Abstract
We reported that VEGF-A induces proliferation of human pulmonary valve endothelial cells (HPVEC) through NFATc1 activation (1). Here we show that VEGF-A increases the migration of human pulmonary valve endothelial cells (HPVEC) through NFATc1 activation, suggesting that VEGF-A/NFATc1 regulates migration of HPVECs. To learn how this pathway may be involved in post-natal valvular repair, HPVECs were treated with VEGF-A, with or without cyclosporine A, to selectively block VEGF-NFATc1 signaling. DSCR1 and HB-EGF were two genes, identified by DNA microarray, whose expression was up-regulated by VEGF-A in a cyclosporine-A-sensitive manner. Silencing DSCR1 increased migration of HPVECs whereas silencing HB-EGF inhibited migration. This differential effect suggests that VEGF-A/NFATc1 signaling might be a crucial coordinator of endothelial cell migration in post-natal valves.
Keywords: valve endothelial cells, NFATc1, VEGF, calcineurin, EMT, DSCR1, HB-EGF
1.Introduction
Developmental heart malformations are the most frequently found congenital anomaly in children (2). Among them, heart valve defects often require replacing the defective valve by a surgical procedure. Thereafter, the integrity and optimal function of valve must be followed carefully. However, little is known about the genes regulating the integrity and maintenance of valve function throughout post-natal and adult life.
The nuclear factor of activated T cells (NFAT) family of transcription factors was first identified in immune cells (3). There are four isoforms, NFATc1, c2, c3, and c4 (alternative names are NFAT1-4). While knockout of other NFAT proteins produces defects in immune systems or other organs (4, 5), knockout of NFATc1 blocks the formation of the pulmonary and aortic valves, which in turn is embryonic lethal at ED 14-15 (6, 7). Cyclosporin A (CsA) is an inhibitor of cacineurin, a phosphatase that activates NFATs and allows NFATs to translocate into the nucleus where the regulate transcription. Thus CsA inhibits the calcinuerin-NFAT signaling pathways (5). Vascular endothelial growth factor-A (VEGF-A), a central regulator of angiogenesis and vasculogenesis, activates NFATc1 in human valve endothelial cells isolated from normally discarded surgical specimens (1). We showed previously that VEGF-A rapidly activates NFATc1 translocation into the nucleus and that this is required for the full mitogenic activity of VEGF-A in HPVECs (1, 8).
The primordial heart valve cushions are swellings in the atrioventricular (AV) junction and outflow tract at ED 9.5 in mice. The cushion is made up of extracellular matrix components initially, but as endocardial endothelial cells are stimulated to undergo endothelial to mesenchymal transdifferentiation (EMT), the cellularity of the cushion increases as a result of the migration of endothelial cells (9). There are ill-defined signals from myocardium that induce EMT of endothelium, but one strongly implicated factor is bone morphogenic protein-2 (BMP-2) (10-13). In the clonal endothelial cells isolated from sheep heart valves, TGF-β induces EMT significantly (14).
A zebrafish mutant embryo, called the jekyll mutant has a disruption in the gene encoding uridine 5′-diphosphate glucose dehydrogenase, which is essential for the synthesis of hyaluronic acid, an abundant component of the extracellular matrix in endocardial cushions (15). The mutant embryo shows regurgitation or toggling of blood between the atrium and ventricle as well as defects in cell differentiation at the AV boundary, indicating a defect in valve formation. Using a chemical approach, we tested the effect of adding a VEGF receptor-selective inhibitor to zebrafish embryos. Blocking VEGF-A signaling for a four hour period of time induced a phenotype remarkably similar to the jekyll mutant (16). This finding suggested that VEGF-A is a significant up-stream regulator of NFATc1 and implicated VEGF-A and VEGFR-2 in valvulogenesis. However, the down-stream targets of VEGF-NFATc1 signaling are not yet identified in valve endothelial cells.
In this study, we investigated the genes up- and down- regulated by VEGF-A-NFATc1 to further elucidate the function of VEGF-NFATc1 signaling in valve endothelial cells. Among up-regulated genes, Down syndrome critical region-1 (DSCR1) and heparin-binding EGF-like growth factor (HB-EGF) showed opposing behavior in the migration of HPVECs. Since increased migration is a key cellular event in EMT, these results suggest that NFATc1 activation by VEGF-A might affect specific cellular functions that are regulated during valvulogenesis.
2.Materials and Methods
2.1. Cell Culture
Human pulmonic valve endothelial cells (HPVEC) and sheep aortic valve endothelial cells (OVEC) were isolated from human pulmonary valve leaflets as described (7,14).
2.2. Immunofluorescent assay
HPVECs were fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and incubated with mouse anti-human NFATc1 monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) followed by FITC-conjugated anti-mouse IgG.
2.3. Western Blot Analysis
Equal amounts of protein extracts in a SDS-lysis buffer were subjected to 10% SDS-PAGE analysis and transferred to nitrocellulose membrane. The anti-NFATc1 antibody was from Santa Cruz. An enhanced chemiluminescence system (Pierce, Woburn, MA) was used for detection.
2.4. Total RNA isolation and DNA microarray analysis
HPVEC cells were treated with VEGF (50 ng/ml) with or without CsA (1 μg/ml) for 3 h. Total RNA was isolated from the treated cells by single-step procedure with Trizol reagent (Invitrogen, Gaithersburg, MD). The quality of total RNA (260/280 nm ratio) was excellent (above 2.0) and used for further cRNA synthesis and chip hybridization procedure with Gene Chip (HU133A, Affymetrix, Inc., Santa Clara, CA). All microarray data were analyzed at the NetAffx Analysis Center in Affymetrix web site, http://www.affymetrix.com/analysis/index.affx.
2.5. Knock-down of DSCR1 and HB-EGF with siRNA
OVEC cells were transfected with double stranded RNA using a MicroPorator (NanoEnTek, Seoul, Korea). After transfection, cells were harvested for the detection of gene expression by semiquantitative RT-PCR. Sequences of siRNA for DSCR1 were 5′-CUGAUUGCCUGUGUGGCAA dTdT-3′ and for HB-EGF, 5′-UGCCGUCGGUGGUGCUGAUGAAdTdT-3′ (Bioneer, Daejeon, Korea).
2.6. Statistical analysis
ANOVA tests were performed to assess significant differences between control and experimental groups. The level of significance was set at p<0.01 or p<0.05. Results are presented as the mean ± SD (standard deviation).
3. Results and Discussion
3.1. VEGF stimulates NFATc1 nuclear translocation and CsA blocks the NFATc1 activation
We previously studied the function of NFATc1 in vavular endothelium using HPVEC isolated from human pulmonary valve leaflets (7). VEGF treatment (50 ng/ml) rapidly induced the NFATc1 translocation into the nucleus within 30 min (Fig. 1Ab) that was sustained for 3-5 h (data not shown). CsA completely blocked the NFATc1 translocation induced by VEGF (Fig. 1A d). Long-term exposure to VEGF for 3 h also sustained NFATc1 activation (Fig. 1A c). To confirm the activation of NFATc1 we performed western blot analysis and found that dephosphorylation of NFATc1 protein was induced by VEGF after 5 h and treatment with CsA blocked the dephosphorylation of NFATc1, which could be seen by slower mobility on an SDS-PAGE gel (Fig. 1B). However, the more sustained treatment of VEGF (10 or 20 h) had no effect on the induction of NFATc1 protein level and its activation, but immunofluorescence staining of cells showed the NFATc1 nuclear shuffling might be occurring at these time points (Fig. 1B). Thus we collected total RNA after VEGF treatment for 3 h, with or without CsA, to identify the genes regulated by NFATc1 activation in HPVEC.
Fig. 1.
VEGF translocates NFATc1 into the nucleus and CsA inhibits the activation. A. HPVECs were stimulated with 50 ng/ml VEGF for 30 min (b) or for 3 h (c) or CsA (1 μM) plus VEGF for 30 min (d). Cells were incubated with a mouse control IgG (a) or with mouse anti-human NFATc1 (b-d), followed by anti-mouse conjugated FITC. In panel d, HPVECs were preincubated with 1 μM CsA for 2 h prior to addition of VEGF. Photographs were taken at 630× magnification. B. Cell lysates from HPVEC in EBM-2 with growth factors (lane 1), EBM-2 without growth factor (lane 2-8) were analyzed for the expression of NFATc1 by western blot. Cells were treated with VEGF 50 ng in the absence (lane 2, 4, 6) or presence (lane 3, 5, 7, 8) of 1 μM CsA for indicated times. Alpha-tubulin levels in each cell lysate are shown as a control.
3.2.Genes regulated by VEGF stimulation
Microarray data was analyzed as follows: to identify genes regulated only by VEGF-NFATc1 pathway, and not by other VEGF-stimulated pathways, we compared gene expression levels in cells treated with VEGF to those treated with VEGF plus CsA. VEGF-NFATc1 regulated genes would be increased or decreased by VEGF but not affected by VEGF plus CsA. We categorized the significantly up-regulated genes with biological functions, cell growth- and differentiation-related, heart development-related, muscle development-related, Wnt signaling-related genes and viral infection-related genes (Table 1). Genes with roles in cardiac development, such as cystein-rich, angiogenic inducer, 61 (CYR61), which regulates cell adhesion, migration, and proliferation through binding to integrin receptors and heparin sulfate proteoglycans were identified. CYR61 deficient mice show severe atrioventricular septal defects (AVSD) accompanying endocardial cushion defects (17). HB-EGF is known to function in cardiac valve morphogenesis, especially in regulating steps of valve remodeling (18). HB-EGF null mutant mice demonstrate enlarged semilunar and atriovenstricular heart valve, indicating its role in inhibition of mesenchymal cell proliferation (18-19). DKK1 is a secreted inhibitor of Wnt signaling and when overexpressed it has an inhibitory effect on cardiac cushion formation in zebrafish (20). From human Alagille syndrome, Jagged-1 mutation reveals pulmonary valve stenosis (21). Among viral infectious cycle-related genes, IL-8, CBP-1 and CCL2 have been suggested to play a role in cardiac function based on the phenotype of the knockouts and the expression patterns in cardiac patients (22-24). However, the other up-regulated genes listed in Table 1 are not fully defined yet in terms of the cardiac development. Insulin-like growth factor binding protein (IGFBP)-2 was recently identified as an angiogenic or cardiogenic molecule in zebrafish (25). IGFBPs bind to IGFs and might differentially regulate biological functions in a tissue-specific manner. IGFBP-3 is increased by serum deprivation and has an anti-proliferative effect in porcine endothelial cells (26).
Table.
Genes regulated in NFATc1 activation in HPVEC categorized by biological function.
| Gene title | Accession no. | Fold Increased |
|---|---|---|
| cell growth, size, differentiation regulation | ||
| insulin-like growth factor binding protein 7 | 201162_at | 4.6 |
| cysteine-rich, angiogenic inducer, 61 | 201289_at | 4.3 |
| SHC (Src homology 2 domain containing) transforming protein 1 | 201469_s_at | 4.3 |
| quiescin Q6 | 201482_at | 4.9 |
| neuroepithelial cell transforming gene 1 | 201829_at | 4.3 |
| deleted in liver cancer 1 | 210762_s_at | 4.9 |
| tumor protein p53 (Li-Fraumeni syndrome) | 211300_s_at | 4.6 |
| insulin-like growth factor binding protein 3 | 212143_s_at | 4.3 |
| heart development | ||
| PDZ and LIM domain 5 | 211681_s_at | 4.6 |
| DSCR-1 (down syndrom critical region 1) | ||
| muscle development | ||
| Muscle blind-like (Drosophila) | 201151_s_at | 4.9 |
| CUG triplet repeat, RNA binding protein 2 | 202158_s_at | 4.6 |
| heparin-binding EGF-like growth factor (HB-EGF) | 203821_at | 4.6 |
| MADS box transcription enhancer factor 2, polypeptide A (myocyte enhancer factor 2A) | 208328_s_at | 4.3 |
| filamin B, beta (actin binding protein 278) | 208613_s_at | 4.6 |
| jagged 1 (Alagille syndrome) | 209098_s_at | 4.3 |
| MADS box transcription enhancer factor 2, polypeptide C (myocyte enhancer factor 2C) | 209199_s_at | 4.6 |
| tropomyosin 4 | 212481_s_at | 4.3 |
| utrophin (homologous to dystrophin) | 213022_s_at | 4 |
| regulation of Wnt receptor signaling pathway | ||
| dickkopf homolog 1 (Xenopus laevis) (DKK1) | 204602_at | 4 |
| transcription factor 7-like 2 (T-cell specific, HMG-box) | 212761_at | 5.3 |
| viral infectious cycle | ||
| CD81 antigen (target of antiproliferative antibody 1) | 200675_at | 4.6 |
| interleukin 8 (IL8) | 202859_x_at | 4.6 |
| C-terminal binding protein 1 | 212863_x_at | 4.9 |
| chemokine (C-C motif) ligand 2 (CCL2) | 216598_s_at | 4.6 |
| upstream binding protein 1 (LBP-1a) | 218082_s_at | 4.6 |
| chromosome 1 open reading frame 139 | 221958_s_at | 4.3 |
| Gene title | Accession no. | Fold decreased |
| regulation of vascular permeability | ||
| angiomotin | 209521_s_at | 13 |
| tube development or cardiac cell differentiation | ||
| bone morphogenetic protein 10 (BMP10) | 208292_at | 16 |
Significantly downregulated genes by VEGF-NFATc1 activation in HPVEC were highly relevant to the vascular permeability and cardiac cell differentiation. Angiomotin, as a receptor for the angiostatin, an angiogenesis inhibitor, decreased VEGF-induced cell migration in vascular endothelial cells, especially at the polarization of endothelial branching (27), therefore, we can speculate that the endocardial cell migration by VEGF might be caused by the inhibition of angiomotin. BMP10 is highly expressed in Notch1 mutant embryos and was shown to increase myocardial proliferation in cardiac chamber formation (28). However, further investigations of the role of BMP10 in endocardial cells of cardiac valve is needed
3.3. VEGF-A increases migration of HPVECs in a calcineurin- and KDR-dependent manner
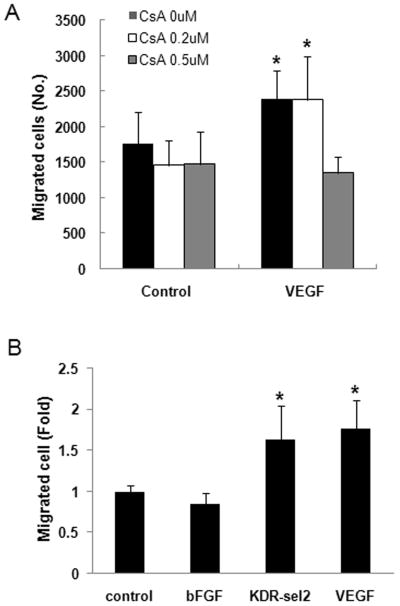
To identify whether VEGF-NFATc1 signaling pathway functions in specific cellular events that occur during EMT, we performed a migration assay because increased migratory ability is feature of EMT (9, 29). VEGF-treated HPVECs showed increased migration (1.4 fold, p<0.01), but cotreatment of CsA 0.5 μM decreased cell migration to the control level (Fig. 2A, p<0.01). This result suggests that calcineurin-dependent NFATc1 contributes to EMT by increasing migration of valve endothelial cells. When cells were treated with KDR receptor-selective variant VEGF (30), migrating cells were almost same as those treated with VEGF, suggesting that this effect of VEGF on migration of HPVEC cells is mediated by VEGFR-2. In contrast, bFGF had no effect on migration of HPVECs (Fig. 2B).
Fig. 2.
VEGF induced migration of valve endothelial cells in calcineurin- and KDR-dependent ways. A. Cell migration assay with HPVECs using Boyden chamber with polycarbonate membrane (8 μM pore size) was performed after VEGF were treated in a lower chamber and cell seeded onto upper chamber with or without CsA (0.2 or 0.5 μM). Migrated cells were counted after stained with hematoxylin and eosin (HE) and removed unmigrated cells. Significant differences from untreated control cells were noted as *p<0.01. B. Cells were treated with bFGF (10 ng/ml), KDR-selective variant VEGF (50 ng/ml), or VEGF (50 ng/ml) and migration assay using Boyden chamber was performed as described in A. Significant differences from untreated control cells were noted as *p<0.01. Data represent mean ± S.D. of a representative experiment (n=3), each performed in triplicate.
3.4. DSCR1 and HB-EGF induced by VEGF demonstrate opposing effects on migration of valve endothelial cells
To further analyze the effect of VEGF on valve EMT, we selected two different genes among those up-regulated by VEGF compared to VEGF plus CsA. DSCR1 is a direct transcriptional target of NFATc1 and a negative feedback inhibitor of calcineurin (31). And HB-EGF k/o mice have thickened valves while NFATc1 k/o mice show absence of semilunar or atrioventricular valves (18, 19, 5). However, HB-EGF was up-regulated by VEGF in our DNA microarray data. Thus we selected these two genes, DSCR1 and HB-EGF.
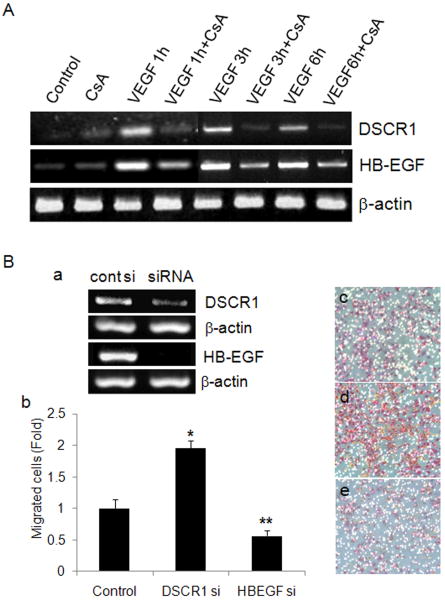
To confirm the DNA microarray data for DSCR1 and HB-EGF, we performed semiquantitative RT-PCR after cells were treated with VEGF in the presence or absence of CsA for 1 h, 3 h or 6 h. The expression of DSCR1 and HB-EGF was significantly up-regulated by VEGF treatment versus cotreatment of CsA until 6 h (Fig. 3A). This confirms the results obtained from the DNA microarray, indicating DSCR1 and HB-EGF are regulated by VEGF-NFATc1 activation. Next, we performed migration assay with ovine cells after knock-down of each gene, individually. siRNA silencing of DSCR1 and HB-EGF worked well, as confirmed by semi-quantitative RT-PCR (Fig. 3Ba). Interestingly, knock-down of DSCR1 significantly increased migration of OVECs (Fig. 3Bb, d) compared to scrambled siRNA control, indicating that DSCR1 might function to inhibit valve EMT. However, knock-down of HB-EGF significantly decreased migration of OVECs (Fig. 3Bb, e). This suggests that HB-EGF has a positive role in the migratory capability of valve endothelial cells.
Fig. 3.
DSCR1 and HB-EGF induced by VEGF have an opposite effect on cardiac valve endothelial cell migration. A. HPVECs were treated with VEGF (50 ng/ml) and/or CsA (1 μM) for 1, 3, or 6 h and collected total RNA for RT-PCR analysis. Beta-actin was used as an internal control. B. OVECs were transfected with siRNA for DSCR1 and HB-EGF and checked mRNA level (a) by RT-PCR. Beta-actin was used as an internal control. Migration assay was performed with knock-down cells with siRNA of DSCR1 and HB-EGF using transwell (8 μM pore size). Migrated cells were stained with HE and counted after unmigrated cells were removed (b). Photographs for migrated cells were taken with control siRNA (c), DSCR1 siRNA (d), or HB-EGF siRNA (e) transfectants (× 200 magnification). Data represent mean ± S.D. of a representative experiment (n=3), each performed in triplicate. Significant differences from control siRNA transfected cells were noted as *p<0.01, from DSCR1 siRNA transfected cells and control siRNA transfected cells as ** p<0.01.
It has been suggested that DSCR1 functions as a negative feedback inhibitor for NFATc1 by inhibiting calcineurin (31). An important question is to define the precise cellular behaviors regulated by this feed back inhibition. For example, VEGF-induced NFATc1 activation enhances proliferation of HPVECs, suggesting that DSCR1 does not have a major effect on proliferation (7). DSCR1 over-expression in OVEC does not inhibit EMT (32). Here, our data suggest that DSCR1 inhibits valve EC migration. Therefore, a mechanism by which VEGF may inhibit or attenuate EMT could be through VEGF-induced DSCR1, which would in turn inhibit cellular migration (31, 33, 34). Alternatively, VEGF might inhibit TGFβ-induced EMT through other genes. The function of HB-EGF in heart development is suggested to be a stimulation of valvulogenesis in some steps through BMP signaling pathway (12, 13). Interestingly, increased activation of BMP signaling molecules, such as Smad1, 5, and 8 was observed in hyperplastic valves of HB-EGF mutants (18), suggesting HB-EGF may oppose BMP signaling. A positive function for VEGF in active cellular migration and EMT (our results, 35) might be mediated by HB-EGF. HB-EGF, therefore, appears to act as a negative regulator for cell proliferation in mesenchymal cells but a positive regulator for cell migration in valve endothelial cells during valve development and remodeling process. Taken together, VEGF-NFATc1 signaling pathway has a role in EMT homeostasis, increasing or inhibiting migration of valve endothelial cells in different contexts of microenvironment of valvulogenesis through the induction of downstream functional genes.
Acknowledgments
This research was supported by a research grant (01-PJ10-PG6-01GN15-0001) from the Ministry of Health and Welfare, Republic of Korea and a grant from the Korean Research Foundation Grant by the Korean Government (MEST) (2009-0066101), and also by NIH R01 HL60490 to JB.
List of abbreviations
- NFAT
nuclear factor in activated T cells
- VEGF
vascular endothelial growth factor
- TGF
transforming growth factor
- EMT
endothelial-mesenchymal transdifferentiation
- DSCR1
Down syndrome critical region 1
- HB-EGF
heparin-binding EGF-like growth factor
- HPVEC
human pulmonary valve endothelial cell
- OVEC
ovine valve endothelial cell
- DKK1
dikkopf 1
- IL-8
interleukin 8
- CBP-1
C-terminal binding protein 1
- CCL2
chemokine ligand 2
- IGFBP
insulin-like growth factor binding protein
- KDR
kinase insert domain receptor
- ED
embryonic day
- CsA
cyclosporine A
- AV
atrioventricular
- αSMA
alpha smooth muscle actin
- FACS
fluorescent activated cell sorter
References
- 1.Johnson EN, Lee YM, Sander TL, Kaushal S, Rabkin E, Schoen JF, Bischoff J. NFATc1 mediates vascular endothelial growth factor-induced proliferation in human pulmonary valve endothelial cells. J Biol Chem. 2003;278:1686–1692. doi: 10.1074/jbc.M210250200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferencz C, Rubin JD, McCarter RJ, Brenner JI, Neill CA, Perry LW, Hepner SI, Downing JW. Congenital heart disease: prevalence at livebirth. The Baltimore-Washington Infant Study. Am J Epidemiol. 1985;121:31–36. doi: 10.1093/oxfordjournals.aje.a113979. [DOI] [PubMed] [Google Scholar]
- 3.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: Regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 4.Crabtree GR. Calcium, calcineurin, and the control of transcription. J Biol Chem. 2001;276:2313–2316. doi: 10.1074/jbc.R000024200. [DOI] [PubMed] [Google Scholar]
- 5.Ranger AM, Grusby MJ, Hodge MR, Gravallese EM, de la Brousse FC, Hoey T, Mickanin C, Baldwin HS, Glimcher LH. The transcription factor NF-ATc is essential for cardiac valve formation. Nature. 1998;392:186–190. doi: 10.1038/32426. [DOI] [PubMed] [Google Scholar]
- 6.de la Pompa JL, Timmerman LA, Takimoto H, Yoshida H, Elia AJ, Samper E, Pooter J, Wakeham A, Marengere L, Langille BL, Crabtree GR, Mak TW. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature. 1998;392:182–186. doi: 10.1038/32419. [DOI] [PubMed] [Google Scholar]
- 7.Shaw JP, Utz PJ, Durand DB, Toole JJ, Emmel EA, Crabtree GR. Identification of a putative regulator of early T cell activation genes. Science. 1988;241:202–205. doi: 10.1126/science.3260404. [DOI] [PubMed] [Google Scholar]
- 8.Combs MD, Yutzey KE. Vascular Endothelial Growth Factor A and RANKL Regulation of NFATc1 in Heart Valve Development. Circ Res. 2009 doi: 10.1161/CIRCRESAHA.109.196469. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramsdell AF, Markwald RR. Induction of endocardial cushion tissue in the avian heart is regulated, in part, by TGFbeta-3-mediated autocrine signaling. Dev Biol. 1997;188:64–74. doi: 10.1006/dbio.1997.8637. [DOI] [PubMed] [Google Scholar]
- 10.Sugi Y, Yamamura H, Okagawa H, Markwald RR. Bone morphogenetic protein-2 can mediate myocardial regulation of atrioventricular cushion mesenchymal cell formation in mice. Dev Biol. 2004;269:505–18. doi: 10.1016/j.ydbio.2004.01.045. [DOI] [PubMed] [Google Scholar]
- 11.Somi S, Buffing AA, Moorman AF, Van Den Hoff MJ. Dynamic patterns of expression of BMP isoforms 2, 4, 5, 6, and 7 during chicken heart development. Anat Rec A Discov Mol Cell Evol Biol. 2004;279:636–51. doi: 10.1002/ar.a.20031. [DOI] [PubMed] [Google Scholar]
- 12.Ma L, Lu MF, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132:5601–11. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- 13.Rivera-Feliciano J, Tabin CJ. Bmp2 instructs cardiac progenitors to form the heart-valve-inducing field. Dev Biol. 2006;295:580–8. doi: 10.1016/j.ydbio.2006.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paranya G, Vineberg S, Dvorin E, Kaushal S, Roth SJ, Rabkin E, Schoen FJ, Bischoff J. Aortic valve endothelial cells undergo transforming growth factor-beta-mediated and non-transforming growth factor-beta-mediated transdifferentiation in vitro. Am J Path. 2001;159:1335–1343. doi: 10.1016/s0002-9440(10)62520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh EC, Stainier DY. UDP-glucose dehydrogenase required for cardiac valve formation in zebrafish. Science. 2001;293:1670–3. doi: 10.1126/science.293.5535.1670. [DOI] [PubMed] [Google Scholar]
- 16.Lee YM, Cope JJ, Ackermann GE, Goishi K, Armstrong EJ, Paw BH, Bischoff J. Vascular endothelial growth factor receptor signaling is required for cardiac valve formation in zebrafish. Developmetnal Dynamics. 2006;235:29–37. doi: 10.1002/dvdy.20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mo FE, Lau LF. The matricellular protein CCN1 is essential for cardiac development. Circ Res. 2006;99:961–9. doi: 10.1161/01.RES.0000248426.35019.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson LF, Qiu TH, Sunnarborg SW, Chang A, Zhang C, Patterson C, Lee DC. Defective valvulogenesis in HB-EGF and TACE-null mice is associated with aberrant BMP signaling. EMBO J. 2003;22:2704–16. doi: 10.1093/emboj/cdg264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwamoto R, Yamazaki S, Asakura M, Takashima S, Hasuwa H, Miyado K, Adachi S, Kitakaze M, Hashimoto K, Raab G, Nanba D, Higashiyama S, Hori M, Klagsbrun M, Mekada E. Heparin-binding EGF-like growth factor and ErbB signaling is essential for heart function. Proc Natl Acad Sci U S A. 2003;100:3221–6. doi: 10.1073/pnas.0537588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurlstone AF, Haramis AP, Wienholds E, et al. The Wnt/beta-catenin pathway regulates cardiac valve formation. Nature. 2003;425:633–7. doi: 10.1038/nature02028. [DOI] [PubMed] [Google Scholar]
- 21.McElhinney DB, Krantz ID, Bason L, et al. Analysis of cardiovascular phenotype and genotype-phenotype correlation in individuals with a JAG1 mutation and/or Alagille syndrome. Circulation. 2002;106:2567–74. doi: 10.1161/01.cir.0000037221.45902.69. [DOI] [PubMed] [Google Scholar]
- 22.Steele IC, Nugent AM, Maguire S, et al. Cytokine profile in chronic cardiac failure. Eur J Clin Invest. 1996;26:1018–22. doi: 10.1046/j.1365-2362.1996.2560587.x. [DOI] [PubMed] [Google Scholar]
- 23.Damås JK, Eiken HG, Oie E, Bjerkeli V, et al. Myocardial expression of CC- and CXC-chemokines and their receptors in human end-stage heart failure. Cardiovasc Res. 2000;47:778–87. doi: 10.1016/s0008-6363(00)00142-5. [DOI] [PubMed] [Google Scholar]
- 24.Gusterson RJ, Jazrawi E, Adcock IM, Latchman DS. The transcriptional co-activators CREB-binding protein (CBP) and p300 play a critical role in cardiac hypertrophy that is dependent on their histone acetyltransferase activity. J Biol Chem. 2003;278:6838–47. doi: 10.1074/jbc.M211762200. [DOI] [PubMed] [Google Scholar]
- 25.Wood AW, Schlueter PJ, Duan C. Targeted knockdown of insulin-like growth factor binding protein-2 disrupts cardiovascular development in zebrafish embryos. Mol Endocrinol. 2005;19:1024–34. doi: 10.1210/me.2004-0392. [DOI] [PubMed] [Google Scholar]
- 26.Delafontaine P, Ku L, Anwar A, Hayzer DJ. Insulin-like growth factor 1 binding protein 3 synthesis by aortic endothelial cells is a function of cell density. Biochem Biophys Res Commun. 1996;222:478–82. doi: 10.1006/bbrc.1996.0769. [DOI] [PubMed] [Google Scholar]
- 27.Aase K, Ernkvist M, Ebarasi L, Jakobsson L, et al. Angiomotin regulates endothelial cell migration during embryonic angiogenesis. Genes Dev. 2007;21:2055–68. doi: 10.1101/gad.432007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grego-Bessa J, Luna-Zurita L, del Monte G, Bolós V, et al. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12:415–29. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armstrong EJ, Bischoff J. Heart valve development: endothelial cell signaling and differentiation. Circ Res. 2004;95:459–70. doi: 10.1161/01.RES.0000141146.95728.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li B, Fuh G, Meng G, Xin X, Gerritsen ME, Cunningham B, de Vos AM. Receptor-selective variants of human vascular endothelial growth factor. Generation and characterization. J Biol Chem. 2000;275:29823–8. doi: 10.1074/jbc.M002015200. [DOI] [PubMed] [Google Scholar]
- 31.Wu H, Kao SC, Barrientos T, Baldwin SH, et al. Down syndrome critical region-1 is a transcriptional target of nuclear factor of activated T cells-c1 within the endocardium during heart development. J Biol Chem. 2007;282:30673–30679. doi: 10.1074/jbc.M703622200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang JH, Wylie-Sears J, Bischoff J. Opposing actions of Notch1 and VEGF in post-natal cardiac valve endothelial cells. Biochem Biophys Res Commun. 2008;374:512–6. doi: 10.1016/j.bbrc.2008.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paruchuri S, Yang JH, Aikawa E, Melero-Martin JM, Khan ZA, Loukogeorgakis S, Schoen FJ, Bischoff J. Human pulmonary valve progenitor cells exhibit endothelial/mesenchymal plasticity in response to vascular endothelial growth factor-A and transforming growth factor-beta2. Circ Res. 2006;99:861–9. doi: 10.1161/01.RES.0000245188.41002.2c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodgers LS, Lalani S, Hardy KM, Xiang X, Broka D, Antin PB, Camenisch TD. Depolymerized hyaluronan induces vascular endothelial growth factor, a negative regulator of developmental epithelial-to-mesenchymal transformation. Circ Res. 2006;99:583–9. doi: 10.1161/01.RES.0000242561.95978.43. [DOI] [PubMed] [Google Scholar]
- 35.Hallaq H, Pinter E, Enciso J, McGrath J, et al. A null mutation of Hhex results in abnormal cardiac development, defective vasculogenesis and elevated Vegfa levels. Development. 2004;131:5197–209. doi: 10.1242/dev.01393. [DOI] [PubMed] [Google Scholar]