The mechanical rigidity of cells and adhesion forces between cells play an important role in a variety of biological processes including cell differentiation, proliferation, and tissue organization. Atomic force microscopy (AFM) has emerged as a powerful tool to quantify these mechanical properties and adhesion forces at the cellular level. Here we demonstrate an instrument that combines AFM with a side-view fluorescent imaging path that enables direct imaging of cellular deformation and cytoskeletal rearrangements along the axis of loading. With this instrument, we were able to directly observe cell shape under load, correlate changes in shape with force-induced ruptures, and image formation of membrane tethers during cell-cell adhesion measurements. Additionally, we observed cytoskeletal reorganization and stress fiber formation while measuring the contractile force of an individual cell. This instrument provides a useful tool for understanding the role of mechanics in biological processes.
Research over the last decade has revealed the importance of the mechanical microenvironment in a variety of biological processes. For example, one study found that substrate rigidity can direct the differentiation of naïve mesenchymal stem cells into muscle, bone, or brain cells, while another study linked substrate stiffness with the malignant phenotype of tumor cells 1,2. A more recent study found that the adhesive forces between ectoderm, mesoderm, and endoderm progenitor cells play a crucial roll in germ layer organization in zebrafish embryos 3. While mechanical loads and properties appear to play an important role in all of these processes, the cellular response to controlled mechanical inputs and the specific molecular mechanisms underlying the observed responses are not well understood, partly due to the lack of appropriate experimental tools.
The mechanical properties of individual cells and the adhesive forces between cells have been characterized using magnetic tweezers, micropipette aspiration, single cell rheometry, optical tweezers, and AFM 4-9. AFM uses a micron-scale cantilever to probe a sample (Fig. 1a) 9. For small deflections, the cantilever behaves like a Hookean spring, exerting a force proportional to its deflection.. By detecting the position of the cantilever with an optical lever, forces exerted on a sample by the cantilever or imposed by the sample on the cantilever can be measured with high resolution (~ pN) over a large dynamic range (hundreds of nN).
Figure 1.
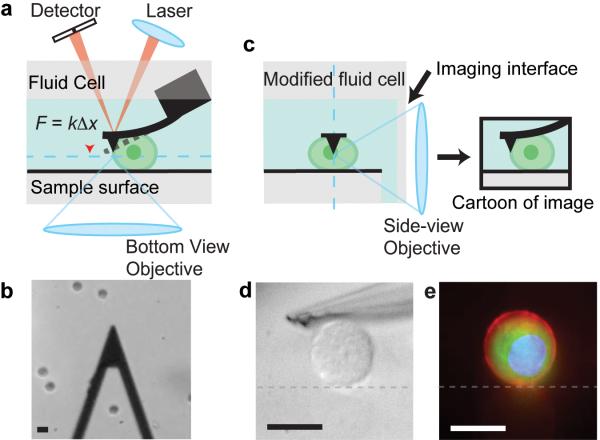
Concept of the Side-view AFM. (a) Cartoon schematic of the typical layout of an AFM. A cantilever (black) is used to probe a sample (green). The undeflected cantilever position is represented by the dotted line. The cantilever base is held rigidly by a fluid cell. A laser is focused onto the tip of the cantilever, and the reflected beam is captured by a detector. The position of the reflected beam on the detector is used to monitor cantilever position. A standard bottom-view imaging path (the image plane is represented by the dotted blue line) is shown. (b) A typical bottom-view image taken in brightfield. The triangular cantilever is seen, as are several trypsinized U2OS cells (circles). (c) Cartoon schematic of side-view imaging path and desired image of sample (image plane is represented by blue dotted line). (d) Side-view image of a U2OS cell and cantilever under oblique white light illumination. Surface is represented by grey dotted line. (e) A merged image consisting of images taken in epi-fluorescence of the cell membrane (red), GFP-actin (green), and the nucleus (blue) of a U2OS cell. The reflection of the sample from the glass sample surface (grey dotted line) can be seen and identifies the surface boundary. Scale bars are all 20 μm.
Because of these capabilities, AFM has been used extensively to study mechanics at the cellular scale. The mechanical properties of cells have been determined by imposing a stress with the cantilever and measuring the resulting strain or by conducting oscillatory microrheology measurements 10,11. In addition, AFM can be used to measure the adhesion between two cells by attaching one cell to a cantilever, pressing it against a second cell solidly attached to a surface, and measuring the force required to pull the two cells apart 12,13.
AFM is often combined with brightfield and fluorescence microscopy to image cellular shape and labeled cellular proteins while making force measurements. Conventional AFMs typically allow straightforward combination with epi-fluorescence imaging systems that provide an image of the sample along a plane parallel to the surface, which we refer to as a “bottom-view” (Fig. 1a,b). However, the most significant cellular deformations and cytoskeletal rearrangements are expected to be aligned with the applied force in a plane perpendicular to the surface. Imaging in this plane via a “side view” would allow one to relate specific cellular responses along the loading direction with the applied load.
One approach to obtaining side-view images of the sample while applying and measuring forces has been to combine AFM with confocal microscopy, which is capable of providing sub-micron resolution images 14. However, the timescale required for high-resolution confocal imaging exceeds that of some typical AFM measurements such as that of cell-cell adhesion, as well as the timescale of many active biological processes of interest 12,13.
A more straightforward approach is to develop a side-view imaging path that aligns the image plane with the axis of loading. In one such instrument, cantilever position is detected with the side-view images instead of optical lever detection, significantly limiting the temporal and spatial resolution of cantilever position 15. In a micropipette-based AFM instrument, the sample is held by a micropipette and can be imaged from the side 16,17. While convenient for holding cells via aspiration, this approach restricts sample geometries and cannot take advantage of large sample areas, such as glass slides and culture dishes used for adherent cells, which are useful for rapid selection and measurement of cell populations. These previously reported instruments have not incorporated epi-fluorescence imaging capabilities of labeled cellular components in the side-view imaging path.
Here we describe the development of an AFM with an optical side-view imaging path to allow direct, high numerical aperture epi-fluorescence imaging of the sample along the loading axis. We demonstrate the utility of this instrument for interpreting the dynamics of adhesion between two cells and analyzing the contractile behavior of an individual cell under tensile forces.
RESULTS
Side-view AFM design
The central design challenge in the development of this instrument was positioning a high-numerical aperture microscope objective in close proximity to a cantilever while retaining thermal noise-limited deflection sensitivity. Furthermore, the cantilever and sample had to be imaged with minimal aberrations while immersed in media. To solve this problem, we designed a glass fluid cell that holds a commercial cantilever chip adjacent to a glass coverslip (Fig. 1c, Supplementary Fig. 1, Online Methods). This coverslip serves as a planar imaging interface through which the side-view objective can image the cantilever and sample under investigation. The minimum distance between the objective and the cantilever is limited by the thickness of the coverslip (~170 μm) and the distance between the cantilever and the edge of the cantilever chip (~0.5 mm for the cantilevers used in our system). Samples can be imaged with oblique white light illumination (Fig. 1d) or in epi-fluorescence (Fig. 1e).
The cantilever position is detected via a calibrated optical lever system similar to conventional AFMs. A collimated laser is focused with a lens onto the cantilever, and the reflected beam from the cantilever surface is collected onto a position sensitive photodiode (Fig. 1a). The position detection resolution of this is thermally limited and is unaffected by the side-view imaging path (Supplementary Fig. 2).
Leukocyte to endothelial cell adhesion
As an initial demonstration of the side-view AFM, we investigated the adhesion between leukocytes and endothelial cells. As part of the immune response to infection or inflammation, leukocytes circulating in the blood vessels must migrate through the endothelial cell layer into the affected tissue 18. The first stage of this response involves the rolling of the leukocytes along the activated endothelial cell layer followed by adhesion of the leukocyte to the endothelial cell 18. After adhesion, the leukocyte may transmigrate through the endothelial cell 18.
The adhesion between leukocytes and endothelial cells has been studied extensively by micropipette aspiration, bulk shear flow measurements, and AFM 6,19-21. However, measurements with AFM have typically been done by allowing cells to touch briefly (~1s) so that only the molecular bonds at the cell surface would be able to form, and thus cytoskeleton recruitment to and reinforcement of the adhesion could be ignored 21. In contrast to those experiments, we probed the adhesion after the leukocyte and endothelial cell had been allowed to form a stronger adhesion over time (~10 minutes). We let leukocytes settle over a monolayer of activated endothelial cells. Within ten minutes, we selected a single leukocyte attached to an endothelial cell and pressed the surface of a Concavalin A (ConA) coated cantilever (just behind the tip) onto the leukocyte, exerting a constant small compressional force (< 1.0 nN) on the leukocyte for ~15 minutes (Fig. 2a, Online Methods). ConA promotes a strong passive attachment between the leukocyte and the cantilever and is commonly used in other leukocyte-endothelial cell adhesion measurements with AFM 21.
Figure 2.
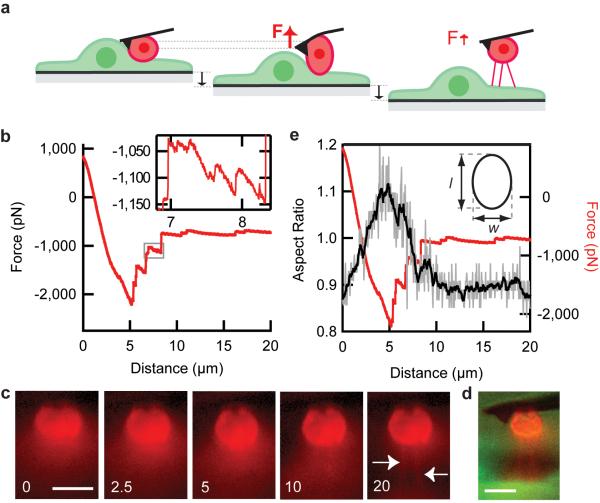
Measurement of adhesion between leukocyte and endothelial cell. (a) Cartoon schematic of experiment design. The cantilever is pressed into a leukocyte (red) attached to an endothelial cell (green), then the sample surface is pulled away at a constant rate. (b) Representative force-distance trace for a single cell [AU: correct?] adhesion experiment where distance refers to the distance the sample surface has moved relative to its initial position, and a positive force indicates a compressive force. The surface was moved away at 500 nm/s. (c) Time series of images of the leukocyte labeled with a membrane dye taken at the selected distances. White arrows point to tethers formed between the leukocyte and endothelial cell. (d) A merged epi-fluorescence side-view image of a leukocyte (red) and an endothelial cell (green) immediately after the adhesion measurement. (e) The aspect ratio of the cell (shown as a cartoon in the inset) is measured for all the images taken with the side-view imaging path (images were taken at 10 Hz) and shown in grey. This trace is smoothed with a Savitzky-Golay 2nd order filter for clarity (black). [AU: Is this a single representative trace? Out of what total n?] Scale bars are 10 μm (c, d).
We then applied a load to the cell-cell adhesion by moving the sample surface away from the cantilever at a constant rate (500 nm/s), which is on the order of the flow speeds experienced physiologically for leukocytes rolling along the endothelium (Fig. 2a) 22. The cantilever was pulled down as the surface was pulled away due to the strength of adhesion between the two cells, so that the cantilever exerted a tensile force on the cell-cell adhesion (Fig. 2b). Eventually, the adhesion between the two cells could not sustain further increases in force, and the adhesive force dropped significantly through a series of discrete ruptures of ~hundreds of pN each. Force ruptures on a smaller scale of ~10-50 pN were also measured. After this de-adhesion phase, only the smaller force ruptures were observed as the two cells were pulled further away from each other, but the total force between the two cells as remained non-zero. Side-view images of the leukocyte, labeled with a membrane dye, revealed the formation of long membrane tethers between the leukocyte and the endothelial cell as the cells were pulled apart (Fig. 2c, Supplementary Video 1, n = 3). Tether formation explains the non-zero steady-state force after leukocyte-endothelial cell de-adhesion because a force (~100 pN/tether) was required to keep a tether from merging back into the cell body, and our side-view images show at least three tethers remain after the two cell bodies have been separated (Fig. 2d) 23.
Next we analyzed the images of leukocyte shape taken with the side-view imaging path. By using a custom thresholding algorithm for each image taken at 10 Hz, we determined the aspect ratio of the leukocyte cell body and compared this to the force trace (Fig. 2e). We found that initially, as some leukocytes were subjected to increasing tensional forces, their aspect ratios increased. This was followed by a decrease in aspect ratio concurrent with the observation of the large force ruptures, after which the aspect ratio reached a steady-state value. The large force ruptures are thus temporally correlated with aspect ratio changes of the leukocyte cell body as the cell returns to a steady-state shape. Since the cytoskeleton is thought to predominantly determine shape of the cell body, these results suggest that the large rupture events represent the de-lamination of the cytoskeleton from the membrane at specific cell-cell adhesion locations. This is consistent with the observation of tethers forming between the two cells, since an unsupported membrane attached at discrete points under tension is expected to form tethers as the most energetically favorable state of attachment at higher forces 23. Thus, the side-view imaging path enabled us to correlate the AFM force measurements and force rupture events with changes in cell shape and provide an interpretation of the large force ruptures.
Adherent cell contraction against a load
As a demonstration of the use of the side-view AFM in probing an active biological process as opposed to a passive mechanical response, we imaged the shape and structure of the actin cytoskeleton during adherent cell contraction of a single cell between two surfaces while measuring the force of contraction. The contractile forces exerted by adherent cells are typically studied for cells on a 2-dimensional soft substrate, where the deformation of the substrate or structures patterned on the substrate can be used to determine the force 24,25. In these cases, the adhesions to the surface are all on the basal surface of the cell, so the contractile forces are all parallel to the adhesions. The geometry of our system allows us to probe the development of cytoskeleton structure and contraction of cells in a quasi-3D environment, where the cell is adhered to two surfaces and is contracting between them. In this case the contractile forces are normal to the adhesions. This geometry has the advantage of allowing a direct application and measurement of force between sets of adhesions on each surface.
Adherent U2OS cells have been previously shown to have a cytoskeleton that is dynamic so that it assembles and remodels quickly but also exhibits a robust structure displaying thick stress fibers26. We investigated the contraction behavior of single adherent U2OS cells under load (Fig. 3a-c). First, we allowed stably transfected U2OS cells expressing GFP-actin to settle on the glass sample surface coated with fibronectin (Fig. 3d). Then, we exerted a small compressional force on an individual cell with the flat portion of a fibronectin-coated cantilever before the cell had time to spread on the sample surface. The cell was observed to spread on the cantilever and sample surfaces as the cell-cantilever interface and cell-surface interface assumed a flat profile (Fig. 3d, n = 11)
Figure 3.
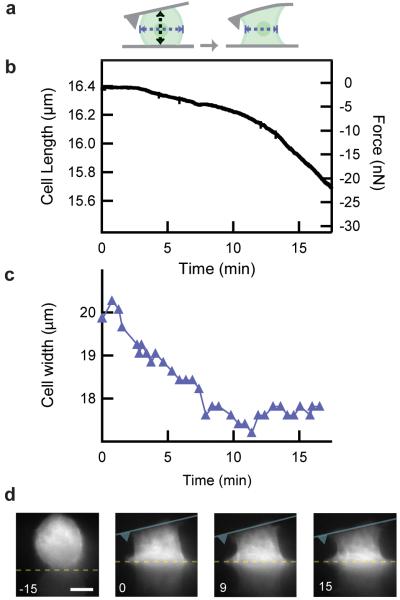
Contraction of U2OS cell against load. (a) Cartoon of experiment design. Cell width, indicated by blue dotted arrows, is measured from side-view images. (b) Cell length and force of contraction are shown for a single representative experiment [AU: correct?]. Cell length was calculated as the difference between the cantilever deflection and the position of the surface, and was calibrated initially by measuring the height of the cell at its midpoint optically with side-view images (a, black dotted arrow). (c) Cell width (a, blue dotted arrows) was measured for every image taken [AU: is this for a single cell?] during the experiment and shown as the blue trace (top) over time. (d) Side-view images taken at the indicated time points of GFP actin in the cell. The location of the surface and cantilever are represented by the yellow dotted line and blue cartoon respectively. Scale bar is 10 μm.
Next, we allowed the cell to contract between the cantilever and the surface while simultaneously imaging the actin cytoskeleton. Since the position of the surface was fixed, the cell contracted against increasing tensile forces as it pulled the cantilever towards the surface. Initially, the rate of cell contraction increases as the cell contracts against increasing force, and then eventually approaches a constant rate of contraction (n = 9). Simultaneously, some actin fibers are observed to grow towards the cantilever in this experiment (Fig 3d, at 15 min, Supplementary Video 2). This suggests that the initial increase in cell contraction rate against increasing loads could be an initiation of a contraction phase, with a rate that increases as the cytoskeleton remodels and connections between the two surfaces in the form of stress fibers are established. Other fibers are observed to remain oriented more parallel to the surface. This type of cytoskeleton architecture is seen in other experiments, though we do not always see the growth of fibers from one surface to the other (see Supplementary Video 3).
As the cell adheres to the two surfaces and begins to contract, we observe a change in morphology from the initial rounded shape to a characteristic “hourglass” shape. To quantify this change in morphology, we measured the width of the cytoskeleton at the midpoint of the cell using a custom thresholding algorithm for each image (Fig. 3c, Supplementary Methods). The lateral contraction completes before the vertical contraction rate increases to reach its maximum.
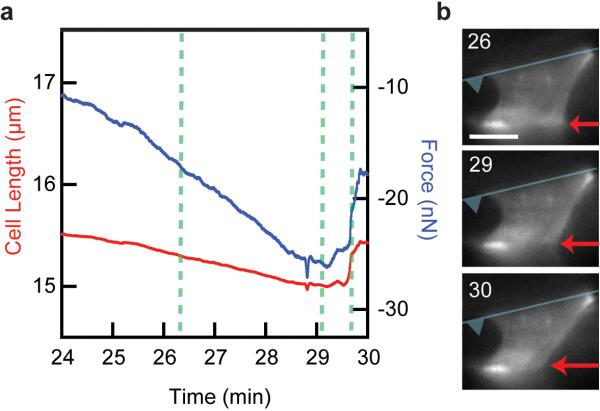
In many of these contraction experiments, the U2OS cells do not reach a steady state length. Instead we observed the contractile force to decrease after reaching a maximum, and the cell length begins to increase again (Fig. 4a, Supplementary Video 3). Interestingly, the side-view images of the cytoskeleton show the rupture of adhesion sites along one of the surfaces to be responsible for this event as opposed to a decrease in contractile force exerted by the cell with the adhesions remaining intact (Fig. 4b). This indicates that the contractile forces that the cytoskeleton can exert across its cell body exceed the strength of the adhesion between the cell and these fibronectin-coated surfaces.
Figure 4.
Contractile force of U2OS cell exceeds force of adhesion to bottom surface. (a) Cell length (red) and contractile force (blue) for a single contracting U2OS cell over time. (b) Series of side-view images taken of GFP actin at the indicated times corresponding to the dotted lines in a. Increase in cell length and decrease in contractile force is coincident with the release of the adhesion (red arrow) to the bottom surface. The location of cantilever surface is represented by the superimposed blue cartoon. Scale bar is 10 μm.
DISCUSSION
In our leukocyte-endothelial cell adhesion experiments, we observed large force rupture events coincident with a return of the leukocyte to a steady-state shape (after the cells had been given time to form a high-strength adhesion), suggesting detachment of the actin cytoskeleton from the membrane is responsible for these large force ruptures. This is consistent with the formation and lengthening of membrane tethers observed following this phase, and the results suggest that the cytoskeleton attachment to the membrane is localized to discrete areas.
Interestingly, we see that the tethers formed in this experiment are extruded from the leukocyte membrane, whereas a previous micropipette-based assay found the tethers to be extruded predominantly from the endothelial cell 19. Several disparities between the two experimental systems may account for this difference. In the micropipette-based experiment, the endothelial cells were rounded in contrast to our experiment in which the endothelial cells were spread out and adopt a configuration that more closely approximates their physiological state. Additionally, the micropipette-based experiments allowed the cells to interact only briefly so that low strength adhesions were formed. These differences highlight the complexity of the adhesion between leukocytes and endothelial cells and point to the importance of boundary conditions and imaging in the interpretation of adhesion behavior.
In the single cell contraction experiments, we find that after spreading onto the cantilever and sample surface, the cell contracts laterally at its midpoint before pulling the cantilever down vertically towards the surface. This inward contraction is perpendicular to the expected direction of contraction between the adhesion sites on the two surfaces and represents an active behavior since a conventional material with a length reducing in one dimension is expected to increase its length in the other two dimensions. Thus the inward contraction of the U2OS cells may be an initial remodeling of the cytoskeleton that enhances the ability of the cell to spread on the two surfaces. Alternatively, it could represent active inward contraction of the cytoskeleton along the midsection of the cell.
On longer timescales, we find that the U2OS cells often do not reach a steady-state contraction length or force, and instead the attachment to one surface sometimes ruptures. This could be due to active sensing of stiffness or force across the cell body, or the U2OS may simply exert contractile forces that the adhesions cannot sustain. This raises the question of what regulates cell attachment to surfaces in a quasi-3D environment. It is also possible that this cellular response may be influenced by the angle of the cantilever relative to the surface, and that the resulting difference in cell height across the cell is actively sensed. Further experiments are needed to determine the underlying mechanism of these behaviors and that of the formation and growth of stress fibers spanning the attachment surfaces.
The combination of side-view imaging with AFM provides insight into mechanical force measurements and reveals interesting mechanical behaviors that raise new questions about cell contractility. We anticipate that this side-view AFM will be useful for future studies of cell mechanics, cell contractility, and cell-cell adhesion, particularly when protein localization or cytoskeletal rearrangement will help to reveal molecular mechanisms underlying the responses. Potential applications of the side-view AFM include probing the deformation of the nucleus and nuclear-cytoskeleton coupling during cell mechanics experiments, observing cytoskeleton recruitment during cell-cell adhesion measurements 27, and quantifying density changes in growing dendritic actin networks under load 28.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank J.W. Shaevitz, M. Van Duijn, M.J. Rosenbluth, A. Crow, and the entire Fletcher lab for helpful discussions, and B. Zuchero (University of California San Francisco) for the gift of GFP transfected U2OS cells used in initial experiments. This work was supported by a National Science Foundation graduate research fellowship (O.C.); a National Institutes of Health National Research Service Award, Hammond research fellowship, and Hartwell biomedical research fellowship (W.A.L.); and a National Institutes of Health R01 (D.A.F.).
Appendix
Online Methods
Instrument components
The essential elements of our side-view imaging path are shown in Supplementary Figure 1. The entire system is mounted on an optical breadboard that is vibrationally isolated with pneumatic support (Technical Manufacturing Corporation) and housed in a custom built acoustic enclosure. The main components of the cantilever position detection system are a 25 mW (used at 5 mW in our system), λ = 785 nm laser (Blue Sky Research), a position sensitive diode as the detector (DL100-7PCBA3, Pacific Silicon Sensors, Inc.), a f = 75 mm lens to focus the laser (Thorlabs, Inc.), a cold mirror (Edmund Optics) to reflect the laser onto to the cantilever, and an uncoated silicon nitride cantilever (MLCT-NO, Veeco Probes). The laser, position sensitive diode, lense, and dichroic mirror are all mounted on a small breadboard (Thorlabs, Inc.) and the breadboard is moved manually with a 3D translation stage (460A, Newport Corp.) at the beginning of every experiment to position the focused laser onto the cantilever. The cantilever is held rigidly in a custom designed and machined fluid cell, and is fixed in space during the experiment. Instead, the sample surface (a 0.5” × 0.5” cover glass mounted on top of a custom machined aluminum piece) is moved during an experiment relative to the cantilever. The sample surface is positioned in XY and coarsely in Z with motorized actuators (CMA-12CCCL, 12.5 mm range, Newport Corporation) mounted in a 3D translation stage (562, 12.5 mm range, Newport Corp.), and then moved finely in Z with a closed loop Z-axis piezoelectric stage (P-620.ZCD, range = 50 μm, resolution = 0.2 nm, Physik Instrumente, LP).
At the beginning of an experiment, 100 μl of fluid/cells is pipetted onto the sample surface. The surface is then moved coarsely to within 25 μm of the cantilever, forming a fluid chamber. The bottom-view imaging path consists of a white lightsource (Fiberlite Fiber optic illuminator, Dolan Jenner Ind.), a 4X objective (Edmund Optics), and a CCD (WAT-902H2 Supreme, Watec). The essential components of the side-view optical path include a water immersion objective for the side-view imaging path (63X/NA = 1.0, WD = 2.0 mm, Carl Zeiss Inc.), a #1 glass coverslip attached to the fluid cell, which serves as the side-view imaging interface, a standard fluorescence imaging suite consisting of a xenon arc lamp/ excitation/ emission filter wheels (Sutter, Inc.), and a cooled, low-light CCD (Retiga-SRV, Q-imaging) to collect images. The side-view objective position is controlled in 3-axes with motorized actuators (Z612B, range = 6.5 mm, Thorlabs, Inc.) mounted in a 3D translation stage (460A, Newport Corp.). This allows us to take image stacks from the side to locate planes in which actin fibers are forming. All other miscellaneous mechanical or optical components were purchased from standard suppliers (i.e. Thorlabs, Edmund Optics, Sutter Instruments) or custom built. The entire system is controlled with software written in LABVIEW (National Instruments).
Cell-cell adhesion experiments
Endothelial cells (HUVECS) were purchased from (Lonza) and cultured in EGM-2 media (Lonza). Human promyelocytic leukemia cells (HL60 line) cells were purchased from ATCC, cultured in RPMI with 10% FBS, and differentiated with 1.5% DMSO for 3-5 days into neutrophil-like cells for experiments. Endothelial cells were plated on a fibronectin-coated glass sample surface to form a monolayer and activated by overnight incubation with TNF-α. These cells were allowed to spread overnight before use. Endothelial cells were labeled with CellTracker Green (Invitrogen), and leukocytes were labeled with the Vybrant DII membrane dye (Invitrogen). Cantilevers were coated with ConA using a procedure described previously. for passive attachment of leukocytes to the cantilever surface21.
Cell contraction experiments
U2OS cells stably transfected with GFP-actin were purchased from Marinpharm GmbH, and cultured in DMEM growth medium (Gibco). A HOECHST 33342 (Invitrogen) nuclear stain and Vybrant DII membrane dye (Invitrogen) were used in Figure 1e. For the experiments shown in Figures 3 & 4, a piranha cleaned cantilever (3:1 mixture of H2SO4 to H2O2) and glass sample surface were incubated with 0.05 mg/ml Fibronectin (Sigma) for at least 1 hour at 37o C in an incubator and then washed in sterile Phosphate Buffered Saline before the experiment. For the experiment itself, U2OS cells were trypsinized from culture and diluted into CO2 independent media at a concentration of 100,000 cells/ml, and immediately added into the fluid chamber.
Data collection and analysis
AFM data was typically collected at 100 Hz. Prior to digitization with a data acquisition card (National Instruments), data was anti-aliased at 50 Hz (Krohn-hite Inc.). Offline data processing was done in Igor Pro (Wavemetrics Inc.). For the analysis of Figure 3 and Figure 4, images were thresholded in Igor Pro, adjusting the threshold until the thresholded image faithfully reproduced the cell shape. The maximum cell height and width were calculated to determine the aspect ratio of the cell in Figure 3. The width of the cell was measured from the thresholded image at a height midway between the cantilever and surface for Figure 3. For the power spectrum shown in Supplementary Figure 2, data was collected at 100 kHz and anti-aliased at 50 kHz.
REFERENCES
- 1.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 2.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Krieg M, et al. Tensile forces govern germ-layer organization in zebrafish. Nature Cell Biology. 2008;10:429–U122. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- 4.Bausch AR, Moller W, Sackmann E. Measurement of local viscoelasticity and forces in living cells by magnetic tweezers. Biophysical Journal. 1999;76:573–579. doi: 10.1016/S0006-3495(99)77225-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shao JY, Hochmuth RM. Micropipette suction for measuring piconewton forces of adhesion and tether formation from neutrophil membranes. Biophysical Journal. 1996;71:2892–2901. doi: 10.1016/S0006-3495(96)79486-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans E, Heinrich V, Leung A, Kinoshita K. Nano- to microscale dynamics of P-selectin detachment from leukocyte interfaces. I. Membrane separation from the cytoskeleton. Biophysical Journal. 2005;88:2288–2298. doi: 10.1529/biophysj.104.051698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desprat N, Richert A, Simeon J, Asnacios A. Creep function of a single living cell. Biophysical Journal. 2005;88:2224–2233. doi: 10.1529/biophysj.104.050278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wottawah F, et al. Optical rheology of biological cells. Physical Review Letters. 2005;94 doi: 10.1103/PhysRevLett.94.098103. [DOI] [PubMed] [Google Scholar]
- 9.Binnig G, Quate CF, Gerber C. Atomic Force Microscope. Physical Review Letters. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 10.Radmacher M, Fritz M, Kacher CM, Cleveland JP, Hansma PK. Measuring the viscoelastic properties of human platelets with the atomic force microscope. Biophysical Journal. 1996;70:556–567. doi: 10.1016/S0006-3495(96)79602-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alcaraz J, et al. Microrheology of human lung epithelial cells measured by atomic force microscopy. Biophysical Journal. 2003;84:2071–2079. doi: 10.1016/S0006-3495(03)75014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benoit M, Gabriel D, Gerisch G, Gaub HE. Discrete interactions in cell adhesion measured by single-molecule force spectroscopy. Nature Cell Biology. 2000;2:313–317. doi: 10.1038/35014000. [DOI] [PubMed] [Google Scholar]
- 13.Panorchan P, et al. Single-molecule analysis of cadherin-mediated cell-cell adhesion. Journal of Cell Science. 2006;119:66–74. doi: 10.1242/jcs.02719. [DOI] [PubMed] [Google Scholar]
- 14.Charras GT, Horton MA. Single cell mechanotransduction and its modulation analyzed by atomic force microscope indentation. Biophysical Journal. 2002;82:2970–2981. doi: 10.1016/S0006-3495(02)75638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canetta E, Duperray A, Leyrat A, Verdier C. Measuring cell viscoelastic properties using a force-spectrometer: Influence of protein-cytoplasm interactions. Biorheology. 2005;42:321–333. [PMC free article] [PubMed] [Google Scholar]
- 16.Heinrich V, Ounkomol C. Biophysics in reverse: Using blood cells to accurately calibrate force-microscopy cantilevers. Applied Physics Letters. 2008;92 [Google Scholar]
- 17.Ounkomol C, Xie H, Dayton PA, Heinrich V. Versatile Horizontal Force Probe for Mechanical Tests on Pipette-Held Cells, Particles, and Membrane Capsules. Biophysical Journal. 2009;96:1218–1231. doi: 10.1016/j.bpj.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vestweber D. Adhesion and signaling molecules controlling the transmigration of leukocytes through endothelium. Immunological Reviews. 2007;218:178–196. doi: 10.1111/j.1600-065X.2007.00533.x. [DOI] [PubMed] [Google Scholar]
- 19.Girdhar G, Shao JY. Simultaneous tether extraction from endothelial cells and leukocytes: Observation, mechanics, and significance. Biophysical Journal. 2007;93:4041–4052. doi: 10.1529/biophysj.107.109298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerszten RE, et al. MCP-1 and IL-8 trigger firm adhesion of monocytes to vascular endothelium under flow conditions. Nature. 1999;398:718–723. doi: 10.1038/19546. [DOI] [PubMed] [Google Scholar]
- 21.Zhang XH, et al. Atomic force microscopy measurement of leukocyte-endothelial interaction. American Journal of Physiology-Heart and Circulatory Physiology. 2004;286:H359–H367. doi: 10.1152/ajpheart.00491.2003. [DOI] [PubMed] [Google Scholar]
- 22.Kunkel EJ, Dunne JL, Ley K. Leukocyte arrest during cytokine-dependent inflammation in vivo. Journal of Immunology. 2000;164:3301–3308. doi: 10.4049/jimmunol.164.6.3301. [DOI] [PubMed] [Google Scholar]
- 23.Derenyi I, Julicher F, Prost J. Formation and interaction of membrane tubes. Physical Review Letters. 2002;88 doi: 10.1103/PhysRevLett.88.238101. [DOI] [PubMed] [Google Scholar]
- 24.Harris AK, Wild P, Stopak D. Silicone-Rubber Substrata - New Wrinkle in the Study of Cell Locomotion. Science. 1980;208:177–179. doi: 10.1126/science.6987736. [DOI] [PubMed] [Google Scholar]
- 25.Chen CS. Mechanotransduction - a field pulling together? Journal of Cell Science. 2008;121:3285. doi: 10.1242/jcs.023507. [DOI] [PubMed] [Google Scholar]
- 26.Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. Journal of Cell Biology. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu YS, et al. Force measurements in E-cadherin-mediated cell doublets reveal rapid adhesion strengthened by actin cytoskeleton remodeling through Rac and Cdc42. Journal of Cell Biology. 2004;167:1183–1194. doi: 10.1083/jcb.200403043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parekh SH, Chaudhuri O, Theriot JA, Fletcher DA. Loading history determines the velocity of actin-network growth. Nature Cell Biology. 2005;7:1219–1223. doi: 10.1038/ncb1336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.