Abstract
Pontine gray neurons of the brain stem are a major source of mossy fiber (MF) afferents to granule cells of the cerebellum. Achieving this connectivity involves an early regionalization of pontine gray neuron cell bodies within the brainstem pontine nuclei, as well as establishing the proper ratio of crossed versus uncrossed MF projections to contralateral versus ipsilateral cerebellar territories. Here, we report expression of the transcription factor Zic1 in newly postmitotic pontine gray neurons and present functional experiments in embryonic and postnatal mice that implicate Zic1 levels as a key determinant of pontine neuron cell body position within the pons and axon laterality. Reducing Zic1 levels embryonically via in utero electroporation of short hairpin RNA interference (shRNAi) vectors shifted the postnatal distribution of pontine neurons from caudolateral to rostromedial territories; by contrast, increasing Zic1 levels resulted in the reciprocal shift, with electroporated cells redistributing caudolaterally. Associated with the latter was a change in axon laterality, with a greater proportion of marked projections now targeting the ipsilateral instead of contralateral cerebellum. Zic1 levels in pontine gray neurons, therefore, play an important role in the development of pontocerebellar circuitry.
Keywords: Pontocerebellar Circuitry, Precerebellar Mossy Fiber Afferents, Cerebellum, Mouse, Transcription Factor
Critical for coordination of skilled movements is a collection of brainstem structures known as precerebellar MF nuclei, aptly named because they provide major cerebellar afferents – MF axons – to granule cells of the cerebellar cortex and output neurons of the cerebellar nuclei (Altman and Bayer, 1997; Brodal and Bjaalie, 1992; Brodal and Bjaalie, 1997; Sotelo, 2004; Taber Pierce, 1966). Through their afferents, brainstem MF neurons transfer information to the cerebellum that is received from neurons residing in the cerebral cortex or spinal cord, the resultant circuitry having powerful means to control cerebellar activity (Altman and Bayer, 1997; Schmahmann and Pandya, 1997; Schwarz and Thier, 1999; Turner, 1941). MFs connect with the cerebellum via a stereotyped ratio of crossed (contralaterally-directed) and uncrossed (ipsilaterally-directed) axon pathways with respect to the brainstem midline (reviewed in Cicirata et al., 2005). While the nature of this ratio suggests an importance for cerebellar computations and motor control (Eccles, 1967; Ito, 1984), little is known about the molecular underpinnings regulating MF laterality. Similarly, little is known about the molecular determinants of MF cell body position within the brain stem and whether soma location and axon laterality are linked. Here we present findings implicating the transcription factor Zic1 in these processes. We focused on assembly and connectivity of the pontine gray nucleus (PGN) because, in mammals, it contributes the greatest number of MFs to the cerebellum, as compared to other classes of MF nuclei, and it appears to do so with a fixed laterality ratio (Brodal and Bjaalie, 1997; Cicirata et al., 2005; Mihailoff et al., 1981; Palay and Chan-Palay, 1974; Rosina and Provini, 1981; Serapide et al., 2001).
The PGN resides in the ventral brain stem (the pons) and is comprised of bilaterally symmetrical lobes, one on each side of the midline (Altman and Bayer, 1987b; Taber Pierce, 1966) (Fig. 1A). Within each lobe are subclasses of MF neurons, categorized by cell body location – rostromedial or caudolateral (Azizi et al., 1981; Brodal and Bjaalie, 1992; Brodal and Bjaalie, 1997; Mihailoff et al., 1981; Rosina and Provini, 1981; Taber Pierce, 1966) – or on laterality of axon projection to the contralateral versus ipsilateral cerebellum (Cicirata et al., 2005). The majority (~80%) of MF axons extend medially from their respective PGN lobe, cross the ventral brainstem midline, and enter the contralateral cerebellum. A smaller cohort of MFs (~20%) project in the opposite direction to neurons situated in the ipsilaterally-located cerebellum, thus their axons do not cross the midline (Cicirata et al., 2005) (Fig. 1A, “contra” and “ipsi” labels are relative to the green lobe of the schematized PGN nucleus, the other lobe of the PGN is indicated in gray). It seems likely that at least some factors involved in determining PGN MF laterality and cell body position exert their effects during intermediate (postmitotic) stages of MF neuron development, after commitment to a generic MF neuron fate has been made but before subtype identity manifests. Specification of generic MF neuron identity occurs early during development, in cycling progenitor cells of the dorsally situated germinal zone called the hindbrain rhombic lip (hRL), and requires the cell-autonomous activity of the basic helix-loop-helix (bHLH) transcription factor Math1 (mAtoh1) (Ben-Arie et al., 1997; Ben-Arie et al., 2000; Farago et al., 2006; Landsberg et al., 2005; Wang et al., 2005). Upon expression of Math1, committed MF cells emerge from the hRL as postmitotic cells that go on to circumnavigate the hindbrain ventrolaterally to take up residence flanking the ventral midline of the pons, from where they extend axons either contralaterally or ipsilaterally (schematized in Figs. 1A-1D). We refer to these early postmitotic cells as MF “precursor” cells because they are not fully differentiated, yet are no longer cycling as progenitor cells. Molecular programs implemented during this intermediate stage of MF neuron development (reviewed in Millen et al., 1999; Sotelo, 2004; Wingate, 2001) may determine, at least in part, where MF neurons settle in the brain stem and the pathway of projection to the cerebellum.
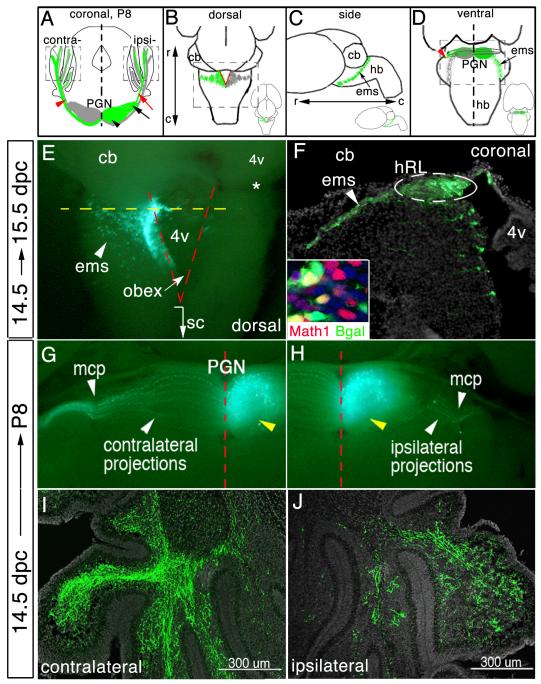
Fig.1.
Visualizing development of a single lobe of the bilateral PGN. (A) Schematic of oblique coronal brain section. The PGN is comprised of rostromedial (black arrowhead) and caudolateral (black arrow) neuronal subpopulations on each side of the brain stem. Green marking represents eGFP/nßgal-transfected neurons and their axons. Most PGN neurons extend axons across brainstem midline toward the contralateral cerebellum (red arrowhead), whereas few project away from the midline toward the ipsilateral cerebellum (red arrow). Dashed boxes, “contra” and “ipsi”, identify area of cerebellar images in (I) and (J), respectively. (B-D) Whole brain schematics. Insets, low-power images of brains. PGN precursor cells emerge from the hRL (red lines) (B) and travel ventrally forming the extramural migratory stream (ems) (C). Postmitotic hRL precursor cells aggregate adjacent to the ventral brainstem midline to form the PGN, extending axons across or away from the midline (D). Dashed boxes in (B) and (D) identify area of images in (E), and (G,H), respectively. (E) Whole brain at 15.5 dpc that was electroporated at 14.5 dpc. eGFP-transfected mitotic cells within the hRL and postmitotic cells within the ems. Dashed yellow line in (E) identifies the ideal axial level of the coronal section in (F). Dashed red lines in (E) demarcate location of the left and right side of the hRL. eGFP+ cells form the ems (arrowhead) (E and F). (F) Immunodetection of eGFP on a high magnification coronal section shows transfected progenitor cells within one side of the hRL and their postmitotic progeny cells within the ems. Inset in (F), co-immunodetection for nßgal and Math1. (G,H) Ventral view of P8 brains that were electroporated at 14.5 dpc. eGFP fluorescence shows labeled cells in the ipsilateral lobe of the PGN (yellow arrowhead) and their contralateral (G) and ipsilateral (H) MF projections. Immunodetection of eGFP shows eGFP+ MF axons throughout the granule cell layer of the contralateral (I) and ipsilateral (J) cerebellum. DAPI staining to highlight cell bodies is shown in gray. r, rostral; c, caudal; cb, cerebellum; sc, spinal cord; 4v, fourth ventricle; mcp, medial cerebellar peduncle.
Molecular programs involved in MF neuron migration, nucleus assembly and MF laterality are just beginning to be characterized. Chemotropic cues secreted by the floor plate, like Netrin-1 (Kennedy et al., 1994), draw young MF neurons ventral (Alcantara et al., 2000; Yee et al., 1999) through activation of the transmembrane receptor deleted in colorectal cancer (Dcc) (Fazeli et al., 1997; Keino-Masu et al., 1996). Rig1/Robo3, a Slit receptor on young MFs, downregulates as leading processes cross the ventral midline (Marillat et al., 2004). Rig1/Robo3-deficiency leads to precocious settling of MF cell bodies laterally, well before reaching the ventral midline; it also results in the extension of processes only ipsilaterally, although it is unclear if these are axons and whether they reach cerebellar targets (Marillat et al., 2004). Rig1/Robo3 is thought to regulate Robo2, another Slit guidance receptor expressed by MF neurons (Marillat et al., 2004). The transcription factor Hoxa2 has recently been shown to regulate directly transcription of Robo2, and in so doing appears critical for maintaining the caudal-to-rostral migration of MF neurons as they traverse from the caudal hRL to the pons (Geisen et al., 2008). Also implicated in postmitotic stages of precerebellar afferent system development is the differential expression of cadherin-type cell adhesion molecules (Taniguchi et al., 2006) as well as intracellular Rho GTPases capable of inducing cytoskeletal changes critical for different migrations (reviewed in Bloch-Gallego et al., 2005). Toward identifying other factors involved in precerebellar afferent system organization as well as molecules involved in MF laterality choice – a process for which little is known – we have analyzed MF lineages for expressed transcription factors and then used gain- and loss-of-function experiments to ascertain how such factors influence MF neuron subtype identity, in particular, site of residence within and axon directionality from the PGN. Here, we present our findings regarding the zinc finger transcription factor Zic1.
Zic family transcription factors, relatives of the Drosophila Opa transcription factor (encoded by the pair rule gene odd-paired) (Aruga et al., 1996; Aruga et al., 1994), are fitting candidates for consideration as regulators of MF axon laterality with respect to the brainstem midline. This is because Zic2 activity in mice has been implicated in directing retinal axon pathway choice at the optic chiasm (García-Frigola et al., 2008; Herrera et al., 2003; Lee et al., 2008) and Zic3 in establishing left-right body asymmetry (Purandare et al., 2002). While the role of Zic genes in early developmental processes such as cerebellar patterning and neuroectoderm differentiation has been well characterized (Aruga, 2004; Aruga et al., 1998; Aruga et al., 2002; Grinberg et al., 2004), investigations into Zic protein functions in postmitotic cells are in their infancy.
Here, we present investigations by which we revealed Zic1 expression in postmitotic MF precursor cells, and then determined the consequences of altered Zic1 levels on MF nucleus formation and axon laterality. We used an electroporation method (Kawauchi et al., 2006; Okada et al., 2007; Saito and Nakatsuji, 2001; Tabata and Nakajima, 2001; Takahashi et al., 2002) to perturb pontine gray MF precursor cells molecularly and acutely by delivering either Zic1-specific RNA interference or overexpression vectors along with reporter plasmids to the pontine gray MF lineage during in utero development. In this way, we bypassed the earlier requirement for Zic1 in neurogenesis and patterning and studied, for the first time, Zic1 functions in later phases of brainstem development. Because the electroporation strategy allowed for directing manipulations to just one side of the embryo, our analyses were also able to include visualization of axon laterality arising from MF neurons situated within a single lobe of the PGN on one side of the brain stem – an axon projection feature otherwise obscured by the passing fibers from homologous PGN neurons of the contralateral side. We found that reducing Zic1 levels shifted the postnatal distribution of the transfected neurons within the PGN, from caudolateral to rostromedial territories. By contrast, increasing Zic1 levels shifted the distribution reciprocally, from rostromedial to caudolateral territories. Associated with this caudolateral shift was a shift in relative proportion of the marked axons projecting to the ipsilateral cerebellum, thus axon laterality was also altered. Zic1, therefore, influences two essential processes during MF system development: where pontine neurons settle in the ventral brain stem and to which cerebellar side they project. Thus, we reveal a new role for Zic1 as a cell autonomous regulator of key aspects of nucleus formation and axon pathway choice in the ventral brain stem.
EXPERIMENTAL PROCEDURES
Animals
Timed pregnant CD1 mice were obtained from Charles River Laboratories (Cambridge, MA). Vaginal plug detection was considered 0.5dpc.
Expression vectors
Complete nlacZ and Zic1 coding regions (Open Biosystems, IMAGE #6813617) were cloned into pCAGGS (Niwa et al., 1991) yielding pCAG-nlacZ and pCAG-Zic1, respectively. Zic1 shRNAi constructs were amplified from oligonucleotides and cloned into pBS/U6 (Sui et al., 2002). Oligonucleotides: Mouse Zic1 (coding region 1407-1429), 5′-TGCTGTTGACAGTGAGCGAGGGCTGGAGCCTTCTTCCGCTTAGTGAAGCCACAGATGTAAGCGGAAGAAGGCTCCAGCCCC TGCCTACTGCCTCGGA-3′; 1866-1888, 5′-TGCTGTTGACAGTGAGCGCACCTTTGCAAGATGTGCGATATAGTGAAGCCACAGATGTATATCGCACATCTTGCAAAGGTATGCCTACTGCCTCGGATGCTGTTGACAGTGAGCGCACCTTTGCAAGATGGCGATATAGTGAAGCCACAGATGTATATCGCACATCTTGCAAAGGTATGCCTACTGCCTCGGA-3′. pCAG-eGFP and an shRNAi vector directed against GAPDH was provided by Dr. C. Cepko (Matsuda and Cepko, 2004).
In utero electroporation
In utero electroporation was performed as previously described (Okada et al., 2007; Saito and Nakasuji, 2001; Tabata and Nakajima, 2001) with modifications. Timed pregnant mice were anesthetized (avertin 0.25 mg/g) at 14.5 dpc and uterine horns exposed. For DNA injection, 3 μl of a stoichiometric plasmid mixture (~ 3 pmol of each construct/μl) plus 0.01% Fast Green in phosphate buffered saline (PBS) was injected into the hindbrain ventricle using pulled glass capillaries. Forceps-type electrodes (7 mm, Harvard Apparatus) were positioned around the caudal hindbrain, anode leftward, and five square electric pulses (50 V, 50 ms) delivered (M830, BTX). Uterine horns were repositioned in the abdomen; excisions closed. Brains were removed at defined developmental stages, and those with unilaterally marked hRL were identified by epifluorescence microscopy and imaged. Embryonic brains were immersion fixed (4% paraformaldehyde (PFA) in PBS pH 7.4) for 4-6 hrs at 4°C; postnatal brains, overnight.
Immunodetection
Fixed whole brains were embedded in Optimal Cutting Temperature (OCT) compound, cryosectioned (20 μm), and mounted on slides (Superfrost Plus). They were fixed in 2% PFA/PBS, washed, blocked with 5% donkey serum/0.1% Triton and incubated in primary antibodies diluted in blocking buffer overnight at 4°C. Slides were washed and incubated in secondary antibodies in blocking buffer for 2 hrs. The following primary antibodies were used: goat polyclonal anti-ßgal antibody (Cappell, 1:2000), chicken polyclonal anti-GFP antibody (1:2000, Abcam), rabbit polyclonal anti-Zic1 (1:2000, Rockland Immunochemicals). The rabbit polyclonal anti-Math1 antibody was provided by Dr. Jane Johnson.
Statistical analyses
Statistical analyses were conducted to determine if the number of neurons shifted from rostromedial to caudolateral PGN territory was significant. Serial sections were collected and immunodetection for nβgal performed. For Zic1 knockdown analysis – On each photographed section, a vertical line and a horizontal line, forming four quadrants, was superimposed at the center of the ipsilateral lobe of the PGN. For both control and experimental cohorts of animals, the total number of βgal+ cells in PGN territory was determined. We then counted the number of cells in the upper left quadrant of the PGN lobe (adjacent to the midline). The number of βgal+ cells in the upper left quadrant was divided by the total number of βgal+ cells in the entire PGN lobe, providing a percentage of total labeled cells. A significantly greater percentage of labeled cells were detected in the upper left quadrant of Zic1 knockdown animals than in control animals (p-value<0.0001). For Zic1 overexpression analysis – On each photographed section, a vertical line was superimposed at the ventral midline and a second vertical line placed in the territory approximately halfway between the midline and the outermost lateral limit of the ipsilateral lobe of the PGN. The distance between the two lines was kept constant. For both control and experimental cohorts of animals, the total number of βgal+ cells in PGN territory was determined. We also counted the total number of cells in the lateral PGN (to the left of the most lateral line). The number of βgal+ cells in the lateral PGN was divided by the total number of βgal+ cells in the entire PGN lobe, providing a percentage of total labeled cells. A significantly greater percentage of labeled cells were detected in the lateral PGN of Zic1 overexpression animals than in control animals (p-value<0.0001). Values from both the knockdown and overexpression studies were subjected to a student’s two tailed paired t-test.
RESULTS
Unilateral gene transfer by electroporation provides a novel means for visualizing PGN formation and axon laterality
The PGN is a bilaterally symmetrical structure with its constituent neurons residing on either side of the brainstem ventral midline (Fig. 1A). Neurons populating the PGN arise from the caudal-most regions of the hRL (Farago et al., 2006; Landsberg et al., 2005; Wang et al., 2005). The hRL is also a bilaterally symmetrical structure, but one that flanks the dorsal midline. MF precursors emerge laterally from both the left and right hRL (Fig. 1B) and stream ventrolaterally (clockwise or counterclockwise, depending on side of origin) along the hindbrain perimeter (Fig. 1C). Pontine gray MF cells ultimately settle adjacent to the ventral midline of the pons (Fig. 1D) and extend axons to target the contralateral or ipsilateral cerebellar hemisphere (Cicirata et al., 2005)(Fig. 1A).
To investigate axon pathway choice and neuron position within the pontine MF system, we employed in utero electroporation of MF progenitor cells in the hRL (Kawauchi et al., 2006; Okada et al., 2007) to manipulate and label only those MF neurons residing on one side of the pons. A stoichiometric mixture of two reporter plasmids, pCAG-eGFP and pCAG-nlacZ, was co-delivered to just those mossy fiber neurons arising from the hRL on the left side of the 14.5 dpc embryo; one encoded a reporter capable of marking cell soma position, being a nuclear-localized ß-galactosidase (nßgal), and the other encoded enhanced green fluorescent protein (eGFP) capable of illuminating axons as well as soma. Electroporations were performed on embryos at ~14.5 dpc because this enabled marking and manipulating the maximal number of pontine gray MF neurons; this is because at 14.5 dpc the greatest number of PGN MF neurons are “born” from the hRL (Taber Pierce, 1966) – that is, undergo terminal mitoses and emerge from the hRL as postmitotic, partially differentiated precursor cells (referred to as precursor cells so as to distinguish them from both cycling progenitor cells as well as fully differentiated postmitotic MF neurons). Furthermore, those MF precursors emerging from the hRL at 14.5 dpc go on to populate the ipsilateral PGN lobe only; by contrast, prior to 14.5 dpc, a small subset of PGN MF precursors appears capable of crossing the ventral midline to settle in the homologous PGN lobe contralateral to the side of origin (Kawauchi et al. 2006), resulting in marked cells in PGN lobes on both sides of the ventral midline, confounding our ability to analyze axon laterality from a single PGN lobe. Because other brainstem mossy fiber neurons, for example those residing in the medulla, emerge from the hRL well before 14.5 dpc, they were not marked using this electroporation protocol; again, thereby offering selective manipulation of just pontine MF neurons.
Plasmid delivery to the caudal portion of the left hRL was verified by collecting whole brains one day (15.5 dpc) after electroporation (n=10 animals). eGFP- and ßgal-marked cells were indeed observed in the caudal hRL, as determined by anatomical position and tissue morphology (Figs. 1E and 1F); moreover labeled cells co-expressed Math1 confirming them as MF progenitor cells (Fig. 1F, inset). In the hRL, progeny cells undergo asymmetric cell division, with one marked daughter cell remaining in, and thus marking, the hRL, while the other migrates ventrolaterally. Marked cells were also observed clustering immediately lateral to the hRL (Figs. 1E and 1F); this reflects a sojourning of postmitotic progeny cells prior to their entering what is referred to as the extramural migratory stream (ems) (Altman and Bayer, 1987b; Engelkamp et al., 1999; Taber Pierce, 1966; Taber Pierce, 1967). Targeting of DNAs to the hRL (versus more ventral territories) was aided because hRL cells are the principal remaining population of proliferating cells in the hindbrain at 14.5 dpc and proliferating cells seem most susceptible to gene transfer by electroporation. Targeting the caudal portion of the hRL was achieved by electrode placement relative to the obex, which can be visualized readily following fast green dye injection into the fourth ventricle (see Methods). Through analyses of brainstem tissue approximately twelve days after electroporation (at postnatal day (P) 8, n=16 animals), we found that the labeled cells come to reside in the PGN and send projections through the medial cerebellar peduncle (mcp) to targets in the contralateral and ipsilateral cerebellum (Fig. 1G and 1H). To more carefully examine laterality of the marked MF axons, we collected coronal sections of the cerebellum and found that from one PGN lobe arose axon projections to both sides of the cerebellum, with the majority projecting contralaterally (Fig. 1I versus 1J, n=16 animals) as expected (Cicirata et al, 2005). Thus, utilizing both the spatial and temporal control provided by the electroporation approach, we were able to selectively introduce DNA plasmids into cells of the pontine gray MF lineage on one side of the brain stem. This permitted a way to readily distinguish between contra- and ipsilateral-projecting populations of PGN MF axons and study aspects of PGN nucleus formation and axon directionality in vivo.
Mossy fiber precursors express Zic1
As part of a larger screen for genes expressed in the mid-to-late gestation hindbrain, we established new aspects of the Zic1 profile. At 14.5 dpc, Zic1 protein was detected in newly emerging PGN precursor cells of the ems immediately as they leave the hRL and begin their traverse dorsal-to-ventral; however, Zic1 protein was not readily detected in the hRL at this stage in development (Fig. 2B). Additional immunohistochemical analyses revealed that this Zic1 expression profile, detectable in newly born postmitotic PGN precursors but not hRL progenitors, was similar at 12.5 dpc, the time at which PGN neurons first begin to leave the hRL (Taber Pierce, 1966) (data not shown). Zic1 expression in MF neurons of the PGN persisted through at least P8 (Figs. 2D and 2E). We observed no differences in the ability to immunodetect Zic1 in PGN MF neurons located rostromedially versus caudolaterally (Fig. 2E). Furthermore, we could immunodetect Zic1 in MF neurons populating other precerebellar nuclei such as the lateral reticular and external cuneate nuclei. By contrast, Zic1 was not detected in neurons of the inferior olivary nucleus, the climbing fiber precerebellar afferent system (data not shown). These data indicate that at these later stages, Zic1 is preferentially expressed in postmitotic precursor cells of the precerebellar MF class and persists even in differentiated MF neurons. Thus, Zic1 is poised to influence events critical to postmitotic MF neurons, possibly migration, nucleogenesis, and/or axon pathfinding.
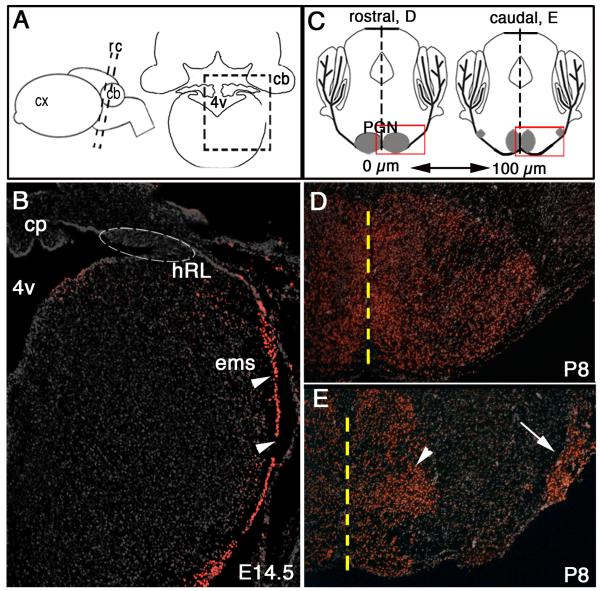
Fig. 2.
Zic1 expression in the postmitotic precerebellar MF lineage. (A) Schematic side view of a mouse brain (left) and a coronal section (right). Boxed area in (A) identifies region in (B). Black dashed lines demarcate idealized axial levels in (D) and (F). (B) Coronal section through the caudal hindbrain at 14.5 dpc showing Zic1+ cells throughout the ems (arrowheads); Zic1+ cells were not detected within progenitor cells of the hRL (located within dashed oval). (C) Schematics of oblique coronal P8 brain sections ~200 μm apart at the axial levels of the rostral (left) and caudal (right) pons, respectively. Boxed area in rostral section identifies region in (D), and boxed area in caudal section identifies region in (E). Black dashed lines demarcate the midline; the PGN is gray. In rostral PGN regions, neurons aggregate medially. Toward more caudal regions, the neurons scatter along the ventral surface and settle into medial (arrowhead) and lateral (arrow) subpopulations. (D,E) Immunodetection for Zic1 on coronal sections through the rostral (D) and caudal (G) PGN at P8 shows Zic1+ cells in medial and lateral populations. Yellow dashed line demarcates the midline. cx, cortex; cp, choroid plexus.
Modulating levels of Zic1 acutely in utero alters the spatial distribution of pontine mossy fiber neurons
RNAi-based knockdown of Zic1 in cells of the pontine mossy fiber lineage
To investigate the role of Zic1 during PGN development, we used individually two different DNA-based shRNAi vectors delivered by electroporation in utero. Each shRNAi vector was directed against a different sequence within the coding portion of the Zic1 transcript. More specifically, the shRNAi construct, either directed against Zic1 (pU6-Zic1sh1866 or pU6-Zic1sh1401) or GAPDH (pU6-shGADPH (Matsuda and Cepko, 2004)) as a control, was electroporated in utero along with the two reporter vectors (pCAG-nlacZ and pCAG-eGFP). Embryos at ~14.5 dpc were employed, with plasmid transfer directed to the hRL situating on the left side of the embryo; brainstem tissue was harvested approximately twelve days later (P8) for analysis. Use of the control plasmid, pU6-shGADPH, addressed the possibility that the RNAi mechanism, in general, might trigger a reaction that would nonspecifically alter MF neuron and PGN development. To account for variances in transfection efficiency among animals, we limited our analyses to only those animals with comparable numbers of transfected cells (control group, average=809 ± 24.7 nβgal+ cells, n=8 animals; knockdown group, average=804 ± 16.9, n=14 animals).
Animals having received shRNAi plasmid directed against Zic1 showed an altered distribution of MF neurons within the PGN as compared to controls. As reference, control animals – recipients of the reporter plasmids along with the shRNAi plasmid directed against GAPDH – showed marked (nßgaland GFP-labeled) cells distributed throughout the rostrocaudal extent of the PGN (Figs. 3A-3C). By contrast, recipients of the reporter plasmids along with Zic1 shRNAi plasmids showed marked cells more localized, having situated predominantly in rostromedial portions of the PGN (Figs. 3E and 3F). Moreover, within this rostromedial subterritory, labeled cells clustered more dorsally. There was near, if not complete, loss of marked cells in the caudolateral PGN (Fig. 3G). Quantitative analyses revealed that ~20% (160.5 ± 29.0) of control-transfected cells located within the rostromedial PGN come to reside in the upper left quadrant (Fig. 3A, asterisk), while ~67% (538.5 ± 39.4) of Zic1 shRNAi-transfected cells in the rostromedial PGN settle in this region (Fig. 3E, asterisk).
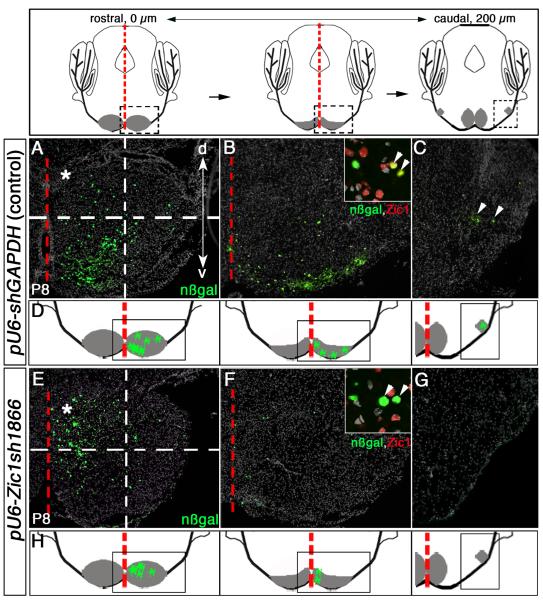
Fig. 3.
Knockdown of Zic1 in the pontine gray MF lineage leads to a shift in neuron distribution to rostromedial brainstem territories. Above, schematics of coronal sections along the rostrocaudal (RC) axis of the PGN at P8. Distance between rostral and caudal sections is ~200 μm; PGN is in gray. Red dashed line demarcates brainstem midline. Boxed area in the rostral schematic identifies neurons in rostromedial PGN in (A) and (E). Boxed area in the middle schematic identifies the PGN territory in (B) and (F). Boxed area in the caudal schematic identifies the caudolateral PGN territory in (C) and (G). (A-C) Immunodetection for nßgal on coronal sections of P8 control animals that were co-transfected with control (pU6-shGAPDH) and reporter vectors at 14.5 dpc. nßgal+ cells along the ventral periphery of the rostromedial (A and B) and caudolateral (C) PGN territories. Inset in (B), co-immunodetection of nßgal (green) and Zic1 (red) shows transfected cells in control animals express relatively high levels of Zic1 protein (arrowheads). (D) Summary schematics of findings from control conditions, showing cells transfected with control and reporter vectors as green markings distributed throughout the rostomedial and caudolateral PGN populations; the PGN is gray. (E-G) Coronal sections through the PGN of P8 animals co-transfected at 14.5 dpc with pU6-Zic1sh1866 and reporter vectors. Immunodetection for nßgal reveals transfected cells scattered primarily in the rostromedial portions of the PGN (E) and (F), with reduction of transfected cells settling in the caudolateral PGN (G). Inset in (F), co-immunodetection of nßgal (green) and Zic1 (red) shows that most transfected cells receiving the Zic1 shRNAi construct expressed little to no detectable levels of Zic1 protein, indicating that this shRNAi construct efficiently suppressed Zic1 protein levels (arrowheads). Dashed-line grids in (A) and (E) clarify distribution differences of transfected cells between control animals and Zic1 knockdown animals (upper left quadrants, asterisks). A significantly greater proportion of nßgal+ cells transfected with the Zic1 shRNAi construct settled in the rostromedial PGN as compared to control animals (student’s t-test, p<0.0001). (H) Summary schematics of findings from Zic1 shRNAi knockdown study, showing transfected cells distributed predominantly in the rostromdial PGN. d, dorsal; v, ventral.
To assess qualitatively the reduction in Zic1 protein following pU6-Zic1sh1866 or pU6-Zic1sh1401 shRNAi vector delivery, we used an antibody to Zic1 to detect the relative level of Zic1 expression in nßgal+ cells (postmitotic descendants of the originally transfected progenitors) in control animals (Fig. 3B, inset) and Zic1 knockdown animals (Fig. 3F, inset). We analyzed Zic1 expression levels in 400 transfected cells for each animal and found that acute suppression of Zic1 levels was achieved in postmitotic MF precursor cells and mature neurons: in control animals ~90% (365.8 ± 10.5) of the GFP+,nßgal+ cells expressed Zic1 at readily detectable levels, whereas after Zic1 knockdown, codetection of Zic1 and nßgal was observed in only ~10% (40 ± 5.2) of the transfected cells. Associated with this reduction in Zic1 expression was a shift in neuron distribution to the rostromedial portion of the pontine brain stem. Thus, our finding revealed that maintaining proper levels of Zic1 expression in postmitotic MF neurons was required for their normal distribution within the PGN.
Zic1 overexpression in cells of the pontine mossy fiber lineage
We next conducted a gain-of-function analysis to investigate whether increased Zic1 levels in MF neurons would drive the reciprocal phenotype, that is neuron redistribution to caudolateral PGN territory. A Zic1 expression vector (pCAG-Zic1) or control vector (pCAGGS) together with reporter vectors (pCAG-lacZ and pCAG-eGFP) were injected into the hindbrain fourth ventricle of 14.5 dpc embryos and electroporated. Brains were harvested at P8, and we analyzed those with comparable numbers of nßgal-transfected cells in the ventral brain stem (for those that received pCAG-Zic1, 800.7 ± 8.6 total nßgal+ cells, n=10 animals; for controls, 803.6 ± 12.9 total nßgal+ cells, n=10 animals). We found that animals transfected with the Zic1 expression construct showed a shift in the spatial distribution of MF neurons, with 30% (255.8 ± 13.7 nßgal+ cells) of the labeled cell population residing in caudolateral aspects of the PGN (Fig. 4Ci-4Cv). By contrast, in control animals, only 15% (118.6 ± 3.4 nßgal+ cells) of the total cell population resides in this caudolateral territory (Figs. 4Ai-4Av). Moreover, there was a trend toward lateral positioning in rostral PGN territory (compare Figs. 4A and 4C). Thus, increasing levels of Zic1 in the MF lineage resulted in an altered spatial distribution of MF neurons within the PGN in a manner reciprocal to that observed following Zic1 reduction.
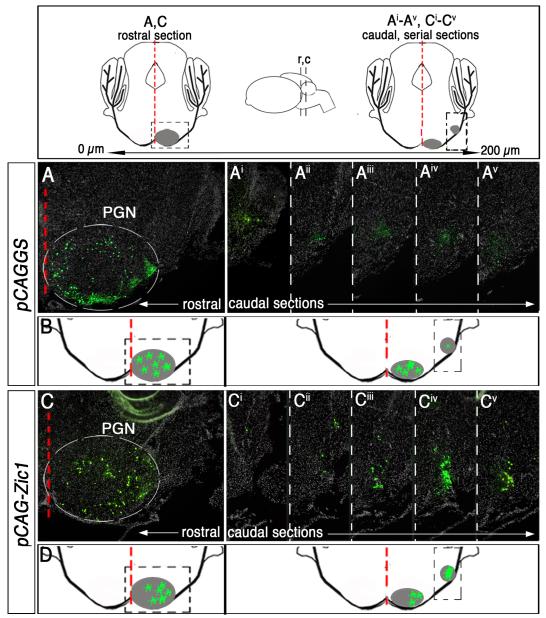
Fig. 4.
Zic1 overexpression in cells of the precerebellar MF lineage leads to a redistribution of neurons to caudolateral territories. Above, schematics of coronal sections along the rostrocaudal axis of the PGN at P8. Inset (center) depicts side view of the brain, and black dashed lines indicate idealized axial levels of coronal sections to the left and right. Distance between rostral and caudal sections is ~200 μm; PGN is in gray. Red dashed line demarcates brainstem midline. Boxed area in the rostral schematic identifies neurons in rostromedial PGN in (A) and (C). Boxed area in the caudal schematic identifies region in caudolateral PGN (Ai-Av) and (Ci-Cv). (A and Ai-v) Immunodetection for nßgal highlights cells co-transfected with control vector (pCAGGS) and reporter vectors. Coronal section of the rostral PGN of a P8 animal that was electroporated at 14.5 dpc (B). Five serial sections of the caudal PGN (A-Av). (B) Summary schematic of labeled cells within the PGN of control animals. (C and Ci-v) Immunodetection for nßgal shows cells co-transfected with pCAG-Zic1 and reporter vectors. Coronal section of the rostral PGN (C). Five serial sections of the caudal PGN (C-Cv). A significantly greater proportion of nßgal+ cells (transfected with pCAG-Zic1) reside in caudolateral PGN as compared to control animals in (Ai-Av) (student’s t-test, p<0.0001). (D) Summary schematic of labeled cells that have shifted to caudolateral aspects of the PGN in Zic1 overexpression animals.
Increased Zic1 levels promoted the ipsilateral axon pathway choice
Increasing Zic1 levels in PGN neurons not only resulted in an increased proportion of transfected neurons settling in caudolateral pontine territories, but also resulted in an increased proportion of labeled MF axons projecting to the ipsilateral cerebellum. In control animals, the majority of PGN MF afferents crossed the ventral midline and projected to the contralateral cerebellum, as reflected by eGFP in MFs throughout the granule cell layer of the contralateral cerebellum (Figs. 5A and 5C) rather than the ipsilateral cerebellum (Figs. 5B and 5D). By contrast, animals having received the Zic1 expression construct showed most eGFP signal in the ipsilateral (Figs. 5F and 5H) rather than contralateral cerbellum (Figs. 5E and 5G).
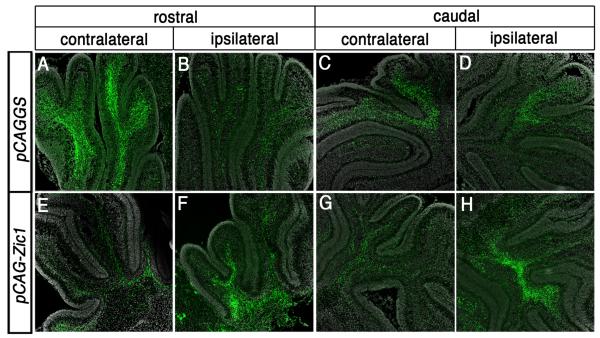
Fig. 5.
Increasing levels of Zic1 in the pontine MF neuron lineage promotes MF axon targeting to the ipsilateral instead of the usual contralateral cerebellum. (A-D) High magnification coronal images of cerebellar sections from P8 control animal that was electroporated at 14.5 dpc. Immunodetection for eGFP at the axial levels of the rostral (A,B) and caudal (C,D) cerebellum shows that a greater proportion of eGFP+ MF axons project throughout the forming granule cell layer of the contralateral cerebellum (A,C) rather than the ipsilateral cerebellum (B,D). (E-H) High magnification coronal images of cerebellar sections from Zic1 overexpression animals. By contrast to control animals, there is a greater proportion of eGFP+ MF axons within the ipsilateral cerebellum (F,H) in comparison to the contralateral cerebellum (E,G).
Predicted by these and the above findings is that knock-down of Zic1 levels would result not only in the observed rostromedial shift in MF cell body position but also a contralateral, rather than ipsilateral, choice in axon projection. While this may be the case, it was technically difficult to discern given the already high ratio of contra-/ipsilateral projections in the wild type.
DISCUSSION
By manipulating Zic1 levels selectively in pontine gray MF lineages in utero, we perturbed, in a cell-autonomous fashion, two steps in the development of the precerebellar MF afferent system, and in so doing revealed two developmental processes regulated by Zic1: (1) determination of pontine gray neuron position within the ventral brain stem and (2) axon (MF) pathway choice, between projecting across versus away from the brainstem ventral midline to target contra- versus ipsilaterally-located cerebellar territories, respectively. Fueling these studies was our discovery that Zic1 is expressed in postmitotic MF precursors during an intermediate stage in their development – during the period in which they migrate, coalesce to form nuclei, and extend axons. Acute reduction of Zic1 levels in pontine gray cells led to a cell-autonomous shift in their distribution toward rostromedial brainstem territories; by contrast, increased levels of Zic1 led to a reciprocal redistribution of transfected neurons toward caudolateral territories. An increased level of Zic1 in the pontine gray MF lineage was also associated with a cell autonomous increase in the relative proportion of ipsilaterally-directed MF projections. We thus revealed that Zic1 levels in postmitotic pontine gray precursor cells are critical for development of pontocerebellar circuitry.
Postmitotic cells of the brainstem mossy fiber lineage express Zic1
Zic1 expression is temporally dynamic and spatially complex within the developing mouse nervous system (Aruga, 2004; Nagai et al., 1997). By mRNA and protein detection on hindbrain tissue sections, we confirmed the well-established expression profile for Zic1 and established a later-stage expressing cell population - the set of postmitotic cells of the brainstem MF lineage. Up to 12 dpc, Zic1 mRNA and protein were detectable in progenitor cells in the dorsal third of the ventricular zone, as previously reported (Nagai et al., 1997); however, at later developmental stages (~12.5 dpc-16.5 dpc), we found that this progenitor zone expression was largely extinguished, with expression shifting to postmitotic MF cells. Zic1 protein and mRNA persisted in young MF neurons during subsequent steps involving their migration to the brainstem ventral midline, aggregation to form structured nuclei (e.g. the PGN), and extension of MF axons (our analyses extended to P8), which suggested previously unappreciated processes in which Zic1 action may be critical.
Laterality choice by mossy fibers at the brainstem ventral midline revealed through unilateral electroporations of the dorsally-located rhombic lip
The expression of Zic1 in newly postmitotic pontine MF precursors suggested a role for Zic1 in an intermediate stage of MF neuron development, after the initial specification of a generic MF neuron fate. Delineating the developmental processes influenced by this later-phase of Zic1 expression necessitated a conditional strategy capable of bypassing the earlier (prior to 12 dpc) requirement for Zic1 for proliferation and patterning of dorsal neuroectoderm. Thus, we needed to develop a means to alter Zic1 levels later in development, specifically in MF lineages in the mid-to-late gestation embryo. Equally critical was an ability to manipulate and label MF neurons on only one embryo side, as this allowed for assaying MF laterality choice at the ventral midline. This is because PGN MF axons that cross the ventral midline ultimately track to the cerebellum alongside ipsilaterally-directed MF axons that arise from the homologous PGN nucleus of the other side (Fig. 1A), making it difficult to study laterality choice by MFs when both PGN lobes are labeled. To address this set of experimental needs, we modified an in utero electroporation technique so as to achieve MF lineage-specific, stage-specific and unilateral delivery of Zic1 overexpression or knockdown plasmids along with reporter-encoding marker (track-tracing) plasmids. We were able to study, for the first time, functions served by Zic1 in late-stages of brain stem development, such as in PGN nucleus formation and MF axon laterality choice at the ventral brainstem midline.
Zic1 levels shape the pontine mossy fiber afferent system through cell autonomous actions at multiple developmental time points
MF neurons were found to distribute aberrantly within the ventral pons in response to cell-autonomous alterations in Zic1 levels. Low Zic1 levels led to a rostromedial distribution, as if the migrating pontine gray precursors failed to sense “stop” signals emanating from the brainstem rostroventral midline or were exuberantly propelled to this midline. By contrast, high Zic1 levels led to a settling of the transfected pontine gray neurons in caudolateral regions of the PGN. In both cases, the altered distributions appeared to occur within the normal rostromedial-to-caudolateral limits of the PGN nucleus. Transfected cells did not appear to distribute aberrantly outside the PGN or the ems, thus, at least grossly, the migration process up to the PGN appeared normal regardless of Zic1 levels. Also associated with high Zic1 levels was a cell autonomous shift in axon laterality at the brainstem ventral midline; rather than sending axons across the ventral midline to the contralateral cerebellum, the majority of transfected cells extended MFs away from the ventral midline to the ipsilateral cerebellum. Taken together, our findings indicate that pontine neuron position within the PGN and its MF connectivity with the cerebellum are dependent, at least in part, upon the levels of Zic1. These phenotypes appeared to restrict to the transfected cells, as we observed no gross perturbations in the non-transfected, yet intermingled neuron populations. Furthermore, because the redistributed transfected cells remained within the confines of the rostromedial-to-caudolateral limits of the PGN and continued to send projections to cerebellar granule cells (even if now ipsilaterally- rather than contralaterally-directed), it seems most likely that Zic1 levels are affecting aspects of subtype identity within the pontine gray MF lineage (e.g. as relates to cell body location) as opposed to causing a more drastic change in fate to a different neuron class.
Because pontine neuron cell body position along the rostromedial-to-caudolateral axis of the PGN correlated with axon laterality (caudolateral position correlating with ipsilateral MF projections and rostromedial position, with contralateral MF projections), it is possible that the primary developmental process controlled by Zic1 is selection of where to settle within the PGN. Site of residence may then secondarily determine or constrain axon pathway choice. Predicted by our findings then is the possibility that the endogenous level of Zic1 in a MF neuron correlates with the localization of that MF neuron within the PGN. Current immunodetection techniques for Zic1 were not sufficiently quantitative to test this prediction; nevertheless, this important question remains a focus of future work.
These Zic1-mediated MF phenotypes likely reflect Zic1 actions in postmitotic pontine gray precursor cells, rather than mitotic progenitor cells in the hRL, even though our electroporation approach drives vector expression in both populations. This is because endogenous Zic1 levels are low or absent in MF progenitor cells at the time of electroporation (Fig. 2B), thus it seems most likely that the delivered shRNA is neutral in the progenitor population but poised to reduce the production of endogenous Zic1 immediately upon its normal onset which occurs in the postmitotic population. Our overexpression results also favor the postmitotic stage of MF neuron development as the venue from which the observed phenotypes are set in motion. First, by multiple assays, transfected MF progenitor cells of the hRL appeared indistinguishable from the untransfected homologous hRL territory on the other embryo side. No differences were observed in the number of proliferating cells nor in the duration of proliferation and production of postmitotic MF progeny cells – plausible phenotypes given that in earlier stage neuroectoderm, where Zic1 is normally expressed, it is required to sustain neurogenesis (Aruga et al., 1998; Aruga et al., 2002; Aruga et al., 1994), with dorsal neural tube hypoplasia resulting in the absence of Zic1. Yet following forced expression of Zic1, we did not observe hyperproliferation of the hRL. Second, the reciprocity between the knockdown and overexpression phenotypes suggests a shared stage of action, with the knockdown experiments anchoring this action to postmitotic MF precursors.
In summary, in the developing brain stem, Zic1 appears to control different processes at different stages. At earlier stages during embryogenesis, Zic1 controls aspects of neuroectodermal proliferation and differentiation (Aruga et al., 1998; Aruga et al., 2002; Aruga et al., 1994), ensuring that the correct number of neurons is generated. Later during embryogenesis, as shown here, modulation of Zic1 levels appears to affect processes related to differentiation of a particular pontine gray neuron subtype identity, in this case, defined by pontine neuron cell body position within the PGN and MF axon laterality.
Zic1 and other molecules employed during pontine mossy fiber development
A generic MF neuron fate is initially specified through the cell-autonomous action of Math1 in progenitor cells of the hRL (Ben-Arie et al., 1997; Ben-Arie et al., 2000; Landsberg et al., 2005; Wang et al., 2005). In parallel and/or in sequence, events must then be enacted that result in the generation of different MF neuron subtypes, the latter defined in part by their final anatomical position of residence within the brain stem and axon trajectory relative to the brainstem ventral midline resulting in projection to different sides of the cerebellum. Our findings revealed aspects of these events; specifically that Zic1 levels are critical to the process of positional identity with in the PGN. The only other transcription factor reported to differentially affect development of subtypes of pontine MF neurons is the E-protein Tcf4 (Flora et al., 2007). Mice null for the Tcf4 gene show PGN MF precursors stopping short of the PGN, but in this case largely remaining in the ems – the trail taken by MF neuron precursors after emerging from the hRL (Altman and Bayer, 1987b; Taber Pierce, 1966). Tcf4-mediated effects on axon extension and laterality remain unclear. Other brainstem MF neuron subtypes appear unaffected by loss of Tcf4, such as those that populate the medulla after their journey from the hRL. Thus, Tcf4, as a heterodimer with Math1, is thought to trigger transcriptional events that set in motion programs required specifically for the development of pontine, but not other, MF neuron subtypes (Flora et al., 2007). During the neurogenetic interval in which PGN MF neurons are born (emerge as postmitotic precursor cells), rhombic lip cells express Math1 and Tcf4 but not Zic1; we showed that at this embryonic stage, Zic1 is restricted to postmitotic MF progeny cells. Thus, it is likely that Math1 and Tcf4 act prior to Zic1 with respect to development of the PGN MF neuron subtype. Transcription factors, like Pax6 (Engelkamp et al., 1999; Landsberg et al., 2005), Barhl1 (Bulfone et al., 2000; Li et al., 2004), and Olig3 (Liu et al., 2008), are also critical for normal MF neuron development, but are required globally by all brainstem MF lineages, not specific subtypes. Thus, to date, only the transcription factors Tcf4 and now Zic1 have been identified as having the ability to differentially affect the development of specific pontine MF neuron subtypes.
Mossy fiber neuron position within the brain stem is also affected by alteration of various axon guidance ligand-receptor pairs such as netrins and Dcc, and Slits and Robos. In mice lacking either Netrin (Serafini et al., 1996) or Dcc (Fazeli et al., 1997), MF neurons emerge from the rhombic lip but fail to reach ventral brain stem territory, instead forming ectopic clusters along the ventrolateral surface of the developing brain stem (Yee et al., 1999). Similarly, mice null for the Rig1/Robo3 receptor show MF precursors enriched in ventrolateral clusters (Marillat et al., 2004) as if MF precursors precociously stopped a journey otherwise intended to extend to more medial brainstem territories. These laterally stalled MF neurons have processes extending toward ipsilateral dorsal territories. If these ipsilateral processes are in fact axons terminating in the ipsilateral cerebellum, rather than lagging processes, then loss of Rig1/Robo3 shares similarities with both phenotypes associated with overexpression of Zic1 – a redistribution of MF neurons to lateral brainstem territories and axon extension ipsilaterally rather than contralaterally. These similarities raise the possibility that high Zic1 levels may act to suppress Rig1/Robo3 activity, either by suppressing its transcription directly or indirectly, or by indirectly regulating the subcellular localization of Rig1/Robo3 and/or its half-life. Interestingly, Rig1 also appears critical to axon laterality for another Math1-descendant neuronal population – dorsal spinal cord commissural neurons. Rig1 expression and floor plate crossing by commissural axons are disrupted following loss of the LIM homeodomain transcription factors Lhx2 and Lhx9 (Wilson et al., 2008). This work is also exciting as it links transcription factor action to changes in axon laterality. Cell adhesion molecules like TAG-1 (Backer et al., 2002) and various cadherins (Kawauchi et al., 2006; Taniguchi et al., 2006) are, like Rig1/Robo3, expressed by MF growth cones and are required for normal MF neuron migration and nucleogenesis. While Zic1 activity and cell adhesion properties have yet to be linked in any system, cell adhesion properties have been linked to the activity of Gli transcription factors (Li et al., 2007), which show strong homology to Zic proteins (Mizugishi et al., 2001). Moreover, certain Zic and Gli proteins dimerize to achieve new activities. Thus, alterations in Zic1 levels could lead to altered Gli activities and ultimately altered cell adhesion. The timing and mechanisms by which these varied classes of molecules may act downstream of Zic1 to coordinate PGN MF neuron position and axon laterality choice is an exciting area for future study; especially important will be an elucidation of the direct transcriptional targets of Zic1 in the MF lineage.
In other neural systems, such as the visual system, Zic2 plays a role in determining axon laterality at the optic chiasm. Recent analyses have revealed that Zic2 is both required (Herrera et al., 2003) and sufficient (García-Frigola et al., 2008) to alter the trajectory of retinal ganglion cell axons from crossed to uncrossed, lending support to our findings that Zic1 mediates MF divergence with respect to the brainstem ventral midline. One of the mechanisms by which Zic2 promotes axon divergence at the optic chiasm is through EphB1 signaling, possibly by transcriptionally regulating EphB1 receptor expression. Thus, in future studies of MF divergence at the brainstem ventral midline, it will also be important to explore Ephs and Ephrins.
In conclusion, here we provide evidence for Zic1 as a cell-autonomous regulator of pontine neuron position and MF laterality choice at the ventral brainstem midline. Thus Zic1 appears to serve critical functions at two stages of pontine gray neuron development: early on, controlling neuorectodermal proliferation and differentiation to ensure generation of the correct number of MF neurons (Aruga et al., 1998; Aruga et al., 2002; Aruga et al., 1994), and later, as shown here, to establish proper pontocerebellar circuitry.
Fig. 6.
Schematic representation of proposed model of Zic1 action in postmitotic precerebellar MF neurons. Under normal conditions, MF neurons of the PGN reside throughout the rostrocaudal extent of the nucleus (right). Variations in Zic1 protein levels in MF neurons determines where in the PGN an individual MF neuron takes up residence and to which side of the cerebellum it projects. MF neurons in which Zic1 levels were reduced settled in rostromedial PGN territories and projected to contralateral target regions in the cerebellum (middle), whereas MF neurons in which Zic1 levels were increased settled in caudolateral PGN territories and projected to ipsilateral targets (right, asterisk).
Acknowledgements
We thank the Dymecki and Cepko labs for critiques, especially Drs. J. Trimarchi and R. Kanadia; Dr. A. Hill for technical advice; Dr. C. Cepko for the pCAG-eGFP and pU6-shGAPDH vectors; and Dr. J. Johnson for the Math1 antibody. This work was supported by predoctoral fellowship F31 HD049320 (H.J.D.) and NIH P01HD036379 and R01HD051936 (S.M.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcantara S, Ruiz M, De Castro F, Soriano E, Sotelo C. Netrin 1 acts as an attractive or as a repulsive cue for distinct migrating neurons during the development of the cerebellar system. Development. 2000;127:1359–72. doi: 10.1242/dev.127.7.1359. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer S. Development of the cerebellar system: in relation to its evolution, structure, and functions. CRC Press; Boca Raton, Florida: 1997. [Google Scholar]
- Altman J, Bayer SA. Development of the precerebellar nuclei in the rat: III. The posterior precerebellar extramural migratory stream and the lateral reticular and external cuneate nuclei. J Comp Neurol. 1987a;257:513–28. doi: 10.1002/cne.902570404. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the precerebellar nuclei in the rat: IV. The anterior precerebellar extramural migratory stream and the nucleus reticularis tegmenti pontis and the basal pontine gray. J Comp Neurol. 1987b;257:529–52. doi: 10.1002/cne.902570405. [DOI] [PubMed] [Google Scholar]
- Aruga J. The role of Zic genes in neural development. Mol Cell Neurosci. 2004;26:205–21. doi: 10.1016/j.mcn.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Aruga J, Minowa O, Yaginuma H, Kuno J, Nagai T, Noda T, Mikoshiba K. Mouse Zic1 is involved in cerebellar development. J Neurosci. 1998;18:284–93. doi: 10.1523/JNEUROSCI.18-01-00284.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruga J, Nagai T, Tokuyama T, Hayashizaki Y, Okazaki Y, Chapman VM, Mikoshiba K. The mouse zic gene family. Homologues of the Drosophila pair-rule gene odd-paired. J Biol Chem. 1996;271:1043–7. doi: 10.1074/jbc.271.2.1043. [DOI] [PubMed] [Google Scholar]
- Aruga J, Tohmonda T, Homma S, Mikoshiba K. Zic1 promotes the expansion of dorsal neural progenitors in spinal cord by inhibiting neuronal differentiation. Dev Biol. 2002;244:329–41. doi: 10.1006/dbio.2002.0598. [DOI] [PubMed] [Google Scholar]
- Aruga J, Yokota N, Hashimoto M, Furuichi T, Fukuda M, Mikoshiba K. A novel zinc finger protein, zic, is involved in neurogenesis, especially in the cell lineage of cerebellar granule cells. J Neurochem. 1994;63:1880–90. doi: 10.1046/j.1471-4159.1994.63051880.x. [DOI] [PubMed] [Google Scholar]
- Azizi SA, Mihailoff GA, Burne RA, Woodward DJ. The pontocerebellar system in the rat: an HRP study. I. Posterior vermis. J Comp Neurol. 1981;197:543–8. doi: 10.1002/cne.901970402. [DOI] [PubMed] [Google Scholar]
- Backer S, Sakurai T, Grumet M, Sotelo C, Bloch-Gallego E. Nr-CAM and TAG-1 are expressed in distinct populations of developing precerebellar and cerebellar neurons. Neuroscience. 2002;113:743–8. doi: 10.1016/s0306-4522(02)00221-x. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–72. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Hassan BA, Bermingham NA, Malicki DM, Armstrong D, Matzuk M, Bellen HJ, Zoghbi HY. Functional conservation of atonal and Math1 in the CNS and PNS. Development. 2000;127:1039–48. doi: 10.1242/dev.127.5.1039. [DOI] [PubMed] [Google Scholar]
- Bloch-Gallego E, Causeret F, Ezan F, Backer S, Hidalgo-Sanchez M. Development of precerebellar nuclei: instructive factors and intracellular mediators in neuronal migration, survival and axon pathfinding. Brain Res Brain Res Rev. 2005;49:253–66. doi: 10.1016/j.brainresrev.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Brodal P, Bjaalie JG. Organization of the pontine nuclei. Neurosci Res. 1992;13:83–118. doi: 10.1016/0168-0102(92)90092-q. [DOI] [PubMed] [Google Scholar]
- Brodal P, Bjaalie JG. Salient anatomic features of the cortico-ponto-cerebellar pathway. Prog Brain Res. 1997;114:227–49. doi: 10.1016/s0079-6123(08)63367-1. [DOI] [PubMed] [Google Scholar]
- Bulfone A, Menguzzato E, Broccoli V, Marchitiello A, Gattuso C, Mariani M, Consalez GG, Martinez S, Ballabio A, Banfi S. Barhl1, a gene belonging to a new subfamily of mammalian homeobox genes, is expressed in migrating neurons of the CNS. Hum Mol Genet. 2000;9:1443–52. doi: 10.1093/hmg/9.9.1443. [DOI] [PubMed] [Google Scholar]
- Cicirata F, Zappala A, Serapide M, Parenti R, Panto M, Paz C. Different pontine projections to the two sides of the cerebellum. Brain Res Brain Res Rev. 2005;49:280–94. doi: 10.1016/j.brainresrev.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Eccles JC. Circuits in the cerebellar control of movement. Proc Natl Acad Sci U S A. 1967;58:336–43. doi: 10.1073/pnas.58.1.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelkamp D, Rashbass P, Seawright A, van Heyningen V. Role of Pax6 in development of the cerebellar system. Development. 1999;126:3585–96. doi: 10.1242/dev.126.16.3585. [DOI] [PubMed] [Google Scholar]
- Farago AF, Awatramani RB, Dymecki SM. Assembly of the brainstem cochlear nuclear complex is revealed by intersectional and subtractive genetic fate maps. Neuron. 2006;50:205–18. doi: 10.1016/j.neuron.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Fazeli A, Dickinson SL, Hermiston ML, Tighe RV, Steen RG, Small CG, Stoeckli ET, Keino-Masu K, Masu M, Rayburn H, et al. Phenotype of mice lacking functional Deleted in colorectal cancer (Dcc) gene. Nature. 1997;386:796–804. doi: 10.1038/386796a0. [DOI] [PubMed] [Google Scholar]
- Flora A, Garcia J, Thaller C, Zoghbi H. The E-protein Tcf4 interacts with Math1 to regulate differentiation of a specific subset of neuronal progenitors. Proc Natl Acad Sci U S A. 2007;104:15382–7. doi: 10.1073/pnas.0707456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Frigola C, Carreres MI, Vegar C, Mason C, Herrera E. Zic2 promotes axonal divergence at the optic chiasm midline by EphB1-dependent and -independent mechanisms. Development. 2008;135:1833–1841. doi: 10.1242/dev.020693. [DOI] [PubMed] [Google Scholar]
- Geisen MJ, Di Meglio T, Pasqualetti M, Ducret S, Brunet JF, Chedotal A, Rijli FM. Hox paralog group 2 genes control the migration of mouse pontine neurons through slit-robo signaling. PLoS Biol. 2008;6:e142. doi: 10.1371/journal.pbio.0060142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinberg I, Northrup H, Ardinger H, Prasad C, Dobyns WB, Millen KJ. Heterozygous deletion of the linked genes ZIC1 and ZIC4 is involved in Dandy-Walker malformation. Nat Genet. 2004;36:1053–1055. doi: 10.1038/ng1420. [DOI] [PubMed] [Google Scholar]
- Herrera E, Brown L, Aruga J, Rachel RA, Dolen G, Mikoshiba K, Brown S, Mason CA. Zic2 patterns binocular vision by specifying the uncrossed retinal projection. Cell. 2003;114:545–57. doi: 10.1016/s0092-8674(03)00684-6. [DOI] [PubMed] [Google Scholar]
- Ito M. The modifiable neuronal network of the cerebellum. Jpn J Physiol. 1984;34:781–92. doi: 10.2170/jjphysiol.34.781. [DOI] [PubMed] [Google Scholar]
- Kawauchi D, Taniguchi H, Watanabe H, Saito T, Murakami F. Direct visualization of nucleogenesis by precerebellar neurons: involvement of ventricle-directed, radial fibre-associated migration. Development. 2006;133:1113–23. doi: 10.1242/dev.02283. [DOI] [PubMed] [Google Scholar]
- Keino-Masu K, Masu M, Hinck L, Leonardo ED, Chan SSY, Culotti JG, Tessier-Lavigne M. Deleted in colorectal cancer (DCC) encodes a Netrin receptor. Cell. 1996;87:175–185. doi: 10.1016/s0092-8674(00)81336-7. [DOI] [PubMed] [Google Scholar]
- Kennedy TE, Serafini T, de la Torre JR, Tessier-Lavigne M. Netrins are diffusible chemotropic factors for commissural axons in the embryonic spinal cord. Cell. 1994;78:425–435. doi: 10.1016/0092-8674(94)90421-9. [DOI] [PubMed] [Google Scholar]
- Landsberg RL, Awatramani RB, Hunter NL, Farago AF, DiPietrantonio HJ, Rodriguez CI, Dymecki SM. Hindbrain rhombic lip is comprised of discrete progenitor cell populations allocated by Pax6. Neuron. 2005;48:933–47. doi: 10.1016/j.neuron.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Lee R, Petros T, Mason C. Zic2 regulates retinal ganglion cell axon avoidance of ephrinB2 through inducing expression of the guidance receptor EphB1. J Neurosci. 2008;28:5910–9. doi: 10.1523/JNEUROSCI.0632-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Qiu F, Xu A, Price SM, Xiang M. Barhl1 regulates migration and survival of cerebellar granule cells by controlling expression of the neurotrophin-3 gene. J Neurosci. 2004;24:3104–14. doi: 10.1523/JNEUROSCI.4444-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Deng W, Lobo-Ruppert S, Ruppert J. Gli1 acts through Snail and E-cadherin to promote nuclear signaling by beta-catenin. Oncogene. 2007;26:4489–4498. doi: 10.1038/sj.onc.1210241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Li H, Hu X, Yu L, Liu H, Han R, Colella R, Mower GD, Chen Y, Qiu M. Control of precerebellar neuron development by Olig3 bHLH transcription factor. J Neurosci. 2008;28:10124–10133. doi: 10.1523/JNEUROSCI.3769-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillat V, Sabatier C, Failli V, Matsunaga E, Sotelo C, Tessier-Lavigne M, Chedotal A. The slit receptor Rig-1/Robo3 controls midline crossing by hindbrain precerebellar neurons and axons. Neuron. 2004;43:69–79. doi: 10.1016/j.neuron.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Cepko C. Electrporation and RNA Interference in the rodent retina in vivo and in vitro. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihailoff GA, Burne RA, Azizi SA, Norell G, Woodward DJ. The pontocerebellar system in the rat: an HRP study. II. Hemispheral components. J Comp Neurol. 1981;197:559–77. doi: 10.1002/cne.901970403. [DOI] [PubMed] [Google Scholar]
- Millen K, Millonig J, Wingate R, Alder J, Hatten M. Neurogenetics of the cerebellar system. J Child Neurol. 1999;14:574–81. doi: 10.1177/088307389901400905. [DOI] [PubMed] [Google Scholar]
- Mizugishi K, Aruga J, Nakata K, Mikoshiba K. Molecular properties of Zic proteins as transcriptional regulators and their relationship to GLI proteins. J Biol Chem. 2001;276:2180–8. doi: 10.1074/jbc.M004430200. [DOI] [PubMed] [Google Scholar]
- Nagai T, Aruga J, Takada S, Gunther T, Sporle R, Schughart K, Mikoshiba K. The expression of the mouse Zic1, Zic2, and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev Biol. 1997;182:299–313. doi: 10.1006/dbio.1996.8449. [DOI] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–9. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Okada T, Keino-Masu K, Masu M. Migration and nucleogenesis of mouse precerebellar neurons visualized by in utero electroporation of a green fluorescent protein gene. Neurosci Res. 2007;57:40–9. doi: 10.1016/j.neures.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Palay SL, Chan-Palay . Cerebellar Cortex: Cytology and Organization. Springer; New York: 1974. [Google Scholar]
- Purandare SM, Ware SM, Kwan KM, Gebbia M, Bassi MT, Deng JM, Vogel H, Behringer RR, Belmont JW, Casey B. A complex syndrome of left-right axis, central nervous system and axial skeleton defects in Zic3 mutant mice. Development. 2002;129:2293–302. doi: 10.1242/dev.129.9.2293. [DOI] [PubMed] [Google Scholar]
- Rosina A, Provini L. Pontine projections to crus I and crus II of the cat cerebellum. A horseradish peroxidase study. Neuroscience. 1981;6:2613–24. doi: 10.1016/0306-4522(81)90107-x. [DOI] [PubMed] [Google Scholar]
- Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD, Pandya DN. The cerebrocerebellar system. Int Rev Neurobiol. 1997;41:31–60. doi: 10.1016/s0074-7742(08)60346-3. [DOI] [PubMed] [Google Scholar]
- Schwarz C, Thier P. Binding of signals relevant for action: towards a hypothesis of the functional role of the pontine nuclei. Trends Neurosci. 1999;22:443–51. doi: 10.1016/s0166-2236(99)01446-0. [DOI] [PubMed] [Google Scholar]
- Serafini T, Colamarino SA, Leonardo ED, Wang H, Beddington R, Skarnes WC, Tessier-Lavigne M. Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system. Cell. 1996;87:1001–14. doi: 10.1016/s0092-8674(00)81795-x. [DOI] [PubMed] [Google Scholar]
- Serapide MF, Panto MR, Parenti R, Zappala A, Cicirata F. Multiple zonal projections of the basilar pontine nuclei to the cerebellar cortex of the rat. J Comp Neurol. 2001;430:471–84. doi: 10.1002/1096-9861(20010219)430:4<471::aid-cne1044>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Sotelo C. Cellular and genetic regulation of the development of the cerebellar system. Prog Neurobiol. 2004;72:295–339. doi: 10.1016/j.pneurobio.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Sui G, Soohoo C, Affar EB, Gay F, Shi Y, Forrester WC, Shi YA. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA. 2002;99:5515–5529. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata H, Nakajima K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience. 2001;103:865–72. doi: 10.1016/s0306-4522(01)00016-1. [DOI] [PubMed] [Google Scholar]
- Taber Pierce E. Histogenesis of the nuclei griseum pontis, corporis pontobulbaris and reticularis tegmenti pontis (Bechterew) in the mouse. An autoradiographic study. J Comp Neurol. 1966;126:219–54. doi: 10.1002/cne.901260205. [DOI] [PubMed] [Google Scholar]
- Taber Pierce E. Histogenesis of the dorsal and ventral cochlear nuclei in the mouse. An autoradiographic study. J Comp Neurol. 1967;131:27–54. doi: 10.1002/cne.901310104. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Saito K, Nomura T, Osumi N. Manipulating gene expressions by electroporation in the developing brain of mammalian embryos. Differentiation. 2002;70:155–62. doi: 10.1046/j.1432-0436.2002.700405.x. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, Kawauchi D, Nishida K, Murakami F. Classic cadherins regulate tangential migration of precerebellar neurons in the caudal hindbrain. Development. 2006;133:1923–31. doi: 10.1242/dev.02354. [DOI] [PubMed] [Google Scholar]
- Turner R, G. WJ. Functional anatomy of the brachium pontis. J. Neurophysiol. 1941;4:196–206. [Google Scholar]
- Wang VY, Rose MF, Zoghbi HY. Math1 Expression Redefines the Rhombic Lip Derivatives and Reveals Novel Lineages within the Brainstem and Cerebellum. Neuron. 2005;48:31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Wingate RJ. The rhombic lip and early cerebellar development. Curr Opin Neurobiol. 2001;11:82–8. doi: 10.1016/s0959-4388(00)00177-x. [DOI] [PubMed] [Google Scholar]
- Wilson SI, Lee KJ, Dodd J. A Molecular Program for Contralateral Trajectory: Rig-1 Control by LIM Homeodomain Transcription Factors. Neuron. 2008;59:413–24. doi: 10.1016/j.neuron.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Yee KT, Simon HH, Tessier-Lavigne M, O’Leary DM. Extension of long leading processes and neuronal migration in the mammalian brain directed by the chemoattractant netrin-1. Neuron. 1999;24:607–22. doi: 10.1016/s0896-6273(00)81116-2. [DOI] [PubMed] [Google Scholar]