Abstract
Copper is an essential micronutrient, but toxic in excess. Sulfolobus solfataricus cells have the ability to adapt to fluctuations of copper levels in their external environment. To better understand the molecular mechanism behind the organismal response to copper, the expression of the cluster of genes copRTA, which encodes the copper-responsive transcriptional regulator CopR, the copper-binding protein CopT, and CopA, has been investigated and the whole operon has been shown to be cotranscribed at low levels from the copR promoter under all conditions, whereas increased transcription from the copTA promoter occurs in the presence of excess copper. Furthermore, the expression of the copper-transporting ATPase CopA over a 27-hour interval has been monitored by quantitative real-time RT-PCR and compared to the pattern of cellular copper accumulation, as determined in a parallel analysis by Inductively Coupled Plasma Optical Emission spectrometry (ICP-OES). The results provide the basis for a model of the molecular mechanisms of copper homeostasis in Sulfolobus, which relies on copper efflux and sequestration.
Keywords: Archaea, Sulfolobus solfataricus, copper resistance, regulation of transcription, metal toxicity
1. Introduction
Copper is a transition metal and an important trace element because of the essential role it plays in a range of biological processes. In contrast, the occurrence of copper levels beyond the physiological range causes serious damage to all molecular components. Studies on yeast have led to the proposition that virtually no free copper ions are present in the cell under normal conditions [1]. The response of cells to copper excess/deficiency is accomplished through the interplay of copper-binding proteins, copper-responsive regulators, transporters for the efflux and uptake of copper, and copper-requiring enzymes. Genetic determinants of copper homeostasis have been described for several bacterial species [2; 3; 4; 5; 6; 7]. In particular, the various components of the Enterococcus irae and Escherichia coli copper-homeostasis systems, their regulation, and interactions have been thoroughly studied [2; 8]. Many sequenced archaeal genomes encode homologs of Cu (I) and Cu (II) transporting ATPases [9]. However, investigations of the response in archaea to changes of copper levels are still limited. Structural studies of individual functional domains of the Cu (I)-transporting ATPase CopA in Archeoglobus fulgidus have provided useful insights into its activities and functions [10; 11; 12; 13; 14]. A. fulgidus also possesses a Cu(II)-transporting ATPase, CopB, that has been biochemically characterized [15]. The transcriptional analysis of a cop locus responsible for survival in the presence of copper has been reported in the extreme acidophilic archaeon “Ferroplasma acidarmanus” strain Fer1, where cotranscription of genes encoding the copper binding protein CopZ and the putative copper transporting ATPase CopB was shown to increase in response to Cu (II) [16]. An interesting mechanism for copper detoxification has been described in Sulfolobus metallicus, which is based on sequestration by organic phosphate, possibly followed by active efflux of the metal-phosphate complex [17]. The Sulfolobus solfataricus genome encodes a cop locus, which includes the three open reading frames (ORFs) Sso2651, Sso2652, and Sso10823, encoding the CopA ATPase, a copper-responsive regulator, and a putative copper-binding protein, respectively [18]. Cotranscription of Sso2652 and Sso10823 has been reported to specifically increase in the presence of copper, while the copper-responsive regulator binds sequences surrounding the putative copA promoter in S. solfataricus strain P2 [19]. In this study, the response of Sulfolobus solfataricus to copper has been further investigated in the strain 98/2. The selection of the genetically tractable strain 98/2 [20] will expand the scope of analyses aimed to the elucidation of archaeal interactions with copper. To gain better insights into the Sulfolobus response to copper levels, the transcription of the three genes of the copRTA operon has been examined under different conditions and in a time course experiment, and the changes in the amount of copper associated with the cells have been monitored over time. Based on the data obtained, a preliminary model for the maintainance of copper homeostasis in Sulfolbus is proposed.
2. Materials and methods
2.1. Growth conditions
Sulfolobus solfataricus strains 98/2 or P2 (DSM 1617) were cultured at 80°C in a defined standard medium (SM) as described in [20]; the medium was supplemented with 0.2% sucrose as the carbon and energy source. Batch cultures were inoculated to obtain a density corresponding to an OD540 of about 0.025, with aliquots withdrawn from mid-log phase cultures. Growth was monitored at a wavelength of 540 nm on a Beckman DU-520 spectrophotometer (Beckman Coulter, USA). All the experiments were carried out on the strain 98/2, unless otherwise stated.
2.2. RNA extraction
Total RNA was isolated from S. solfataricus cultures in their exponential phase of growth (OD540=0.3–0.6). Before centrifugation at 3500 g for 15 min, cells were mixed with two volumes of RNA Protect (Qiagen, USA). RNA was extracted from the cell pellets using the RNAeasy Mini kit (Qiagen, USA) and treated with DNase (Ambion, USA), as recommended by the manufacturer. DNA contamination was excluded by PCR using primers targeting the 16S rRNA gene. The quantity and quality of the RNA obtained was evaluated both spectrophotometrically on a NanoDrop ND-1000 spectrophotometer (NanoDrop, USA) and by agarose gel electrophoresis [21].
2.3. Reverse-transcription PCR (RT-PCR) and real-time quantitative RT-PCR (qRT-PCR) analyses
For RT-PCR analysis, total RNA (0.5 μg) was analyzed in 25-μl reactions using the Enhanced Avian HS RT-PCR Kit (Sigma-Aldrich, USA). The amplification products were separated on a 1.2% agarose gel by electrophoresis, and the gel images were acquired using a GelLogic 440 Imaging System (Eastman Kodak, USA). Specific transcripts were quantified by qRT-PCR using the iScript One-Step RT-PCR kit with SYBR Green (Bio-Rad, USA) and the real-time detection system iCycler iQ (Bio-Rad, USA). Reactions, in triplicate, were assembled according to the protocol of the manufacturer, and using 0.2 μg of total RNA in a 25-μl reaction. Primers were designed using the OligoPerfect Designer software (Invitrogen, USA) to have a composition that was suitable for use in both RT-PCR and qRT-PCR (Table 1). Specificity of each pair of primers was confirmed by sequencing. The efficiency of the PCR amplifications was determined from the slopes of the dilution curves of the target RNA. The cycle threshold (Ct) values obtained were used in the “2−ΔΔCt Method” to calculate the relative changes in gene expression [22]. Expression of the target RNAs of interest was normalized to the level of the Sso0067 transcript, detected using the primers 0067-F and 0067-R (Table 1). Sso0067 encodes a ribosomal protein and its expression is not affected by copper exposure, as determined by microarray analysis (unpublished).
Table 1.
Oligonucleotides used in this work.
| ID | Sequence (5'-3') | Target |
|---|---|---|
| q2651-F | GAATAGTTGGGATGCATTGT | |
| q2651-R | ACTACCCCCTTAACGTTTTC | copA |
| q2652-F | TTTATTGCCTTCGCCATTTC | |
| q2652-R | GTTGCGTGCAAATTTTTCCT | copR |
| 2652-F | TGCAATTCTTGCTTGTCTGG | cop operon (paired with q2651-R) |
| q10823-F | ATGATAATCGATCCGGTTTG | |
| q10823-R | ATTCCTTAAATACTCTTCCGGA | copT |
| q0067-F | TACCAATTGTCGCTTTTGCT | |
| q0067-R | CAAATCACCATCTGGAGGAA | reference transcript |
2.4. Analysis of copper content
Cell samples were harvested from exponentially growing S. solfataricus cultures. Cell pellets were washed with 10 mM EDTA to remove the copper adsorbed to the cell wall, then rinsed with SM without added trace metals (Zn, Cu, Mo, V, and Co). Before centrifugation at 3500 g for 15 min, aliquots were removed from each sample for determination of protein concentration using the BCA Protein Assay (Pierce, USA). Cell pellets were resuspended in 50% nitric acid, and digested for 16 hrs at 25°C. The total copper content was analyzed using a Vista Pro radial view Inductively Coupled Plasma Optical Emission Spectrometer (ICP-OES, Varian Inc., USA). Controls included non-inoculated SM medium and untreated cultures.
3. Results and discussion
3.1. Physiological response to copper
To establish the optimal concentration of copper to be used in this study, cells were exposed to CuCl2 at concentrations ranging from 0 mM to 2.5 mM. The MIC, defined as the lowest concentration that completely inhibits cell growth immediately after exposure, was determined to be 1.5 mM (Fig.1a). In response to copper concentrations that were equal or greater than 1.5-mM CuCl2, a lag phase of variable duration was observed. The duration of the lag phase was directly proportional to the metal concentration, and growth resumed thereafter, indicating a slow adaptation to levels of copper that were above the MIC. To rule out the possibility that this effect was due to the appearance of copper-resistant mutants, cells adapted to 2-mM copper-containing medium were subcultured in fresh medium in the absence of the metal. After 4 cycles of cell divisions, the cells were re-inoculated into fresh medium containing 2 mM copper. A lag phase was observed again, indicating that the response to copper concentrations above the MIC was the result of physiological adaptation (unpublished). To limit the manifestation of extensive stress responses that would overshadow the response to copper, only the sublethal concentration of 0.75 mM copper was used for transcript measurements. The level of tolerance to copper observed in S. solfataricus 98/2 was within the range observed for most microorganisms. It was previously observed that the carbon source affected the copper sensitivity of microbes [23], probably because of the binding of copper to the thiol groups of some amino acids and other molecules. Therefore, all experiments were carried out using cells grown on 0.2% sucrose as the sole carbon source. Interestingly, the S. solfataricus strain P2 showed a higher copper sensitivity than the strain 98/2, displaying a growth rate on 0.2 mM copper comparable to that observed for strain 98/2 grown in the presence of 2.5 mM copper (Fig.1b). The two strains were originally isolated in Italy and USA, respectively. It was speculated that their physical separation might have led to different functionality of the copper responsive system. Thus, genetic diversity either at the level of the transcriptional regulator CopR or within the region of the putative copTA promoter was hypothesized to be responsible for the different copper sensitivities of the two strains. However, the sequencing of the segment of the cop operon that includes copR, copT, and their intergenic space in the strain 98/2 (GenBank accession no. EU544670), showed that this region was identical to the corresponding sequence in strain P2, indicating that other factors probably contribute to these differences.
Fig. 1.
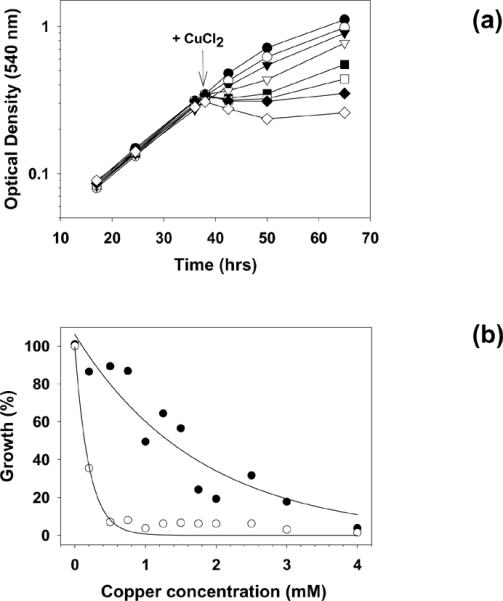
Effect of CuCl2 on growth of S. solfataricus. (a) An exponentially growing culture of strain 98/2 was used as inoculum for 8 subcultures. Copper was added at the time indicated by the arrow, at the following final concentrations: (●) = 0 mM, (○) = 0.75 mM, (▾) = 1 mM, (▿) = 1.25 mM, (∎) = 1.5 mM, (◻) = 1.75 mM, (◆) = 2 mM, (◇) = 2.5 mM. (b) Growth of P2 (○) or 98/2 (●). Copper was added at the time of inoculum and cell growth is expressed as percentage of the untreated control. Best fit curves were obtained by nonlinear regression applied to both sets of data.
3.2. Cotranscription of copRTA
The cop operon consists of three genes oriented in the same direction, represented by ORF's Sso2652, Sso10823, and Sso2651. With the possible exception of the Cu (I) transporter CopA, and the Cu (II) transporter CopB, there are no unique identifiers for functional homologs of copper responsive sequences in prokaryotes. Herein, it is proposed the designation “CopR” for the product of ORF Sso2652, in agreement with the nomenclature applied to metal responsive transcriptional regulators: ArsR, NikR, MerR [24], and CopR [25; 26]. “CopT” is suggested for Sso10823, which corresponds to a stand alone TRASH domain [27]. In an attempt to elucidate the molecular mechanisms behind the regulation of the copRTA operon (Fig. 2a), RT-PCR was used to test whether the genes copR, copT, and copA were cotranscribed. Using the primers pair 2652-F and q2651-R, designed to amplify across the genes copR and copA, a unique band of the expected size was obtained (Fig. 2a, b); sequencing of the amplicon confirmed its specificity and indicated the cotranscription of all the three genes. Cotranscription of the copper-binding protein and the ATPase-coding genes was previously reported to be detected by primer extension [19], but the cotranscription of the copR gene was not addressed in that analysis. By specifically targeting the region encompassing the three genes, the regulator-encoding copR was also shown to be a part of the transcript. This is an interesting finding because it provides additional insight into the regulation of copper homeostasis in Sulfolobus, as discussed later in this section. In the next step, the induction of individual genes was investigated by qRT-PCR. The primer pairs used for the detection of copR and copT are listed in Table 1. In cells treated with copper, the levels of copT mRNA exceeded the amounts of the same transcript in untreated cultures, displaying a greater than 20-fold induction after 120 min, thus behaving as copA (see section 3.3); however, the level of copR expression was unaffected by copper (Fig. 2c). This trend agreed with the data obtained by primer extension [19], and, moreover, this method provided an accurate quantitation of the fold induction. The fact that the genes of the operon copRTA are cotranscribed, that copR is constitutively expressed (Fig. 2c), and that copTA transcription is considerably affected by excess copper, suggests a model where, in the absence of excess copper, copT and copA are transcribed together with copR, constitutively, from the copR promoter, whereas the supplementary transcription of copTA in the presence of copper results from the induction of a second promoter upstream of copTA promoter. The constitutive expression of copTA probably provides a constant and low-level supply of the proteins CopT and CopA that maintain homeostasis, allowing the cell to adjust to small fluctuations in copper levels under normal conditions.
Fig. 2.
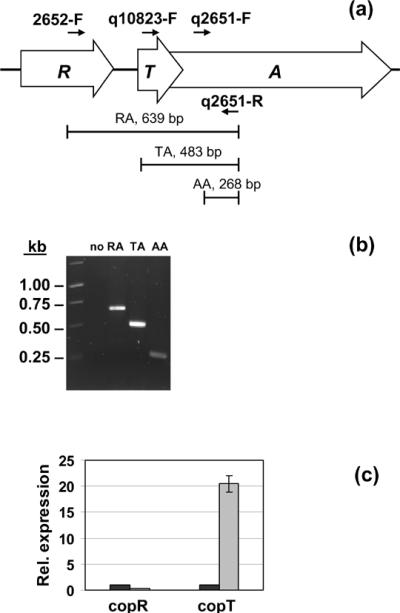
Co-transcription of copR, copT and copA. (a) Diagram of the cop operon and location of the primers. (b) Agarose gel electrophoresis of the amplicons corresponding to the regions encompassing copR/copA (RA), copT/copA (TA) and targeting copA (AA); (no) negative control without reverse transcriptase. (c) Expression of copR and copT in the presence of copper (gray bars) relative to untreated controls (black bars). Results are reported as means ± standard errors.
3.3. Transcription of copA occurs transiently in response to copper
To better understand the regulation of cell response, the expression of copA was monitored in a time-course experiment. Total RNA samples, isolated at different times after treating cell cultures with 0.75 mM CuCl2 were subjected to qRT-PCR analysis using primers targeting the copA transcript (Table 1). Sulfolobus responded to exposure to a sub-lethal copper excess by the transient active transcription of the copA gene. The copA transcript reached a peak 1 hr after treatment, corresponding to approximately 35-fold the uninduced level; thereafter, the transcript level decreased until a steady-state level of 2-3-fold induction is reached in the following 16 hours (Fig. 3). Moreover, in cells cultured for several generations in the presence of 0.75 mM copper, the amount of copA transcript was maintained at 2-3-fold the uninduced level (unpublished), indicating that its rate of expression was maintained constant during long-term exposures, provided the concentration of copper did not change. The above observation can be explained as follows. A high rate of CopA synthesis is necessary to reestablish homeostasis during early copper exposure. Thereafter, the drop in intracellular concentration of copper causes a decrease in copA transcription, which leads to the establishment of a rate of CopA synthesis sufficient to maintain the internal equilibrium at the new cytoplasmic copper concentration. It has been previously reported [19] that treating S. solfataricus P2 with 5 mM copper causes an accumulation of the copA transcript over a 2-hrs period. This might be explained by a higher demand for CopA, extended over time, in the presence of copper concentrations above the MIC. This observation is also consistent with the behavior of S. solfataricus 98/2 for copper levels above 0.75 mM (Fig. 1a). Furthermore, the fact that copA is transiently induced after copper challenge and that its transcription subsequently declines but is maintained at a low basal induced level for the duration of exposure might indicate an accumulation of the CopA transporter. The persistence of a stable CopA protein would reduce the demand for further copA transcription and translation.
Fig. 3.
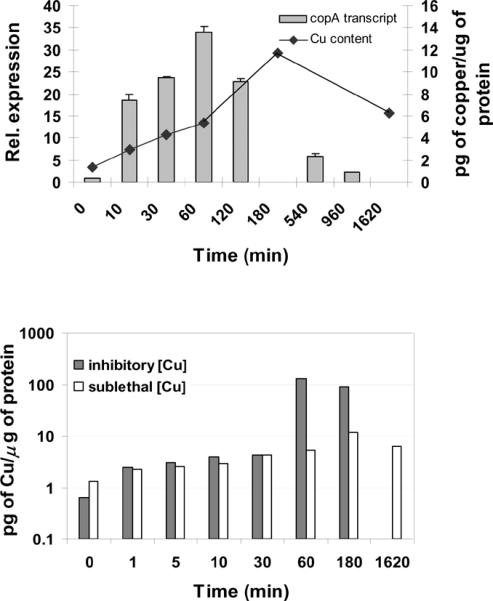
Time course analysis of copper content and copA induction in response to copper. (a) Total RNA was isolated at different times after treatment of the cultures with 0.75 mM copper. Samples were not collected at 180 and 1620 min. Results are expressed as means ± standard errors. Bars represent the normalized ratio between amounts of transcript in copper treated and untreated cultures. For comparison, changes in copper content (◆) after exposure to 0.75 mM copper are superimposed. (b) Copper content in cultures treated with 0.75 mM (white bars) or 1.25 mM copper (dark bars).
3.4. Determination of copper content
To test if the pattern of expression of copA depended on internal fluctuations of copper, the copper associated with the cells was analyzed in a time course experiment by ICP-OES spectrometry, and was observed to slowly increase during the first 3 hrs of monitoring. This might be explained with the sequestration and consequent accumulation of intracellular excess copper by CopT. Alternatively, although the cell pellets were washed with EDTA, these measurements might reflect some residual copper adsorption to the cell surface. After prolonged exposure, the amount of copper returned to the level measured 1 hr after treatment (Fig. 3a), probably due to efflux mediated by CopA. The amount of copper in a 0.75 mM solution is 47.6 × 103 μg/L. However, the copper associated with the cells in a similar medium, 30 min after treatment, is three orders of magnitude lower, being approximately 11.6 μg/L (or 183 nM). These data suggest that active efflux has a major role in maintainance of copper homeostasis, and that sequestration also contributes to it, possibly by fine tuning the system. Cells treated with inhibitory copper concentrations showed a similar pattern of copper accumulation, except for a large increase 60 min after exposure (Fig. 3b). Bioremediation studies have established that dead cells, i.e. algae, have a higher capacity of binding metal ions [28]. Therefore the large increase in copper content may indicate cell death.
4. Conclusions
When S. solfataricus cells are challenged with copper, their rate of growth slows down, and this decrease is proportional to the concentration of copper tested. Although immediately after copper challenge a MIC of 1.5 mM copper has been determined, cells become resistant to environs containing up to 2.5 mM of CuCl2 after prolonged exposure. An increased level of the transcripts of genes directly involved in copper detoxification, namely copA and copT, which are co-transcribed, is observed after treatment with 0.75 mM copper. One hour after copper challenge, the level of copA transcript declines until a steady-state level corresponding to an approximately 2-fold induction is reached. Similarly, after treatment with excess copper, the amount of the metal associated with the cells is maintained constant, after a slight accumulation followed by a decrease. These results are consistent with the existence of a feedback-like mechanism of action, where accumulation of the CopA transporter, derived from the burst of copA transcription, causes a drop in the levels of cytoplasmic copper ions. Sequestration of the metal by CopT contributes to limit the availability of free copper. The lowered copper level results in a reduced effect on CopR and consequently a decline of copA transcription. The different copper sensitivity of the two Sulfolobus strains P2 and 98/2, both having identical copR and copT genes and copTA promoter sequences, supports the hypothesis that additional factors probably have a role in the response of the cells to copper concentrations, and that these factors might be different in the two strains studied. Further research is required to elucidate the steps involved and to identify the additional genetic determinants of the response of the cell to varying levels of copper in the surroundings.
Acknowledgements
This research was supported by Hatch Project NJ01136 and a SEBS Pre-Tenure Award to EB, a GAANN fellowship to AV, and NIH Bridge Award GM58389 to MC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O'Halloran TV. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284:805–8. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- [2].Rensing C, Grass G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol Rev. 2003;27:197–213. doi: 10.1016/S0168-6445(03)00049-4. [DOI] [PubMed] [Google Scholar]
- [3].Chintalapati S, Al Kurdi R, van Scheltinga AC, Kuhlbrandt W. Membrane structure of CtrA3, a copper-transporting P-type-ATPase from Aquifex aeolicus. J Mol Biol. 2008;378:581–95. doi: 10.1016/j.jmb.2008.01.094. [DOI] [PubMed] [Google Scholar]
- [4].Sitthisak S, Knutsson L, Webb JW, Jayaswal RK. Molecular characterization of the copper transport system in Staphylococcus aureus. Microbiology. 2007;153:4274–83. doi: 10.1099/mic.0.2007/009860-0. [DOI] [PubMed] [Google Scholar]
- [5].Magnani D, Barre O, Gerber SD, Solioz M. Characterization of the CopR regulon of Lactococcus lactis IL1403. J Bacteriol. 2008;190:536–45. doi: 10.1128/JB.01481-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Smaldone GT, Helmann JD. CsoR regulates the copper efflux operon copZA in Bacillus subtilis. Microbiology. 2007;153:4123–8. doi: 10.1099/mic.0.2007/011742-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Espariz M, Checa SK, Audero ME, Pontel LB, Soncini FC. Dissecting the Salmonella response to copper. Microbiology. 2007;153:2989–97. doi: 10.1099/mic.0.2007/006536-0. [DOI] [PubMed] [Google Scholar]
- [8].Solioz M, Stoyanov JV. Copper homeostasis in Enterococcus hirae. FEMS Microbiol Rev. 2003;27:183–95. doi: 10.1016/S0168-6445(03)00053-6. [DOI] [PubMed] [Google Scholar]
- [9].Bini E. Archaea-metal interactions: metabolism and strategies of resistance. In: Blum P, editor. Archaea: New models for prokaryotic biology. Caister Academic Press; 2008. pp. 27–43. [Google Scholar]
- [10].Sazinsky MH, Agarwal S, Arguello JM, Rosenzweig AC. Structure of the actuator domain from the Archaeoglobus fulgidus Cu(+)-ATPase. Biochemistry. 2006;45:9949–55. doi: 10.1021/bi0610045. [DOI] [PubMed] [Google Scholar]
- [11].Sazinsky MH, Mandal AK, Arguello JM, Rosenzweig AC. Structure of the ATP binding domain from the Archaeoglobus fulgidus Cu+-ATPase. J Biol Chem. 2006;281:11161–6. doi: 10.1074/jbc.M510708200. [DOI] [PubMed] [Google Scholar]
- [12].Rice WJ, Kovalishin A, Stokes DL. Role of metal-binding domains of the copper pump from Archaeoglobus fulgidus. Biochem Biophys Res Commun. 2006;348:124–31. doi: 10.1016/j.bbrc.2006.07.012. [DOI] [PubMed] [Google Scholar]
- [13].Mandal AK, Yang Y, Kertesz TM, Arguello JM. Identification of the transmembrane metal binding site in Cu+-transporting PIB-type ATPases. J Biol Chem. 2004;279:54802–7. doi: 10.1074/jbc.M410854200. [DOI] [PubMed] [Google Scholar]
- [14].Cattoni DI, Flecha FL, Arguello JM. Thermal stability of CopA, a polytopic membrane protein from the hyperthermophile Archaeoglobus fulgidus. Arch Biochem Biophys. 2008;471:198–206. doi: 10.1016/j.abb.2007.12.013. [DOI] [PubMed] [Google Scholar]
- [15].Mana-Capelli S, Mandal AK, Arguello JM. Archaeoglobus fulgidus CopB is a thermophilic Cu2+-ATPase: functional role of its histidine-rich-N-terminal metal binding domain. J Biol Chem. 2003;278:40534–41. doi: 10.1074/jbc.M306907200. [DOI] [PubMed] [Google Scholar]
- [16].Baker-Austin C, Dopson M, Wexler M, Sawers RG, Bond PL. Molecular insight into extreme copper resistance in the extremophilic archaeon `Ferroplasma acidarmanus' Fer1. Microbiology. 2005;151:2637–46. doi: 10.1099/mic.0.28076-0. [DOI] [PubMed] [Google Scholar]
- [17].Remonsellez F, Orell A, Jerez CA. Copper tolerance of the thermoacidophilic archaeon Sulfolobus metallicus: possible role of polyphosphate metabolism. Microbiology. 2006;152:59–66. doi: 10.1099/mic.0.28241-0. [DOI] [PubMed] [Google Scholar]
- [18].She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, De Moors A, Erauso G, Fletcher C, Gordon PM, Heikamp-de Jong I, Jeffries AC, Kozera CJ, Medina N, Peng X, Thi-Ngoc HP, Redder P, Schenk ME, Theriault C, Tolstrup N, Charlebois RL, Doolittle WF, Duguet M, Gaasterland T, Garrett RA, Ragan MA, Sensen CW, Van der Oost J. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci U S A. 2001;98:7835–40. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ettema TJ, Brinkman AB, Lamers PP, Kornet NG, de Vos WM, van der Oost J. Molecular characterization of a conserved archaeal copper resistance (cop) gene cluster and its copper-responsive regulator in Sulfolobus solfataricus P2. Microbiology. 2006;152:1969–79. doi: 10.1099/mic.0.28724-0. [DOI] [PubMed] [Google Scholar]
- [20].Worthington P, Hoang V, Perez-Pomares F, Blum P. Targeted disruption of the alpha-amylase gene in the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol. 2003;185:482–8. doi: 10.1128/JB.185.2.482-488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sambrook J, Fritsch EF, Maniatis T. Molecular cloning : a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y.: 1989. [Google Scholar]
- [22].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- [23].Miller KW, Risanico SS, Risatti JB. Differential tolerance of Sulfolobus strains to transition metals. FEMS Microbiology Letters. 1992;93:69–73. [Google Scholar]
- [24].Pennella MA, Giedroc DP. Structural determinants of metal selectivity in prokaryotic metal-responsive transcriptional regulators. Biometals. 2005;18:413–28. doi: 10.1007/s10534-005-3716-8. [DOI] [PubMed] [Google Scholar]
- [25].Cantini F, Banci L, Solioz M. The copper-responsive repressor CopR of Lactococcus lactis is a `winged helix' protein. Biochem J. 2009;417:493–9. doi: 10.1042/BJ20081713. [DOI] [PubMed] [Google Scholar]
- [26].Mills SD, Lim CK, Cooksey DA. Purification and characterization of CopR, a transcriptional activator protein that binds to a conserved domain (cop box) in copper-inducible promoters of Pseudomonas syringae. Mol Gen Genet. 1994;244:341–51. doi: 10.1007/BF00286685. [DOI] [PubMed] [Google Scholar]
- [27].Ettema TJ, Huynen MA, de Vos WM, van der Oost J. TRASH: a novel metal-binding domain predicted to be involved in heavy-metal sensing, trafficking and resistance. Trends Biochem Sci. 2003;28:170–3. doi: 10.1016/S0968-0004(03)00037-9. [DOI] [PubMed] [Google Scholar]
- [28].Kadukova J, Vircikova E. Comparison of differences between copper bioaccumulation and biosorption. Environ Int. 2005;31:227–32. doi: 10.1016/j.envint.2004.09.020. [DOI] [PubMed] [Google Scholar]


