, Lincoln, NE). The confirmed mutants were subcloned into expression vector pMT5.
2.2. Expression and spectral analyses of visual pigments (in vitro assay)
The opsin cDNAs of full-length were subcloned into the EcoRI and SalI restriction sites of the expression vector pMT5 and these plasmids were expressed in COS1 cells by transient transfection (e.g., Yokoyama, 2000b). In short, the pigments were regenerated by incubating the opsins with 11-cis-retinal (Storm Eye Institute, Medical University of South Carolina) and purified using immobilized 1D4 (Industry Liaison Office, University of British Columbia) in buffer W1 (50 mM N-(2-hydroxyethyl)piperazine-N′-2-ethanesulfonic acid (HEPES) (pH 6.6), 140 mM NaCl, 3 mM MgCl2, 20% (w/v) glycerol and 0.1% dodecyl maltoside). UV visible spectra were recorded at 20 °C using a Hitachi U-3000 dual beam spectrophotometer. Visual pigments were bleached for 3 min using both 366 nm UV light illuminator and a 60 W standard light bulb equipped with a Kodak Wratten #3 filter at a distance of 20 cm. Data were analyzed using Sigmaplot software (Jandel Scientific, San Rafael, CA).
2.3. Statistical analyses and chemical modelling of amino acid sequences
So far, both amino acid sequences and in vitro assay-based absorption spectra have been characterized for 17 SWS1 pigments (Table 1). Using these pigments (Table 1), the nucleotide sequences of ancestral pigments were inferred using a likelihood-based Bayesian method (Yang, 1997).
Table 1.
The λmax values (in parentheses) of SWS1 pigments that are determined by in vitro assay
| Pigment | Reference for λmax | GenBank |
|---|---|---|
| Goldfish (P359) | Yokoyama (2000a) | D85863 |
| Zebrafish (P355) | Chinen et al. (2003) | AB087810 |
| Frog (P423) | Yokoyama (2000a) | U23463 |
| Salamander (P356) | Ma et al. (2001) | AF038948 |
| Chameleon (P358) | Yokoyama (2000a) | AF134192 |
| Gecko (P360) | Yokoyama and Takenaka (unpublished) | AY024356 |
| Pigeon (P393) | Yokoyama (2000a) | AF149234 |
| Zebra finch (P359) | Yokoyama (2000a) | AF222331 |
| Chicken (P415) | Yokoyama (2000a) | M92039 |
| Budgerigar (P363) | Yokoyama (2000a) | Y11787 |
| Human (P414) | Shi et al. (2001) | M13295 |
| Mouse (P359) | Yokoyama (2000a) | U49720 |
| Rat (P358) | Yokoyama (2000a) | U63972 |
| Bovine (P438) | Fasick et al. (2002) | U92557 |
| Guinea pig (P420) | Parry et al. (2004) | AY552608 |
| Elephant (P419) | Yokoyama et al. (2005) | AY686753 |
| Wallaby (P420) | Deeb et al. (2003) | AY286017 |
Bovine, Bos taurus; Budgerigar, Melopsittacus undulatus; Chameleon, Anolis carolinensis; Chicken, Gallus gallus; Elephant, Loxodonta africana; Frog, Xenopus laevis; Goldfish, Carassius auratus; Gecko, Gekko gekko; Guinea pig, Cavia porcellus; Human, Homo sapiens; Mouse, Mus musculus; Pigeon, Columba livia; Rat, Rattus norvegicus; Salamander, Ambystoma tigrinum; Wallaby, Macropus eugenii; Zebra finch, Taeniopygia guttata; Zebrafish, Danio rerio.
At present, the bovine rhodopsin has been crystallized (Palczewski et al., 2000; Okada et al., 2002, 2004). Using these crystal structures of the rhodopsins as templates, we modelled specific structural changes that are associated with the transition between absorption spectra of UVand violet pigments using the SWISS MODEL program (www.expasy.ch/swissmod), which adopt energy minimization procedure, with Pdb Viewer (HYPERLINK http://www.expasy.ch/spdbv).
3. Results and discussion
3.1. Spectral tuning of visual pigments
The molecular basis of spectral tuning of SWS1 pigments has been studied first considering avian species and then mammalian species. The initial site-directed mutagenesis results have provided a rather simple picture of the molecular genetic basis of UV vision; the violet pigments of chicken and pigeon become UV-sensitive by S90C (Yokoyama et al., 2000), whereas the UV pigments of zebra finch (Yokoyama et al., 2000) and budgerigar (Wilkie et al., 2000) become violet-sensitive by C90S. When the mouse UV and human blue pigments were analyzed next, the picture of spectral tuning became far more complicated. That is, the λmax value of the mouse pigment was transformed into that of the human pigment by making the amino acid changes, F46T, F49L, T52F, F86L, T93P, A114G, and S118T in TM I–III. Likewise the seven reverse mutations in the human pigment resulted in a λmax value virtually identical to that of the mouse pigment (Shi et al., 2001). The transformations between the two pigments are characterized by strong synergistic interactions among the seven amino acids. In particular, the synergistic interactions were so intense that the λmax-shifts from the mouse pigment to the human pigment had been detected only when multiple amino acids were replaced (Shi et al., 2001). Today, we also know that some amino acid changes at site 86 can cause significant λmax-shifts individually. For example, F86Y, F86V, and F86S increase the λmax value by 17–66 nm depending on the background of a specific visual pigment (Fasick et al., 2002; Cowing et al., 2002; Shi and Yokoyama, 2003; Parry et al., 2004; Yokoyama and Tada, in press; Yokoyama et al., 2005). The effects of these amino acid changes on the λmax-shift are also subjected to strong synergistic interactions with other amino acids.
In addition, amino acid changes at site 97 in bovine (P438) pigment (Fasick et al., 2002), at site 113 in frog (P423) pigment (Babu et al., 2001), and at site 116 in the avian ancestral pigment (Shi and Yokoyama, 2003) and elephant (P419) pigment (Yokoyama et al., 2005) shift the λmax value of SWS1 pigments significantly. At present, therefore, a total of 11 critical amino acid sites only in TM I–III are known to be involved in the spectral tuning of SWS1 pigments. As noted earlier, this argument ignores another observation that the magnitude of λmax-shift caused by specific amino acid changes in UV pigment can be very different from that caused by the reverse mutations in violet pigments. Such non-symmetric effects of amino acid changes in UV and violet pigments must be caused by interactions with other amino acids. Then, where are these interactions coming from? This motivates the question: where are these amino acids located?
To study the effects of such synergistic interactions of different amino acids on the λmax-shifts, it is helpful to construct various chimeric pigments between UV and violet pigments (e.g., Shi et al., 2001). When the TM I of the mouse pigment is replaced by that of the human pigment, the chimeric pigment is unstable and its λmax value cannot be evaluated. Therefore, using frog (P423) pigment and its ancestral amphibian pigment with a λmax value of 359 nm (pigment c, Shi and Yokoyama, 2003), we constructed two sets of seven chimeras: 1) those containing TM I, II, III, I+II, I+III, II+III, and I+II+III of the frog pigment with the background of the ancestral pigment and 2) those containing the corresponding segments of the ancestral pigment with the background of the frog pigment.
Table 2 shows the λmax values of the ancestral, frog, and 14 chimeric pigments measured using in vitro assay. The ancestral pigment with TM II+III of the frog pigment (chimera 6) has a λmax value to 421 nm, whereas the frog pigment with TM II +III of the ancestral pigment (chimera 9) has a λmax to 361 nm. Thus, just like the human (P414) pigment (Shi et al., 2001), it appears that only TM I–III, or more specifically TM II and III, are solely responsible for the functional differentiation of the frog pigment. However, the ancestral pigment with TM III of the frog pigment (chimera 3) increases the λmax value by 51 nm, whereas the frog pigment with TM III of the ancestral pigment (chimera 12) reduces the λmax value only by 15 nm, again showing strong non-symmetric effects of amino acid changes in the UV and violet pigments. We can also see that the ancestral pigment with TM II of the frog pigment (chimera 2) and that with TM II and TM IV–VII of the frog pigment (chimera 10) increase the λmax value by 24 and 45 nm, respectively, demonstrating that TM IV–VII are involved in the spectral tuning of the frog pigment. This is totally unexpected from our current understanding of the molecular basis of spectral tuning of SWS1 pigments.
Table 2.
The λmax values for the chimeras between the frog and its ancestral pigments
| Visual pigment | TMa |
λmax (nm) | |||
|---|---|---|---|---|---|
| I | II | III | IV–VII | ||
| Ancestor | A | A | A | A | 359 |
| Chimera 1 | F | A | A | A | 359 |
| Chimera 2 | A | F | A | A | 383 |
| Chimera 3 | A | A | F | A | 410 |
| Chimera 4 | F | F | A | A | 389 |
| Chimera 5 | F | A | F | A | 417 |
| Chimera 6 | A | F | F | A | 421 |
| Chimera 7 | F | F | F | A | 422 |
| Chimera 8 | A | A | A | F | 360 |
| Chimera 9 | F | A | A | F | 361 |
| Chimera 10 | A | F | A | F | 404 |
| Chimera 11 | A | A | F | F | 404 |
| Chimera 12 | F | F | A | F | 408 |
| Chimera 13 | F | A | F | F | 404 |
| Chimera 14 | A | F | F | F | 418 |
| Frog | F | F | F | F | 423 |
A — Ancestral segment; F — Frog segment.
In order to evaluate all individual and synergistic effects of different TM segments on the λmax-shift, we define 1) Z and θI, θII, θIII, and θIV–VII as the λmax of the ancestral pigment and the effects of replacing TM I, II, III, and IV–VII of the ancestral pigment by the corresponding segments of the frog pigment on the λmax-shift, respectively, and 2) their synergistic effects θI × II, θI × III, θI × IV–VII, θII × III, θII × IV–VII, θIII × IV–VII, θI × II × III, θI × II × IV–VII, θI × III × IV–VII, θII × III × IV–VII, and θI × II × III × IV–VII on the λmax-shift. Then, all λmax values in Table 2 can be expressed as functions of Z and combinations of different θ values. Solving the 16 equations, we can show that 1) the ancestral pigment has a λmax value of 359 nm and 2) interactions II×IV–VII and I×II×III×IV–VII increase the λmax value by 20 and 14 nm, respectively, and interactions III×IV–VII, I×II×IV–VII, I×III×IV–VII, and II×III×IV–VII decrease the λmax by 7, 3, 8, and 17 nm, respectively. Thus, although their individual effects on the λmax-shift are nil (see chimeras 1 and 8), TM I and IV–VII are also involved in the spectral tuning of the frog pigment by interacting with other TM segments. The difference between the λmax values of chimeras 2 and 10 can be easily explained by considering the interaction between TM II and TM IV–VII. For unknown reasons, when all of these synergistic effects involving TM IV–VII are combined together, the total effect on the λmax-shift is abolished. It seems most likely that this negligible overall effect of TM IV–VII on the λmax-shift is caused by some structural requirement of visual pigment, providing a new dimension in exploring structure and function of visual pigments. The negligible overall effect is the reason why the spectral tuning of SWS1 pigments appears to be modulated solely by amino acid differences in TM I–III.
The comparative analyses of the ancestral and frog pigment illustrate that the differential λmax-shifts caused by forward and reverse mutations in UV and violet pigments are due not only to amino acids in TM I–III but also to interactions of amino acids within and among TM I–VII. Thus, the molecular basis of spectral tuning of SWS1 pigments is far more complicated than previously thought. Such interactions will become particularly important in studying the evolutionary processes of spectral adjustments (or adaptations) of visual pigments to certain ecological environments. The frog experiment does not specify which of TM IV–VII are actually involved in the synergistic interaction and the molecular basis of the spectral tuning of SWS1 pigments remains to be clarified.
3.2. Evolution of UV and violet (or blue) pigments
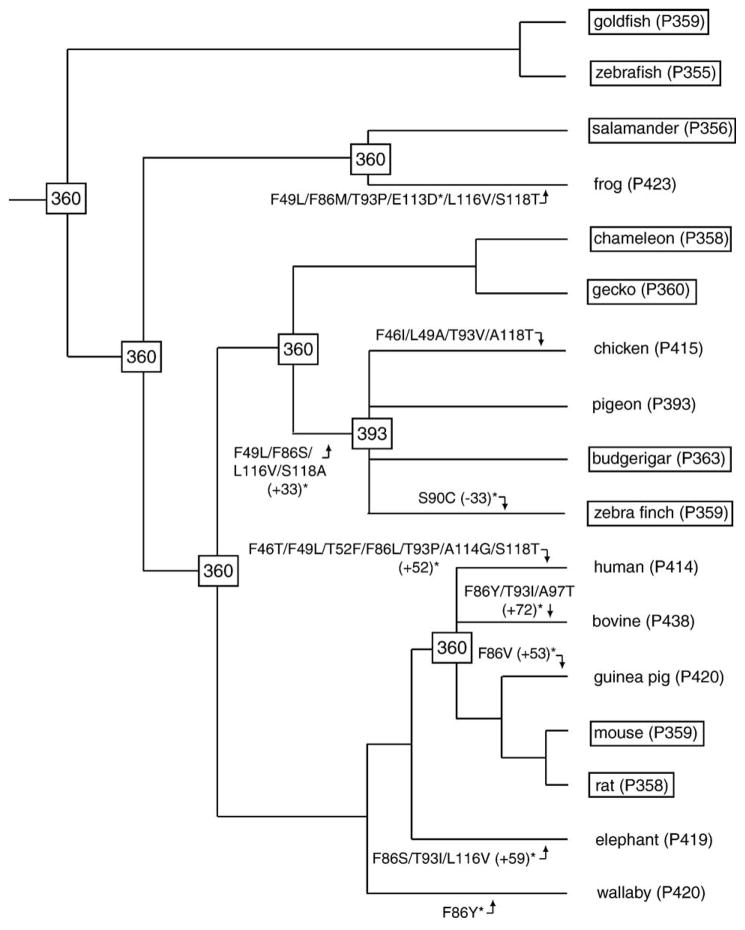
Despite our ignorance of the molecular basis of spectral tuning in the SWS1 pigments, it is still possible to explore the evolutionary processes of UV and violet vision by engineering SWS1 pigments of ancestral organisms. Fig. 1 shows a composite evolutionary tree of the 17 SWS1 pigments, which is consistent with the organismal tree based on molecular and paleontological data (e.g., Yokoyama and Radlwimmer, 2001; Yokoyama and Takenaka, 2005). Given the phylogenetic tree topology in Fig. 1, the ancestral amino acid sequences have been inferred by a likelihood-based Bayesian method by using a modified version of the Jones, Taylor, and Thornton model (Yang, 1997). Using Dayhoff and equal-input models, the same amino acid replacements at these sites are inferred (see also Shi and Yokoyama, 2003). Then, visual pigments at major nodes have been engineered and their λmax values are evaluated (Shi and Yokoyama, 2003). From the inferred amino acid sequences, we can trace amino acid replacements at the 11 critical sites in the eight violet pigments (Fig. 1). So far, the effects of these changes on the λmax-shift have been examined experimentally for the avian ancestral UV and violet pigments (Shi and Yokoyama, 2003) and the contemporary violet pigments of frog (Babu et al., 2001), chicken, pigeon, zebra finch (Yokoyama et al., 2000), budgerigar (Wilkie et al., 2000), human (Shi et al., 2001), bovine (Fasick et al., 2002; see also Cowing et al., 2002), guinea pig (Parry et al., 2004), elephant (Yokoyama and Takenaka, 2005), and wallaby (Yokoyama and Tada, in press) (Fig. 1).
Fig. 1.
The composite phylogenetic tree and amino acid replacements at sites 46, 49, 52, 86, 90, 93, 97, 114, 116, and 118 for 18 SWS1 pigments. The numbers in boxes are the λmax values of engineered ancestral pigments (Shi and Yokoyama, 2003). The UV pigments are also boxed. Stars (*) indicate that the effects of amino acid replacements have been actually tested using in vitro assay. The branch lengths are not in scale.
The inferred amino acid sequences reveal that several identical amino acid replacements have occurred independently multiple times; all F49L, F86S, F86Y, T93P, T93I, and S118T occurred twice and L116V three times (Fig. 1). In the future, such parallel amino acid replacements may be used to test whether or not these sites have undergone positive selection (e.g., Zhang and Kumar, 1997; Briscoe, 2002). Indeed, using such a method, we have shown that positive evolution of triple amino acid replacements (S180, Y277F, and T285) has occurred in the M/LWS pigment group (Yokoyama and Takenaka, 2005). As more data accumulate, statistical tests based on the numbers of synonymous and nonsynonymous substitutions per codon site will also be used (e.g., Spady et al., 2005). It should be stressed, however, that the effects of all potentially important amino acid changes on the λmax-shift must be examined experimentally (Yokoyama and Takenaka, 2005).
3.3. E113 and spectral tuning of violet pigments
As noted earlier, the protonated and unprotonated Schiff base-linked chromophores detect light at 440 and 365 nm, respectively. This observation casts an interesting possibility that the spectral tuning of SWS1 pigments may depend on whether or not their Schiff base-linked chromophores are protonated. At present, four chemical hypotheses that explain the modulation of absorption spectra of various visual pigments have been proposed: 1) twisting about the single and double bonds of the polyene chain of the 11-cis-retinal; 2) electrostatic interactions between the Schiff base-linked chromophore and the counterion (E113); 3) interactions between the chromophore with other charged, polar, or polarized groups; and 4) unprotonated Schiff-base chromophore in UV pigments (Ebrey and Takahashi, 2002). The protonation of the chromophore is caused by the counterions E113 and E181 (e.g., Ebrey and Takahashi, 2002). Thus, E113 can have a significant role in the spectral tuning of SWS1 pigments.
Biochemical roles of the counterion E113 in the spectral tuning of SWS1 pigments have been studied experimentally by Shi et al. (2001), Babu et al. (2001), and Fasick et al. (2002). For example, when E113Q is introduced into mouse (P359) pigment, the mutant pigment has a λmax value of 352 nm and is still UV-sensitive, having little effect on the spectral sensitivity in the mouse UV pigment. Since Q113 does not work as a counterion, this result implies that E113 is not working as the counterion. When E113Q is introduced into the human blue opsin, the mutant opsin fails to bind to 11-cis-retinal (Shi et al., 2001; see also Fasick et al., 2002). However, when E113Q is introduced into the mouse pigment with seven mutations, F46T/F49L/T52F/F86L/T93P/A114G/S118T, the mutant pigment shifts its λmax value from 411 to 369 nm. Since the λmax value and tertiary structure of the mouse UV pigment with the seven specific amino acid changes and those of the human blue pigment are very close to each other (see below), it is most likely that the human blue pigment has a protonated Schiff base-linked chromophore (see also Yokoyama and Shi, 2000). Therefore, the drastic decrease in the λmax value seems to occur because the Schiff base-linked chromophore was originally protonated (see also Sakmar et al., 1989; Zhukovsky and Oprian, 1989; Nathans, 1990a,b; Nakayama and Khorana, 1991). The same suggestion has been made by Babu et al. (2001) and Fasick et al. (2002) using frog (P423) and bovine (P438), respectively.
If E113 is not used for the protonation of the Schiff base, why do the mouse UV pigment, and virtually all other pigments in that matter, still have E113? It is most likely that E113 has another function in phototransduction. For example, when exposed to UV light, it takes the mouse pigment with E113Q about 10 min to shift its λmax from 352 to 380 nm. Compared with this, the wild-type mouse UV pigment achieves a λmax value of 380 nm even after 2 min of UV exposure. Thus, although it has very little effect on the λmax-shift, E113 in the mouse UV pigment seems important in photobleaching and possibly subsequent phototransduction, as suggested by Shi et al. (2001).
3.4. Spectral tuning and tertiary structure of UV and violet pigments
In order to understand the molecular basis of spectral tuning of visual pigments, it is essential to relate the λmax-shifts caused by certain amino acid changes to the changes in the tertiary and chemical structures of visual pigments. For that purpose, it would be ideal to have the crystal structures of both UV and violet pigments. At present, however, only the bovine rhodopsin has been crystallized (Palczewski et al., 2000; Okada et al., 2002, 2004). Therefore, we will evaluate two sets of closely related distance measures: 1) the distance between the oxygen in the backbone of a particular amino acid and that of E113 in UV pigment (d1) and that between those in violet pigment (d2) and 2) the distance between the Schiff base nitrogen and the oxygen in the backbone of a particular amino acid in UV pigment (e1) and that in violet pigment (e2). Since the distances between the two charged atoms of E113 and the Schiff base nitrogen are very similar (3.3 and 3.8 Å) for all visual pigments considered here, d and e values represent two distance measures from slightly different angles. To distinguish the structures of UV and violet pigments, we shall simply search for amino acid sites where either the relationship |d1 − d2| or |e1 − e2| is larger than a certain value, t. In the following, we set t=0.05 Å and identify all structural changes of visual pigments caused by critical amino acid changes.
3.4.1. Mouse UV pigment and human blue pigment
When TM II, III, I+II, II+III, and I+II+III of the human (P414) pigment are introduced into the mouse (P359) pigment, the chimeric pigments increase their λmax values from 359 to 381, 363, 396, 405, and 414 nm, respectively (Shi et al., 2001). As noted earlier, when the TM I of human pigment is introduced into the mouse pigment, the chimeric pigment is unstable and its λmax value cannot be evaluated. However, the λmax-shifts caused by the TM I, II, and III of the human pigment can be traced to amino acid changes F46T/F49L/T52F, F86L/T93P, and A114G/S118T, respectively (Shi et al., 2001).
When the tertiary structures of the mouse and human pigments are compared, |d1 − d2| > 0.05 Å holds only for amino acid sites 42, 43, and 45 in TM I and 89, 91, 92, and 93 in TM II (Table 3). Note that these sites and the seven critical amino acid sites that are involved in the spectral tuning of the mouse and human pigments do not necessarily coincide. In fact, out of the seven critical amino acid sites that are involved in the functional differentiation, only site 93 is included in the structural change between the two pigments. Comparisons of the seven pairs of d1 and d2 values show that the open spaces between E113 in TM III and TM I and between E113 and TM II are often narrower in the mouse UV pigment than in the human blue pigment (Table 3).
Table 3.
Distances (in Å) between amino acids and E113 in the mouse and human pigments, where the corresponding distances between the amino acid and the two wild-type (WT) pigments are >0.05 Å
| Amino acid | Mouse (P359)a |
Human (P414) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| WT | F86L | TM III | TM II | TM I– II | TM II– III | 7 ch. | WT | 7 ch. | |
| (359) | (359) | (363) | (381) | (395) | (403) | (411) | (414) | (360) | |
| A42 | 15.86 | 15.86 | 15.94 | 15.89 | 16.16 | 15.96 | 16.22 | 16.22 | 15.90 |
| F43 | 13.87 | 13.87 | 13.95 | 13.88 | 13.74 | 13.94 | 13.80 | 13.80 | 13.88 |
| G45 | 15.19 | 15.18 | 15.26 | 15.21 | 15.39 | 15.27 | 15.45 | 15.44 | 15.22 |
| F89 | 9.26 | 9.72 | 9.31 | 9.64 | 9.64 | 9.68 | 9.68 | 9.69 | 9.17 |
| V91 | 5.63 | 5.62 | 5.71 | 5.52 | 5.52 | 5.58 | 5.58 | 5.57 | 5.62 |
| F92 | 8.55 | 8.52 | 8.63 | 8.42 | 8.42 | 8.48 | 8.48 | 8.30 | 8.44 |
| T93 | 9.83 | 9.83 | 9.91 | 9.84 | 9.84 | 9.90 | 9.90 | 9.91 | 9.84 |
TM I, TM II, and TM III contain AA changes F46T/F49L/T52F, F86L/ T93P, and A114G/S118T, respectively. Note that the λmax-shifts caused by these amino acid changes can differ slightly from those caused by replacing each TM segment (Shi et al., 2001). The 7 ch. in the mouse UVand human blue pigments contain F46T/F49L/T52F/F86L/T93P/A114G/S118T and the reverse changes, respectively (Shi et al., 2001). The numbers in parentheses refer to λmax values of visual pigments.
To study the transitional steps between the UV and blue pigments, we have also evaluated d1 values with six sets of amino acid changes: 1) F86L in TM II; 2) A114G/S118T in TM III; 3) F86L/T93P in TM II; 4) F46T/F49L/T52F/F86L/T93P in TM I–II; 5) F86L/T93P/A114G/S118T in TM II–III; and 6) F46T/F49L/T52F/F86L/T93P/A114G/S118T in TM I–III. The respective changes are known to increase the λmax of mutant pigment by 0, 4, 22, 36, 44, and 52 nm (Shi et al., 2001). Table 3 shows that as the λmax value of a mutant pigment gets closer to that of the human pigment, their tertiary structure becomes more similar; on the other hand, the structure of the human pigment with the seven reverse amino acid changes also becomes virtually identical to that of the mouse pigment.
3.4.2. Mouse UV pigment and bovine blue pigment
The mouse pigments with F86Y and F86Y/T93I/A97T have λmax values of 424 and 430 nm, respectively (Fasick et al., 2002). When the e1 and e2 distances are measured, |e1 − e2| > 0.05 Å holds only for C87 and F89 (Table 4). Again, as the λmax value of a mutant gets closer to that of the bovine pigment, their tertiary structure becomes similar. However, for C87, the e1 is still shorter than e2 by 0.2 Å. The incomplete change in the tertiary structure of the mouse pigment with the triple mutations reflects the fact that the λmax of the mutant pigment is still 8 nm lower than that of the bovine (P438) pigment. The structural difference can again be characterized by the narrower space between F89 and the Schiff base nitrogen in the mouse pigment than that in the bovine pigment.
Table 4.
Distances between amino acids and the Schiff base nitrogen in the mouse and bovine (P438) pigments, where the corresponding distances between the amino acid and the Schiff base nitrogen of the two wild-type (WT) pigments are >0.05 Å
| Mouse (P359) | Bovine (P438) | |||
|---|---|---|---|---|
| AA | WT | F86Ya | F86Y/T93I/A97Ta | WT |
| (359) | (424) | (430) | (438) | |
| C87 | 11.45 | 10.56 | 10.57 | 10.79 |
| F89 | 11.98 | 12.05 | 12.23 | 12.23 |
The λmax values (nm) are taken from Fasick et al. (2002).
3.4.3. UV and violet pigments in birds
When d1 values for zebra finch (P358) pigment and d2 values for chicken (P415) and pigeon (P393) pigments are evaluated, we cannot find any amino acid sites with |d1 − d2| > 0.05 Å. Thus, it is most likely that the λmax differences in the avian UV and violet pigments are caused solely by the structural change at the amino acid site 90 or by other factors affected by it. In fact, if we consider the molecular structural difference between S90 and C90, now including their side chains, the closest distance from C90 to E113 in zebra finch (P358) pigment is 7.16 Å, while that in zebra finch (P358) pigment with C90S is 8.85 Å. On the other hand, the corresponding distances for the pigeon (P393) pigment and mutant pigment with S90C are 8.89 and 7.68 Å, respectively. These results show that amino acids C90 in the UV pigments are actually closer to E113 than S90 in the violet pigments.
All of these structural analyses show that a pocket exists between the counterion E113 (or the Schiff base nitrogen) and amino acid sites 87–93 in TM II. When we inspect the tertiary structures of these visual pigments, we find a channel-like structure at this region of TM II and III. With the narrower opening of the channel, the access of water molecules to this region may be limited. This may result in displacement of positive charge away from the Schiff base nitrogen, leading to deprotonation of the Schiff base (Rafferty and Shichi, 1981; Deng et al., 1994; Harosi and Sandorfy, 1995; Nagata et al., 1997).
3.5. Quantum chemical computations of chicken (P415) pigment
Upon absorbing a photon, the 11-cis-retinal chromophore in the ground electronic state (S0) is transformed into an excited state (S1) or excited states with higher energy. Then S1-chromophore goes through a conical intersection region and return to S0-chromophore (Gonzalez-Luque et al., 2000). The excitation energies and oscillator strength obtained by quantum chemical computations can be related to the λmax value of visual pigments. The most widely applied computational procedures for calculation of a large protein system is the QM:MM method that combines a quantum mechanical (QM) method and a molecular mechanical (MM) method (Field et al., 1990).
To make computational calculations of the visual pigments both executable and reliable, theoretical chemists often combine basically two computational methods: 1) a highly accurate and computationally intensive method for the neighborhood of the 11-cis-retinal and 2) a less accurate but less intensive computational method for the rest of pigments. Morokuma and his coworkers have developed a more general hybrid method, ONIOM (e.g., Dapprich et al., 1999; Vreven and Morokuma, 2003). Here, we shall introduce the results of an application of such quantum chemical computations to chicken (P415) pigment, which has a λmax value of 415 nm. Note that when S90C is introduced into this chicken pigment, the mutant pigment has a λmax value of 369 nm (Yokoyama et al., 2000). Using the ONIOM method, we have evaluated the effect of S90C on the λmax-shift in chicken (P415) pigment (Ohmiya et al., 2005). The results show that when water molecules specified by Okada et al. (2002) are included in the computation, the λmax values of the pigments with protonated and unprotonated chromophores for chicken (P415) pigment have λmax values of 512 and 398 nm, respectively, while the corresponding values for the mutant chicken pigment with S90C are 506 and 397 nm (Table 5). The absolute λmax values evaluated by the ONIOM method differ significantly from those of chicken (P415) and its mutant pigments. To improve the accuracy of these estimates, more time-consuming computations are needed. Despite these drawbacks, it is still clear that the protonation status of the chromophore strongly influences the λmax value of the chicken pigment. The S90C mutation decreases the λmax values of both protonated and unprotonated chromophores, but its direct effect on the λmax-shift is small. When water molecules are excluded in the calculation, the λmax values decrease slightly for the unprotonated pigments, but not for the protonated pigments. These theoretical results are compatible with the experimental prediction that UV pigments have the unprotonated chromophore (Babu et al., 2001; Shi et al., 2001; Fasick et al., 2002).
Table 5.
The λmax values (nm) of chicken (P415) pigment, evaluated by ONIOM (TD-B3LYP/6-31G*: Amber94)/ONIOM(B3LYP/6-31G) method
| The Schiff base | Wild type |
Mutant (S90C) |
||
|---|---|---|---|---|
| H2O | No H2O | H2O | No H2O | |
| Unprotonated | 398 | 386 | 397 | 380 |
| Protonated | 512 | 514 | 506 | 521 |
Taken from Ohmiya et al. (2005).
4. Conclusions and perspectives
Recent mutagenesis analyses of UV and violet pigments have dramatically improved our understanding of the roles of key amino acids in the spectral tuning of SWS1 pigments. Until very recently, it is widely accepted that amino acid sites only in TM I–III are involved in the differentiation of the absorption spectra of SWS1 pigments. Here we have seen that the absorption spectra of SWS1 pigments are modulated not only by amino acids in TM I–III but also by those in TM IV–VII. Today, we know neither critical amino acid sites in TM IV–VII nor their exact roles in modulating the absorption spectra of SWS1 pigments. In order to explore how UV and violet vision of different organisms have adapted to various ecological and physiological environments, the nature of the synergistic effects of TM I–VII has to be clarified. For that purpose, extensive site-directed mutagenesis experiments will be needed.
The crystal structure of the bovine rhodopsin shows that TM I–III and TM VII are located near the Schiff base nitrogen of the chromophore (Palczewski et al., 2000; Okada et al., 2002, 2004), which is generally protonated by E113 in TM III and possibly by E181 in the extracellular IV–V loop. The mutagenesis results of E113Q strongly suggest that UV pigments have unprotonated Schiff base-linked chromophore. The Schiff base pocket around E113 tends to be narrower in UV pigments than in violet pigments, which may limit the access of water molecules in that region and deprotonate the chromophore. Indeed, the ONIOM calculations of the chicken violet pigment also show that the deprotonation of the Schiff base-linked chromophore is the major cause of UV-sensitivity. In this specific calculation, water molecules had only minor influences on the differentiation of UV and violet pigments. It should be noted, however, that a rather small number of water molecules have been considered and may not detect the effects of removal of water molecules in the UV pigments on the λmax-shift. Thus, the cause for the deprotonation of the Schiff base nitrogen remains elusive. In the future, chemical computational analyses should also be applied to different sets of UV pigment, violet pigment, and intermediate forms and relate their results to experimentally evaluated λmax values. To make the predictions more accurate, it is highly desirable to obtain the crystal structures of UV and violet pigments.
Acknowledgments
We thank Drs. K. Morokuma, K. Ohmiya, R. Yokoyama, and anonymous reviewers for their comments on the manuscript. This work was supported by a grant from the National Institutes of Health and start-up fund from Emory University.
Abbreviations
- λmax
wavelength of maximal absorption
- SWS1
short wavelength-sensitive type 1
References
- Babu KR, Dukkipati A, Birge RR, Knox BE. Regulation of phototransduction in short-wavelength cone visual pigments via the retinylidene Schiff base counterion. Biochemistry. 2001;40:13760–13766. doi: 10.1021/bi015584b. [DOI] [PubMed] [Google Scholar]
- Briscoe AD. Homology modeling suggests a functional role for parallel amino acid substitutions between bee and butterfly red- and green-sensitive opsins. Mol Biol Evol. 2002;19:983–986. doi: 10.1093/oxfordjournals.molbev.a004158. [DOI] [PubMed] [Google Scholar]
- Chinen A, Hamaoka T, Yamada Y, Kawamura S. Gene duplication and spectral diversification of cone visual pigments of zebrafish. Genetics. 2003;163:663–675. doi: 10.1093/genetics/163.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowing JA, Poopalasundaram S, Wilkie SR, Robinson PR, Bowmaker JK, Hunt DM. The molecular mechanism for the spectral shifts between vertebrate ultraviolet- and violet-sensitive cone visual pigments. Biochem J. 2002;367:129–135. doi: 10.1042/BJ20020483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapprich S, Komaromi K, Bynn KS, Morokuma M, Frisch MJ. A new ONIOM implementation in Gaussian 98: Part I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives. J Mol Struct (Theorchem) 1999;1:461–462. [Google Scholar]
- Deeb SS, Wakefield MJ, Tada T, Marotte L, Yokoyama S, Marshall Graves JA. The cone visual pigments of an Australian marsupial, the Tammar wallaby (Macropus eugenii): sequence, spectral tuning, and evolution. Mol Biol Evol. 2003;20:1642–1649. doi: 10.1093/molbev/msg181. [DOI] [PubMed] [Google Scholar]
- Deng H, Huang L, Callender R, Ebrey T. Evidence for a bound water molecule next to the retinal Schiff base in bacteriorhodopsin and rhodopsin: a resonance Raman study of the Schiff base hydrogen/deuterium exchange. Biophys J. 1994;66:1129–1136. doi: 10.1016/S0006-3495(94)80893-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrey T, Koutalos Y. Vertebrate photoreceptors. Prog Retin Eye Res. 2001;20:49–94. doi: 10.1016/s1350-9462(00)00014-8. [DOI] [PubMed] [Google Scholar]
- Ebrey TG, Takahashi Y. Photobiology of retinal proteins. In: Coohill TP, Valenzeno DP, editors. Photobiology for the 21st Century. Valdenmar; Overland Park, KA: 2002. pp. 101–133. [Google Scholar]
- Fasick JI, Applebury ML, Oprian DD. Spectral tuning in the mammalian short wavelength sensitive cone pigments. Biochemistry. 2002;41:6860–6865. doi: 10.1021/bi0200413. [DOI] [PubMed] [Google Scholar]
- Field MJ, Bash PA, Karplus MJ. A combined quantum mechanical and molecular mechanical potential for molecular dynamics simulation. J Comp Chem. 1990;11:700. [Google Scholar]
- Gonzalez-Luque R, Garavelli M, Bernardi F, Merchan M, Robb MA. Computational evidence in favor of a two-state, two-mode model of the retinal chromophore photoisomerization. Proc Natl Acad Sci U S A. 2000;97:9379–9384. doi: 10.1073/pnas.97.17.9379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harosi FI, Sandorfy C. Retinylidene-opsin Schiff base chromophores and their accessibility to water. Photochem Photobiol. 1995;61:510–517. [Google Scholar]
- Jacobs GH. Ultraviolet vision in vertebrates. Am Zool. 1992;32:544–554. [Google Scholar]
- Kochendoerfer GG, Lin SW, Sakmar TP, Mathies RA. How color visual pigments are tuned. Trends Biochem Sci. 1999;24:300–305. doi: 10.1016/s0968-0004(99)01432-2. [DOI] [PubMed] [Google Scholar]
- Ma JX, et al. Salamander UV cone pigment: sequence, expression, and spectral properties. Vis Neurosci. 2001;18:393–399. doi: 10.1017/s0952523801183057. [DOI] [PubMed] [Google Scholar]
- Nagata T, Terakita A, Kandori H, Kojima D, Shichida Y. Water and peptide backbone structure in the active center of bovine rhodopsin. Biochemistry. 1997;36:6164–6170. doi: 10.1021/bi962920t. [DOI] [PubMed] [Google Scholar]
- Nakayama TA, Khorana HG. Mapping of the amino acids in membrane-embedded helices that interact with the retinal chromophore in bovine rhodopsin. J Biol Chem. 1991;266:4269–4275. [PubMed] [Google Scholar]
- Nathans J. Determinants of visual pigment absorbance: role of charged amino acids in the putative transmembrane segments. Biochemistry. 1990a;29:937–942. doi: 10.1021/bi00456a013. [DOI] [PubMed] [Google Scholar]
- Nathans J. Determinants of visual pigment absorbance: identification of the retinylidene Schiff’s base counterion in bovine rhodopsin. Biochemistry. 1990b;29:9746–9752. doi: 10.1021/bi00493a034. [DOI] [PubMed] [Google Scholar]
- Ohmiya K, Morokuma K, Yokoyama S. ONIOM (QM:MM) Calculation of Chicken SWS1 Visual Pigment and its S90C Mutant. Am Chem Soc Abstract. 2005 http://www.chemistry.org/portal/a/c/s/1/acsdisplay.html?DOC=meetings_index.html.
- Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc Natl Acad Sci U S A. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation in rhodopsin in light of a new 2.2 Å crystal structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- Palczewski K, et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Parry JWL, Poopalasundaram S, Bowmaker JK, Hunt DM. A novel amino acid substitution is responsible for spectral tuning in a rodent violet-sensitive visual pigment. Biochemistry. 2004;43:8014–8020. doi: 10.1021/bi049478w. [DOI] [PubMed] [Google Scholar]
- Rafferty CN, Shichi H. The involvement of water at the retinal binding site in rhodopsin and early light-induced intramolecular protein transfer. Photochem Photobiol. 1981;33:229–234. doi: 10.1111/j.1751-1097.1981.tb05329.x. [DOI] [PubMed] [Google Scholar]
- Sakmar TP, Franke RR, Khorana HG. Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc Natl Acad Sci U S A. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Yokoyama S. Molecular analysis of the evolutionary significance of ultraviolet vision in vertebrates. Proc Natl Acad Sci U S A. 2003;100:8308–8313. doi: 10.1073/pnas.1532535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Radlwimmer FB, Yokoyama S. Molecular genetics and the evolution of ultraviolet vision in vertebrates. Proc Natl Acad Sci U S A. 2001;98:11731–11736. doi: 10.1073/pnas.201257398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spady TC, Seehausen O, Loew ER, Jordan RC, Kocher TD, Carleton KL. Adaptive molecular evolution in the opsin genes of rapidly speciating cichlid species. Mol Biol Evol. 2005;22:1412–1422. doi: 10.1093/molbev/msi137. [DOI] [PubMed] [Google Scholar]
- Vreven T, Morokuma K. Investigation of the S0 S1 excitation in bacteriorhodopsin with the ONIOM (MO:MM) hybrid method. Theor Chem Acc. 2003;109:125–132. [Google Scholar]
- Wilkie SE, Robinson PR, Cronin TW, Poopalasundaram S, Bowmaker JK, Hunt DM. Spectral tuning of avian violet- and ultraviolet-sensitive visual pigments. Biochemistry. 2000;39:7895–7901. doi: 10.1021/bi992776m. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Molecular evolution of vertebrate visual pigments. Prog Retin Eye Res. 2000a;19:385–419. doi: 10.1016/s1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Phylogenetic analysis and experimental approaches to study color vision in vertebrates. Methods Enzymol. 2000b;315:312–325. doi: 10.1016/s0076-6879(00)15851-3. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Molecular evolution of color vision in vertebrates. Gene. 2002;300:69–78. doi: 10.1016/s0378-1119(02)00845-4. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Radlwimmer FB. The molecular genetics of red and green color vision in vertebrates. Genetics. 2001;158:1697–1710. doi: 10.1093/genetics/158.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Shi Y. Genetics and evolution of ultraviolet vision in vertebrates. FEBS Lett. 2000;486:167–172. doi: 10.1016/s0014-5793(00)02269-9. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Tada T. Evolution of ultraviolet vision in vertebrates. In: Bullock TH, editor. Evolution of Nervous Systems. Elsevier; Amsterdam: in press. [Google Scholar]
- Yokoyama S, Takenaka N. Statistical and molecular analyses of evolutionary significance of red-green color vision and color blindness in vertebrates. Mol Biol Evol. 2005;22:968–975. doi: 10.1093/molbev/msi080. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Yokoyama R. Adaptive evolution of photoreceptors and visual pigments in vertebrates. Annu Rev Ecol Syst. 1996;27:534–567. [Google Scholar]
- Yokoyama S, Radlwimmer FB, Blow NS. Ultraviolet pigments in birds evolved from violet pigments by a single amino acid change. Proc Natl Acad Sci U S A. 2000;97:7366–7371. doi: 10.1073/pnas.97.13.7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Takenaka N, Agnew DW, Shoshani J. Elephants and human color-blind deuteranopes have identical sets of visual pigments. Genetics. 2005;170:335–344. doi: 10.1534/genetics.104.039511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kumar S. Detection of convergent and parallel evolution at the amino acid sequence level. Mol Biol Evol. 1997;14:527–536. doi: 10.1093/oxfordjournals.molbev.a025789. [DOI] [PubMed] [Google Scholar]
- Zhukovsky EA, Oprian DD. Effect of carboxylic acid side chains on the absorption maximum of visual pigments. Science. 1989;246:928–930. doi: 10.1126/science.2573154. [DOI] [PubMed] [Google Scholar]