Abstract
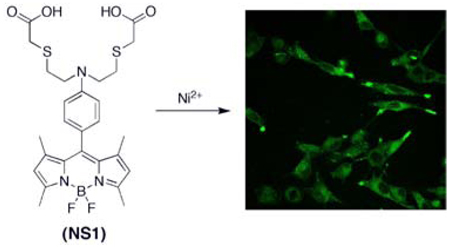
We present the synthesis and properties of Nickelsensor-1 (NS1), a new water-soluble, turn-on fluorescent sensor that is capable of selectively responding to Ni2+ in aqueous solution and in living cells. NS1 combines a BODIPY chromophore and a mixed N/O/S receptor to provide good selectivity for Ni2+ over a range of biologically abundant metal ions in aqueous solution. In addition to these characteristics, confocal microscopy experiments further show that NS1 can be delivered into living cells and report changes in intracellular Ni2+ levels in a respiratory cell model.
Nickel is an essential metal nutrient for supporting life, but loss of nickel homeostasis is harmful to prokaryotic and eukaryotic organisms alike.1 Elegant studies continue to elucidate mechanisms for Ni2+ uptake, regulation, and efflux,2–10 as well as to define the redox and non-redox roles of nickel biochemistry in microbial and plant systems.11–17 However, the contributions of nickel homeostasis to mammalian health and disease remain largely unexplored.18 In this context, excess nickel accumulation can aberrantly affect respiratory and immune systems, but mechanisms of nickel imbalance are insufficiently understood.19,20
To help elucidate the roles of nickel in living systems, we are developing Ni2+-selective fluorescent indicators as part of a larger program aimed at studying metals in biology by molecular imaging.21,22 Such chemical tools, in principle, can be used to monitor exchangeable nickel pools with spatial and temporal resolution and provide a complement to standard bulk techniques for measuring total nickel content such as atomic absorption or inductively coupled plasma mass spectrometry. A major chemical challenge to this end is designing systems with Ni2+-specific responses over other biologically relevant metal ions in water. Examples of Ni2+-responsive fluorescent probes remain rare; Ni2+-selective peptide,23,24 protein,25 polymer,26,27 and small-molecule based sensors28–30 have been reported but have not been utilized for cellular imaging, whereas the commercial Zn2+ sensor Newport Green DCF also responds to Ni2+ and Ti3+ and has been used to detect their accumulation in cells.31–34 In this report, we present the synthesis and properties of Nickelsensor-1 (NS1, 5), a new turn-on fluorescent sensor for the selective detection of Ni2+ in water and in biological samples. NS1 features visible wavelength spectral profiles and a ca. 25-fold fluorescence increase upon Ni2+ binding. Confocal microscopy experiments show that this indicator can reliably monitor changes in Ni2+ levels within living mammalian cells.
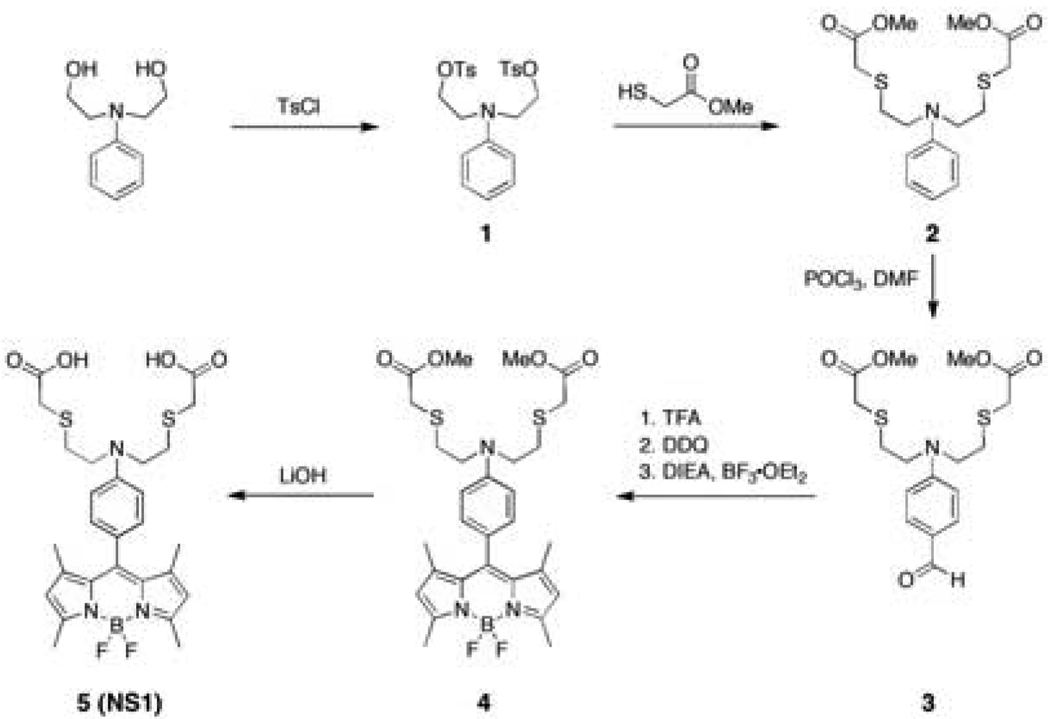
Our design for NS1 combines a BODIPY dye reporter with a mixed N/O/S receptor to satisfy the Ni2+ cation (Scheme 1). Addition of ditosylate 1 to Cs2CO3 and methyl thiogylcolate affords diester 2 in 41% yield. Vilsmeier formylation of 2 using POCl3/DMF followed by basic workup furnishes aldehyde 3 in 60% yield. BODIPY 4 is obtained in a one-pot, three-step procedure via condensation of 3 with 2,4-dimethylpyrrole, followed by DDQ oxidation and boron insertion with BF3•OEt2 (38% overall yield for three steps). Ester hydrolysis of 4 under basic conditions gives NS1 (5) in 71% yield.
Scheme 1.
Synthesis of Nickelsensor-1 (NS1)
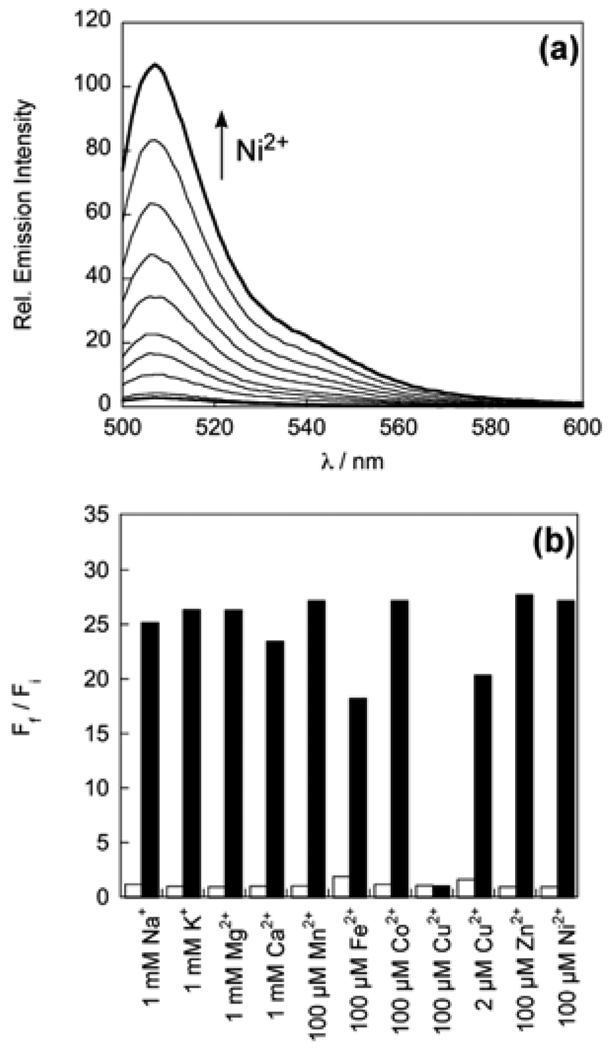
Spectroscopic evaluation of NS1 was performed in 20 mM HEPES buffered to pH 7.1. The optical features of the probe are characteristic of the BODIPY platform. Apo NS1 displays one visible region absorption band centered at 495 nm (ε = 5.8 × 103 M−1 cm−1) and an emission maximum at 507 nm (Φ = 0.002). Addition of 50 equiv of Ni2+ triggers a ca. 25-fold fluorescence turn-on (Φ = 0.055, Figure 1a) with no shifts in absorption (λabs = 495 nm, ε = 5.5 × 103 M−1 cm−1) or emission maxima (λem = 507 nm) compared to the apo probe. The turn-on response is reversible; treatment of Ni2+-loaded NS1 with the divalent metal ion chelator TPEN restores NS1 fluorescence back to baseline levels. A Hill plot indicates a simple binding process with no cooperativity (Figure S1a), and the apparent Kd for Ni2+ binding to NS1 is 193 ± 5 µM (Figure S1b).
Figure 1.
(a) Fluorescence response of 2 µM NS1 to Ni2+. Spectra shown are for Ni2+ concentrations of 0, 2, 5, 10, 15, 25, 35, 50, 75, 100 µM. Spectra were acquired in 20 mM HEPES, pH 7.1, with 488 nm excitation. (b) Fluorescence responses of 2 µM NS1 to various metal ions. Bars represent the final (Ff) over the initial (Fi) integrated emission. Spectra were acquired in HEPES, pH 7.1. White bars represent the addition of the competing metal ion to a 2 µM solution of NS1. Black bars represent addition of 100 µM Ni2+ to the solution. Excitation was provided at 488 nm, with emission integrated over 498—700 nm.
NS1 exhibits a selective turn-on fluorescence response to Ni2+ in water. Responses of 2 µM NS1 to the presence of various biologically relevant metal ions are shown in Figure 1b. The fluorescence profiles of apo or Ni2+-bound NS1 are unchanged in the presence of 1 mM Na+, K+, Mg2+, and Ca2+, indicating excellent selectivities for Ni2+ over these alkali and alkaline earth cations. Moreover, a series of 3d divalent metal cations, including 100 µM Mn2+, Fe2+, Co2+, and Zn2+, do not trigger NS1 fluorescence enhancements or interfere with the Ni2+ response. Of the first-row divalent transition metal ions, Cu2+ at 100 µM can mute the turn-on Ni2+ response of NS1, but lower Cu2+ levels (2 µM) minimize this interference. As expected, Cu2+ binds the sensor due to Irving-Williams series considerations, but the paramagnetic d9 ion quenches fluorescence, suggesting that the fluorescence increase for Ni2+ is due to a diamagnetic d8 state.
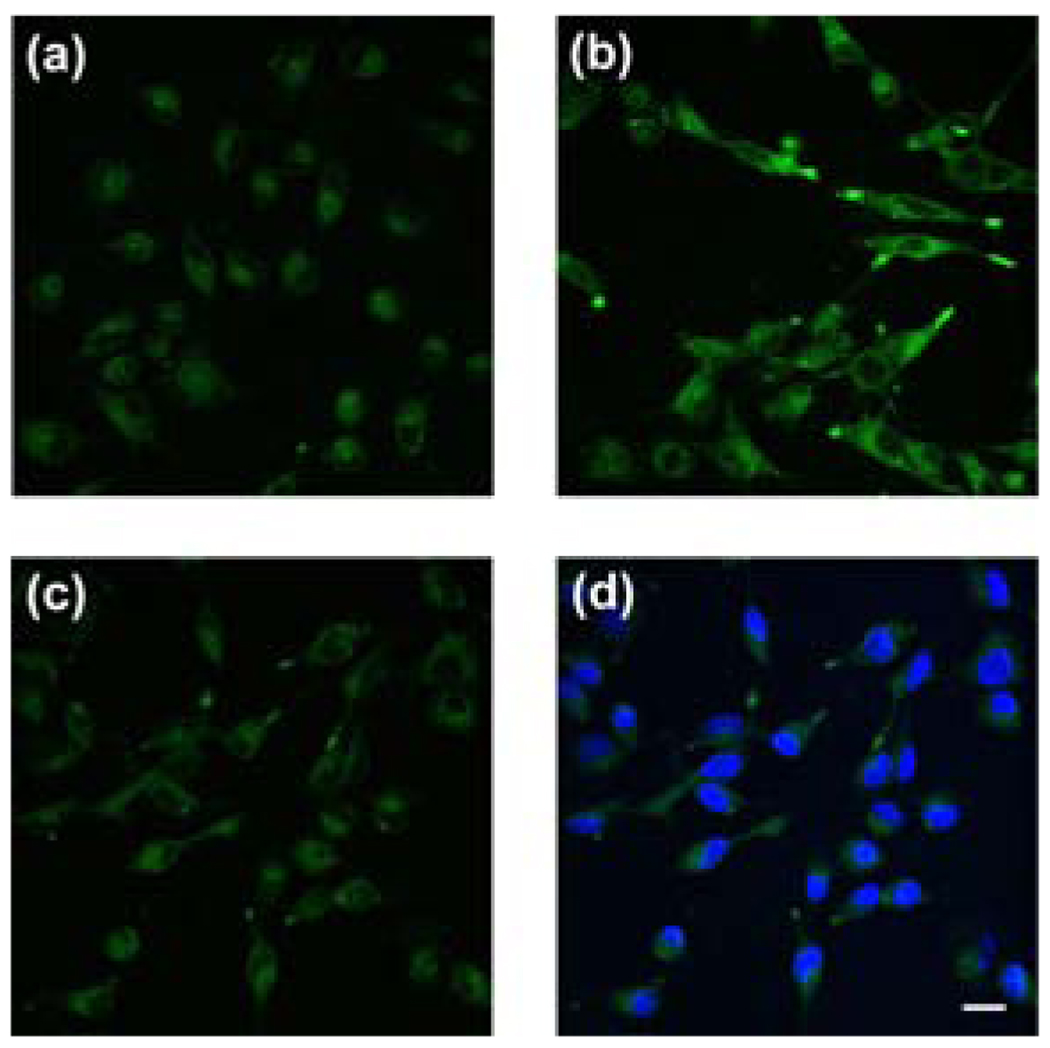
We next established the ability of NS1 to track Ni2+ levels in living cells using a model for respiratory nickel exposure. Live-cell confocal microscopy imaging experiments utilized the acetoxymethyl ester form of NS1 (NS1-AM) to enhance membrane permeability. Live human lung carcinoma A549 cells loaded with a 1:1 (v/v) mixture of NS1-AM and F-127 Pluronic acid (10 µM) for 35 min at 37 °C show weak intracellular fluorescence (Figure 2a). A549 cells supplemented with 1 mM NiCl2 in the growth medium for 18 h at 37 °C and then staining with NS1-AM under the same loading conditions results in an increase in observed intracellular fluorescence intensity (Figure 2b); previous experiments establish that this exposure level of nickel is not lethal to lung carcinoma cells, whereas 2–10 mM Ni2+ causes widespread cell death.35 Treatment of cells loaded with NS1-AM and Ni2+ with the divalent metal chelator TPEN (1 mM) for 1 min at 25 °C reverses the observed fluorescence increases (Figure 2c). Finally, Hoescht-3342 staining confirms that the cells are viable throughout the imaging studies (Figure 2d). These data establish that NS1 can respond to changes in intracellular Ni2+ levels within living cells.
Figure 2.
Live-cell imaging of intracellular Ni2+ levels by confocal microscopy. (a) Control A549 cells incubated with a 1:1 mixture of 10 µM NS1-AM and F-127 Pluronic acid for 35 min at 37 °C. (b) Cells supplemented with 1 mM NiCl2 in the growth medium for 18 h at 37 °C and stained with 10 µM NS1-AM and F-127 Pluronic acid for 35 min at 37 °C. (c) NS1-loaded, 1 mM Ni2+-supplemented cells treated with 1 mM of the divalent metal chelator TPEN for 1 min at 25 °C. (D) NS1-loaded, 1 mM Ni2+-supplemented cells treated with 1 mM TPEN, stained with 5 µM Hoescht-3342 to show cell viability. Scale bar = 20 µm.
In closing, we have described the synthesis, spectroscopy, and application of NS1, a new fluorescent sensor for Ni2+ in biological samples. NS1 is a unique Ni2+-responsive small-molecule indicator that features visible excitation and emission profiles and a selective turn-on response to Ni2+ compared to other biological metal ions. Confocal microscopy experiments show that NS1 can be used for detecting changes in Ni2+ levels within living cells. Future plans will focus on improving the optical brightness and binding affinities of this first-generation probe as well as applying NS1 and related chemical tools to probe the cell biology of nickel.
Supplementary Material
Acknowledgment
We thank the Dreyfus, Packard, and Sloan Foundations, the Hellman Faculty Fund, Amgen, NSF (CAREER CHE-0548245), NIH (GM 79465), and HHMI for providing funding for this work. We thank Holly Aaron (UCB Molecular Imaging Center) and Ann Fischer and Michelle Yasukawa (UCB Tissue Culture Facility) for expert technical assistance.
Footnotes
Supporting Information Available: Synthetic and experimental details (PDF). This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Sigel A, Sigel H, Sigel RKO, editors. Nickel and Its Surprising Impact in Nature. Vol. 2. England: John Wiley & Sons Ltd.; 2007. [Google Scholar]
- 2.Chivers PT, Sauer RT. Protein Sci. 1999;8:2494–2500. doi: 10.1110/ps.8.11.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dosanjh NS, Michel SL. Curr. Opin. Chem. Biol. 2006;10:123–130. doi: 10.1016/j.cbpa.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 4.Giedroc DP, Arunkumar AI. Dalton Trans. 2007:3107–3120. doi: 10.1039/b706769k. [DOI] [PubMed] [Google Scholar]
- 5.Hausinger RP, Zamble DB. In: Molecular Microbiology of Heavy Metals. Nies DH, Silver S, editors. Heidelberg, Germany: Springer; 2007. pp. 287–320. [Google Scholar]
- 6.Wang SC, Dias AV, Zamble DB. Dalton Trans. 2009:2459–2466. doi: 10.1039/b818167p. [DOI] [PubMed] [Google Scholar]
- 7.Carrington PE, Chivers PT, Al-Mjeni F, Sauer RT, Maroney MJ. Nat. Struct. Biol. 2003;10:126–130. doi: 10.1038/nsb890. [DOI] [PubMed] [Google Scholar]
- 8.Schreiter ER, Sintchak MD, Guo Y, Chivers PT, Sauer RT, Drennan CL. Nat. Struct. Biol. 2003;10:794–799. doi: 10.1038/nsb985. [DOI] [PubMed] [Google Scholar]
- 9.Phillips CM, Schreiter ER, Guo Y, Wang SC, Zamble DB, Drennan CL. Biochemistry. 2008;47:1938–1946. doi: 10.1021/bi702006h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwig JS, Leitch S, Herbst RW, Maroney MJ, Chivers PT. J. Am. Chem. Soc. 2008;130:7592–7606. doi: 10.1021/ja710067d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maroney MJ. Curr. Opin. Chem. Biol. 1999;3:188–199. doi: 10.1016/S1367-5931(99)80032-5. [DOI] [PubMed] [Google Scholar]
- 12.Drennan CL, Doukov TI, Ragsdale SW. J. Biol. Inorg. Chem. 2004;9:511–515. doi: 10.1007/s00775-004-0563-y. [DOI] [PubMed] [Google Scholar]
- 13.Evans DJ. Coord. Chem. Rev. 2005;249:1582–1595. [Google Scholar]
- 14.Fontecilla-Camps JC, Volbeda A, Cavazza C, Nicolet Y. Chem. Rev. 2007;107:4273–4303. doi: 10.1021/cr050195z. [DOI] [PubMed] [Google Scholar]
- 15.Lindahl PA. Angew. Chem., Int. Ed. Engl. 2008;47:4054–4056. doi: 10.1002/anie.200800223. [DOI] [PubMed] [Google Scholar]
- 16.Ragsdale SW. J. Biol. Chem. 2009;284:18571–18575. doi: 10.1074/jbc.R900020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia W, Li H, Sze KH, Sun H. J. Am. Chem. Soc. 2009;131:10031–10040. doi: 10.1021/ja900543y. [DOI] [PubMed] [Google Scholar]
- 18.Goodman JE, Prueitt RL, Dodge DG, Thakali S. Crit. Rev. Toxicol. 2009;39:365–417. doi: 10.1080/10408440902762777. [DOI] [PubMed] [Google Scholar]
- 19.Costa M, Davidson TL, Chen H, Ke Q, Zhang P, Yan Y, Huang C, Kluz T. Mutat. Res. 2005;592:79–88. doi: 10.1016/j.mrfmmm.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Nemec AA, Leikauf GD, Pitt BR, Wasserloos KJ, Barchowsky A. Am. J. Respir. Cell Mol. Biol. 2009;41:69–75. doi: 10.1165/rcmb.2008-0409OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domaille DW, Que EL, Chang CJ. Nat. Chem. Biol. 2008;4:168–175. doi: 10.1038/nchembio.69. [DOI] [PubMed] [Google Scholar]
- 22.Que EL, Domaille DW, Chang CJ. Chem. Rev. 2008;108:1517–1549. doi: 10.1021/cr078203u. [DOI] [PubMed] [Google Scholar]
- 23.Torrado A, Walkup GK, Imperiali B. J. Am. Chem. Soc. 1998;120:609–610. [Google Scholar]
- 24.Pearce DA, Walkup GK, Imperiali B. Bioorg. Med. Chem. Lett. 1998;8:1963–1968. doi: 10.1016/s0960-894x(98)00354-0. [DOI] [PubMed] [Google Scholar]
- 25.Salins LL, Goldsmith ES, Ensor CM, Daunert S. Anal. Bioanal. Chem. 2002;372:174–180. doi: 10.1007/s00216-001-1169-7. [DOI] [PubMed] [Google Scholar]
- 26.Wang BY, Hu YL, Su ZX. React. Funct. Polym. 2008;68:1137–1143. [Google Scholar]
- 27.Wang BY, Liu XY, Hu YL, Su ZX. Polym. Int. 2009;58:703–709. [Google Scholar]
- 28.Fabbrizzi LLM, Pallavicini P, Perotti A, Taglietti A, Sacchi D. Chem. Eur. J. 1996;2:75–82. [Google Scholar]
- 29.Bolletta F, Costa I, Fabbrizzi L, Licchelli M, Montalti M, Pallavicini P, Prodi L, Zaccheroni N. J. Chem. Soc., Dalton Trans. 1999:1381–1385. [Google Scholar]
- 30.Jiang LJ, Luo QH, Wang ZL, Liu DJ, Zhang Z, Hu HW. Polyhedron. 2001;20:2807–2812. [Google Scholar]
- 31.Ke Q, Davidson T, Kluz T, Oller A, Costa M. Toxicol. Appl. Pharmacol. 2007;219:18–23. doi: 10.1016/j.taap.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 32.Thierse HJ, Helm S, Pink M, Weltzien HU. J. Immunol. Methods. 2007;328:14–20. doi: 10.1016/j.jim.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 33.Zhao J, Bertoglio BA, Devinney MJ, Jr, Dineley KE, Kay AR. Anal. Biochem. 2009;384:34–41. doi: 10.1016/j.ab.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cadosch D, Meagher J, Gautschi OP, Filgueira L. J. Neurosci. Methods. 2009;178:182–187. doi: 10.1016/j.jneumeth.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Chen H, Costa M. Exp. Biol. Med. 2006;231:1474–1480. doi: 10.1177/153537020623100905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.