Abstract
We identify the helicase-SANT–associated (HSA) domain as the primary binding platform for nuclear actin-related proteins (ARPs) and actin. Individual HSA domains from chromatin remodelers (RSC, yeast SWI-SNF, human SWI-SNF, SWR1 and INO80) or modifiers (NuA4) reconstitute their respective ARP–ARP or ARP–actin modules. In RSC, the HSA domain resides on the catalytic ATPase subunit Sth1. The Sth1 HSA is essential in vivo, and its omission causes the specific loss of ARPs and a moderate reduction in ATPase activity. Genetic selections for arp suppressors yielded specific gain-of-function mutations in two new domains in Sth1, the post-HSA domain and protrusion 1, which are essential for RSC function in vivo but not ARP association. Taken together, we define the role of the HSA domain and provide evidence for a regulatory relationship involving the ARP–HSA module and two new functional domains conserved in remodeler ATPases that contain ARPs.
Transcriptional regulation involves the concerted action of chromatin regulators, transcription factors and the basal transcription machinery. The two general types of chromatin regulators are chromatin remodelers, which reposition and restructure nucleosomes1, and chromatin modifiers, which add or remove covalent marks from the histone proteins2. These chromatin regulatory complexes work together to mark and move nucleosomes, which can either help silence or activate transcription, depending on the context. Remodelers bear a catalytic ATPase subunit required for ATP-dependent nucleosome repositioning1,3,4, whereas modifier complexes bear one or more subunits with histone-modification potential2,5,6. However, in both cases, most of the subunits of chromatin regulatory complexes are nonenzymatic. These attendant subunits are specialized for diverse tasks: targeting the complex to particular nucleosomes, enabling complex association with particular DNA binding proteins or other complexes, or helping to regulate enzymatic activity.
Intriguingly, actin and ARPs are among the associated subunits of certain chromatin-remodeling and chromatin-modifying complexes7–9. ARPs have been studied extensively in the budding yeast Saccharomyces cerevisiae, which contains ten ARPs10. Four ARPs (ARPs 1, 2, 3 and 10) reside mainly in the cytoplasm and have clear roles in actin filament nucleation and cytoskeletal regulation. In contrast, the remaining six ARPs (ARPs 4, 5, 6, 7, 8 and 9) are nuclear. Remarkably, all nuclear ARPs are associated exclusively with chromatin-regulating complexes, including ATP-dependent nucleosome remodelers and certain histone acetyltransferase (HAT) complexes. ARPs and actin are stable and stoichiometric members of SWI-SNF11,12, SWR113 and INO80 family14 remodelers but are absent in ISWI and CHD family remodelers. Actin and ARPs are also present in HAT complexes related to the yeast NuA4 complex, which is essential for histone H4 acetylation15,16. In addition, human complexes that combine both HAT and remodeler activities, such as p400, also bear ARPs and actin17. However, beyond these notable exceptions, most HAT complexes (such as SAGA, SAS and ADA) lack ARPs18,19.
Each chromatin complex associates with particular nuclear ARP proteins. For example, both the yeast SWI-SNF complex and the paralogous RSC complex contain Arp7 and Arp9, but lack actin11,12. Human orthologs bear the ARP BAF53 as well as actin itself 20. Thus, SWI-SNF complexes can contain either two ARPs or an ARP–actin pair. Interestingly, remodelers of the SWR1 and INO80 families bear actin and multiple ARPs; SWR1 contains Arp4 and Arp6 (ref. 21), whereas INO80 contains Arp4, Arp5 and Arp8 (ref. 14). Thus, remodelers show both selectivity and diversity in their association with ARPs, but the basis for these properties has not been elucidated.
For yeast RSC and SWI-SNF, Arp7 and Arp9 function as obligate heterodimers22, and in SWI-SNF they are bound to the catalytic subunit Snf2 (ref. 23). Likewise, in human SWI-SNF, BAF53 and actin are tightly associated with the ATPase subunit BRG1 (ref. 20). For Ino80, omission of a large region of the N terminus leads to the loss of Arp4, Arp8 and actin, as well as other subunits, raising the possibility of an ARP nucleation domain in this region24. In addition, a large insertion in the ATPase domain of Swr1 (unique to INO80 and SWR1 family remodelers) is required for the association of Arp6, as well as five other subunits25. These results established the association of ARPs with the ATPase subunit; however, the domain(s) that enables the association with particular ARPs was not defined. Two previous experiments suggested that ARPs or actin regulate the ATPase domain. First, actin binding drugs moderately reduce the DNA-dependent ATPase activity of human SWI-SNF20. Second, the omission of ARP proteins from INO80 prevented DNA-dependent ATPase activity and DNA binding24. However, the ARP proteins in the RSC complex show only modest levels of ATPase regulation22, raising the possibility that different remodeler families might, to different extents, rely on ARP regulation. Regardless, these studies did not address the mechanism or domain relationships that enable ARP docking or ATPase regulation.
Our work addresses two central questions: how do ARPs and actin selectively associate with chromatin regulators, and what domain relationships are involved in enabling ATPase regulation by ARPs? We provide several lines of evidence that the HSA domain is the general binding platform for nuclear ARPs and actin. We also provide genetic evidence that the ARP–HSA module cooperates with two domains (termed post-HSA and protrusion 1) that are present together only in chromatin-remodeling ATPases that contain ARPs, providing a mechanistic framework for the regulation of the ATPase subunit.
RESULTS
Remodelers with ARPs have an HSA and a post-HSA domain
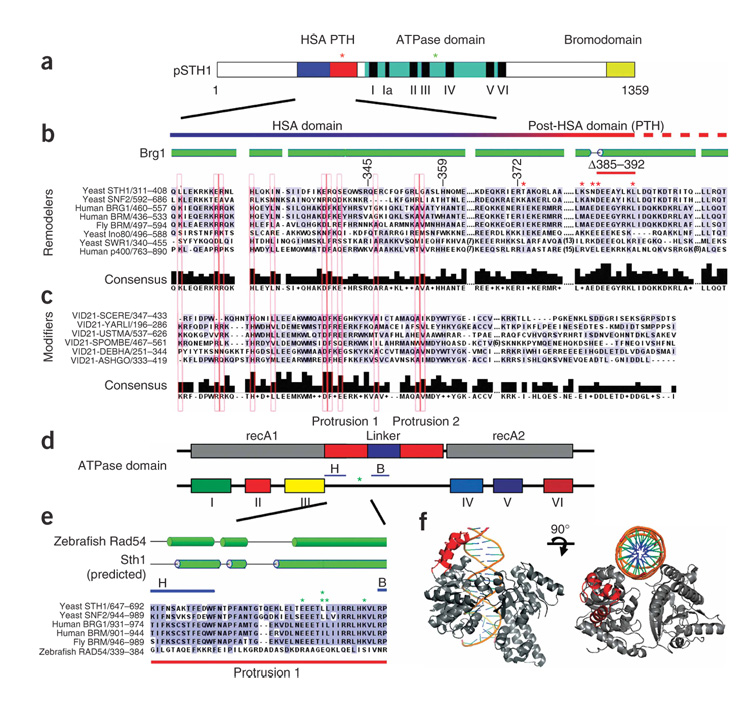
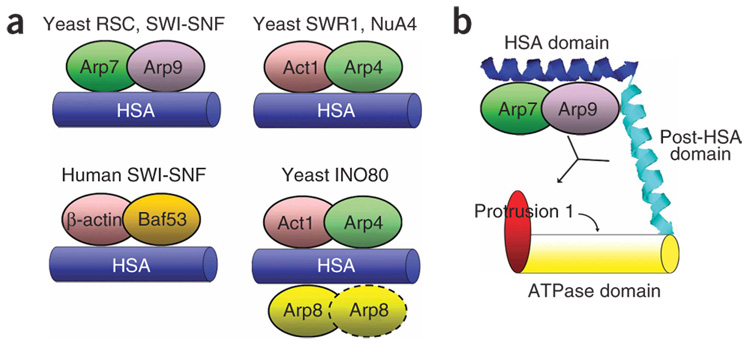
Extensive BLAST and pairwise alignments26 among remodeler ATPases reveal an HSA domain in several remodeler families: SWI-SNF, SWR1, INO80 and p400 (Fig. 1). Although it was not previously recognized, an HSA domain is present adjacent to the ATPase domain in Sth1, the catalytic subunit of RSC (Fig. 1a,b). Notably, all remodeler complexes with an HSA domain contain ARPs and/or actin. In contrast, all remodelers that lack an HSA domain (for example, ISWI and CHD1) likewise lack ARPs and actin. In keeping with this concept, histone-modifying complexes that contain ARPs or actin (for example, NuA4) contain a protein with an HSA domain (Eaf1, also known asVid21, in NuA4) (Fig. 1c). In contrast, chromatin complexes lacking ARPs (SAGA, SLIK, SAS and TFIID) lack an HSA domain (data not shown). Taken together, our bioinfomatic analyses reveal a notable correlation: all protein complexes that contain an HSA domain bear at least two ARPs or an ARP–actin pair.
Figure 1.
HSA, post-HSA (PTH) and protrusion 1 domains in ARP-containing remodeling complexes are conserved; arpΔ suppressors cluster in Sth1. (a) A schematic depicting the domains of Sth1. Clusters of arpΔ suppressor mutations are depicted with asterisks. (b) Alignment of HSA and post-HSA domains of remodelers. Secondary-structure predictions used PSIPRED. Locations of mutations that suppress arpΔ lethality are indicated with asterisks (red, within the HSA; green, within protrusion 1). (c) Alignment of HSA domains of Eaf1 (also known as Vid21)–related proteins, with residues conserved within remodelers extended to b and boxed in red. The post-HSA domain is not present in Eaf1-related proteins from Saccharomyces cerevisiae (SCERE), Yarrowia lipolytica (YARL), Ustilago maydis (USTMA), Schizosaccharomyces pombe (SPOMBE), Debaryomyces hansenii (DEBHA), Ashbya gossypii (ASHGO). (d) Schematic of SNF2 family ATPase domains, which bear a unique insertion between domains III and IV, containing two protrusions and two highly conserved subdomains (H and B)29. sth1 ATPase mutations that suppress arpΔ lethality cluster between these regions (green asterisk). recA1 and recA2 refer to the regions in helicases/translocases that share homology to recA. (e) Mutations that suppress arpΔ mutations (green asterisks) cluster tightly within protrusion 1, between the H-B region. (f) Structural depiction of protrusion 1 of Sulfolobus solfataricus Rad54 (in red). DNA and ATP binding regions are not localized to protrusion 1.
Our alignments also revealed a highly conserved region directly adjacent to the HSA domain in remodeler ATPases, which we term the post-HSA domain (Fig. 1a,b). The precise boundary between the HSA and the post-HSA domains is not clear, with the alignments suggesting a transition between them in the region around amino acid 370 in Sth1. The post-HSA domain is present exclusively in remodelers that contain ARPs; it is absent in HAT complexes (Fig. 1b,c and data not shown). These results prompted tests for links between the ARPs and these domains by genetic and biochemical methods.
Isolation of ten arpΔ suppressors
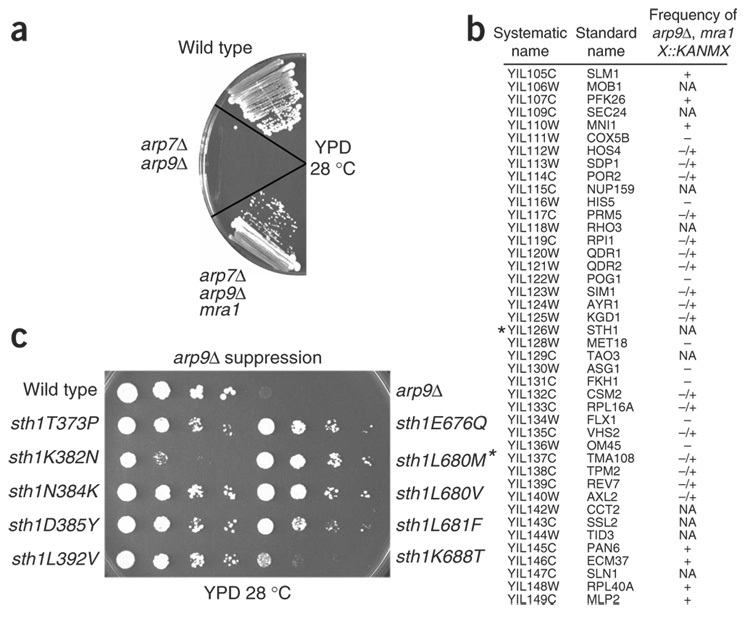
A connection between ARPs, the HSA domain and the post-HSA domain emerged from genetic studies involving selections for arp mutant suppressors. arp7Δ or arp9Δ mutants are barely viable in the W303 background and are inviable in the S288C background. However, spontaneous arpΔ suppressors arose at low frequency in the W303 background during growth on rich media plates22 (Fig. 2a). In total, seven independent arp9Δ suppressors and three independent arp7Δ suppressors were isolated; these were termed mra mutants (as they modified the requirement for actin-related proteins). One arp7Δ suppressor (mra1-1) suppressed an arp7Δ arp9Δ double mutation, showing that mra1 can restore growth ability in the absence of ARP function (Fig. 2a and data not shown). Although it showed rapid growth on rich medium, the mra1-1 arpΔ strain was inviable under various stress conditions (Supplementary Table 1 online). These defects are conferred by the arpΔ mutation and not by the mra mutation, as the isolated mra mutations lacked these or other strong phenotypes (data not shown), which prevented the cloning of mra gene(s) by complementation.
Figure 2.
Particular sth1 mutations suppress arpΔ mutations. (a) Suppression of the arp7Δ arp9Δ lethality by mra1-1. (b) Linkage analysis maps arpΔ suppressor mutations to the STH1 locus. Genes flanking STH1 (marked with an asterisk) on chromosome IX are depicted, and the scoring reflects the frequency of obtaining triple-mutant spores (mra1 arp9Δ XD::KANMX): +, spore patches; −/+, infrequent punctate spore colonies; –, no spores; NA; not applicable (essential genes). Shown is the data for mra1-1, and linkage to STH1 was also demonstrated for the nine other mra mutations (not shown). (c) arp9Δ suppression by sth1 alleles. Reference strains were wild type and arp9Δ. An asterisk indicates the arp9Δ arp7Δ background. All strains are the original spontaneous suppressors.
arpΔ suppressor mutants are alleles of STH1
We identified mra genes and mutations using a relatively new linkage mapping approach27 involving a synthetic genetic array (SGA)—a library of yeast strains in which each nonessential gene is knocked out and replaced with a marker (XΔ∷KANMX). Briefly, a MATα mra1 arp9Δ strain was crossed to the complete set of haploid knockouts (4,891 MATa XΔ∷KANMX strains). Following sporulation, we identified sporulated diploids that failed to produce haploid XΔ∷KANMX mra1 arp9Δ progeny, demonstrating linkage of XΔ∷KANMX to mra1 (Methods). Remarkably, this procedure revealed the linkage of mra1 to STH1 (Fig. 2b). The other nine mra mutants also showed tight linkage to STH1, and sequencing of the STH1 locus from each of the ten mra strains revealed that each encoded a single amino acid substitution in Sth1: sth1T373P, sth1K382N, sth1N384K, sth1D385Y, sth1L392V, sth1E676Q, sth1L680V, sth1L680M, sth1L681F and sth1K688T (Fig. 2c). Furthermore, sth1N384K and sth1L680M mutations suppressed arpΔ mutations when tested in isolation on plasmids (data not shown), verifying that arpΔ suppression relied solely on these alleles. Thus a genome-wide screen for arpΔ suppression yielded only sth1 alleles.
sth1 and mra mutations cluster within two Sth1 domains
The mra mutations clustered tightly to two small regions of Sth1 (Fig. 1a). Five sth1 mutations clustered tightly within the post-HSA domain, providing an initial link between ARP and post-HSA function (red asterisks, Fig. 1a,b). The other five mutations also clustered tightly within a special insertion in the catalytic ATPase domain of Sth1, termed protrusion 1 (green asterisks, Fig. 1a,d,e). Chromatin-remodeling ATPases are a subfamily within the Super-family 2 (SF2) family of helicases and translocases, and all Snf2 family ATPases contain an insertion located between ATPase domains III and IV28,29 (Fig. 1a,d,e). In Rad54, the only Snf2 family ATPase that has been crystallized, this insertion consists of two protrusions (1 and 2) connected by a linker29,30 (Fig. 1d). These five mutations (green asterisks, Fig. 1d,e) cluster within 13 amino acids inside protrusion 1. Protrusion 1 from Rad54 is a three-helix domain (shown in red, Fig. 1f) and is not directly adjacent to either the DNA binding or the ATP binding regions. At present, nothing is known about the function of protrusion 1, although our isolation of arpΔ suppressors in this region raises the possibility that protrusion 1 might work together with the ARPs to help regulate ATPase activity. Protrusion 1 in Rad54 consists of three helices, and structural predictions suggest a similar three-helix relationship in Snf2 family remodelers (Fig. 1e). Chromatin remodelers that contain ARPs share high sequence identity in protrusion 1, whereas those that lack ARPs, such as Rad54, diverge. This further suggests connections between protrusion 1 and ARPs.
Next, we addressed whether these mutations in the post-HSA domain or in protrusion 1 were gain-of-function alleles or loss-of-function alleles. Notably, whereas the original alleles L392V and L681F conferred suppression, L392P and L681S substitutions did not (Fig. 2c and data not shown). Furthermore, the sth1 alleles that conferred arpΔ suppression were semidominant (data not shown). This establishes that these sth1 mutations are dominant gain-of-function alleles.
We were surprised that our genetic suppression experiments yielded many dominant gain-of-function mutations in both the post-HSA domain and in protrusion 1, but none was obtained in the central HSA domain itself. Here we speculated that the HSA domain might be the crucial binding platform for ARPs, whereas the other two domains might communicate that binding to the ATPase domain, with the mutations we isolated mimicking the bound state, thus suppressing an arpΔ genotype.
The HSA domain of Sth1 binds Arp7 and Arp9
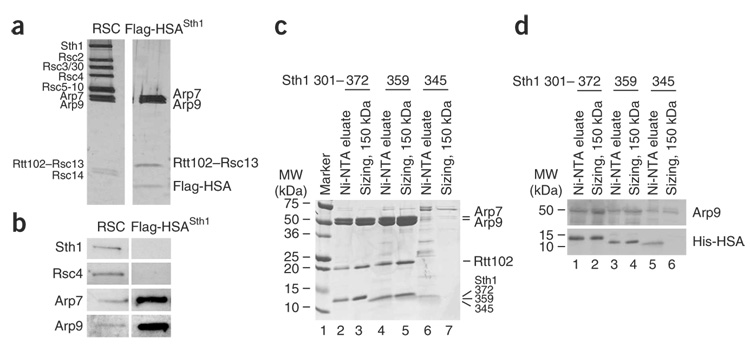
To determine whether the HSA domain is the primary binding platform for ARPs, we first tested the HSA of Sth1. We initially tagged versions of either the entire Sth1 protein (Flag-Sth1) or the Sth1 HSA region (amino acids 301–383; Flag-HSASth1−10 × His–tagged), and assessed ARP association by co-immunoprecipitation with anti-Flag antibodies. All RSC members tested associated with full-length Sth1, whereas the HSA domain associated with Arp7 and Arp9 but not with other RSC members (Supplementary Fig. 1a online). As a definitive test, we expressed a Flag- and His-tagged HSA domain in yeast and purified it to near homogeneity using a combination of nickel–nitrilotriacetic acid (Ni-NTA) and anti-Flag chromatography. This procedure yielded four proteins: Arp7, Arp9, Rtt102 (also known as Rsc13) and the HSA domain of Sth1 (Fig. 3a), determined by western analysis (Fig. 3b) and MS sequencing. Yeast actin was not associated with these complexes (Fig. 3a and data not shown), consistent with earlier studies of RSC composition. Rtt102, a nonessential gene with no known homologs31,32, is not required for the association of the ARPs with the HSA, as Arp7 and Arp9 co-precipitated with the HSA domain in a strain lacking Rtt102 (Supplementary Fig. 1b). Furthermore, biochemical reconstitution experiments show that Rtt102 binds only the ARPs and not the HSA (Supplementary Fig. 1c). Thus, the HSA region of Sth1 interacts directly with Arp7 and Arp9.
Figure 3.
The HSA domain of Sth1 is sufficient to bind Arp7 and Arp9. (a) Tagged Sth1 HSA (amino acids 301–383) copurifies Arp7, Arp9 and Rtt102 (Methods). Proteins were revealed by silver staining and identified by MS sequencing (see text). (b) Western analysis of RSC and the HSA–Arp7–Arp9–Rtt102 complexes. (c) Reconstitution of the complex and determination of the minimal ARP binding HSA domain. Sth1 HSA truncation derivatives (Sth1301–372, Sth1301–359 or Sth1301–345 tagged with 10 × His) were expressed along with untagged Arp7, Arp9 and Rtt102. Extracts were purified using Ni-NTA agarose and subjected to gel filtration for sizing (Methods). (d) Western analysis of recombinant complexes.
Reconstitution of an HSA–Arp7–Arp9–Rtt102 complex
To further establish the interaction of the HSA with ARPs and to better define the region required for ARP association, we reconstituted a recombinant HSA–Arp7–Arp9–Rtt102 complex. To achieve this, the Sth1 HSA region (amino acids 301–372, 10 × His–tagged) was coexpressed in bacteria along with untagged full-length Arp7, Arp9 and Rtt102. The Sth1301–372 derivative was purified by Ni-NTA chromatography and subjected to gel filtration chromatography, yielding a four-protein complex (Fig. 3c, lanes 2 and 3). To determine the minimal HSA region sufficient for ARP association, we expressed truncated HSA derivatives and checked for complex reconstitution. One derivative, Sth1301–359, encompasses the region of the HSA domain that is most highly conserved between remodelers and modifier proteins (Fig. 1) but lacks the post-HSA domain. Notably, this minimal HSA domain afforded the complete four-protein reconstituted complex (Fig. 3c, lane 5). In contrast, a shorter derivative lacking a portion of the HSA domain, Sth1301–345, was stably produced but behaved poorly, with minimal ARP recovery and severe aggregation on the sizing column (Fig. 3c, lanes 6 and 7). However, we note that the small percentage that eluted at the proper size (150 kDa) binds low levels of ARPs (Fig. 3d). Taken together, we define a minimal stable HSA domain that is sufficient to reconstitute a complex containing Arp7, Arp9 and Rtt102.
HSA domains mediate selective ARP–actin association
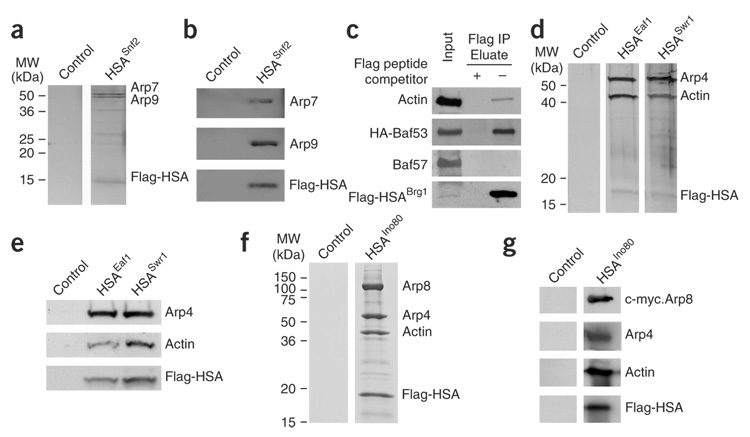
To test the generality and specificity of HSA–ARP–actin association, we examined the ARP and actin associations of other HSA domains. Yeast SWI-SNF and RSC both contain Arp7 and Arp9 (ref. 11,12). To test whether the HSA domain found in the SWI-SNF catalytic subunit Snf2 binds Arp7 and Arp9, we expressed a Flag-tagged and 10 × His–tagged Snf2 HSA domain (Snf2578–659) in a wild-type yeast strain, and purified this domain to near homogeneity using Ni-NTA and anti-Flag chromatography. The purification yielded three proteins, Arp7, Arp9 and the Snf2 HSA domain, visualized by silver staining (Fig. 4a), and their identities were confirmed by western analysis (Fig. 4b). Human SWI-SNF complex contains one actin-related protein (BAF53) and β-actin20. We expressed a tagged BRG1 HSA domain (Flag-HSABRG1) in RKO cells. Immunopurification using anti-Flag antibodies followed by immunoblot analysis revealed co-precipitation of BAF53 and actin (Fig. 4c). Here the small percentage of actin associated with the HSA is expected, as the vast majority performs cytoskeletal roles. Also, the BAF57 subunit of human SWI-SNF was not co-precipitated, showing that the entire SWI-SNF complex was not co-precipitated with the isolated HSA. Thus, the HSA domain of a human SWI-SNF remodeler is sufficient for ARP and actin association.
Figure 4.
Individual HSA domains are sufficient to bind actin and particular ARPs. (a) The yeast Snf2 HSA domain nucleates Arp7 and Arp9. The HSA region of Snf2 (578–659, Flag-tagged and 10 × His-tagged) was expressed in a wild-type strain (YBC928). Complexes were purified with Ni-NTA, then anti-Flag beads (Methods) and visualized by silver staining. (b) Western analysis confirms the protein identities in a. (c) The BRG1 HSA associates with BAF53 and actin. The BRG1 HSA (462–531, Flag-tagged) and HA-tagged BAF53 were coexpressed by transient transfection in RKO cells. Immunoprecipitation analysis with anti-Flag antibody followed by western blotting reveals specific associations with BAF53 and actin. (d) The Swr1 and Eaf1 HSA domains nucleate Arp4 and actin. The HSA regions (Flag-tagged and 10 × His–tagged) of Swr1 or Eaf1 (p1871 and p1870) were expressed, along with untagged Arp4 and actin (p1883) in a wild-type strain (YBC928). Complexes were purified using Ni-NTA and anti-Flag beads (Methods) and visualized by silver staining. (e) Western analysis confirms protein identities in d. (f) The Ino80 HSA nucleates Arp4, Arp8 and actin. The HSA region of Ino80 (462–598, Flag-tagged and His-tagged, (p2441)), along with untagged Arp4 and actin (p1883), V5-Arp5 and c-myc–Arp8 (p2436), were expressed in a wild-type strain. Complexes were purified with Ni-NTA, then anti-Flag beads (Methods) and visualized by Coomassie. (g) Western analysis confirms the protein identities shown in f.
Other yeast complexes that contain ARPs and actin are the remodelers SWR1 (ref. 25) and INO80 (ref. 24), and the HAT complex NuA4 (ref. 15). To test for ARP association, we expressed their HSA domains (Swr1340–411; Ino80462–598; Eaf1287–406) in yeast as tagged derivatives bearing both 10 × His and Flag tags. Swr1 and Eaf1 HSA domains were purified to near homogeneity through a combination of Ni-NTA and anti-Flag chromatography, which yielded three proteins (Fig. 4d). Sequencing by MS and immunoblot analysis identified Arp4, actin and their respective HSAs (Fig. 4e and data not shown). Staining and immunoblot analyses of the purified Ino80 HSA domain revealed associated Arp4, actin and Arp8, (Fig. 4f,g) but not Arp5 (data not shown). Our results with Ino80 support earlier work by others showing that the large N terminus of Ino80 is necessary for Arp4, Arp8 and actin association24. We extend that work by identifying an HSA domain in Ino80 and by establishing the HSA as the associating domain, although the particular ARP(s) that associates directly with the HSA is not distinguishable. We note that each HSA was highly selective; no actin was found in association with the HSA from Sth1, and no Arp7 or Arp9 is found in association with the HSA domains from Eaf1, Swr1 or Ino80 (Fig. 4 and data not shown). Furthermore, although remodeler complexes interact with nucleosomes, histone H3 was not present at detectable levels either by staining or western analysis in the HSA–ARP–actin modules purified from yeast (data not shown). Taken together, each HSA is sufficient for its selective ARP–ARP or ARP–actin nucleation.
We next determined whether the Eaf1 HSA domain is required for ARP association. An Eaf1 derivative lacking residues 347–418 (Eaf1ΔHSA) was stably produced but no longer associated with Arp4 and actin, or with the Epl1 or Esa1 (HAT) subunits of NuA4 (Fig. 5a,b). These results suggest that the HSA–ARP module of Eaf1 is required for two functions, Arp4–actin association and assembly of Eaf1 into the NuA4 complex.
Figure 5.
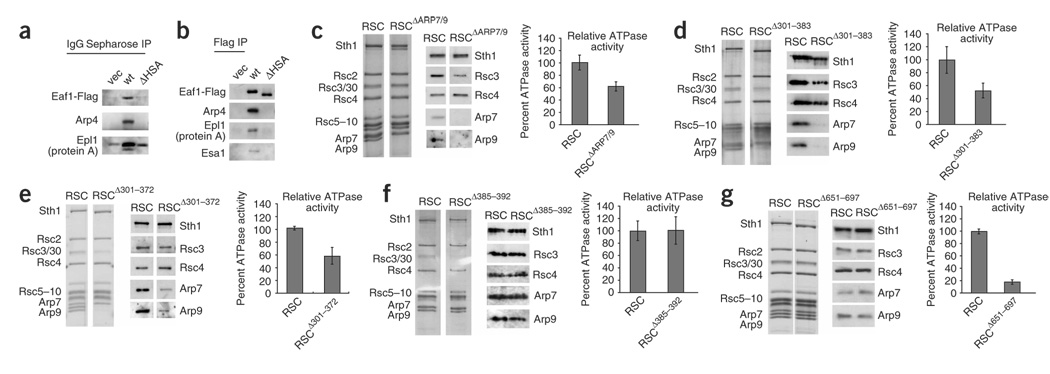
The HSA domains of Sth1 and NuA4 are required for Arp and actin binding, and, in RSC, full ATPase activity. (a) The Eaf1 HSA region is necessary for Epl1 association with Eaf1, Arp4 and actin. Flag-tagged Eaf1 (p2345), or a derivative lacking the HSA domain (Eaf1ΔHSA (p2348)) was expressed in an eaf1Δ strain (YBC3068) that also expressed TAP-tagged Epl1 from its genomic locus. Proteins associated with Epl1 (by pull-down with IgG sepharose) were examined by western analysis. (b) The Eaf1 HSA region is necessary for association of Arp4 and actin with Eaf1. Flag-tagged Eaf1 (p2345), or a derivative lacking the HSA domain (Eaf1ΔHSA (p2348)) was expressed in an eaf1Δ strain. Eaf1 was pulled down with anti-Flag agarose, and associated proteins were examined by western analysis. (c–g) RSC complex derivatives containing Sth1 domain mutations, or arp mutations, were purified and examined for composition by silver staining (left) and western analysis (middle), and then monitored for ATPase activity, relative to a wild-type derivative purified alongside the mutant complex (right). For each, the error bar indicates s.d. (c) RSC complex lacking Arp7 and Arp9. (d) RSC complex lacking the Sth1 HSA region (RSCΔ301–383). (e) RSC complex lacking the Sth1 region sufficient for ARP binding (RSCΔ301–372). (f) RSC complex lacking the Sth1 post-HSA domain (RSCΔ385–392). (g) RSC complex lacking the Sth1 protrusion 1 domain (RSCΔ651–697).
We then explored whether particular regions of the Sth1 HSA are responsible for selective association with Arp7 or Arp9. We performed extensive mutagenesis of the Sth1 HSA domain, replacing conserved sets of consecutive amino acids (Supplementary Fig. 2 online). Notably, we did not observe the selective loss of either Arp7 or Arp9 (Supplementary Fig. 2). In the rare instances where ARPs were partially lost, Arp7 and Arp9 were both released, supporting the idea that they interact with the HSA as a dimer. Thus, the Arp7–Arp9 dimer seems to interact with many locations along the HSA.
ARP association with RSC requires the HSA domain
To determine whether the HSA or post-HSA domain is necessary for ARP association with RSC, we purified RSC complexes with domain deletions to near homogeneity (Fig. 5). A stable Sth1 protein derivative could be produced if we deleted the entire HSA along with a portion of the post-HSA (sth1Δ301–383), or the regions sufficient for ARP binding (sth1Δ301–359 and sth1Δ301–372), or all of protrusion 1 (sth1Δ651–697). In the case of the post-HSA domain, its highly conserved central portion could be deleted without causing Sth1 instability (sth1Δ385–392; Fig. 1b). We expressed these Flag-tagged Sth1 mutant derivatives with the conditional MET25 promoter in cells containing a wild-type STH1 allele (for viability) and purified them, generating RSCΔ301–383, RSCΔ301–372, RSCΔ385–392 and RSCΔ651–697 (Fig. 5d–g). By a similar strategy, using plasmid-borne wild-type Sth1, we isolated RSC complex lacking ARPs (RSCΔARP7/9) from a strain rendered viable owing to a genomic mra mutation (Fig. 5c). We found that RSCΔ301–383 lacked Arp7 and Arp9 entirely (Fig. 5d), consistent with our results that amino acids 301–383 or 301–372 alone were sufficient for robust ARP binding (note that Arp7 often co-migrates at or near Rsc10). Notably, a small fraction of the ARPs remained present in the RSCΔ301–372 derivative, suggesting that the post-HSA domain may interact weakly with the ARPs (Fig. 5e). RSCΔ385–392 and RSCΔ651–697 both show full ARP association (Fig. 5f,g), showing that deletions involving the highly conserved regions of the post-HSA or protrusion 1 domains do not lead to the loss of ARPs in a RSC complex that has an intact HSA domain. Thus, the HSA domain of Sth1 is necessary and sufficient for the robust association of ARPs with RSC, with a section of the post-HSA domain providing a minor contribution.
Regulation of RSC ATPase activity
Although the low yields of the mutant RSC complexes prevented detailed tests of nucleosome remodeling, we obtained sufficient amounts to test DNA-dependent ATPase activity. To provide an accurate comparison, we purified wild-type RSC (using Flag-tagged wild-type Sth1) and the mutant derivatives side by side each time with identical solutions and conditions (Fig. 5). The DNA-dependent ATPase activity (Vmax) of the RSCΔ301–383 complex was reduced about two-fold from that of the wild-type complex (Fig. 5d), with some variability. In previous work, we purified RSC complex lacking Arp7 or Arp9 and found a slight reduction in ATPase activity (~85% of wild type)22, although the derivative that was tested is now known to have contained an sth1N384K replacement, which may have slightly restored ATPase activity resulting from the loss of ARP proteins. To address this, we tested RSCΔARP7/9 (which bears wild-type Sth1) and found it two-fold less active than wild-type RSC (Fig. 5c), establishing the full impact of ARPs on ATPase activity. The ATPase activity of RSCΔ301–372, which has low levels of Arp7 and Arp9, is reduced to ~60% of the activity observed with the wild type (Fig. 5e). Notably, the loss of protrusion 1 reduced ATPase activity substantially (to ~20% of wild-type), even though it retains ARP proteins (Fig. 5g). Thus, the HSA, ARP proteins and protrusion 1 are required for full ATPase activity. Notably, the activity of the RSCΔ385–392 complex was not reduced (Fig. 5f), raising the possibility that the post-HSA domain regulates the ATPase in a manner not revealed in this assay.
The HSA, post-HSA and protrusion 1 domains are essential
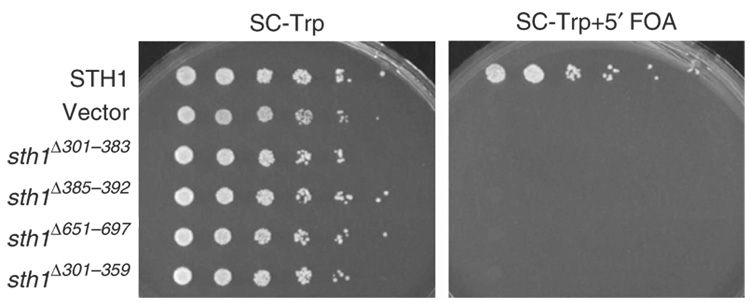
To determine the importance of the HSA, post-HSA and protrusion 1 domains for RSC function in vivo, we expressed derivatives lacking all or portions of these domains and tested for complementation of sth1Δ. All three derivatives were stably expressed and assembled into RSC complexes (Fig. 5 and data not shown). Sth1 absolutely requires all three of these domains to function, as the sth1 alleles bearing deletions in each of these domains fail to complement sth1Δ under all conditions tested (Fig. 6 and data not shown). Taken together, we show that the three domains characterized here are required for viability and RSC function in vivo.
Figure 6.
The HSA, post-HSA and protrusion 1 domains are essential for viability. TRP1-marked plasmids bearing wild-type STH1, sth1Δ301–383, sth1Δ385–392, sth1Δ651–697 or sth1Δ301–359 (p976, p1724, p1683, p2500 or p2465, respectively) were transformed into an sth1Δ strain (YBC943) covered by wild-type STH1 on a URA3-marked plasmid (p114). Loss of the URA3-marked plasmid is enforced on medium containing 5′ fluoroorotic acid (FOA), which prevents the growth of Ura3+ cells. Inability to lose the URA3-marked plasmid bearing wild-type STH1 demonstrates the lack of complementation by the sth1 domain deletion derivative. Vector, empty vector control.
DISCUSSION
Our work provides three important findings. First, we define the HSA domain as a general binding platform for nuclear ARPs and show that the individual HSA domains can reconstitute particular ARP–ARP or ARP–actin modules (Fig. 7a). Second, we identify a new essential domain, the post-HSA domain, and connect this domain to ARP function through genetic suppression relationships. Third, we provide the first functional evidence for the protrusion 1 domain: it is necessary for Sth1 function and viability in vivo and full ATPase activity in vitro, and is linked to ARP function through genetic suppression. Notably, these dominant mutations in the post-HSA and protrusion 1 domains of Sth1 (ten total) were isolated in an unbiased genome-wide screen for arpΔ suppressors, and were the only mutations obtained. Taken together, we define the role of the HSA domain, and reveal a three-domain regulatory relationship network in the remodeler ATPase consisting of an ARP–HSA module, the post-HSA domain and protrusion 1 of the ATPase.
Figure 7.
Summary of HSA domain complexes and a model for HSA–ARP regulation of ATPase activity via the adjacent post-HSA domain and protrusion 1. (a) The HSA domains from yeast modifying complexes RSC (Sth1), SWI-SNF (Snf2), SWR1 (Swr1), NuA4 (Eaf1), INO80 (Ino80) and human SWI-SNF (BRG1) nucleate binding of their respective ARP–ARP or ARP–actin members. (b) The HSA–ARPs, post-HSA and protrusion 1 domains interact to regulate the function of the ATPase. Actin-related proteins bind strongly to the HSA, with a weak interaction with the post-HSA also detected. The ARPs and protrusion 1 positively regulate ATPase activity, and the ability of the ARPs to influence (or bind) protrusion 1 may be regulated by the post-HSA domain.
Here we define HSA domains as both necessary and sufficient to assemble specific ARP–actin modules. Thus, some attribute of the domain must select a particular combination of ARPs and/or actin. We note that cytoplasmic Arp2 and Arp3 bind an α-helical platform on the p38 protein33. In keeping with this idea, secondary-structure predictions suggest that HSA domains are composed of long α-helices (Fig. 1). Taken together, these data lead us to speculate that the HSA–ARP–actin modules present on nuclear chromatin remodelers might have evolved from the p38–Arp2–Arp3 module resident in the cytoplasm. Furthermore, particular residues along the helical surface may have evolved for selective ARP complementarity. Here our mutagenesis data suggest that the HSA may have evolved to bind ARP–ARP or ARP–actin dimer units, rather than selecting them individually (Supplementary Fig. 2).
SWR1 and INO80 are shown here to have an HSA domain that is sufficient for the association of actin and Arp4, which are members of both complexes, but not Arp5 and Arp6, which are unique to each complex (only INO80 contains Arp5, and only SWR1 has Arp6). Other work showed that association with Arp5 and Arp6 requires the ‘major insertion’ domain, located in the III–IV loop just adjacent to protrusion 2, within their respective ATPase subunits24,34. This insertion also mediates the association of the Ruv helicases, which are required for Arp5 and Arp6 association. Taken together, there are two modes by which ARPs can bind to chromatin remodeling ATPases: (i) a primary interaction via the HSA domain, which is found in all ARP-containing remodelers and modifier complexes; and (ii) through proteins bound to the major insertion domain found in a subset of remodelers. We note that the association of actin with these remodelers is solely dependent on the HSA domain.
We define the post-HSA domain as a domain adjacent to the HSA that is conserved in remodelers that bear ARPs but absent in other remodeler families such as CHD and ISWI. Likewise, modifier proteins such as those of the NuA4 family contain an HSA but lack a post-HSA domain. Although adjacent to the HSA, the post-HSA is not required for robust ARP association or selectivity, but it makes a small contribution to full ARP interaction. We suggest that the post-HSA functions with the HSA and the ARPs to help regulate an aspect of remodeler ATPase function, as specific post-HSA mutations confer arp suppression. However, mutations in (or deletions within) the post-HSA did not diminish the activity of the ATPase domain in our in vitro assay; thus, additional factors or assay systems are probably required to observe this regulation in vitro.
One interesting question is the extent to which alterations in ATPase activity underlie the inviability of arpΔ mutants. Previous work on INO80 clearly showed elimination of ATPase activity following the loss of ARP components24. In contrast, our previous examination of a RSC complex lacking ARPs revealed only ~15% reduction in ATPase activity22, and thus we questioned whether this reduction was of a magnitude sufficient to confer inviability. However, the RSC derivative examined also contained an sth1N384K replacement, which may have partially restored the reduction in ATPase activity that results from the loss of ARP proteins. Here we resolved this issue by isolating a RSC complex that contained wild-type Sth1 but lacked ARPs, and this complex showed a two-fold reduction in ATPase activity, nearly identical to the results we obtained from a RSC derivative lacking the HSA (RSCΔ301–383), which also lacked ARPs. Thus, RSC and INO80 differ considerably in the extent to which they rely on ARPs for ATPase regulation.
Protrusion 1 is part of a larger insertion residing in the III–IV loop and is a distinguishing feature of Snf2 family ATPases. Here we show that protrusion 1 is essential for Sth1 function. Protrusion 1 in Rad54 consists of three helices, and structural predictions suggest a similar three-helix relationship in Sth1 (Fig. 1e). Chromatin remodelers that contain ARPs share high sequence identity in protrusion 1, whereas those that lack ARPs are divergent. We speculate that protrusion 1 is highly similar in ARP-containing remodelers in order to interact functionally with the HSA–ARP module and/or the post-HSA domain; protrusion 1 may be the target within the ATPase domain that is regulated by the ARP–HSA module (Fig. 7b). As protrusion 1 is located on the opposite side of the structure from the DNA binding or ATP binding domains, its impact on ATPase activity is likely to occur through an allosteric mechanism.
Although the precise relationships remain to be defined, our work, along with that of others20,24, suggests a unifying concept: ARPs influence remodeler function by interacting with the ATPase III–IV loop. For SNF2 family remodelers, the regulatory interaction occurs between ARPs and protrusion 1, and is enabled by the ARPs docking on the adjacent HSA and post-HSA domains. For INO80 and SWR1 family remodelers, a subset of their ARPs (Arp4, Arp8 and actin) also dock using the adjacent HSA domain, and one additional ARP docks on the major insertion region adjacent to protrusion 2. In both cases, the crucial feature is HSA docking and interaction with protrusions in the ATPase III–IV loop. Future work, both biochemical and structural, should more precisely define how ARPs and these newly identified domains help to regulate the remodeler.
METHODS
Media, genetic methods and strains
Standard procedures were used for media preparations, transformations, sporulation and tetrad analysis. All strains are derivatives of S288C or W303. Strain identities are provided in the figure legends, and full genotypes are listed in Supplementary Table 2 online.
Linkage analysis
YBC251 was constructed by placing HIS3 under control of the MFA1 promoter by integrating the HIS3 open reading frame at MFA1 in YBC425 as described previously35. Mating type was converted to MATα by introducing a pRSGalI.HO plasmid, growth in 1.5% (w/v) galactose, plating and mating type screening. For a linkage assessment, we generated a haploid tester MATα strain (MATα mra1-1 arp9Δ) also bearing MFA1∷HIS3, which confers histidine prototrophy only in MATa cells. This strain was crossed to the complete set of haploid knockouts (4,891 MATa strains) with a kanamycinresistance marker (KANMX) replacing the open reading frame (denoted XΔ∷KANMX) of nearly all the nonessential genes in S. cerevisiae. We then sporulated the resulting diploids and monitored the ability to obtain triple-mutant (XΔ∷KANMX mra1-1 arp9Δ) haploid progeny (His+ due to MATa MFA1∷HIS3). Linkage was determined by the inability to obtain XΔ∷KANMX mra1-1 arp9Δ triple-mutant haploids.
Plasmids
The pRS314.sth1Δ385–392 (p1683) and MET25∷sth1Δ385–392 (p1682) plasmids were constructed by homologous recombination using a PCR fragment and a gapped pRS314.STH1 (p976) and Met25∷STH1 plasmid (p1170), respectively. Primers used were BC3719 and BC3721. A primer sequence table is available upon request. Likewise, MET25∷sth1Δ301–383 and/or pRS314.sth1Δ301–383 (p1684 and p1724), sth1Δ301–359 (p2463 and p2465) and sth1Δ301–372 (p2524) were constructed similarly or by site-directed mutagenesis using primers BC3722 and BC3723, BC3836 and BC3837, and BC3877 and BC3878, respectively. Yeast expression constructs MET25∷Flag.Sth1HSA.10 × His (amino acids 301–383) (p1684), GAL1∷Flag.Snf2HSA.10 × His (amino acids 578–659)(p2535), GAL1∷Flag.Eaf1HSA.10 × His (amino acids 287–406)(p1870), GAL1∷Flag.Ino80HSA.10 × His (amino acids 462–598)(p2441) and GAL1∷Flag.Swr1HSA.10 × His (amino acids 340–411)(p1687) were synthesized by PCR and cloned into either the MET25-inducible (p521) or GAL1-inducible (p533) expression vector. The primers used to construct the expression vectors were BC3843 and BC3844 (Sth1), BC3699 and BC3700 (Eaf1), BC3708 and BC3709 (Ino80) and BC3692 and BC3701 (Swr1). The Flag-tagged BRG1 HSA (amino acids 462–531) expression vector (p1877) was constructed by PCR amplification (using primers BC3702 and BC3703) of human cDNA and subcloning into pcDNA3 (Invitrogen). The HA-BAF53 expression vector was a kind gift of Michael Cole36. The V5-Arp5, cmyc-Arp8 Gal-inducible expression vector (p2436) was constructed from a GAL1–10 divergent promoter construct (p1999) using PCR amplification of genomic DNA with primers BC3712, BC3713, BC3714 and BC3715. MET25∷sth1Δ651–697 and pRS314.sth1Δ651–697 (p2499 and p2500) were constructed by site-directed mutagenesis, deleting amino acids 651–697 with primers BC3836 and BC3837. The constructs were sequence verified.
For recombinant expression, Sth1 amino acids 301–372, 301–359 or 301–345 were N′-terminally 10 × His tagged using PCR (with primers BC3704, 3705, 3706 and 3707) and subcloned into the pCDF-Duet expression vector (Novagen). Rtt102 was constructed by PCR (using primers BC3834 and BC3835) and subcloned into the same pCDF-Duet vector as the Sth1 truncation constructs, creating plasmids p2443, p2444 and p2445, respectively.
Recombinant expression
Constructs were transformed into BL21 CodonPlus competent cells (Stratagene), autoinduced and harvested. The cells were lysed in lysis buffer (200 mM NaCl, 50 mM Tris-HCl (pH 7.5), 10% (v/v) glycerol, 1 mM β-mercaptoethanol and protease inhibitors) by sonication and precleared, and the supernatant was bound to Ni-NTA beads (Qiagen). The beads were washed in lysis buffer with 20 mM imidazole, and proteins were eluted with buffer containing 250 mM imidazole. The eluate was subjected to sizing by gel filtration on a Superdex 200 column (GE Healthcare Inc.)
Extract preparation and immunoprecipitation
Yeast extracts were prepared as described previously37. For co-immunoprecipitation experiments, anti-Flag beads (Sigma) were incubated with whole-cell extracts (400 µg) derived from transformants of YBC405 grown in synthetic complete (SC) medium, containing methionine, washed twice with IP buffer (250 mM NaCl, 50 mM Tris-HCl (pH 7.7), 1 mM EDTA, 0.05% (v/v) Tween-20, 10% (v/v) glycerol) and eluted with SDS sample buffer or Flag peptide as indicated. For purification experiments, cellular extracts were prepared as described previously37, incubated with Ni-NTA beads, and washed and then eluted with buffer containing 250 mM imidazole. The nickel eluate was bound to anti-Flag agarose beads (Sigma), washed with IP buffer and eluted using 3 × Flag peptide. Antibodies used were anti–protein A (Sigma), anti-Esa1 (Abcam), anti–6 × His (BD Biosciences), anti-Arp4 (our rabbit polyclonal), anti–histone H3 C-terminal (Abcam) and anti-actin (yeast; gift from J. Shaw, University of Utah).
For BRG1 HSA expression, RKO cells were cultured in DMEM media (Invitrogen) with 10% (v/v) FBS (Hyclone). Cells were transfected with 1 µg of each expression vector using Lipofectamine (Invitrogen) as per the manufacturer’s protocol. Cells were harvested at 72 h after transfection (150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 10% (v/v) glycerol, 0.1% (v/v) NP-40, 1 mM PMSF and 1 × protease inhibitor cocktail (Roche)) and precleared, and extracts were immunoprecipitated using anti-Flag agarose beads. Precipitants were eluted with 3 × Flag peptide and subjected to western blotting using antibodies specifically recognizing Flag, HA (HCI Core Laboratories), actin (MPBio) and BAF57 (a gift from W. Wang, US National Institutes of Health (NIH)).
RSC purification and ATPase assays
BCY211 or BCY262 (wild-type and mra Sth1) harboring an integrated RSC2.TAP and Flag-tagged Sth1 (or derivative) on a plasmid was used to purify RSC as described previously4,38, except that column elutions were performed with 300 mM potassium acetate and an additional purification step using anti-Flag agarose beads was performed, and eluted using 3 × Flag peptide. ATPase activity was measured as described previously4.
Alignments and structures
The HSA domain was characterized using Pfam39. The alignments were performed in Jalview40 using the MAFFT41. The structure figures were made with PyMOL (http://www.pymol.org), and secondary-structure predictions were performed in PSIPRED42.
Supplementary Material
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
ACKNOWLEDGMENTS
We thank J. Wittmeyer and A. Saha (Cairns laboratory) for plasmids, strains and expertise on protein purification. We thank J. Shaw, D. Close, M. Kasten, T. Parnell and J. Lenkart (all University of Utah) for reagents and advice. We thank W. Wang (US National Institutes of Health (NIH)) and M. Cole (Dartmouth University) for antibodies and plasmids, P. Hollenhorst and C. Foulds (University of Utah) for cell culture reagents and advice. This work was supported by the NIH (GM60415 to B.R.C., and support of H.S.), CA24014 for core facilities, and the Howard Hughes Medical Institute (support of B.R.C. and K.H.).
Footnotes
Published online at http://www.nature.com/nsmb/
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Havas K, Whitehouse I, Owen-Hughes T. ATP-dependent chromatin remodeling activities. Cell. Mol. Life Sci. 2001;58:673–682. doi: 10.1007/PL00000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 3.Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown CE, Lechner T, Howe L, Workman JL. The many HATs of transcription coactivators. Trends Biochem. Sci. 2000;25:15–19. doi: 10.1016/s0968-0004(99)01516-9. [DOI] [PubMed] [Google Scholar]
- 6.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olave IA, Reck-Peterson SL, Crabtree GR. Nuclear actin and actin-related proteins in chromatin remodeling. Annu. Rev. Biochem. 2002;71:755–781. doi: 10.1146/annurev.biochem.71.110601.135507. [DOI] [PubMed] [Google Scholar]
- 8.Blessing CA, Ugrinova GT, Goodson HV. Actin and ARPs: action in the nucleus. Trends Cell Biol. 2004;14:435–442. doi: 10.1016/j.tcb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 9.Chen M, Shen X. Nuclear actin and actin-related proteins in chromatin dynamics. Curr. Opin. Cell Biol. 2007;19:326–330. doi: 10.1016/j.ceb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Poch O, Winsor B. Who’s who among the Saccharomyces cerevisiae actin-related proteins? A classification and nomenclature proposal for a large family. Yeast. 1997;13:1053–1058. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1053::AID-YEA164>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 11.Peterson CL, Zhao Y, Chait BT. Subunits of the yeast SWI/SNF complex are members of the actin-related protein (ARP) family. J. Biol. Chem. 1998;273:23641–23644. doi: 10.1074/jbc.273.37.23641. [DOI] [PubMed] [Google Scholar]
- 12.Cairns BR, Erdjument-Bromage H, Tempst P, Winston F, Kornberg RD. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol. Cell. 1998;2:639–651. doi: 10.1016/s1097-2765(00)80162-8. [DOI] [PubMed] [Google Scholar]
- 13.Krogan NJ, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 14.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 15.Galarneau L, et al. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell. 2000;5:927–937. doi: 10.1016/s1097-2765(00)80258-0. [DOI] [PubMed] [Google Scholar]
- 16.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 17.Ikura T, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 18.Grant PA, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 19.Osada S, et al. The yeast SAS (something about silencing) protein complex contains a MYST-type putative acetyltransferase and functions with chromatin assembly factor ASF1. Genes Dev. 2001;15:3155–3168. doi: 10.1101/gad.907201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao K, et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 21.Mizuguchi G, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 22.Szerlong H, Saha A, Cairns BR. The nuclear actin-related proteins Arp7 and Arp9: a dimeric module that cooperates with architectural proteins for chromatin remodeling. EMBO J. 2003;22:3175–3187. doi: 10.1093/emboj/cdg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Zaurin R, Beato M, Peterson CL. Swi3p controls SWI/SNF assembly and ATP-dependent H2A–H2B displacement. Nat. Struct. Mol. Biol. 2007;14:540–547. doi: 10.1038/nsmb1238. [DOI] [PubMed] [Google Scholar]
- 24.Shen X, Ranallo R, Choi E, Wu C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell. 2003;12:147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- 25.Wu WH, et al. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat. Struct. Mol. Biol. 2005;12:1064–1071. doi: 10.1038/nsmb1023. [DOI] [PubMed] [Google Scholar]
- 26.Letunic I, et al. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 2002;30:242–244. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jorgensen P, et al. High-resolution genetic mapping with ordered arrays of Saccharomyces cerevisiae deletion mutants. Genetics. 2002;162:1091–1099. doi: 10.1093/genetics/162.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanya HS, Bird LE, Brannigan JA, Wigley DB. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 29.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thoma NH, et al. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat. Struct. Mol. Biol. 2005;12:350–356. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- 31.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 32.Fiori A, et al. Disruption of six novel genes from chromosome VII of Saccharomyces cerevisiae reveals one essential gene and one gene which affects the growth rate. Yeast. 2000;16:377–386. doi: 10.1002/1097-0061(20000315)16:4<377::AID-YEA537>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 33.Hitchcock-DeGregori SE. Now, swing your partner! 3D-domain switching of WASP activates Arp2/3 complex. Nat. Struct. Biol. 2003;10:583–584. doi: 10.1038/nsb0803-583. [DOI] [PubMed] [Google Scholar]
- 34.Jonsson ZO, Jha S, Wohlschlegel JA, Dutta A. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol. Cell. 2004;16:465–477. doi: 10.1016/j.molcel.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 35.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 36.Park J, Wood MA, Cole MD. BAF53 forms distinct nuclear complexes and functions as a critical c-Myc-interacting nuclear cofactor for oncogenic transformation. Mol. Cell. Biol. 2002;22:1307–1316. doi: 10.1128/mcb.22.5.1307-1316.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cairns BR, et al. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- 38.Puig O, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 39.Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 41.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.