Abstract
The Type I Diabetes Genetics Consortium (T1DGC) has collected thousands of multiplex and simplex families with type I diabetes (T1D) with the goal of identifying genes involved in T1D susceptibility. These families have all been genotyped for the HLA class I and class II loci and a subset of samples has been typed for an major histocompatibility complex (MHC) single-nucleotide polymorphism (SNP) panel. In addition, the T1DGC has genotyped SNPs in candidate genes to evaluate earlier reported T1D associations. Individual SNPs and SNP haplotypes in IL4R, which encodes the α-chain of the IL4 and IL13 receptors, have been associated with T1D in some reports, but not in others. In this study, 38 SNPs in IL4R were genotyped using the Sequenom iPLEX Gold MassARRAY technology in 2042 multiplex families from nine cohorts. Association analyses (transmission-disequilibrium test and parental-disequilibrium test) were performed on individual SNPs and on three-SNP haplotypes. Analyses were also stratified on the high-risk HLA DR3/DR4-DQB1*0302 genotype. A modest T1D association in HBDI families (n = 282) was confirmed in this larger collection of HBDI families (n = 424). The variant alleles at the non-synonymous SNPs (rs1805011 (E400A), rs1805012 (C431R), and rs1801275 (Q576R)), which are in strong linkage disequilibrium, were negatively associated with T1D risk. These SNPs were more associated with T1D among non-DR3/DR4-DQB1*0302 genotypes than DR3/DR4-DQB1*0302 genotypes. This association was stronger, both in terms of odds ratio and P-values, than the initial report of the smaller collection of HBDI families. However, the IL4R SNPs and the three-SNP haplotype containing the variant alleles were not associated with T1D in the total data. Thus, in the overall families, these results do not show evidence for an association of SNPs in IL4R with T1D.
Keywords: polymorphism, genotype, haplotype
Introduction
Type I diabetes (T1D), an autoimmune disease involving destruction of the insulin-producing cells of the pancreas, has a significant genetic component (λs = 15).1 On the basis of linkage and association analyses, the strongest contribution (40–50%) comes from the HLA region.2,3 Although alleles at the HLA-DR and -DQ-encoding loci are the major determinants of genetic risk for T1D,4,5 association studies have revealed that multiple genes within the HLA region contribute to T1D risk.6–12 In addition, many T1D susceptibility regions and genes outside the HLA region have been identified by linkage and by associations analyses.13–15 Of the genes identified by association, some have been detected by hypothesis-free genome-wide approaches,15 whereas others have been identified in candidate gene studies, on the basis of biological plausibility of the gene and/or of the specific polymorphism. Many of the candidate gene studies investigated a limited number of single-nucleotide polymorphisms (SNPs) and samples and, consequently, had very modest statistical power. The results of many of the reported candidate genes studies of T1D association are discordant. To address the issue of limited statistical power in the published candidate gene-association studies, the Type I Diabetes Genetics Consortium (T1DGC), an international collaboration that has collected thousands of multiplex and simplex T1D families,16 has performed genotyping and association analysis of multiple SNPs in 21 T1D candidate genes.
One of the 21 candidate genes studied in the Rapid Response Project is IL4R, which encodes the α-chain of the IL4 and IL13 receptors. Polymorphisms in IL4R have been reported to be associated with allergy and asthma17 and cervical cancer,18 as well as with T1D;19,20 several IL4R non-synonymous SNPs have been associated with differences in signaling.21 A study of the multiplex HBDI families (n = 282) investigated eight SNPs in IL4R and, on the basis transmission–disequilibrium test (TDT) analysis, reported a modest protective effect of the variant allele at several tightly linked non-synonymous SNPs.19 SNP rs1805015 (S503P) was the only individual SNP that exhibited nominal significance in this small study. This significance emerged only after stratification on families in which neither affected sib had the high-risk DR3/DR4-DQB1*0302 genotype. The percent transmission for the 503P variant allele was 44.6% (P = 0.06) for all families. In contrast, transmission of the 503P allele was 47.8% (P = 0.61) in families with a DR3/DR4-DQB1*0302-affected child and 42% (P = 0.03) in families in which neither affected sib had DR3/DR4-DQB1*0302. TDT analysis of an eight-SNP haplotype that included the variant alleles at rs1805011 (E400A), rs2234898 (L414L), rs1805012 (C431R), rs1805015 (S503P), and rs1801275 (Q576R) head a 33% transmission and an odds ratio (OR) of 0.49 (95% confidence interval (CI) = 0.28–0.81).
A small study of Filipino T1D cases and controls also reported an association of IL4R SNPs with T1D.20 The association analysis in this study was performed without stratification on the DR3/DR4-DQB1*0302 genotype because this high-risk genotype is very rare in the Filipino population. Consistent with the earlier results,19 this study reported a modest protective effect of a seven-SNP haplotype that included the same five variant alleles (400A 414L 431R, 503P, and 576R) (OR = 0.4; 95% CI = 0.2–0.8; P = 0.005). Four of the five SNPs are non-synonymous.
Two studies, one in Caucasian multiplex families and one in Filipino cases and controls, suggested that the variant alleles at a series of linked SNPs in IL4R were associated with protection from T1D. Small sample sizes and multiple testing, however, limited statistical power. An earlier report found no association with IL4R (only rs1801275 and Q576R were genotyped).22 More recently, a much larger study consisting of 3475 T1D families, including 1244 Finnish families, genotyped eight IL4R SNPs and found no significant evidence for association with T1D.23 Subsequent to this report, another large study examined the IL4R SNPs and T1D in large family and case/control datasets and earlier published data and found no single-SNP association with T1D.24
In addition to the analysis of IL4R SNPs, the study of Filipino T1D cases and controls also investigated the possible association of SNPs in IL4 and IL13, genes that encode the ligands of the IL4 receptor, and possible gene–gene interaction between SNPs in these genes and SNPs in IL4R. On the basis of the earlier small case–control data, risk for T1D might be determined by specific combinations of genotypes at IL4R, IL4, and IL13. However, the large study found no evidence for association or interaction between SNPs in IL4R, IL4, and IL13.24
The T1DGC carried out SNP genotyping on 38 SNPs in IL4R, 10 SNPs in IL4, and 5 SNPs in IL13 on a collection of 2042 multiplex families from nine different populations. The data were subjected to TDT and parental TDT (PDT) analyses. An association analysis of the T1DGC genotyping data for 19 candidate genes, including the IL4R, IL4, and IL13, found no evidence of association for any of the SNPs in these three genes in the overall dataset.25 Here, we present association analyses on the results from the individual cohorts in the IL4R T1DGC dataset.
Results
A panel of 38 SNPs in IL4R was genotyped using the Sequenom iPLEX platform on the nine cohorts (multiplex family collections from nine geographic locations) listed in Table 1. IL4R SNPs, their positions, and minor allele frequencies are shown in Table 2. Although 38 tagging SNPs in IL4R were attempted for genotyping in this study, two of the SNPS for which T1D associations were reported earlier, rs2234898 (L414L) and rs1805015 (S503P),19 were not genotyped. The association analyses were performed using the TDT and PDT methods that used meiotic transmissions to each affected sib compared with expectation under the hypothesis of ‘no association’. In addition, the analyses were stratified on the high-risk DR3/DR4-DQB1*0302 haplotype.
Table 1.
Cohorts used for the study (proportions of DR3/DR4 vs non-DR3/DR4 are shown, as well as average age of onset in each cohort)
| Region | Cohort | N pedigrees | N pedigrees MHC |
Not DR3/DR4 trios |
DR3/DR4 trios |
Age of onset mean |
s.d. | n T1D patients age of onset |
Num peds with age of onset |
|---|---|---|---|---|---|---|---|---|---|
| 1 | AP | 169 | 118 | 130 | 109 | 10.34 | 7.94 | 366 | 169 |
| 2 | DAN | 130 | 94 | 106 | 85 | 14.99 | 11.23 | 293 | 130 |
| 2 | EUR | 428 | 329 | 429 | 232 | 11.83 | 8.26 | 893 | 428 |
| 4 | HBDI | 424 | 413 | 540 | 342 | 12.28 | 8.67 | 937 | 415 |
| 4 | JOS | 71 | 53 | 56 | 52 | 11.83 | 7.53 | 148 | 71 |
| 4 | NA | 295 | 217 | 263 | 172 | 8.76 | 6.62 | 637 | 295 |
| 5 | BDA | 393 | 0 | 0 | 0 | 12.60 | 9.76 | 853 | 391 |
| 5 | SAR | 74 | 52 | 51 | 52 | 12.75 | 8.71 | 150 | 74 |
| 5 | UK | 108 | 91 | 93 | 90 | 8.22 | 5.29 | 242 | 108 |
| Total | 2092 | 1367 | 1668 | 1134 |
Abbreviations: MHC, major histocompatibility complex; T1D, type I diabetes.
Table 2.
IL4R SNPs tested, their locations, alleles, and functions
| Marker number |
SNP ID | Reference assembly chromosome position |
Major allele | Minor allele |
Minor allele frequency |
SNP functiona |
|---|---|---|---|---|---|---|
| 1 | rs2057768 | 27229596 | C | T | 0.2874 | Not reportedb |
| 2 | rs2107356 | 27230905 | G | A | 0.4125 | 5′ near gene |
| 3 | rs6498012 | 27239475 | C | G | 0.379 | Intron |
| 4 | rs1110470 | 27243928 | C | T | 0.4733 | Intron |
| 5 | rs4787948 | 27248560 | A | G | 0.2933 | Intron |
| 6 | rs2283563 | 27253855 | G | A | 0.3121 | Intron |
| 7 | rs3024537 | 27260320 | G | A | 0.1438 | Intron |
| 8 | rs1805010 | 27263704 | A | G | 0.4466 | I75V |
| 9 | rs3024560 | 27264168 | T | G | 0.3536 | Intron |
| 10 | rs3024571 | 27265428 | C | T | 0.0891 | N167N |
| 11 | rs2301807 | 27265599 | G | T | 0.0499 | Intron |
| 12 | rs3024578 | 27265852 | G | A | 0.0826 | Intron |
| 13 | rs2239347 | 27266522 | A | C | 0.4548 | Intron |
| 14 | rs3116578 | 27267337 | C | T | 0.0201 | Intron |
| 15 | rs3024613 | 27271754 | G | A | 0.4816 | Intron |
| 16 | rs3024614 | 27271846 | A | G | 0.0562 | Intron |
| 17 | rs3024622 | 27272954 | C | G | 0.3448 | Intron |
| 18 | rs4787423 | 27274835 | T | C | 0.1368 | Intron |
| 19 | rs3024668 | 27279450 | C | T | 0.0524 | Intron |
| 20 | rs2234897 | 27281113 | T | C | 0.0244 | F313F |
| 21 | rs1805011 | 27281373 | A | C | 0.1137 | E400A |
| 22 | rs1805012 | 27281465 | T | C | 0.1053 | C431R |
| 23 | rs1801275 | 27281901 | A | G | 0.2114 | Q576R |
| 24 | rs1805016 | 27282428 | A | C | 0.0558 | S752A |
| 25 | rs2074570 | 27282658 | A | G | 0.0405 | 3′ UTR |
| 26 | rs8832 | 27283288 | G | A | 0.4423 | 3′ UTR |
| 27 | rs3024685 | 27284411 | A | G | 0.3923 | Not reportedb |
| 28 | rs12102586 | 27285554 | C | T | 0.0914 | Not reportedb |
| 29 | rs4787956 | 27285750 | T | C | 0.3439 | Not reportedb |
| 30 | rs16976728 | 27289213 | G | A | 0.3831 | Not reportedb |
| 31 | rs4787426 | 27292232 | T | G | 0.1271 | Not reportedb |
| 32 | rs12445135 | 27293007 | C | T | 0.0298 | Not reportedb |
| 33 | rs4787427 | 27293895 | C | G | 0.324 | Not reportedb |
| 34 | rs7191188 | 27296912 | C | T | 0.2386 | Not reportedb |
| 35 | rs6498015 | 27299125 | C | T | 0.1287 | Not reportedb |
| 36 | rs6498016 | 27299289 | G | A | 0.1842 | Not reportedb |
| 37 | rs2382722 | 27300127 | A | G | 0.4611 | Not reportedb |
| 38 | rs9944340 | 27301092 | T | C | 0.2699 | Not reportedb |
| 39 | rs6498017 | 27302359 | G | A | 0.1396 | Not reportedb |
Single-nucleotide polymorphism (SNP) location relative to the IL4R gene and function in the context of changes to the peptide sequence of the IL4R gene, as described in the GeneView ‘Function’ entries of the relevant entry for each rs number in the NCBI’s Entrez dbSNP database.
No data was available in the GeneView entry describing this SNP as resulting in either a synonymous or non-synonymous replacement, or describing a change in an untranslated region as being in a 5′ UTR, 3′ UTR, or intron.
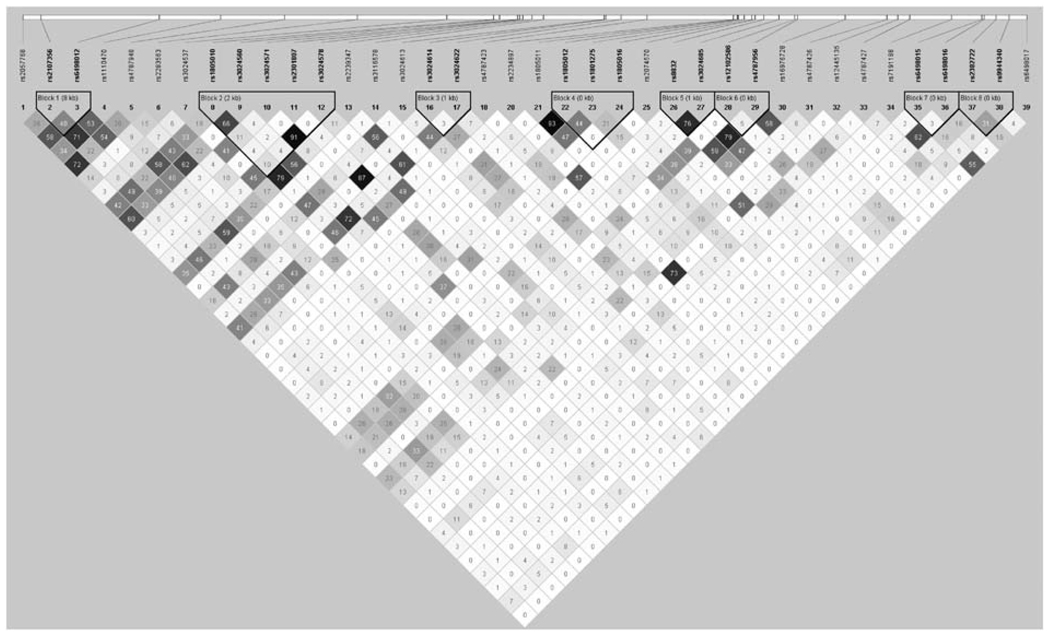
The patterns of LD among 38 IL4R SNPs are shown in Figure 1. None of the individual SNPs showed a statistically significant association with T1D in the overall dataset. All the IL4R SNPs that exhibited a nominally significant (<0.05) association with T1D by either TDT or PDT analysis in one or more of the different cohorts (before or after stratification) are shown in Table 3. No consistent pattern of association with T1D was observed across all cohorts.
Figure 1.
Linkage disequilibrium (R2) values between all SNPs across all cohorts.
Table 3.
RR in IL4R SNPs with significant TDT or PDT P-values
| Cohort | Stratification | Marker number |
SNP | Over- transmitted allele |
IS | TU | % Trans | TDT P-value |
PDT P-value |
RR minor |
CI_minor |
|---|---|---|---|---|---|---|---|---|---|---|---|
| EUR | DR3/DR4-DQB1*0302+ | 1 | rs2057768 | C | Major | 110:82 | 0.5729 | 0.0429 | 0.2782 | 0.75 | (0.56–0.99) |
| EUR | No stratification | 1 | rs2057768 | C | Major | 244:198 | 0.5520 | 0.0285 | 0.0365 | 0.81 | (0.67–0.98) |
| EUR | DR3/DR4-DQB1*0302+ | 2 | rs2107356 | A | Minor | 154:118 | 0.5662 | 0.0288 | 0.0652 | 1.31 | (0.60–0.97) |
| EUR | No stratification | 2 | rs2107356 | A | Minor | 361:300 | 0.5461 | 0.0176 | 0.0186 | 1.20 | (0.71–0.97) |
| NA | DR3/DR4-DQB1*0302− | 2 | rs2107356 | A | Minor | 123:92 | 0.5721 | 0.0342 | 0.0204 | 1.34 | (0.57–0.98) |
| SAR | No stratification | 2 | rs2107356 | G | Major | 66:44 | 0.6000 | 0.0353 | 0.0972 | 0.67 | (0.45–0.98) |
| EUR | DR3/DR4-DQB1*0302+ | 3 | rs6498012 | C | Major | 136:102 | 0.5714 | 0.0273 | 0.0975 | 0.75 | (0.58–0.97) |
| EUR | No stratification | 3 | rs6498012 | C | Major | 304:245 | 0.5537 | 0.0117 | 0.0158 | 0.81 | (0.68–0.95) |
| EUR | No stratification | 4 | rs1110470 | T | Minor | 325:277 | 0.5399 | 0.0503 | 0.0285 | 1.17 | (1.00–1.38) |
| SAR | No stratification | 4 | rs1110470 | C | Major | 71:48 | 0.5966 | 0.0344 | 0.0985 | 0.68 | (0.47–0.97) |
| AP | DR3/DR4-DQB1*0302− | 5 | rs4787948 | A | Major | 54:29 | 0.6506 | 0.0057 | 0.0851 | 0.54 | (0.34–0.84) |
| AP | No stratification | 5 | rs4787948 | A | Major | 106:65 | 0.6199 | 0.0016 | 0.2415 | 0.61 | (0.45–0.83) |
| DAN | DR3/DR4-DQB1*0302− | 5 | rs4787948 | A | Major | 29:23 | 0.5577 | 0.4049 | 0.0411 | 0.79 | (0.46–1.37) |
| NA | DR3/DR4-DQB1*0302− | 6 | rs2283563 | A | Minor | 104:72 | 0.5909 | 0.0156 | 0.0010 | 1.44 | (0.51–0.93) |
| NA | No stratification | 6 | rs2283563 | A | Minor | 159:130 | 0.5502 | 0.0878 | 0.0017 | 1.22 | (0.65–1.03) |
| AP | DR3/DR4-DQB1*0302− | 7 | rs3024537 | G | Major | 35:13 | 0.7292 | 0.0012 | 0.3410 | 0.37 | (0.20–0.70) |
| AP | DR3/DR4-DQB1*0302+ | 7 | rs3024537 | G | Major | 47:16 | 0.7460 | 0.0001 | 0.0006 | 0.34 | (0.19–0.60) |
| AP | No stratification | 7 | rs3024537 | G | Major | 82:29 | 0.7387 | 0.0000 | 0.0009 | 0.35 | (0.23–0.54) |
| HBDI | DR3/DR4-DQB1*0302− | 7 | rs3024537 | G | Major | 134:103 | 0.5654 | 0.0437 | 0.0274 | 0.77 | (0.60–0.99) |
| HBDI | No stratification | 7 | rs3024537 | G | Major | 239:188 | 0.5597 | 0.0135 | 0.0088 | 0.79 | (0.65–0.95) |
| JOS | DR3/DR4-DQB1*0302+ | 7 | rs3024537 | A | Minor | 17:7 | 0.7083 | 0.0382 | 0.0772 | 2.43 | (0.17–0.99) |
| NA | DR3/DR4-DQB1*0302− | 7 | rs3024537 | G | Major | 51:31 | 0.6220 | 0.0264 | 0.1452 | 0.61 | (0.39–0.95) |
| BDA | DR3/DR4-DQB1*0302− | 8 | rs1805010 | G | Minor | 169:132 | 0.5615 | 0.0327 | 0.1089 | 1.28 | (1.02–1.61) |
| JOS | DR3/DR4-DQB1*0302− | 8 | rs1805010 | A | Major | 23:13 | 0.6389 | 0.0934 | 0.0254 | 0.56 | (0.29–1.11) |
| NA | DR3/DR4-DQB1*0302− | 8 | rs1805010 | A | Major | 115:81 | 0.5867 | 0.0149 | 0.0166 | 0.70 | (0.53–0.93) |
| EUR | DR3/DR4-DQB1*0302+ | 9 | rs3024560 | T | Major | 130:100 | 0.5652 | 0.0476 | 0.2078 | 0.77 | (0.59–1.00) |
| NA | DR3/DR4-DQB1*0302− | 10 | rs3024571 | C | Major | 40:20 | 0.6667 | 0.0091 | 0.0327 | 0.50 | (0.29–0.85) |
| NA | No stratification | 10 | rs3024571 | C | Major | 71:48 | 0.5966 | 0.0344 | 0.0416 | 0.68 | (0.47–0.97) |
| NA | DR3/DR4-DQB1*0302− | 12 | rs3024578 | G | Major | 36:18 | 0.6667 | 0.0134 | 0.0631 | 0.50 | (0.28–0.88) |
| JOS | DR3/DR4-DQB1*0302− | 13 | rs2239347 | A | Major | 20:12 | 0.6250 | 0.1551 | 0.0455 | 0.60 | (0.29–1.23) |
| NA | DR3/DR4-DQB1*0302− | 13 | rs2239347 | A | Major | 129:83 | 0.6085 | 0.0015 | 0.0017 | 0.65 | (0.49–0.85) |
| BDA | DR3/DR4-DQB1*0302− | 15 | rs3024613 | G | Major | 163:125 | 0.5660 | 0.0249 | 0.0396 | 0.77 | (0.61–0.97) |
| BDA | No stratification | 15 | rs3024613 | G | Major | 304:253 | 0.5458 | 0.0306 | 0.0614 | 0.83 | (0.70–0.98) |
| DAN | DR3/DR4-DQB1*0302− | 15 | rs3024613 | A | Minor | 43:29 | 0.5972 | 0.0979 | 0.0483 | 1.48 | (0.42–1.08) |
| AP | DR3/DR4-DQB1*0302− | 17 | rs3024622 | G | Minor | 64:43 | 0.5981 | 0.0417 | 0.0189 | 1.49 | (1.01–2.19) |
| EUR | DR3/DR4-DQB1*0302+ | 17 | rs3024622 | C | Major | 131:101 | 0.5647 | 0.0486 | 0.1021 | 0.77 | (0.60–1.00) |
| AP | No stratification | 18 | rs4787423 | T | Major | 64:37 | 0.6337 | 0.0069 | 0.0977 | 0.58 | (0.39–0.87) |
| DAN | DR3/DR4-DQB1*0302− | 20 | rs2234897 | T | Major | 4:0 | 1.0000 | 0.0185 | 0.0578 | 0.11 | (0.01–2.08) |
| AP | DR3/DR4-DQB1*0302− | 21 | rs1805011 | C | Minor | 30:16 | 0.6522 | 0.0375 | 0.0587 | 1.88 | (1.02–3.44) |
| HBDI | DR3/DR4-DQB1*0302− | 21 | rs1805011 | A | Major | 111:76 | 0.5936 | 0.0103 | 0.0153 | 0.68 | (0.51–0.92) |
| HBDI | No stratification | 21 | rs1805011 | A | Major | 184:132 | 0.5823 | 0.0034 | 0.0071 | 0.72 | (0.57–0.90) |
| HBDI | DR3/DR4-DQB1*0302− | 22 | rs1805012 | T | Major | 112:79 | 0.5864 | 0.0167 | 0.0208 | 0.70 | (0.53–0.94) |
| HBDI | No stratification | 22 | rs1805012 | T | Major | 183:133 | 0.5791 | 0.0048 | 0.0094 | 0.72 | (0.58–0.91) |
| AP | DR3/DR4-DQB1*0302− | 23 | rs1801275 | G | Minor | 47:27 | 0.6351 | 0.0193 | 0.0239 | 1.74 | (1.08–2.79) |
| DAN | DR3/DR4-DQB1*0302+ | 23 | rs1801275 | G | Minor | 38:22 | 0.6333 | 0.0377 | 0.0104 | 1.73 | (1.02–2.92) |
| HBDI | DR3/DR4-DQB1*0302− | 23 | rs1801275 | A | Major | 169:133 | 0.5596 | 0.0381 | 0.0309 | 0.79 | (0.63–0.99) |
| HBDI | No stratification | 23 | rs1801275 | A | Major | 287:238 | 0.5467 | 0.0323 | 0.0338 | 0.83 | (0.7–0.98) |
| AP | DR3/DR4-DQB1*0302− | 24 | rs1805016 | A | Major | 8:4 | 0.6667 | 0.2437 | 0.0285 | 0.50 | (0.15–1.67) |
| DAN | DR3/DR4-DQB1*0302+ | 24 | rs1805016 | C | Minor | 15:6 | 0.7143 | 0.0459 | 0.0311 | 2.50 | (0.97–6.44) |
| DAN | No stratification | 24 | rs1805016 | C | Minor | 28:16 | 0.6364 | 0.0687 | 0.0222 | 1.75 | (0.95–3.23) |
| JOS | DR3/DR4-DQB1*0302− | 24 | rs1805016 | C | Minor | 3:0 | 1.0000 | 0.0414 | 0.1573 | 7.00 | (0.36–135.52) |
| DAN | DR3/DR4-DQB1*0302− | 25 | rs2074570 | A | Major | 8:2 | 0.8000 | 0.0496 | 0.1167 | 0.25 | (0.05–1.18) |
| UK | DR3/DR4-DQB1*0302+ | 25 | rs2074570 | A | Major | 14:4 | 0.7778 | 0.0153 | 0.0222 | 0.29 | (0.09–0.87) |
| UK | No stratification | 25 | rs2074570 | A | Major | 22:9 | 0.7097 | 0.0177 | 0.0097 | 0.41 | (0.19–0.88) |
| UK | DR3/DR4-DQB1*0302+ | 27 | rs3024685 | G | Minor | 60:40 | 0.6000 | 0.0448 | 0.1365 | 1.50 | (1.01–2.24) |
| DAN | DR3/DR4-DQB1*0302+ | 28 | rs12102586 | T | Minor | 27:11 | 0.7105 | 0.0084 | 0.0040 | 2.45 | (1.22–4.95) |
| SAR | DR3/DR4-DQB1*0302− | 28 | rs12102586 | T | Minor | 6:1 | 0.8571 | 0.0465 | 0.2059 | 6.00 | (0.72–49.84) |
| UK | No stratification | 28 | rs12102586 | C | Major | 41:23 | 0.6406 | 0.0235 | 0.0378 | 0.56 | (0.34–0.93) |
| EUR | DR3/DR4-DQB1*0302+ | 29 | rs4787956 | T | Major | 117:86 | 0.5764 | 0.0293 | 0.0617 | 0.74 | (0.56–0.97) |
| EUR | No stratification | 29 | rs4787956 | T | Major | 280:237 | 0.5416 | 0.0585 | 0.0465 | 0.85 | (0.71–1.01) |
| UK | DR3/DR4-DQB1*0302+ | 29 | rs4787956 | C | Minor | 57:34 | 0.6264 | 0.0153 | 0.0489 | 1.68 | (0.39–0.91) |
| UK | DR3/DR4-DQB1*0302+ | 30 | rs16976728 | A | Minor | 55:36 | 0.6044 | 0.0456 | 0.1181 | 1.53 | (0.43–1.00) |
| EUR | DR3/DR4-DQB1*0302+ | 31 | rs4787426 | T | Major | 61:40 | 0.6040 | 0.0360 | 0.2243 | 0.66 | (0.44–0.98) |
| EUR | No stratification | 31 | rs4787426 | T | Major | 147:112 | 0.5676 | 0.0294 | 0.0868 | 0.76 | (0.60–0.97) |
| DAN | DR3/DR4-DQB1*0302− | 32 | rs12445135 | C | Major | 6:0 | 1.0000 | 0.0039 | 0.0348 | 0.08 | (0.004–1.37) |
| JOS | No stratification | 32 | rs12445135 | T | Minor | 6:1 | 0.8571 | 0.0465 | 0.0833 | 6.00 | (0.72–49.84) |
| UK | DR3/DR4-DQB1*0302− | 33 | rs4787427 | G | Minor | 32:17 | 0.6531 | 0.0308 | 0.0469 | 1.88 | (1.05–3.39) |
| UK | DR3/DR4-DQB1*0302− | 34 | rs7191188 | C | Major | 38:22 | 0.6333 | 0.0377 | 0.0652 | 0.58 | (0.34–0.98) |
| EUR | DR3/DR4-DQB1*0302− | 35 | rs6498015 | C | Major | 100:65 | 0.6061 | 0.0062 | 0.2798 | 0.65 | (0.48–0.88) |
| EUR | No stratification | 35 | rs6498015 | C | Major | 148:107 | 0.5804 | 0.0101 | 0.2535 | 0.72 | (0.56–0.93) |
| UK | DR3/DR4-DQB1*0302− | 35 | rs6498015 | T | Minor | 27:14 | 0.6585 | 0.0406 | 0.0478 | 1.93 | (1.01–3.68) |
| UK | No stratification | 35 | rs6498015 | T | Minor | 51:32 | 0.6145 | 0.0362 | 0.0637 | 1.59 | (1.02–2.48) |
| EUR | DR3/DR4-DQB1*0302− | 36 | rs6498016 | A | Minor | 127:96 | 0.5695 | 0.0376 | 0.0148 | 1.32 | (0.58–0.99) |
| EUR | No stratification | 36 | rs6498016 | A | Minor | 201:150 | 0.5726 | 0.0064 | 0.0070 | 1.34 | (0.60–0.92) |
| SAR | DR3/DR4-DQB1*0302− | 36 | rs6498016 | G | Major | 17:7 | 0.7083 | 0.0382 | 0.0643 | 0.41 | (0.17–0.99) |
| UK | DR3/DR4-DQB1*0302− | 36 | rs6498016 | G | Major | 34:18 | 0.6538 | 0.0253 | 0.0388 | 0.53 | (0.30–0.93) |
| HBDI | No stratification | 37 | rs2382722 | A | Major | 386:345 | 0.5280 | 0.1293 | 0.0492 | 0.89 | (0.78–1.03) |
| UK | DR3/DR4-DQB1*0302− | 37 | rs2382722 | G | Minor | 38:21 | 0.6441 | 0.0258 | 0.0182 | 1.81 | (1.06–3.08) |
| AP | DR3/DR4-DQB1*0302+ | 38 | rs9944340 | T | Major | 56:29 | 0.6588 | 0.0031 | 0.0172 | 0.52 | (0.33–0.81) |
| EUR | DR3/DR4-DQB1*0302+ | 38 | rs9944340 | T | Major | 109:77 | 0.5860 | 0.0187 | 0.0074 | 0.70 | (0.53–0.94) |
Abbreviations: CI, confidence interval; PDT, parental-disequilibrium test; RR, relative risk; SNP, single-nucleotide polymorphism; TDT, transmission-disequilibrium test.
Significant P-values are shown in bold.
A pattern of association consistent with the initial reports of T1D association was observed, however, in the HBDI cohort (Table 4).19,20 The variant alleles for IL4R SNP rs1805011 (400A), rs1805012 (431R), and rs1801275 (576R) (a series of three linked non-synonymous SNPs) are associated with a reduced risk for T1D in the HBDI families, both in the absence of stratification and in those families with no DR3/DR4-DQB1*0302-affected siblings (Table 4). SNP rs1805011 (E400A) shows the strongest effect with a relative risk (RR) of 0.72 (95% CI = 0.57–0.9, P = 0.0034) in all HBDI families and an RR = 0.68 (0.51–0.92, P = 0.010) in the subset of families with no DR3-DR4-DQB1*0302-affected sibs.
Table 4.
RR in IL4R SNPs with significant TDT or PDT P-values in the HBDI families
| Cohort | Stratification | Marker number |
SNP | Over- transmitted allele |
IS | TU | % Trans | TDT P-value |
PDT P-value |
RR minor |
CI_minor |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HBDI | DR3/DR4-DQB1*0302− | 7 | rs3024537 | G | Major | 134:103 | 0.5654 | 0.0437 | 0.0274 | 0.77 | (0.60–0.99) |
| HBDI | No stratification | 7 | rs3024537 | G | Major | 239:188 | 0.5597 | 0.0135 | 0.0088 | 0.79 | (0.65–0.95) |
| HBDI | DR3/DR4-DQB1*0302− | 21 | rs1805011 | A | Major | 111:76 | 0.5936 | 0.0103 | 0.0153 | 0.68 | (0.51–0.92) |
| HBDI | No stratification | 21 | rs1805011 | A | Major | 184:132 | 0.5823 | 0.0034 | 0.0071 | 0.72 | (0.57–0.90) |
| HBDI | DR3/DR4-DQB1*0302− | 22 | rs1805012 | T | Major | 112:79 | 0.5864 | 0.0167 | 0.0208 | 0.70 | (0.53–0.94) |
| HBDI | No stratification | 22 | rs1805012 | T | Major | 183:133 | 0.5791 | 0.0048 | 0.0094 | 0.72 | (0.58–0.91) |
| HBDI | DR3/DR4-DQB1*0302− | 23 | rs1801275 | A | Major | 169:133 | 0.5596 | 0.0381 | 0.0309 | 0.79 | (0.63–0.99) |
| HBDI | No stratification | 23 | rs1801275 | A | Major | 287:238 | 0.5467 | 0.0323 | 0.0338 | 0.83 | (0.70–0.98) |
| HBDI | No stratification | 37 | rs2382722 | A | Major | 386:345 | 0.5280 | 0.1293 | 0.0492 | 0.89 | (0.78–1.03) |
Abbreviations: CI, confidence interval; PDT, parental-disequilibrium test; RR, relative risk; SNP, single-nucleotide polymorphism; TDT, transmission-disequilibrium test.
Significant P-values are shown in bold.
The results of the analysis of three-marker haplotypes are provided in Table 5. A three-marker haplotype (consisting of the variant allele at rs1805011, rs1805012, and rs1801275) has a transmission of 41%, with an RR of 0.71, an OR of 0.54 (0.39–0.74), and a marginally significant TDT (P = 0.043) for this haplotype. The transmissions to each affected child in these multiplex families were all analyzed, so that the dataset is equivalent to 540 trio families with a DR3/DR4-DQB1*0302 and 342 trio families without a DR3/DR4-DQB1*0302-affected child. This is in contrast to the initial study of 242 HBDI families in which transmission only to the proband was evaluated.19 In addition, SNP rs3024537 also exhibited a modest protective effect in all HBDI families (RR = 0.79; 0.65–0.95) as well as in those without DR3/DR4-DQB1*0302 (RR = 0.77; CI 0.60–0.99) (Table 4). This SNP also was nominally protective in the North American and Asia-Pacific cohorts, but seemed to be positively associated with T1D in the Joslin cohort, although the sample size for this latter cohort was very small.
Table 5.
Significant three-locus haplotypes in the HBDI families
| Marker numbers |
SNP 1 | SNP 2 | SNP 3 | Stratification | TDT P-value |
Haplotype | Number transmitted |
Number not transmitted |
% Transmitted |
RR | OR | OR CI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3-4-5 | rs6498012 | rs1110470 | rs4787948 | DR3/DR4-DQB1*0302+ | 0.039 | C-T-A | 120 | 94 | 56.07 | 1.23 | 1 | 1–1 |
| rs6498012 | rs1110470 | rs4787948 | 0.039 | G-C-G | 92 | 88 | 51.11 | 1.19 | 0.82 | 0.55–1.22 | ||
| rs6498012 | rs1110470 | rs4787948 | 0.039 | C-C-A | 60 | 71 | 45.79 | 1.00 | 0.66 | 0.43–1.02 | ||
| rs6498012 | rs1110470 | rs4787948 | 0.039 | G-C-A | 25 | 42 | 37.32 | 0.65 | 0.47 | 0.27–0.82 | ||
| rs6498012 | rs1110470 | rs4787948 | 0.039 | C-T-G | 0 | 2 | 0.00 | 2.00E-08 | 0 | 0–0 | ||
| 7–8–9 | rs3024537 | rs1805010 | rs3024560 | None | 0.014 | G-G-G | 251 | 232 | 51.96 | 1.73 | 0.95 | 0.74–1.22 |
| rs3024537 | rs1805010 | rs3024560 | 0.014 | G-A-T | 290 | 255 | 53.21 | 1.72 | 1 | 1–1 | ||
| rs3024537 | rs1805010 | rs3024560 | 0.014 | A-G-T | 91 | 98 | 48.15 | 1.57 | 0.82 | 0.59–1.14 | ||
| rs3024537 | rs1805010 | rs3024560 | 0.014 | G-G-T | 18 | 21 | 46.15 | 1.42 | 0.75 | 0.39–1.45 | ||
| rs3024537 | rs1805010 | rs3024560 | 0.014 | A-G-G | 55 | 99 | 35.72 | 1.00 | 0.49 | 0.34–0.71 | ||
| 7-8-9 | rs3024537 | rs1805010 | rs3024560 | DR3/DR4-DQB1*0302− | 0.019 | A-G-T | 52 | 48 | 52.01 | 2.07 | 0.99 | 0.63–1.55 |
| rs3024537 | rs1805010 | rs3024560 | 0.019 | G-G-G | 149 | 130 | 53.40 | 2.01 | 1.05 | 0.76–1.45 | ||
| rs3024537 | rs1805010 | rs3024560 | 0.019 | G-A-T | 161 | 147 | 52.27 | 1.91 | 1 | 1–1 | ||
| rs3024537 | rs1805010 | rs3024560 | 0.019 | G-G-T | 9 | 12 | 42.86 | 1.41 | 0.68 | 0.28–1.67 | ||
| rs3024537 | rs1805010 | rs3024560 | 0.019 | A-G-G | 31 | 65 | 32.29 | 1.00 | 0.44 | 0.27–0.71 | ||
| 19-20-21 | rs3024668 | rs2234897 | rs1805011 | None | 0.021 | C-T-A | 203 | 142 | 58.84 | 1.11 | 1 | 1–1 |
| rs3024668 | rs2234897 | rs1805011 | 0.021 | C-C-A | 24 | 26 | 48.00 | 1.00 | 0.65 | 0.36–1.17 | ||
| rs3024668 | rs2234897 | rs1805011 | 0.021 | T-T-C | 56 | 70 | 44.44 | 0.88 | 0.56 | 0.37–0.84 | ||
| rs3024668 | rs2234897 | rs1805011 | 0.021 | C-T-C | 77 | 117 | 39.69 | 0.74 | 0.46 | 0.32–0.66 | ||
| rs3024668 | rs2234897 | rs1805011 | 0.021 | T-T-A | 4 | 9 | 30.77 | 0.51 | 0.31 | 0.09–1.03 | ||
| 20-21-22 | rs2234897 | rs1805011 | rs1805012 | None | 0.008 | T-A-T | 193 | 135 | 58.84 | 1.16 | 1 | 1–1 |
| rs2234897 | rs1805011 | rs1805012 | 0.008 | C-A-T | 23 | 26 | 46.94 | 1.00 | 0.62 | 0.34–1.13 | ||
| rs2234897 | rs1805011 | rs1805012 | 0.008 | T-C-C | 118 | 168 | 41.26 | 0.83 | 0.49 | 0.36–0.68 | ||
| rs2234897 | rs1805011 | rs1805012 | 0.008 | T-C-T | 2 | 7 | 22.22 | 0.29 | 0.20 | 0.04–0.98 | ||
| 20-21-22 | rs2234897 | rs1805011 | rs1805012 | DR3/DR4-DQB1*0302− | 0.019 | T-A-T | 116 | 75 | 60.73 | 1.30 | 1 | 1–1 |
| rs2234897 | rs1805011 | rs1805012 | 0.019 | C-A-T | 13 | 16 | 44.83 | 1.00 | 0.53 | 0.24–1.15 | ||
| rs2234897 | rs1805011 | rs1805012 | 0.019 | T-C-C | 68 | 101 | 40.24 | 0.88 | 0.44 | 0.29–0.66 | ||
| rs2234897 | rs1805011 | rs1805012 | 0.019 | T-C-T | 2 | 7 | 22.22 | 0.32 | 0.18 | 0.04–0.91 | ||
| 21-22-23 | rs1805011 | rs1805012 | rs1801275 | None | 0.043 | A-T-A | 231 | 181 | 56.07 | 1.00 | 1 | 1–1 |
| rs1805011 | rs1805012 | rs1801275 | 0.043 | A-T-G | 94 | 97 | 49.21 | 0.97 | 0.76 | 0.54–1.07 | ||
| rs1805011 | rs1805012 | rs1801275 | 0.043 | C-C-G | 103 | 149 | 40.87 | 0.71 | 0.54 | 0.39–0.74 | ||
| rs1805011 | rs1805012 | rs1801275 | 0.043 | C-T-G | 2 | 3 | 40.00 | 0.54 | 0.52 | 0.09–3.16 | ||
| 23–24–25 | rs1801275 | rs1805016 | rs2074570 | None | 0.049 | G-C-G | 2 | 0 | 100.00 | 77740 | Inf | 0–infinity |
| rs1801275 | rs1805016 | rs2074570 | 0.049 | A-A-A | 237 | 190 | 55.50 | 1.00 | 1 | 1–1 | ||
| rs1801275 | rs1805016 | rs2074570 | 0.049 | G-C-A | 65 | 65 | 50.00 | 0.98 | 0.80 | 0.54–1.19 | ||
| rs1801275 | rs1805016 | rs2074570 | 0.049 | G-A-G | 34 | 36 | 48.57 | 0.98 | 0.76 | 0.46–1.26 | ||
| rs1801275 | rs1805016 | rs2074570 | 0.049 | G-A-A | 114 | 161 | 41.45 | 0.74 | 0.57 | 0.42–0.77 | ||
| 25-26-27 | rs2074570 | rs8832 | rs3024685 | DR3/DR4-DQB1*0302− | 0.032 | A-G-G | 6 | 4 | 59.99 | 2.89 | 1.19 | 0.33–4.31 |
| rs2074570 | rs8832 | rs3024685 | 0.032 | G-A-G | 2 | 2 | 50.00 | 2.12 | 0.80 | 0.11–5.72 | ||
| rs2074570 | rs8832 | rs3024685 | 0.032 | A-G-A | 191 | 152 | 55.68 | 2.12 | 1 | 1–1 | ||
| rs2074570 | rs8832 | rs3024685 | 0.032 | A-A-G | 152 | 159 | 48.88 | 1.88 | 0.76 | 0.56–1.04 | ||
| rs2074570 | rs8832 | rs3024685 | 0.032 | G-G-A | 21 | 25 | 45.65 | 1.64 | 0.67 | 0.36–1.24 | ||
| rs2074570 | rs8832 | rs3024685 | 0.032 | A-A-A | 27 | 57 | 32.14 | 1.00 | 0.38 | 0.23–0.62 | ||
| 26-27-28 | rs8832 | rs3024685 | rs12102586 | None | 0.027 | G-G-C | 13 | 8 | 61.91 | 2.57 | 1.42 | 0.58–3.47 |
| rs8832 | rs3024685 | rs12102586 | 0.027 | A-G-T | 56 | 43 | 56.57 | 1.99 | 1.14 | 0.74–1.74 | ||
| rs8832 | rs3024685 | rs12102586 | 0.027 | G-A-C | 327 | 285 | 53.43 | 1.62 | 1 | 1–1 | ||
| rs8832 | rs3024685 | rs12102586 | 0.027 | A-G-C | 257 | 273 | 48.49 | 1.48 | 0.82 | 0.65–1.04 | ||
| rs8832 | rs3024685 | rs12102586 | 0.027 | G-A-T | 45 | 55 | 45.00 | 1.25 | 0.71 | 0.47–1.09 | ||
| rs8832 | rs3024685 | rs12102586 | 0.027 | A-A-C | 55 | 89 | 38.19 | 1.00 | 0.54 | 0.37–0.78 | ||
| 26-27-28 | rs8832 | rs3024685 | rs12102586 | DR3/DR4-DQB1*0302− | 0.020 | G-G-C | 6 | 4 | 60.00 | 2.87 | 1.15 | 0.32–4.15 |
| rs8832 | rs3024685 | rs12102586 | 0.020 | A-G-T | 33 | 25 | 56.90 | 2.40 | 1.01 | 0.58–1.78 | ||
| rs8832 | rs3024685 | rs12102586 | 0.020 | G-A-C | 188 | 144 | 56.63 | 2.08 | 1 | 1–1 | ||
| rs8832 | rs3024685 | rs12102586 | 0.020 | A-G-C | 143 | 164 | 46.59 | 1.74 | 0.67 | 0.49–0.91 | ||
| rs8832 | rs3024685 | rs12102586 | 0.020 | G-A-T | 25 | 32 | 43.86 | 1.42 | 0.60 | 0.34–1.05 | ||
| rs8832 | rs3024685 | rs12102586 | 0.020 | A-A-C | 26 | 52 | 33.34 | 1.00 | 0.38 | 0.23–0.64 | ||
| 28-29-30 | rs12102586 | rs4787956 | rs16976728 | DR3/DR4-DQB1*0302+ | 0.041 | C-C-A | 121 | 87 | 58.17 | 1.00 | 1.49 | 1.04–2.14 |
| rs12102586 | rs4787956 | rs16976728 | 0.041 | C-T-G | 135 | 145 | 48.21 | 0.78 | 1 | 1–1 | ||
| rs12102586 | rs4787956 | rs16976728 | 0.041 | T-T-A | 7 | 8 | 46.66 | 0.71 | 0.94 | 0.33–2.66 | ||
| rs12102586 | rs4787956 | rs16976728 | 0.041 | T-T-G | 31 | 37 | 45.59 | 0.68 | 0.90 | 0.53–1.53 | ||
| rs12102586 | rs4787956 | rs16976728 | 0.041 | C-T-A | 37 | 42 | 46.84 | 0.67 | 0.95 | 0.57–1.56 | ||
| rs12102586 | rs4787956 | rs16976728 | 0.041 | C-C-G | 6 | 18 | 25.00 | 0.25 | 0.36 | 0.14–0.93 | ||
| 34-35-36 | rs7191188 | rs6498015 | rs6498016 | DR3/DR4-DQB1*0302+ | 0.004 | C-C-A | 12 | 5 | 70.59 | 1.00 | 2.87 | 0.98–8.37 |
| rs7191188 | rs6498015 | rs6498016 | 0.004 | C-T-G | 63 | 48 | 56.75 | 0.62 | 1.57 | 1–2.45 | ||
| rs7191188 | rs6498015 | rs6498016 | 0.004 | T-C-A | 79 | 70 | 53.02 | 0.53 | 1.35 | 0.9–2.02 | ||
| rs7191188 | rs6498015 | rs6498016 | 0.004 | T-C-G | 29 | 28 | 50.88 | 0.50 | 1.24 | 0.7–2.2 | ||
| rs7191188 | rs6498015 | rs6498016 | 0.004 | C-C-G | 118 | 141 | 45.56 | 0.45 | 1 | 1–1 | ||
| rs7191188 | rs6498015 | rs6498016 | 0.004 | T-T-G | 0 | 9 | 0.00 | 5.16E-10 | 0 | 0–0 | ||
| 36-37-38 | rs6498016 | rs2382722 | rs9944340 | DR3/DR4-DQB1*0302+ | 0.041 | G-G-C | 2 | 0 | 1.00 | 1.33E+09 | Inf | 0–infinity |
| rs6498016 | rs2382722 | rs9944340 | 0.041 | G-A-T | 48 | 37 | 56.48 | 1.04 | 1.59 | 0.96–2.62 | ||
| rs6498016 | rs2382722 | rs9944340 | 0.041 | A-A-T | 84 | 65 | 56.37 | 1.00 | 1.58 | 1.04–2.4 | ||
| rs6498016 | rs2382722 | rs9944340 | 0.041 | G-A-C | 90 | 96 | 48.38 | 0.79 | 1.15 | 0.78–1.69 | ||
| rs6498016 | rs2382722 | rs9944340 | 0.041 | G-G-T | 103 | 126 | 44.98 | 0.74 | 1 | 1–1 | ||
| rs6498016 | rs2382722 | rs9944340 | 0.041 | A-G-T | 0 | 3 | 0.00 | 2.05E-08 | 0 | 0–0 |
Abbreviations: OR, odds ratio; RR, relative risk; SNP, single-nucleotide polymorphism; TDT, transmission-disequilibrium test.
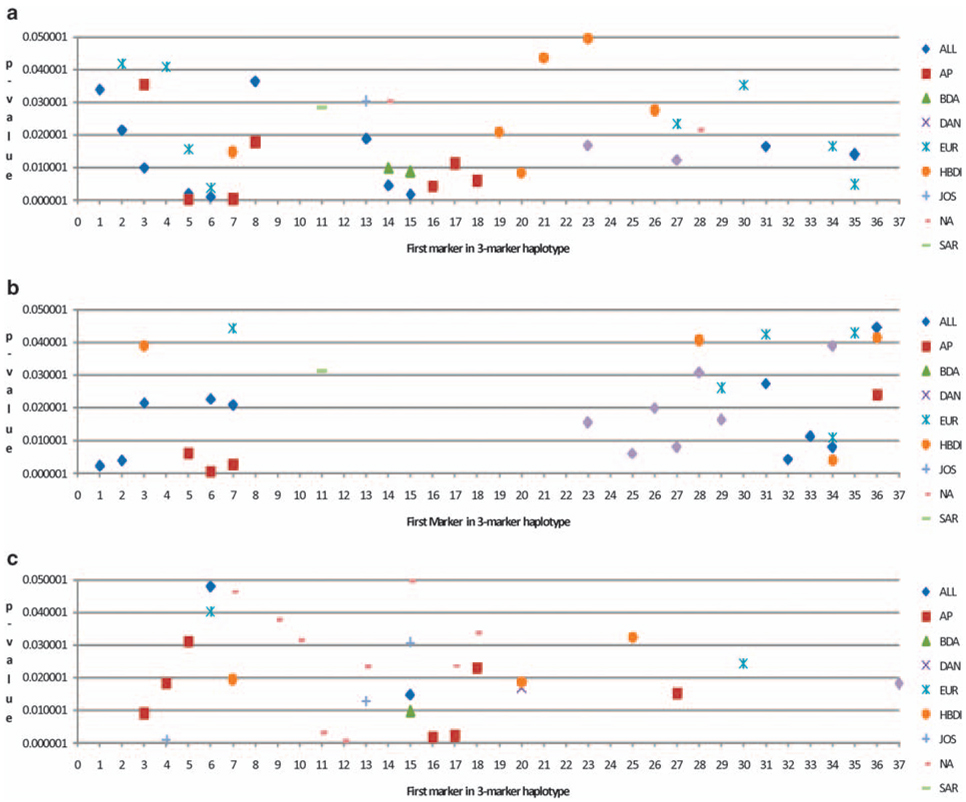
The strength of significance for the association of three-SNP haplotypes with T1D in the individual cohorts as well as the overall dataset is shown in Figure 2a–c. In the HBDI families, all three-SNP haplotypes containing the rs1805011 400A variant are associated with T1D in the unstratified analysis (Figure 2a); the haplotype with 400A as the middle SNP is also associated in the families without DR3/DR4-DQB1*0302 (Figure 2c). In the total data, haplotypes including the first three SNPs (rs2057768, rs2107356, and rs6498012) were associated in the unstratified data and in families with DR3/DR4-DQB1*0302. In these data, haplotypes with rs1805010 (I75V), rs2239347, rs3116578, and rs3024613 (all intronic) and rs4787426 and rs6498015 were associated in the unstratified data. Haplotypes with SNP rs4787426, rs12445135, rs4787427, rs7191188, and rs6498016 were also associated in the families with DR3/DR-DQB1*0302.
Figure 2.
TDT significance in IL4R three-SNP haplotypes: (a) TDT significance in three locus haplotypes, no stratification; (b) TDT significance in three locus haplotypes with DR3/DR4-DQB1*0302; and (c) TDT significance in three locus haplotypes without DR3/DR4-DQB1*0302.
Discussion
Many published association studies of candidate gene polymorphisms examine only a few SNPs per gene and have relatively small sample sizes, resulting in limited statistical power to identify disease-associated alleles. Consequently, reports of association for candidate genes with T1D are often discordant. The extensive set of multiplex families, collected by the T1DGC, has enabled replication analyses of a variety of reported associations (the T1DGC Rapid Response Project). Although the association of three linked SNPs in IL4R observed in this large set of HBDI families was nominally significant and consistent with the initial reports in a subset of HBDI families and a small case/control study among Filipinos, no evidence for association of any individual SNPs was seen in the overall dataset.
The simplest interpretation of the lack of association of any of the individual IL4R SNPs in the overall data is that the earlier reported associations,19,20 as well as the nominally significant observations within the HBDI cohort, are spurious and simply reflect false positive results (type I error). It may be premature, however, to definitively exclude IL4R as a T1D candidate gene. The HBDI families represent the largest individual cohort in the T1DGC dataset and the protective association of the three linked non-synonymous SNPs observed, after stratification, in the families without DR3/DR4-DQB1*0302 replicate earlier findings.19 The set of HBDI families in the T1DGC collection include those studied in the initial report;19 however, two meiotic transmissions, rather than one transmission per family, were analyzed in this larger T1DGC HBDI family collection. As a result, the association of IL4R SNPs with T1D became more significant with this expanded dataset. The question remains why IL4R SNPs seem to be associated in the HBDI cohort, but not in the overall dataset. It is conceivable that heterogeneity among these cohorts, in terms of environmental triggers or interacting genetic effects or linkage disequilibrium patterns, could account for these differences. Some three-SNP haplotypes show nominally significant association in the overall dataset, although, unlike rs180511 and flanking SNPs, there is no prior hypothesis associated with them. In conclusion, this large study of IL4R SNPs provides no clear and consistent evidence of T1D association, but the complex pattern of data suggests that some SNPs should not be definitively excluded as T1D-associated polymorphisms.
Materials and methods
Study population
The T1DGC has created a resource base of well-characterized families from multiple ethnic groups to characterization of T1D susceptibility genes (http://www.t1dgc.org). Genotyping for the Rapid Response project was performed on 2295 families in nine cohorts. The families selected for these analyses consisted mainly of nuclear families with an affected sib pair. This study reports the results for the IL4R gene in 9251 individuals, including 5580 children (4445 (80%) affected, 840 (15%) unaffected, and 295 (5%) unknown status) and 3671 parents (157 (4%) affected, 2576 (70%) unaffected, and 938 (26%) unknown status). The majority of the subjects were Caucasian (81%) with 18% unknown ethnicity and 1% other (Asian, African American, and Pacific Islander).
Genotyping
SNP genotyping was performed by the Broad Institute Center for Genotyping and Analysis (http://www.broad.mit.edu/gen_analysis/genotyping/). Aliquots of the T1DGC source 96-well plates were adjusted to 5–10 ng ml−1 in water in new 96-well plates. The iPLEX Gold chemistry of Sequenom’s MassARRAY platform (San Diego, CA, USA) was used for genotyping of all SNPs as part of the larger set of T1DGC Rapid Response Project. Sequenom’s SpectroDesigner software was used for SNP assay design, and SpectroTyper 4.0 was used to call genotypes automatically, followed by manual review.
Statistical genetic analysis
A TDT26 was performed on each of the markers as implemented in Haploview software (version 4.1).27 The transmission proportions were used to compute ORs and 95% CIs as described.28 The PDT method, as implemented in Haploview 4.1, was also used as a family based test of genetic association.29 This method incorporates parental phenotypes and, specifically, the parental genotype–phenotype correlation terms. The model is based on the between-within-sibship-association model using a liability-threshold-model approach. The incorporation of parental phenotypes can considerably increase power, as compared with the standard TDT and equivalent quantitative tests, whereas providing both significant protection against stratification and a means of evaluating the contribution of stratification to positive results. This methodology enables the extraction of more information from existing family based collections that are currently being genotyped and analyzed by use of standard approaches.
For pedigrees, full DRB1-DQB1 typing was available.4 T1D patients were stratified into those carrying DR3/DR4, defined here as carrying one DRB1*0301-DQB1*0201 haplotype and one DRB1*0401/02/04/05/08-DQB1*0302/04 or DQB1*0201 haplotype. All other participants were categorized as non-DR3/DR4.
Acknowledgements
We are grateful to the family members who contributed samples and to all the participating T1DGC investigators and sites listed at www.t1dgc.org. This research uses resources provided by the Type I Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Human Genome Research Institute (NHGRI), National Institute of Child Health and Human Development (NICHD), and Juvenile Diabetes Research Foundation International (JDRF) and supported by U01 DK062418. Genotyping was performed at the Broad Institute Center for Genotyping and Analysis is supported by grant U54 RR020278 from the National Center for Research Resources.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Risch N. Assessing the role of HLA-linked and unlinked determinants of disease. Am J Hum Genet. 1987;40:1–14. [PMC free article] [PubMed] [Google Scholar]
- 2.Concannon P, Erlich HA, Julier C, Morahan G, Nerup J, Pociot F, et al. Type 1 diabetes: evidence for susceptibility loci from four genome-wide linkage scans in 1,435 multiplex families. Diabetes. 2005;54:2995–3001. doi: 10.2337/diabetes.54.10.2995. [DOI] [PubMed] [Google Scholar]
- 3.Noble JA, Valdes AM, Cook M, Klitz W, Thomson G, Erlich HA. The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. Am J Hum Genet. 1996;59:1134–1148. [PMC free article] [PubMed] [Google Scholar]
- 4.Erlich H, Valdes AM, Noble J, Carlson JA, Varney M, Concannon P, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57:1084–1092. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nejentsev S, Howson JM, Walker NM, Szeszko J, Field SF, Stevens HE, et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450:887–892. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noble JA, Valdes AM, Bugawan TL, Apple RJ, Thomson G, Erlich HA. The HLA class I A locus affects susceptibility to type 1 diabetes. Hum Immunol. 2002;63:657–664. doi: 10.1016/s0198-8859(02)00421-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noble JA, Valdes AM, Thomson G, Erlich HA. The HLA class II locus DPB1 can influence susceptibility to type 1 diabetes. Diabetes. 2000;49:121–125. doi: 10.2337/diabetes.49.1.121. [DOI] [PubMed] [Google Scholar]
- 8.Valdes AM, Erlich HA, Noble JA. Human leukocyte antigen class I B and C loci contribute to type 1 diabetes (T1D) susceptibility and age at T1D onset. Hum Immunol. 2005;66:301–313. doi: 10.1016/j.humimm.2004.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cucca F, Dudbridge F, Loddo M, Mulargia AP, Lampis R, Angius E, et al. The HLA-DPB1-associated component of the IDDM1 and its relationship to the major loci HLA-DQB1, -DQA1, and -DRB1. Diabetes. 2001;50:1200–1205. doi: 10.2337/diabetes.50.5.1200. [DOI] [PubMed] [Google Scholar]
- 10.Erlich HA, Rotter JI, Chang JD, Shaw SJ, Raffel LJ, Klitz W, et al. Association of HLA-DPB1*0301 with IDDM in Mexican-Americans. Diabetes. 1996;45:610–614. doi: 10.2337/diab.45.5.610. [DOI] [PubMed] [Google Scholar]
- 11.Lie BA, Todd JA, Pociot F, Nerup J, Akselsen HE, Joner G, et al. The predisposition to type 1 diabetes linked to the human leukocyte antigen complex includes at least one non-class II gene. Am J Hum Genet. 1999;64:793–800. doi: 10.1086/302283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nejentsev S, Gombos Z, Laine AP, Veijola R, Knip M, Simell O, et al. Non-class II HLA gene associated with type 1 diabetes maps to the 240-kb region near HLA-B. Diabetes. 2000;49:2217–2221. doi: 10.2337/diabetes.49.12.2217. [DOI] [PubMed] [Google Scholar]
- 13.Bottini N, Gloria-Bottini F, Borgiani P, Antonacci E, Lucarelli P, Bottini E. Type 2 diabetes and the genetics of signal transduction: a study of interaction between adenosine deaminase and acid phosphatase locus 1 polymorphisms. Metabolism. 2004;53:995–1001. doi: 10.1016/j.metabol.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 14.Marron MP, Raffel LJ, Garchon HJ, Jacob CO, Serrano-Rios M, Martinez Larrad MT, et al. Insulin-dependent diabetes mellitus (IDDM) is associated with CTLA4 polymorphisms in multiple ethnic groups. Hum Mol Genet. 1997;6:1275–1282. doi: 10.1093/hmg/6.8.1275. [DOI] [PubMed] [Google Scholar]
- 15.Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, Smink LJ, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 16.Rich SS, Concannon P, Erlich H, Julier C, Morahan G, Nerup J, et al. The Type 1 Diabetes Genetics Consortium. Ann N Y Acad Sci. 2006;1079:1–8. doi: 10.1196/annals.1375.001. [DOI] [PubMed] [Google Scholar]
- 17.Chatila TA. Interleukin-4 receptor signaling pathways in asthma pathogenesis. Trends Mol Med. 2004;10:493–499. doi: 10.1016/j.molmed.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Ivansson EL, Gustavsson IM, Magnusson JJ, Steiner LL, Magnusson PK, Erlich HA, et al. Variants of chemokine receptor 2 and interleukin 4 receptor, but not interleukin 10 or Fas ligand, increase risk of cervical cancer. Int J Cancer. 2007;121:2451–2457. doi: 10.1002/ijc.22989. [DOI] [PubMed] [Google Scholar]
- 19.Mirel DB, Valdes AM, Lazzeroni LC, Reynolds RL, Erlich HA, Noble JA. Association of IL4R haplotypes with type 1 diabetes. Diabetes. 2002;51:3336–3341. doi: 10.2337/diabetes.51.11.3336. [DOI] [PubMed] [Google Scholar]
- 20.Bugawan TL, Klitz W, Alejandrino M, Ching J, Panelo A, Solfelix CM, et al. The association of specific HLA class I and II alleles with type 1 diabetes among Filipinos. Tissue Antigens. 2002;59:452–469. doi: 10.1034/j.1399-0039.2002.590602.x. [DOI] [PubMed] [Google Scholar]
- 21.Kruse S, Japha T, Tedner M, Sparholt SH, Forster J, Kuehr J, et al. The polymorphisms S503P and Q576R in the interleukin-4 receptor alpha gene are associated with atopy and influence the signal transduction. Immunology. 1999;96:365–371. doi: 10.1046/j.1365-2567.1999.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reimsnider SK, Eckenrode SE, Marron MP, Muir A, She JX. IL4 and IL4Ralpha genes are not linked or associated with type 1 diabetes. Pediatr Res. 2000;47:246–249. doi: 10.1203/00006450-200002000-00016. [DOI] [PubMed] [Google Scholar]
- 23.Maier LM, Twells RC, Howson JM, Lam AC, Clayton DG, Smyth DJ, et al. Testing the possible negative association of type 1 diabetes and atopic disease by analysis of the interleukin 4 receptor gene. Genes Immun. 2003;4:469–475. doi: 10.1038/sj.gene.6364007. [DOI] [PubMed] [Google Scholar]
- 24.Maier LM, Chapman J, Howson JM, Clayton DG, Pask R, Strachan DP, et al. No evidence of association or interaction between, the IL4RA, IL4, and IL13 genes in type 1 diabetes. Am J Hum Genet. 2005;76:517–521. doi: 10.1086/428387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howson JMM, Walker NM, Smyth DJ, Todd JA the Type I Diabetes Genetics Consortium. Analysis of 19 genes for association with type I diabetes in the Type I Diabetes Genetics Consortium families. Genes Immun. 2009;10 Suppl 1:S74–S84. doi: 10.1038/gene.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 28.Kazeem GR, Farrall M. Integrating case-control and TDT studies. Ann Hum Genet. 2005;69(Pt 3):329–335. doi: 10.1046/j.1529-8817.2005.00156.x. [DOI] [PubMed] [Google Scholar]
- 29.Purcell S, Sham P, Daly MJ. Parental phenotypes in family-based association analysis. Am J Hum Genet. 2005;76:249–259. doi: 10.1086/427886. [DOI] [PMC free article] [PubMed] [Google Scholar]