Abstract
Purpose
Fifteen independent genetic variants have been implicated in prostate cancer risk by recent genome-wide association studies. However, their association with clinicopathologic features of prostate cancer is uncertain.
Experimental Design
We systematically evaluated these 15 variants in 1,563 prostate cancer patients undergoing radical prostatectomy, taking advantage of the uniform tumor stage and grade information available for each of these cases. Associations of these variants with aggressiveness, pathologic Gleason scores, pathologic stage, age at diagnosis, or serum PSA levels were tested.
Results
After adjusting for multiple testing, none of the SNPs was individually or cumulatively associated with aggressiveness or individual clinicopathologic variables of prostate cancer such as Gleason scores, pathologic stage, or age at diagnosis of prostate cancer. The reported risk allele (G) for SNP rs2735839 in the KLK3 gene at 19q13 was more frequent in less aggressive prostate cancer patients (0.89) than in more aggressive prostate cancer patients (0.86), nominal P = 0.03, or in controls (0.86), nominal P = 0.04. Considering that this allele was also significantly associated with higher serum PSA levels among controls (nominal P = 0.003), the observed trend of higher frequency of this risk allele between less and more aggressive prostate cancer, or between less aggressive and controls may be due to detection bias of PSA screening.
Conclusions
Prostate cancer risk variants recently discovered from genome-wide case-control association studies are not associated with clinicopathologic variables in this population. Case-case studies are urgently needed in order to discover genetic variants that predict tumor aggressiveness.
Keywords: association, prostate cancer, genetics, aggressiveness, Gleason score, stage, KLK3
Introduction
Recently, several single nucleotide polymorphisms (SNPs) have been implicated in prostate cancer risk by genome-wide association studies (1–9). In contrast to the difficulty in replicating association for previously reported prostate cancer risk variants (10), these novel risk variants can be consistently replicated in multiple study populations (11–20). However, results on associations of these risk variants with clinicopathologic features of prostate cancer were inconsistent. While there is a moderate trend of risk alleles observed more often in prostate cancer patients with early age at diagnosis, higher Gleason grade and/or stage or more aggressive form of the disease in some reports (1,3,5,11–14,15), no such associations were found in other studies (2,4,16–17). These inconsistent results may be partially due to the heterogeneous nature in evaluating clinical and pathologic features of prostate cancer.
To better understand associations of these variants with clinicopathologic variables of prostate cancer, we systematical evaluated all prostate cancer risk variants reported to date from recent genome-wide association studies in a population of 1,563 cases who underwent radical prostatectomy for treatment of prostate cancer at Johns Hopkins Hospital. Because each patients’ cancer was extensively and accurately graded and staged using uniform criteria in this case series, it offers an excellent ability to assess associations of genetic variants with clinicopathologic characteristics of prostate cancer.
Materials and Methods
Study subjects
The JHH study population was described in detail elsewhere (12). Briefly, the prostate cancer patients were 1,563 men of European descent (by self report) who underwent radical prostatectomy for treatment of prostate cancer at The Johns Hopkins Hospital from January 1, 1999, through December 31, 2006. Because the prostate gland was removed entirely for each patient, each tumor was accurately and systematically graded using the Gleason scoring system (21) and staged using the TMN (tumor–node–metastasis) system (22), as described previously (23). We defined more aggressive and less aggressive disease based on pathologic tumor stage and Gleason score (Table 1). Tumors with pathologic Gleason Scores of 7 or higher, or pathologic stage T3 or higher, or N+ or M+ (i.e., either high-grade or non-organ-confined disease) were over-sampled from this patient population and defined as more aggressive disease (N=1,017). Tumors with pathologic Gleason score of 6 or lower and pathologic stage T2/N0/M0 (i.e., cancer confined to the prostate) were defined as less aggressive disease (N=546). In a recent study with postoperative follow-up for more than 2,500 patients with pathologically organ-confined, Gleason score 6 or less prostate cancer, biochemical recurrence and local recurrence after radical prostatectomy were extremely rare, and no patients experienced distant metastases or prostate cancer specific mortality (24). Normal seminal vesicle tissue that was obtained and frozen at the time of surgery was used to isolate DNA for genotyping of case patients. As a reference group, men undergoing screening for prostate cancer at The Johns Hopkins Hospital and The Johns Hopkins University Applied Physics Lab (Columbia, MD) during the same time period were asked to participate as control subjects. Serum prostate-specific antigen (PSA) levels, digital rectal examination (DRE) results, and demographic information were available for these subjects. A total of 482 men of European descent (by self report) met our inclusion criteria as control subjects for this study: normal DRE, PSA levels less than 4.0 ng/mL, and age older than 55 years.
Table 1.
Clinicopathologic variables of prostate cancer patients
| More aggressive cases | Less aggressive cases | |
|---|---|---|
| Number of subjects | 1,017 | 528 |
| Mean age (SD), year | 60.1 (6.89) | 56.8 (6.46) |
| Age at diagnosis, No. (%) | ||
| < 65 years | 708 (72) | 463 (88) |
| ≥ 65 years | 274 (28) | 64 (12) |
| Serum PSA levels, No. (%) | ||
| < 4.0 ng/mL | 76 (10) | 191 (36) |
| ≥ 4.0 ng/mL | 660 (90) | 333 (64) |
| Pathologic stage, No. (%) | ||
| T2N0 | 186 (27) | 528 (100) |
| T3 or N+ or M+ | 502 (73) | 0 (0) |
| Pathological Gleasone score, No. (%) | ||
| ≤ 6 | 72 (8) | 528 (100) |
| 7 | 606 (63) | 0 (0) |
| 8 | 148 (15) | 0 (0) |
| ≥ 9 | 131 (14) | 0 (0) |
Selection of SNPs and SNP genotyping
We selected 15 SNPs that were implicated in four genome-wide association studies and were replicated in at least one independent study population in the original papers (1–9). They included one SNP each from 8q24 (three separate sub regions), 17q12, 17q24.3, 3p12, 6q25, 7p15, 7q21, 9q33, 10q11, 10q26, 11q13, 19q13, and Xp11. The SNP at 2q15 was not included because no evidence for prostate cancer association was found in this study population in the initial report (8).
These 15 SNPs were genotyped using a MassARRAY QGE iPLEX system (Sequenom, Inc. San Diego, CA). Polymerase chain reaction (PCR) and extension primers for these SNPs were designed using MassARRAY Assay Design 3.0 software (Sequenom, Inc). The primer information is available at http://www.wfubmc.edu/genomics. PCR and extension reactions were performed according to the manufacturer’s instructions, and extension product sizes were determined by mass spectrometry using the Sequenom iPLEX system. Duplicate test samples and two water samples (PCR negative controls) that were blinded to the technician were included in each 96-well plate. The average genotype call rate for these SNPs was 98.1% and the average concordance rate was 99.8% among 100 duplicated quality control samples. Each of the SNPs in the autosomal chromosomes was in Hardy-Weinberg equilibrium (P ≥ 0.05).
Statistical analyses
Allele frequency differences between two groups of patients (more vs. less aggressive, early vs. late age at diagnosis, or organ-confined vs. non-organ-confined) were tested for each SNP using a chi-square test with 1 degree of freedom. Associations of increasing Gleason scores (≤ 6, 7, 8, or ≥ 9) with risk genotypes of each SNP, assuming a dominant or recessive model, were tested using the Cochran-Armitage test for trend with 1 degree of freedom. Associations of serum PSA levels with each SNP were tested separately in patients (pre-operative PSA) or in controls (at the time of sampling) using an additive model. PSA levels were logarithm transformed before tests.
We tested the cumulative effects of prostate cancer risk SNPs on aggressiveness of prostate cancer by counting the number of prostate cancer associated alleles (based on the best-fitting genetic model from single SNP analysis) of these 15 SNPs in each subject. The OR for more aggressive prostate cancer for men carrying more prostate cancer risk alleles (2nd, 3rd, 4th, and 5th quintiles) was estimated by comparing to men carrying the lowest quintile (1st) using logistic regression analysis. The Cochran-Armitage test for trend was also performed to test increasing risk for aggressive prostate cancer with increasing quintile of number of risk alleles.
All reported P-values were based on a two sided test.
Results
As a primary analysis, we tested associations of these 15 SNPs with aggressiveness of prostate cancer defined using tumor stage and grade as determined for the radical prostatectomy specimen (Table 2). Three SNPs reached a nominal P < 0.05, including rs9364554 at 6q25 (P = 0.04), rs2735839 at 19q13 (P = 0.03), and rs5945619 at Xp11 (P = 0.02). The reported risk alleles were more frequent among more aggressive prostate cancer patients for SNPs at 6q25 and Xp11. In contrast, the reported risk allele (G) for SNP at 19q13 was more frequent in less aggressive prostate cancer patients (0.89) than in more aggressive prostate cancer patients (0.86), P = 0.03, and in controls (0.86), P = 0.04. However, when a Bonferroni correction was applied to account for 15 independent tests, none of the SNPs reached an adjusted P of 0.003 that is required for a 5% study-wise significance level. Results were similar when we removed cases who have Gleason score of 7 and localized disease from the group of more aggressive prostate cancer.
Table 2.
Association of 15 reported prostate cancer risk SNPs with clinicopathologic variables in prostate cancer patiients
| Freq. of risk allele in controls | Aggressiveness |
Stage |
Age at diagnosis (year) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Freq. of risk alleles |
Freq. of risk alleles |
Freq. of risk alleles |
||||||||||||
| Alleles§ |
More | Less | P | T3/N+/M+ | T2/N0/M0 | P | ≥ 65 | <65 | P | |||||
| SNPs | Chr | Position* | Ref | Risk | N=486 | N=1,017 | N=528 | N=714 | N=528 | N= | N= | |||
| rs2660753 | 3p12 | 87,193,364 | C | T | 0.12 | 0.14 | 0.13 | 0.53 | 0.13 | 0.14 | 0.75 | 0.14 | 0.14 | 0.72 |
| rs9364554 | 6q25 | 160,804,075 | C | T | 0.27 | 0.31 | 0.27 | 0.04 | 0.30 | 0.28 | 0.35 | 0.28 | 0.30 | 0.47 |
| rs10486567 | 7p15 | 27,749,803 | T | C | 0.80 | 0.78 | 0.78 | 0.65 | 0.78 | 0.78 | 0.79 | 0.76 | 0.79 | 0.11 |
| rs6465657 | 7q21 | 97,654,263 | T | C | 0.46 | 0.48 | 0.49 | 0.57 | 0.48 | 0.48 | 0.79 | 0.46 | 0.49 | 0.22 |
| rs16901979 | 8q24 (2) | 128,194,098 | C | A | 0.04 | 0.05 | 0.05 | 0.63 | 0.05 | 0.06 | 0.34 | 0.04 | 0.06 | 0.26 |
| rs6983267 | 8q24 (3) | 128,482,487 | T | G | 0.50 | 0.56 | 0.58 | 0.40 | 0.56 | 0.58 | 0.22 | 0.59 | 0.56 | 0.20 |
| rs1447295 | 8q24 (1) | 128,554,220 | C | A | 0.08 | 0.14 | 0.12 | 0.27 | 0.14 | 0.13 | 0.15 | 0.12 | 0.13 | 0.50 |
| rs1571801 | 9q33 | 121,506,927 | G | T | 0.23 | 0.26 | 0.26 | 0.64 | 0.26 | 0.27 | 0.55 | 0.27 | 0.26 | 0.42 |
| rs10993994 | 10q11 | 51,219,502 | C | T | 0.43 | 0.47 | 0.46 | 0.59 | 0.49 | 0.46 | 0.18 | 0.45 | 0.47 | 0.29 |
| rs4962416 | 10q26 | 126,686,862 | A | G | 0.28 | 0.31 | 0.33 | 0.38 | 0.32 | 0.32 | 0.81 | 0.32 | 0.32 | 0.89 |
| rs10896449 | 11q13 | 68,751,243 | A | G | 0.52 | 0.57 | 0.57 | 0.86 | 0.57 | 0.58 | 0.66 | 0.57 | 0.57 | 0.79 |
| rs4430796 | 17q12 | 33,172,153 | C | T | 0.51 | 0.58 | 0.58 | 0.74 | 0.57 | 0.58 | 0.70 | 0.56 | 0.58 | 0.33 |
| rs1859962 | 17q24.3 | 66,620,348 | T | G | 0.48 | 0.51 | 0.51 | 0.95 | 0.53 | 0.51 | 0.28 | 0.50 | 0.52 | 0.45 |
| rs2735839 | 19q13 | 56,056,435 | A | G | 0.86 | 0.86 | 0.89 | 0.03 | 0.86 | 0.89 | 0.02 | 0.87 | 0.88 | 0.56 |
| rs5945619 | Xp11 | 51,074,708 | A | G | 0.33 | 0.42 | 0.36 | 0.02 | 0.43 | 0.37 | 0.05 | 0.40 | 0.40 | 0.98 |
Build35
Risk alleles are based on the previously published studies
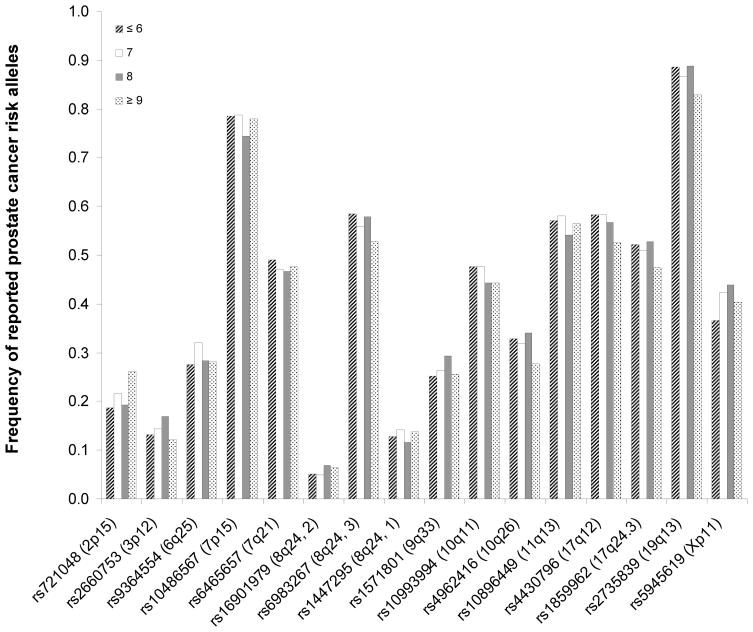
As a secondary analysis, we tested associations of these 15 SNPs with individual clinicopathologic characteristics of prostate cancer (Table 2). For pathological stage, only rs2735839 at 19q13 reached a nominal P < 0.05; the reported risk allele (G) was more frequent in organ-confined than non-organ-confined prostate cancer patients (P = 0.02). For age at diagnosis, none of the 15 SNPs reached a nominal P of 0.05. Similarly, we did not find any significant trend of association between reported risk genotypes (using either dominant or recessive model) and each increasing Gleason score (≤ 6, 7, 8, or ≥ 9). As shown in Figure 1, we observed no consistent increase or decrease in the patterns of the reported risk alleles with increasing Gleason scores.
Figure 1.
Frequencies of reported risk alleles of 15 SNPs in patients with Gleason scores of ≤ 6, 7, 8, or ≥ 9. No consistent increase or decrease in the patterns of the reported risk alleles with increasing Gleason scores was found.
We also tested association of these 15 SNPs with pre-operative serum PSA levels among all patients. Not unexpectedly, no significant association, at a nominal P < 0.05, was found for any of these 15 SNPs (Table 3). To assess the association of these 15 SNPs with serum PSA levels among men not thought to have prostate cancer, we tested the associations in 482 undiagnosed control subjects. Even though this group was preselected to have PSA values below 4ng/ml, one SNP rs2735839 at 19q13 was significantly associated with PSA levels in these men (P = 0.003); men who are homozygous for the reported risk allele (GG) had higher mean PSA levels (1.23 ng/mL) than carriers of GA (0.86 ng/mL) and AA (0.79 ng/mL). This association is biologically plausible because this SNP is in the 3′ of the KLK3 gene that encodes for a PSA precursor.
Table 3.
Association of 15 SNPs with serum PSA levels in cases (preoperative) and controls
| Alleles§ |
Cases |
Controls |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | Chr | Position* | W | R | WW | WR | RR | P | WW | WR | RR | P |
| rs2660753 | 3p12 | 87,193,364 | C | T | 8.80 | 8.66 | 13.09 | 0.35 | 1.09 | 1.17 | 1.78 | 0.55 |
| rs9364554 | 6q25 | 160,804,075 | C | T | 8.70 | 8.62 | 11.15 | 0.18 | 1.14 | 1.08 | 1.19 | 0.64 |
| rs10486567 | 7p15 | 27,749,803 | T | C | 10.04 | 8.78 | 8.84 | 0.77 | 1.15 | 1.07 | 1.14 | 0.72 |
| rs6465657 | 7q21 | 97,654,263 | T | C | 8.37 | 8.88 | 9.30 | 0.61 | 1.13 | 1.08 | 1.17 | 0.66 |
| rs16901979 | 8q24 (2) | 128,194,098 | C | A | 8.86 | 8.49 | 16.01 | 0.40 | 1.10 | 1.16 | 2.77 | 0.20 |
| rs6983267 | 8q24 (3) | 128,482,487 | T | G | 8.75 | 8.78 | 9.03 | 0.95 | 1.04 | 1.12 | 1.16 | 0.57 |
| rs1447295 | 8q24 (1) | 128,554,220 | C | A | 8.77 | 9.43 | 7.29 | 0.52 | 1.10 | 1.14 | 0.55 | 0.27 |
| rs1571801 | 9q33 | 121,506,927 | G | T | 8.88 | 9.09 | 7.68 | 0.52 | 1.06 | 1.13 | 1.42 | 0.10 |
| rs10993994 | 10q11 | 51,219,502 | C | T | 9.80 | 8.56 | 8.47 | 0.23 | 1.09 | 1.07 | 1.29 | 0.12 |
| rs4962416 | 10q26 | 126,686,862 | A | G | 9.29 | 8.31 | 8.95 | 0.35 | 1.11 | 1.10 | 1.10 | 0.99 |
| rs10896449 | 11q13 | 68,751,243 | A | G | 10.15 | 8.49 | 8.73 | 0.19 | 1.05 | 1.17 | 1.07 | 0.34 |
| rs4430796 | 17q12 | 33,172,153 | C | T | 8.75 | 9.23 | 8.40 | 0.50 | 1.11 | 1.06 | 1.20 | 0.34 |
| rs1859962 | 17q24.3 | 66,620,348 | T | G | 9.23 | 8.73 | 8.86 | 0.83 | 1.21 | 1.08 | 1.03 | 0.19 |
| rs2735839 | 19q13 | 56,056,435 | A | G | 9.09 | 8.34 | 8.99 | 0.69 | 0.79 | 0.86 | 1.23 | 2.9E-03 |
| rs5945619 | Xp11 | 51,074,708 | A | G | 8.76 | 8.94 | 0.93 | 1.10 | 1.00 | 1.12 | 0.82 | |
Build35
W (Wild type) and R (Risk) alleles are based on the previously published studies
Finally, we tested the cumulative effect of these 15 risk variants on aggressiveness of prostate cancer. The ORs for aggressive prostate cancer were estimated among patients who have 2nd, 3rd, 4th, or 5th quintile numbers of prostate cancer risk alleles, respectively, compared with patients who have the lowest quintile of prostate cancer risk alleles (Table 4). There was no significant trend of increasing risk for aggressive prostate cancer with increasing quintile of number of risk alleles, P-trend = 0.33.
Table 4.
Association of cumulative effect of risk SNPs with aggressiveness of prostate cancer
| Quintile of # risk alleles | Aggressiveness of prostate cancer |
|||
|---|---|---|---|---|
| OR | 95% CI | P | P-trend | |
| 1st | 1.00 | |||
| 2nd | 1.13 | 0.81–1.57 | 0.49 | |
| 3rd | 1.41 | 1.01–1.97 | 0.04 | |
| 4th | 1.07 | 0.77–1.50 | 0.68 | |
| 5th | 1.15 | 0.81–1.64 | 0.43 | 0.33 |
Discussion
The systematic evaluation of all reported prostate cancer risk variants in this large and uniformly evaluated case series from a single hospital provides compelling evidence that the prostate cancer risk variants discovered to date from genome-wide association studies are not strongly associated with clinicopathologic characteristics of prostate cancer. The null findings from this study were unlikely due to lack of statistical power to detect association. To increase our ability to detect SNP effects on more aggressive disease, we over-sampled men with cancers with high grade or high stage disease as determined after radical prostatectomy. With 1,017 more aggressive and 546 less aggressive prostate cancer patients in our study, we have more than 80% power to detect an allele that is 0.2 (0.1) in the population that confers OR of 1.3 (1.4) for more aggressive prostate cancer. Since all the cases examined in this study were candidates for curative surgery, these associations should be also assessed in large populations of men presenting with clinically non-organ-confined (i.e. metastatic) disease.
The lack of association between these prostate cancer risk variants and aggressiveness of prostate cancer was also found in the data from National Cancer Institute Cancer Genetic Markers of Susceptibility Initiative (CGEMS) (4). The CGEMS study subjects, including 1,172 prostate cancer patients and 1,157 control subjects of European Americans were selected from the Prostate, Lung, Colon and Ovarian (PLCO) Cancer Screening Trial. Among the prostate cancer patients, 737 were classified as having more aggressive prostate cancer, defined as clinical stage T3/T4 or Gleason Score ≥7 based on biopsy specimen, and 624 were classified as having less aggressive disease, defined as clinical Gleason Score < 7 and Stage < III. We downloaded individual genotype data from http://cgems.cancer.gov/data/ and tested association of these 15 SNPs with aggressiveness of prostate cancer. Fourteen of these 15 SNPs were directly genotyped as part of the ~528,000 SNPs in the CGEMS genome-wide association study. One SNP (rs16901979) was imputed from the adjacent genotyped SNPs at 8q24 using the computer program IMPUTE (25). Among these 15 SNPs, only the SNP rs2735839 at 19q13 was significantly associated with aggressiveness of prostate cancer in the CGEMS study, nominal P = 0.003. The association remained significant after adjusting for 15 independent tests using a Bonferroni correction. Interestingly, similar to the findings in our study, the reported risk allele (G) of this SNP was also more frequent in less aggressive prostate cancer (0.89) than in more aggressive prostate cancer (0.85) and unaffected controls (0.84).
The lack of association between prostate cancer risk variants and clinicopathologic characteristics observed in this study has important implications. From a mechanistic perspective, this lack of association with stage or grade may indicate that the initiating events for both more and less aggressive prostate cancers are more similar rather than disparate, and that other factors are more important in determining the aggressive nature of individual prostate cancers. Whether these other factors are genetic, environmental, or stochastic in nature will need to be determined by appropriately designed studies. From a clinical perspective, the unfortunate implication is that these SNPs only provide information about who may be at risk for any prostate cancer, as opposed to a cancer with a predilection for more or less aggressive behavior. Considering that a significant number, yet only ~20% of all prostate cancer patients, die of the disease, it is important to identify markers that predict risk for more aggressive prostate cancer. This subset of patients need to be diagnosed earlier and treated aggressively.
It is interesting to observe that prostate cancer risk allele (G) of rs2735839 in the KLK3 gene was consistently higher among less aggressive (0.89) prostate cancers than more aggressive prostate cancers (0.85–0.86) and unaffected controls (0.84–0.86) in both ours and the CGEMS study. While the basis for this difference is unknown, it may be due to the association of this allele with higher PSA levels per se, rather than with prostate cancer risk. Since the vast majority of patients in this study were diagnosed as a result of elevated PSA levels, any alleles which contribute to such elevated levels will tend to be present in higher frequency among cases, regardless if they are directly associated with prostate cancer risk or not. It is possible that patients with less aggressive prostate cancer are more likely to carry such PSA elevating - alleles than are non-organ confined patients whose PSA levels are elevated as a direct result of more invasive prostate cancer,, rather than the allele associated with higher PSA level. If these speculations are correct, the association of this SNP with aggressiveness of prostate cancer merely represents a detection bias due to PSA screening. Based on the same argument, this PSA related detection bias may also account for the different allele frequencies of this SNP between prostate cancer patients as a group and controls. Obviously, further studies will be necessary to dissect these relationships.
It is important to note that failure to detect association between these prostate cancer risk SNPs and clinicopathologic variables of prostate cancer does not imply lack of such genetic variants in the genome. These 15 prostate cancer risk variants were all discovered using a case-control design. Different study designs, such as case-case studies that compare genetic variants among more or less aggressive prostate cancer cases should be more efficient to identify genetic variants that predict aggressive characteristics of prostate cancer.
Acknowledgments
The authors thank all the study subjects who participated in this study. The authors also thanks for the National Cancer Institute Cancer Genetic Markers of Susceptibility Initiative (CGEMS) for making the data available publicly. The authors take full responsibility for the study design, data collection, analysis and interpretation of the data, the decision to submit the manuscript for publication, and the writing of the manuscript.
The study is partially supported by National Cancer Institute (CA106523, CA106523, and CA95052 to J.X., CA112517 and CA58236 to W.B.I., CA86323 to AWP, and by Department of Defense grant PC051264 to J.X. The support of William T Gerrard, Mario A Duhon, John and Jennifer Chalsty, and David Koch to W.B.I is gratefully acknowledged.
Footnotes
Statement of Clinical Relevance
Since most prostate cancers have a favorable outcome, molecular markers capable of providing prognostic information are urgently needed to assist in the identification of the subset of patients who will benefit from aggressive treatment. In this study we assessed the possible association between recently identified genetic markers of prostate cancer risk and pathologic indicators of cancer aggressiveness. Using pathologic stage and grade to divide men undergoing radical prostatectomy into more or less aggressive cancer groups, we compared the frequencies of alleles at 15 single nucleotide polymorphisms previously confirmed to be associated with risk of prostate cancer. While two SNPs showed different frequencies between the groups with more or less aggressive prostate cancer, no SNPs were significantly different when the results were adjusted for multiple tests. While these markers appear to be informative for the identification of men who may be at elevated risk for a prostate cancer diagnosis, they do not appear to be helpful in identifying men at risk for developing a more aggressive prostate cancer. Additional studies directly comparing cases with more or less aggressive disease in the discovery phase should be pursued to identify genetic variants that predict aggressive characteristics of prostate cancer.
References
- 1.Amundadottir LT, Sulem P, Gudmundsson J, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–8. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 2.Freedman ML, Haiman CA, Patterson N, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103:14068–73. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 4.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 5.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–83. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 6.Duggan D, Zheng SL, Knowlton M, et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99:1836–44. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 7.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 8.Gudmundsson J, Sulem P, Rafnar T, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–3. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eeles RA, Kote-Jarai Z, Giles GG, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–21. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 10.Schaid DJ. The complex genetic epidemiology of prostate cancer. Hum Mol Genet. 2004;(Spec No 1):R103–21. doi: 10.1093/hmg/ddh072. [DOI] [PubMed] [Google Scholar]
- 11.Haiman CA, Patterson N, Freedman ML, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–44. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng SL, Sun J, Cheng Y, et al. Additive effects of two unlinked loci at 8q24 are associated with a considerable fraction of prostate cancer among European Americans. JNCI. 2007;99:1525–33. doi: 10.1093/jnci/djm169. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, McDonnell SK, Slusser JP, et al. Two common chromosome 8q24 variants are associated with increased risk for prostate cancer. Cancer Res. 2007;67:2944–50. doi: 10.1158/0008-5472.CAN-06-3186. [DOI] [PubMed] [Google Scholar]
- 14.Suuriniemi M, Agalliu I, Schaid DJ, et al. Confirmation of a positive association between prostate cancer risk and a locus at chromosome 8q24. Cancer Epidemiol Biomarkers Prev. 2007;16:809–14. doi: 10.1158/1055-9965.EPI-06-1049. [DOI] [PubMed] [Google Scholar]
- 15.Robbins C, Torres JB, Hooker S, et al. Confirmation study of prostate cancer risk variants at 8q24 in African Americans identifies a novel risk locus. Genome Res. 2007;17:1717–22. doi: 10.1101/gr.6782707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Severi G, Hayes VM, Padilla EJ, et al. The common variant rs1447295 on chromosome 8q24 and prostate cancer risk: results from an Australian population-based case-control study. Cancer Epidemiol Biomarkers Prev. 2007;16:610–2. doi: 10.1158/1055-9965.EPI-06-0872. [DOI] [PubMed] [Google Scholar]
- 17.Schumacher FR, Feigelson HS, Cox DG, et al. A common 8q24 variant in prostate and breast cancer from a large nested case-control study. Cancer Res. 2007;67:2951–6. doi: 10.1158/0008-5472.CAN-06-3591. [DOI] [PubMed] [Google Scholar]
- 18.Cheng I, Plummer SJ, Jorgenson E, et al. 8q24 and prostate cancer: association with advanced disease and meta-analysis. Eur J Hum Genet. 2008 Jan 30; doi: 10.1038/sj.ejhg.5201959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun J, Purcell L, Gao Z, et al. Association between sequence variants at 17q12 and 17q24.3 and prostate cancer risk in European and African Americans. Prostate. 2008 doi: 10.1002/pros.20754. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–9. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 21.Epstein JI, Allsbrook WC, Jr, Amin MB, Egevad LL. ISUP Grading Committee. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am J Surg Pathol. 2005;29:1228–42. doi: 10.1097/01.pas.0000173646.99337.b1. [DOI] [PubMed] [Google Scholar]
- 22.Hoedemaeker RF, Vis AN, Van Der Kwast TH. Staging prostate cancer. Microsc Res Tech. 2000;51:423–9. doi: 10.1002/1097-0029(20001201)51:5<423::AID-JEMT4>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 23.Epstein JI. Pathologic assessment of the surgical specimen. Urol Clin North Am. 2001 Aug;28(3):567–94. doi: 10.1016/s0094-0143(05)70164-6. [DOI] [PubMed] [Google Scholar]
- 24.Hernandez DJ, Nielsen ME, Han M, Trock BJ, Partin AW, Walsh PC, Epstein JI. Natural History of Pathologically Organ-Confined (pT2), Gleason Score 6 or Less, Prostate Cancer After Radical Prostatectomy. Urology. 2008 Feb 25; doi: 10.1016/j.urology.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]