Abstract
LC-MS/MS has demonstrated potential for detecting plant pathogens. Unlike PCR or ELISA, LC-MS/MS does not require pathogen-specific reagents for the detection of pathogen-specific proteins and peptides. However, the MS/MS approach we and others have explored does require a protein sequence reference database and database-search software to interpret tandem mass spectra. To evaluate the limitations of database composition on pathogen identification, we analyzed proteins from cultured Ustilago maydis, Phytophthora sojae, Fusarium graminearum, and Rhizoctonia solani by LC-MS/MS. When the search database did not contain sequences for a target pathogen, or contained sequences to related pathogens, target pathogen spectra were reliably matched to protein sequences from nontarget organisms, giving an illusion that proteins from nontarget organisms were identified. Our analysis demonstrates that when database-search software is used as part of the identification process, a paradox exists whereby additional sequences needed to detect a wide variety of possible organisms may lead to more cross-species protein matches and misidentification of pathogens.
Keywords: Pathogen detection, Plant pathology, Protein biomarker, Protein identification
1 Introduction
Disease diagnosis is critical to protecting agricultural crops and preventing pathogen spread. For example, damage by soybean rust in the US could be far worse if not for the cooperative efforts for disease surveying and pathogen screening [1]. In this case, scouting teams are deployed to survey cropland and surrounding areas. Symptomatic plants are evaluated in diagnostic labs where microscopes are used to assess potential infections and PCR is used to amplify soybean rust-specific DNA [2]. Reports of positive findings are sent back to scientists and growers who work together to develop disease progression models and prepare a response plan that may include spraying fungicides. A correct diagnosis is required for the fungicide application to be cost-effective. A misdiagnosis, such as mistaking soybean rust for brown spot, could cost a grower any profit for the year.
Plant diseases tend to be particularly challenging to diagnose quickly and efficiently. Typically, pathogens are characterized by morphological identification and symptom evaluation. However, these visual methods are slow and prone to subjectivity. It is easy to mistake a rust pustule for a brown spot and incorrectly assume that Phakopsora pachyrhizi, the fungus causing soybean rust, is present rather than Septoria glycines, the fungus causing brown spot. To be certain, it may take many months to conclusively identify a pathogen by symptom evaluation, culturing, and microscopy.
By contrast, molecular methods of detection such as PCR and ELISA are objective and can be fast, precise, and economical. However, these methods can only identify the pathogen for which the assays were designed. Because several thousands of possible pathogen-specific PCR primers or antiserum reagents might be needed to identify a pathogen causing a yellow leaf spot symptom on a tomato leaf, it is often difficult to decide which set of primers to test first and which set of primers to use next if the first fail. It is equally difficult to obtain an arsenal of antibodies sufficient for detecting a wide range of pathogens.
Consequently, plant pathologists would benefit by having advanced techniques that may be better suited for the identification of an unknown causal agent of a disease. MS holds such potential for pathogen screening since no pathogen-specific reagents are required to measure the mass of a molecule or the masses of its fragments [3]. Such an advantage brings versatility to the disease screening process and could save time, money, and resources. Although a wide variety of characteristic molecules such as lipids, phospholipids, carbohydrates, or metabolites can be detected by MS, proteins are well-suited for being highly specific biomarkers because they confer indirect species–specific genetic information in their sequences. Specific proteins and their post-translational products identified by MS or MS/MS have been shown by some researchers to be excellent indicators of the presence of animal and plant viruses and bacterial pathogens [4, 5]. We also have experimented with using MS/MS as a detection tool and have used it to characterize a virus previously unidentifiable by symptom evaluation or PCR [6]. The MS/MS detection of distinct peptides belonging to the viral coat protein of potato virus X demonstrated the capability of MS/MS methods to identify the unknown cause of a disease.
As a result of these successful studies, we wish to extend this diagnostic application of MS/MS to identify fungal plant pathogens which are wide-spread, detrimental to a vast array of crops, and sometimes difficult to diagnose. In our prior efforts, we used 2-D PAGE for separating proteins from an infected plant [6]. Proteins were cut from the gels and their peptides were analyzed by MS/MS. Because gels are labor intensive and not suited for quick diagnostic turnaround, we have turned to a more amenable high-throughput protein separation method where we first digest proteins and then separate their peptides by LC before analyzing them by MS/MS (LC-MS/MS [7–12]). This high throughput workflow has been used by other groups to identify characteristic peptides from bacterial pathogens [13–15]. Unlike the 2-D PAGE method where select proteins are analyzed, LC-MS/MS allows for the screening of a complex number of pathogen, host and background proteins in a sample in a single assay. The resulting MS/MS spectra are interpreted with database-search software that compares experimentally acquired spectra to a database of protein sequences [16]. Matches of spectra to peptides from pathogen proteins has been interpreted to imply the presence of the pathogen in a sample [6, 13–15].
To test the feasibility of using LC-MS/MS applications for the identification of multicellular plant pathogens, we have performed a proteomic analysis on three cultured fungi and one oomycete plant pathogen with varying amounts of protein sequence representation in the database used to interpret the spectra. Theoretically, if we can successfully match spectra obtained from pure cultures to the protein sequences of the respective pathogens, we then have reason for further examination of real samples containing complex mixtures of both plant and pathogen proteins. Instead, our results show that the varying degrees of publicly available genome and protein sequence information available for each organism greatly impact and confound our ability to specifically map the resolved spectra back to the target cultured organisms. These results cast doubt on the extended use of database-search programs to interpret tandem mass spectra collected from samples infected with unknown pathogens for which little protein sequence information exists.
2 Methods
2.1 Pathogens and growth conditions
Fusarium graminearum isolate CBS 110246 and Phytophthora sojae isolate CBS 418.91 were obtained from the CBS Fungal Biodiversity Center (Utrecht, The Netherlands). Rhizoctonia solani and Ustilago maydis strain 521 were generously provided by Dr. Dilip Lakshman, Floral & Nursery Plants Research Unit, USDA-ARS (Beltsville, MD) and Dr. Scott Gold, Department of Plant Pathology, University of Georgia (Athens, GA), respectively. F. graminearum and R. solani were both grown on sterile 0.45 μm Protran BA85 NC membranes (Whatman Schleicher & Schuell, Florham Park, NJ) that had been placed on the surface of potato dextrose agar (PDA) media. P. sojae was grown under the same conditions except that V8 tomato juice agar was used in place of PDA. U. maydis 521 was initially streaked across a PDA plate. After allowing U. maydis 521 to grow sufficiently, a transfer was made from the plate to a plastic vial containing 5 mL of potato dextrose broth (PDB) (BD, Franklin Lakes, NJ) using a sterile inoculating loop. After 48 h, the culture was used to inoculate 500 mL PDB. U. maydis 521 was purified from the PDB medium by centrifugation. The pathogenicity of F. graminearum, P. sojae, and R. solani was confirmed by infecting Triticum aestivum cv. Norm spikelets, Glycine max cv. Williams roots and G. max cv. Williams germinating seedlings, respectively, and evaluating characteristic disease symptoms.
2.2 Protein purification
Three experimental replicates were evaluated for each of the four pathogens. Each plant pathogen sample was cooled using liquid N2 and then pulverized with a mortar and pestle. Proteins from each pathogen were precipitated in acetone/TCA, resolubilized in 8 M urea/100 mM Tris-HCl pH 8.5, reduced in trichloroethylphosphine (TCEP) and carboxyamidomethylated in iodoacetamide [17]. The Micro BCA Protein Assay Reagent Kit (Pierce Biotechnology, Rockford, IL) was used to determine protein concentration. Before digesting 1 mg soluble protein with Porozyme immobilized trypsin (Applied Biosystems, Foster City, CA) at 37°C for 12 h, the protein solution was diluted to 2 M urea with 100 mM Tris-HCl pH 8.5 and adjusted to 2 mM CaCl2[17]. Immobilized trypsin was removed by centrifugation. All peptides were separated from debris using 0.45 μm PVDF filters (Millipore, Bedford, MA) and were concentrated by SPE using SPEC-PLUS PT C18 columns (Varian, Lake Forest, CA) followed by centrifugal vacuum evaporation.
2.3 Peptide separation
Columns were prepared from 365 μm od×75 μm id fused-silica capillaries (Polymicro Technologies, Phoenix, AZ). Each capillary was drawn to a 5 μm tip using a P-2000 laser puller (Sutter Instrument Company, Novato, CA) [18]. Then, 9cm of 5 μm Aqua RP C18 resin (Phenomenex, Torrance, CA) followed by 4 cm of 5 μm Luna strong cation exchange resin (Phenomenex) were packed under 600 psi using a helium pressure cell [19]. The peptides were also loaded onto the packed column using the same pressure cell. The loaded column was placed in-line with a Surveyor HPLC pump that is part of the ProteomeX workstation (Thermo Fisher Scientific, Waltham, MA). Peptides were eluted in a 12-step process that included increasing concentrations of salt followed by an increasing gradient of mobile-phase at each step [17]. The eluent was introduced directly into the LCQ-Deca XP IT mass spectrometer via an ESI source. Electrospray voltage (1.8 kV) was applied at a liquid junction before the column via a gold electrode.
2.4 MS/MS and spectra extraction
A parent ion scan was performed over the range 400–1600 m/z. Automated peak recognition, dynamic exclusion, and MS/MS ion scanning of the top three most intense parent ions were controlled by Xcalibur 1.3 (Thermo Fisher Scientific). The tandem mass spectra were extracted from the Xcalibur .raw files and converted into .dta files using Bio-works 3.1 (Thermo Fisher Scientific). Parameters were set at: 400 minimum mass, 3500 maximum mass; 15 minimum ion count; 100 000 minimum TIC; 1.4 Da precursor mass tolerance; and 1 group scan. All spectra not calculated as being singly charged were extracted as both doubly- and triply-charged spectra.
The MS/MS spectra represented as .dta files were analyzed using PepNovo to identify spectra potentially representing peptides from those of insufficient quality (for example, those from nonpeptide ions). PepNovo uses a probabilistic model to assess peptide fragmentation and assigns high-scoring short peptide sequences called tags to the peaks most likely to represent peptide fragmentation [20, 21]. PepNovo was set to evaluate spectra for three amino-acid tags. Spectra having a top-scoring tag exceeding the probability threshold of 0.80 were used for subsequent analysis. A Perl script, merge.pl that is part of the MASCOT 2.1 software package (Matrix Science, London, UK) was used to convert subset .dta files into a single file suitable for searching.
2.5 Peptide sequence identification
The MS/MS spectra were evaluated with MASCOT (version 2.1, Matrix Science) [16] and searched against sequences in the 07/06/2006 release of the NCBI nonredundant (NCBInr) protein database (3 782 570 sequences; ftp://ftp.ncbi.nih.gov/blast/db/FASTA/). Additional protein sequence records for P. sojae (19 027; http://genome.jgi-psf.org/Physo1_1/Physo1_1.download.ftp.html), P. ramorum (15 743; http://genome.jgi-psf.org/Phyra1_1/Phyra1_1.download.ftp.html), and F. graminearum (14 021 sequences; ftp://ftpmips.gsf.de/fusarium/), were appended to NCBInr as described in Section 3. Searches were performed on a seven node 3.2 GHz Dell server with Xeon processors. The search was configured to assume a tryptic digest, and one missed cleavage per peptide was permitted; carboxyamidomethylation was selected as a fixed mass modification; oxidation (M), was set as a variable mass modification. Monoisotopic mass values were used, and peptide mass tolerance and fragment mass tolerance were set at ±1.5 Da and ±0.8 Da, respectively.
2.6 Protein assembly
MASCOT.dat files containing results from MASCOT searches were uploaded to PANORAMICS, a web-based C program that assembles proteins from shotgun proteomics data by making use of peptide assignments acquired from MASCOT output files [22, 23]. By applying a rigorous probability model, PANORAMICS derives consistent confidence measures indicating that protein assemblies from peptides given a search database are correct. The probabilities can be used to determine the false positive rate of protein identification. When computing the probability of identifying a protein, the program takes into account both distinct peptides and shared peptides in a coherent manner by distributing the probabilities of shared peptides among all related proteins. By applying an iterative approach, the distribution of probabilities benefits those proteins that are more likely to be present in the sample. The MASCOT Ions score, the database size, number of candidate peptides in the database with a similar m/z ratio, the length of the peptide sequence and charge state of the ion are incorporated into the probability model. Proteins exceeding a 99.9% probability threshold were kept for evaluation. Peptide sequences, associated MASCOT Ions scores, protein groups and their probabilities are available as Supporting Information for each data set.
3 Results
3.1 Choice of plant pathogens, search databases, and chosen procedures
We were interested in evaluating LC-MS/MS as a method for identifying fungal and oomycete plant pathogens. For this pilot project, we focused our attention on U. maydis, P. sojae, F. graminearum, and R. solani. These pathogens were selected because: (i) pure cultures could be obtained, (ii) they are taxonomically distinct, (iii) they attack economically important crops, (iv) they infect different parts of plants, and (v) there is variation in the amounts of available genome and protein sequence information available for these pathogens. Specifically, U. maydis is a Basidiomycetes fungus that infects maize kernels [24] and its genome has been sequenced and well-annotated (http://www.broad.mit.edu/annotation/genome/ustilago_maydis/Home.html). F. graminearum is an Ascomycetes fungus that causes wheat head blight [25] and P. sojae is an Oomycetes pathogen that causes root and stem rot on soybean [26]. The whole-genome sequence for both F. graminearum and P. sojae is quite extensive and can be considered nearly-complete (http://www.broad.mit.edu/annotation/genome/fusarium_grami nearum/Home.html and [27], respectively). The Basidiomycetes R. solani is a soil-borne pathogen that causes root rot in a broad host range of plants that includes maize, rice, bean, and sorghum [28]. We are not aware of any ongoing genome sequencing efforts for this organism.
To initiate our studies, we extracted proteins from three separate cultures for each organism. The proteins were digested and the peptides analyzed by LC-MS/MS. We then compared MS/MS spectra to a database of protein sequences using MASCOT, a common database-search algorithm. We searched the large NCBInr database rather than a subset database of specific target pathogen sequences because a sizeable database like NCBInr would be needed for real applications in order to detect a wide range of possible pathogens or differentiate between pathogen proteins and other proteins present in the background environment. Within our version of NCBInr, there were 6664 U. maydis and 11 830 F. graminearum protein records to which exact matches could be made (Table 1). Despite the genome sequence coverage for P. sojae [27], only 133 protein records existed for it, which meant that the protein sequences from the gene models from the sequencing effort had not been released when we acquired our version of NCBInr (Table 1). Finally, very little sequence data exists for R. solani and only 37 protein records for this pathogen were in NCBInr (Table 1). We will show in the following subsections the impact of these records on protein identification from the target pathogens. For our controlled study, there theoretically should be no plant proteins and few background proteins other than contaminating human proteins and trypsin that we experimentally introduced. Ideally, matches should be specific to proteins from the target organism.
Table 1.
Summary of the number of nonredundant proteins that were identified from four plant pathogens
| Target plant pathogen | Database | Database records for target plant pathogen | Total NR proteins (99.9% confidence) | NR proteins matching target pathogen | NR proteins matching another species of target pathogen | NR proteins matching nontarget species | # Matching several species (ambiguous for target pathogen identification) |
|---|---|---|---|---|---|---|---|
| U. maydis | NCBInr | 6664 (6662 RefSeq) | 434 | 417 | 0 | 10 | 7 |
| P. sojae | NCBInr | 133 | 79 | 4 | 36 | 23 | 16 |
| P. sojae | NCBInr + JGI | 19 160 | 457 | 433 | 8 | 4 | 12 |
| P. sojae proteins | |||||||
| P. sojae | NCBInr + JGI | 19 160 | 471 | 369 | 28 | 4 | 70 |
| P. sojae + 15 743 JGI | |||||||
| P. ramorum proteins | |||||||
| F. graminearum | NCBInr | 11 830 (11 638 PH-1 RefSeq) | 26 | 23 | 0 | 0 | 3 |
| F. graminearum | NCBInr + MIPS | 25 851 | 27 | 24 | 0 | 0 | 3 |
| F. graminearum | |||||||
| PH-1 proteins | |||||||
| R. solani | NCBInr | 37 | 13 | 1 | 0 | 7 | 5 |
As was previously described, it is current practice to match an MS/MS spectrum to a peptide sequence and scan the descriptive information linked to the associated protein record to determine the organism of origin for the protein record [6, 13–15]. Typically, this information can be used to imply which organisms’ proteins are present in a sample. However, there are three constraints to this workflow that must be addressed. First, while there are distinct peptide sequences that uniquely identify one organism's protein, there are peptides sequences that are shared among proteins from varied organisms [29]. Thus, distinct and shared proteins must be distinguished because the detection of shared peptides can be falsely interpreted to imply proteins from multiple organisms were present when in fact there were proteins from only one organism present in a sample.
Second, several steps in the database searching process are subject to generating misleading information. Statistically significant false spectrum/peptide sequence matches can be made, even when a database contains sequences specific to a target protein [30, 31]. Since a large database composed of proteins from a wide variety of organisms needs to be searched in order to detect as many potential pathogen and background proteins as possible, false spectrum/peptide matches have the chance of occurring more frequently [32]. Such false matches may affect downstream target pathogen identification. Nevertheless, the rates of false matches can be modeled [16, 31, 33], and a probability or score can be assigned to the resulting peptide sequence indicating the likelihood that a spectrum was generated by a peptide from the database [16, 34–36]. Thus, this information must be considered to make spectrum/peptide sequence matches reliably.
Third, database-search algorithms like MASCOT can effectively match MS/MS spectra from one organism to distinct peptide sequences from related organisms. In some cases, protein homology has been exploited to identify proteins in nonmodel organisms using model organism protein sequence reference databases [17]. This feature has been referred as cross-species protein matching [37]. Thus, statistically significant matches can be made to distinct peptides from organisms other than the target and this could negatively impact target pathogen identification.
We have made some recent advances to counteract some of these issues which influence our ultimate goal of determining whether a protein that is specific to a target pathogen is present in a sample. To simplify data analysis, we used a software program that effectively distinguishes distinct and shared peptides and then organizes the matched sets of peptides into a parsimonious set of protein complements [22, 23]. Distinct peptides act as anchors for protein identification and a probability model distributes the contribution that shared peptides make to any associated proteins. In the event that the same set of matched peptide sequences is associated with several proteins, the protein records are grouped. The end result of using this program is the assembly of a set of matched peptides belonging to a protein or set of proteins, and a probability score indicates the likelihood that the assembly is correct with respect to the records in the database. Proteins with a high probability are the most likely to be have been identified by the given peptides. Subsequently, the record information from probable proteins can be used as a starting point for indicating the presence of a target pathogen in a sample. A beneficial side-effect of this program is that probability scores also reflect the false-positive rates. A high threshold can be set such that false spectrum/peptide matches have little impact on the probability-based assembly of proteins.
Consequently, the main consideration to address experimentally is the effect of varying amounts of protein sequence information in the database needed to sufficiently and correctly identify spectra from different pathogens. We are particularly interested in knowing whether homology between related proteins from different organisms will influence spectrum/peptide sequence matches, and how this will affect protein identification and target pathogen detection.
3.2 Potential to identify fungal plant pathogens with well-characterized genomes
For U. maydis, a total of 105 500 MS/MS spectra were collected from three separate experiments. We sought to create a “high-quality” subset of these spectra and did so by selecting spectra with the property of having attributes of being derived from peptides. The purpose for this was two-fold. First, even when peptides are analyzed by MS/MS, many of the tandem mass spectra that are collected are products of other ionized nonpeptide molecules (e.g., polymers from the tubing) or poorly fragmented peptides [38]. Thus, by removing spectra that are poor candidates for resolving peptide sequences, we can improve peptide and protein identifications across the board. The second reason for choosing peptide-centric spectra is to ensure quality across datasets. The importance for this will be made apparent in the later sections.
While there are a number of other different algorithms that can be used to assess mass spectral data quality [39–43], we used the PepNovo algorithm to evaluate whether the mass spectral dataset comprised spectra derived from peptides. PepNovo examines each spectrum and reports a probability for a three-amino-acid-residue sequence tag inferred from the fragment ions [20, 21]. To test this application, we used PepNovo to score all of the primary U. maydis spectra. 59 555 out of 105 500 spectra (56.5%) scored 0.80 or greater. We then searched the primary set and the peptide-centric subset against NCBInr using MASCOT and assembled proteins from the peptide sequence information with PANORAMICS. For the primary set, 796 proteins were assembled at the 95% confidence level and 428 proteins were assembled at the 99.9% confidence level. By comparison, 41 fewer proteins were assembled at the 95% confidence level for the subset, but six more proteins were detected at the 99.9% confidence level. Fewer proteins were assembled from the subset at the 95% level because peptide tag filtering eliminated some of the lower quality spectra that lead to lower confidence matches to peptide sequences. For the same reason, a few more proteins were detected at the higher confidence level from the peptide-centric subset spectra because PANORAMICS protein group probabilities received higher scores since there were fewer low scoring peptide matches to suppress the probability for the protein group. These results indicate that PepNovo can sufficiently identify peptide-centric tandem mass spectra as evidenced by the high-quality peptide sequence matches made by MASCOT and the consistent number of proteins identified even after filtering.
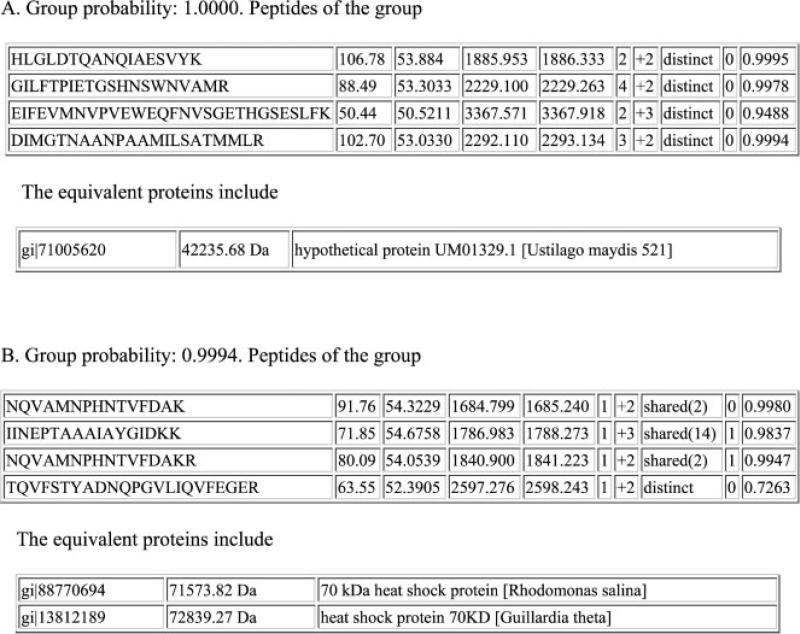
When performing typical proteomic identification, assembled proteins equal to or exceeding a 95% probability are kept for analysis [23]. We believe that a much higher threshold is required for diagnostic specificity. So, a very high probability threshold of 99.9% was chosen, meaning the error-rate for an incorrect protein assembly from the given peptides is less than or equal to 0.1%. This strict criterion ensured that proteins were assembled from two or more distinct peptides or (in a few cases) assembled from a distinct peptide with a very high MASCOT Ions score in relation to the MASCOT Identity score. As a result, 434 protein groups were identified from the filtered U. maydis dataset at the 99.9% level (Table 1; Supporting Information S1). An example of the data used in a U. maydis protein assembly is shown in Fig. 1A. In this example, four distinct peptides from protein record gi|71005620 were resolved by MASCOT. PANORAMICS grouped these peptides together to show with certainty that these peptides identify protein gi|71005620. The information in the header of the record indicates that the protein from which this record was derived came from U. maydis. Thus, this finding implies that the spectra collected were derived from U. maydis and that this U. maydis protein was present in the sample.
Figure 1.
Sample protein assembly data from PANORAMICS. (A) Peptides identified by MASCOT, all from a single U. maydis record (gi|71005620) from NCBInr. The probability for this match is 1.0000. (B) Peptides identified by MASCOT, all found in the same two records from NCBInr matched with equal probability of 0.9994. The columns are arranged as follows: peptide sequence, MASCOT Ions score, MASCOT Identity score, computed peptide mass, observed precursor mass, number of tandem mass spectra assigned to same peptide sequence, the charge states observed for the peptide, whether the peptide is shared between protein groups with different probabilities (number in parentheses is number of other groups peptides are shared with) or is distinct (considered a unique identifier for proteins grouped with the same probability), the number of missed tryptic cleavages in this sequence, and the probability for a particular peptide sequence.
Interestingly, 417 (96.1%) of the proteins detected were assembled using information from U. maydis records (Table 1), while ten proteins were matched to records from other organisms. Two of those were to bovine trypsin, a protein added during sample processing. However, matches were made to proteins specific to other nontarget Basidiomycetes fungi such as Cryptococcus neoformans and Phanerochaete chrysosporium. In addition, there were seven sets of proteins from multiple organisms identified by the same sets of matched peptides (Fig. 1B). Because these peptides could not be mapped to a protein from any one organism, we classified these protein groups as ambiguous for pathogen identification. For both the ambiguous groups and the nontarget matches, it could be argued that principles enabling cross-species protein matching led to the correlation between U. maydis peptide tandem mass spectra and protein database records from nontarget organisms.
On the positive side, 417/434 proteins matched records for the target pathogen, meaning that it is possible that these could be used with a high degree of confidence for indicating the presence of U. maydis in a sample. However, since diagnosis depends on highly confident identifications, we are concerned that ~3.0% of the protein identifications were made using protein records from nontarget organisms. It is possible that this information could be mistakenly interpreted to mean that these nontarget organisms were present in the sample.
Perplexing as it seems, there are several explanations why peptide tandem mass spectra from U. maydis match proteins to nontarget organisms. One reason is that the mass spectrometer that we used achieves high-throughput at the cost of low resolution and mass accuracy. While the former property makes these instruments very sensitive for screening, the latter can reduce the confidence of identification relative to instruments that achieve tens-of-ppm resolution (such as TOF/TOF analyzers). Since the NCBInr database contains many orthologous, paralogous, and homologous sequences to the U. maydis proteins, there are ample opportunities for an incorrect sequence to out-compete the true sequence when evaluating a spectrum with low mass resolution [44]. Spectra acquired when peptides are present at low concentrations or that represent nonrandom peptide fragmentation (despite PepNovo filtering) can also result in spurious peptide identifications from organisms other than the true one [45].
Mutations and modifications may also produce peptides that are misidentified by database-search software. Natural genetic variation may yield variant proteins, potentially generating peptides whose spectra match better to sequences from related organisms. For the U. maydis study, this is probably a negligible effect since we examined the strain whose genome was sequenced. Meanwhile, a protein modification that is left out of the search configuration may cause a spectrum to not match its true sequence in the database [46]. Instead, a match is made to another organism's peptide sequence whose ion fragments are similar to the set produced from the true peptide. These changes may also lead to species misidentification. All together, we suspect that spectral variation and large search databases such as NCBInr containing multiple candidate sequences inevitably resulted in viable matches being made to proteins from nontarget organisms.
Some of these suspicions were confirmed when we examined P. sojae. Although the genome for P. sojae is sequenced [27], only 133 protein records existed for this pathogen in the version of NCBInr that we tested. Consequently, only 79 protein identifications were made at the 99.9% confidence level for 97 736 tagged MS/MS spectra (Supporting Information S2). More importantly, only 5% of those matches (4/79) were made to P. sojae protein sequence records (Table 1). This is in stark contrast to U. maydis where ~96% of the matches were made to target U. maydis records. Surprisingly, more spectra matched sequences from nontarget but related Phytophthora species. Twenty-seven protein records from P. infestans and nine from P. cinnamomi, P. nicotianae, P. palmivora, P. parasitica, and P. megasperma were matched. Finding more matches to Phytophthora spp. other than P. sojae might be explained by the fact that in NCBInr there were 640 records for other Phytophthora spp. in contrast to 133 records for the target P. sojae. We suspect that the additional homologous records afforded more opportunity for MASCOT to make cross-species protein matches. Likewise, 23 matches were made to pathogens other than Phytophthora spp., such as Pythium spp. Taken at face value, these initial results mistakenly imply that many different Phytophthora spp. or other Oomycetes pathogen proteins were in the samples.
It appeared from these results that the lack of target sequences for P. sojae allowed MASCOT to make matches to protein sequences from the other species. To test this hypothesis, we added to NCBInr 19 027 P. sojae protein records derived from the P. sojae genome sequence. This resulted in many more total protein matches being made, 457 at a 99.9% confidence level to be exact, of which 433 were specific to the P. sojae protein records (Table 1; Supporting Information S3). Also, 21 fewer matches were made to P. infestans protein records, indicating the benefits of having information specific to the target pathogen in the database. However, because prior results suggested that a high number of nontarget matches could be made to organisms whose records were more abundant than the target, we added to NCBInr an additional 15 743 nontarget P. ramorum records (derived from its genome sequence [27]) alongside the 19 027 P. sojae records. Using this database, 471 matches were made at a 99.9% confidence level but only 369 were specific to P. sojae (Table 1; Supporting Information S4). The number of specific matches to P. ramorum records increased to 21 and the number of matches ambiguous for identification jumped to 70, most of which were a product of detecting the same sets of matched peptide sequences from homologous P. sojae and P. ramorum protein records. Thus, we conclude that the additional P. ramorum sequences led to a reduction in matches being specifically made to P. sojae records. These results suggest that the addition of nontarget pathogen sequences to a database can increase the rate of nontarget protein identifications.
3.3 Plant pathogens that were difficult to detect
For the next pathogen, F. graminearum, a total of 114 605 MS/MS spectra were collected from three replicate experiments. Peptide tag filtering reduced the set to 62 968 spectra. At the 99.9% confidence level, only 26 protein identifications were made, of which 23 were made to records for F. graminearum (anamorph Gibberella zeae; Table 1, Supporting Information S5). Three of the protein identifications were classified as ambiguous for pathogen identification. Although most of the matches were specifically made to F. graminearum records, we were surprised that so few proteins were identified especially since NCBInr contains a whole complement of RefSeq protein records derived from the F. graminearum genome. By comparison, we identified several hundred proteins from U. maydis and P. sojae samples when genome-derived protein records were present in the search database. Since our spectral quality analysis suggested that nearly the same number of peptide-centric spectra were in the F. graminearum dataset as in the U. maydis dataset, it does not appear that there was any direct correlation between the number of proteins identified and the availability of peptide-centric MS/MS spectra for analysis.
Most likely there are other explanations such as incorrect database records, unique protein variation between the sequenced strain and our isolate, or unpredicted peptide/protein modifications that account for our inability to identify many proteins from F. graminearum. A recent report suggests that there is variation between the 11 638 Broad Institute-derived F. graminearum RefSeq gene models in NCBInr compared to another set generated by the Munich Information Center for Protein Sequences (MIPS) [47]. It is possible that incorrect exon calls could greatly impact the ability of MASCOT to make matches between F. graminearum spectra and these records. To evaluate this further, we appended 14 021 MIPS gene models (ftp://ftpmips.gsf.de/fusarium/Fgraminearum_valid, 8/29/06 release) to NCBInr, and repeated the MASCOT searches and assembly of proteins. Only one additional protein was identified (Table 1). This suggests that differences between the gene models had very little bearing on protein identification. Therefore, other factors must account for the low resolution of F. graminearum proteins. It is possible that there is a significant amount of variation between our F. graminearum isolate belonging to lineage 1 and the sequenced strain, PH-1, which belongs to lineage 7 [48]. It appears that limited gene flow exists in the F. graminearum clade and that reproductive isolation of these lineages could portend allopatric speciation [48]. These differences could partially explain why we were not able to make a larger number of protein identifications using the sequences from PH-1.
Nevertheless, some spectra from our lineage 1 strain were matched using sequences from PH-1, lineage 7. Taken at face value, this data could be mistakenly interpreted to mean that the sequenced strain was detected. Consequently, it appears that using another isolate's reference sequences is problematic for strain identification because mismatches and misidentifications can be made. In order, to confidently identify strains, increased sequence coverage for all strains is needed. However, we suspect trouble using database-search algorithms to unambiguously match spectra to protein records without having sequence representation for all strains and taxonomic groups. Our prior observation that nontarget matches were made between P. sojae spectra and P. ramorum sequences supports our suspicion.
Problems with cross-species protein matching became more apparent when we examined R. solani, a pathogen poorly represented in NCBInr. We managed to identify only 13 proteins at the 99.9% confidence level from the 59 889 tagged MS/MS spectra collected. Only one protein matched a record specific to R. solani (anamorph Thanatephorus cucumeris; Table 1, Supporting Information S6). Of the remaining proteins, seven cross-species protein matches were made to protein records from nontarget but related Basidiomycetes fungi such as the plant pathogens U. maydis and Puccinia graminis, as well as other nonplant pathogens such as C. neoformans, Lentinula edodes, and Volvariella volvacea. Matches were also made to records from the fungi Aspergillus fumigatus and Candida albicans. There were also five protein matches ambiguous for pathogen identification.
Tag-filtering indicated that the R. solani spectral dataset contained as many peptide-centric MS/MS as the U. maydis, P. sojae and F. graminearum spectral subsets. Thus, it is reasonable to believe that it was the extremely limited representation of the R. solani proteome in the NCBInr database that led to matches being made to protein sequences from nontarget organisms. Certainly, it is reasonable to believe that it should be difficult to identify proteins for organisms poorly represented in NCBInr, especially given the dependent nature of database-search algorithms on these databases. However, it appears that valid matches can also be made to proteins from nontarget organisms, which means that data can be misconstrued to suggest that nontarget pathogens are present when they really are not.
4 Discussion
Our initial goal was to determine whether LC-MS/MS could be used to identify peptides and proteins from complex plant pathogens as part of a screening process. We were successful in detecting proteins from target pathogens with sequenced genomes. However, we discovered that database-search algorithms can make cross-species protein matches to records from multiorganism databases. We have shown how the addition of records from a nontarget but related pathogen influenced the interpretation of spectra from P. sojae, a pathogen with a sequenced genome, and how the lack of specific records negatively impacted the identification of proteins from R. solani, a pathogen for which little sequence information exists. This points to trouble when analyzing a sample containing a pathogen with poor sequence representation—database-search programs may reliably match spectra from one organism to sequences from another organism thereby leading to false-positive identification. Simply increasing the numbers of protein records represented for target pathogens will likely not ameliorate the problem since this increases database record variation which throws off spectrum matches to protein records of target organisms. We suspect that LC-MS/MS analysis on a real-world sample that includes pathogen, host and background proteins will become ever more complicated if identification is predicated on the use of database-search software.
Other laboratories have evaluated LC-MS/MS for the detection of peptides unique to biological warfare agents. We are all pursuing the common goal of being able to detect the broadest-range of threatening organisms that might be present in an unknown environmental sample without having to use pathogen-derived reagents. Using a workflow similar to ours, Dworzanski et al. [14] made the case that spectra from a nonpathogenic bacteria could be distinguished using a database containing a variety of bacterial protein records. VerBerkmoes et al. [15] extended this technological evaluation by examining sensitivity versus protein concentration of the target organism. Each group examined the effects of a limited database size and showed that matches to proteins from nontarget organisms were possible. However, the design of the experiments and the interpretation of the results favored the successful detection of the target organisms, thereby indirectly supporting the continued use of database-search software to make peptide sequence-spectrum matches.
By comparison, our study addresses some of the more realistic scenarios not considered in the other studies. First, we differentiate ourselves by searching all of NCBInr, a very large database, because such a large database would have to be searched if one were really trying to identify the unknown cause of a disease. Second, we examine the effect of having a microorganism in the sample whose proteome is not well represented in the database. Third, we examine a much more diverse group of microorganisms. Given our detailed analysis using different microorganisms with varied levels of sequence coverage and examining the outcomes of peptide-spectrum matching using a large, but incomplete database, we do not believe that database-search algorithms are entirely sufficient for interpreting tandem mass spectra when the goal is pathogen identification because cross-species protein matches potentially lead to ambiguous or misidentification of organisms even when the peptide matches and protein identifications are deemed to be very accurate. In other words, results can be easily misinterpreted to suggest a protein from an organism was present in a sample when really a protein record just contributed a sequence that was matched to a spectrum. In conclusion, our findings contradict those of other researchers who claim to have successfully identified microorganisms using an LC-MS/MS workflow that depends on database-search algorithms for the interpretation of spectra.
Clearly, when database-search software is part of the LC-MS/MS scheme, there is a paradox whereby a taxonomically diverse database that is needed to identify a broad range of target pathogens allows for more cross-species protein matches and misidentifications. However, this does not mean that LC-MS/MS cannot be accurately used for pathogen identification. Rather, spectra derived from pathogens should be utilized differently. Instead of using protein databases to infer peptide sequence information from spectra, we propose allowing uninterpreted tandem mass spectra to stand alone as biomarkers for pathogen identification. We have already shown here that peptide-centric tandem mass spectra can be generated from pathogens. These spectral datasets could be assembled into searchable reference libraries. Direct spectrum/spectrum comparisons between spectra from a sample and those in pathogen-specific MS/MS reference libraries may enable more reliable identification and fewer false-positive or nontarget matches. This method would be similar to the one used throughout the world to identify chemical compounds or akin to other proposed spectral matching methods for the identification of peptides [49–53]. Hence, conversion to a spectral library generation and spectral searching system may eventually extend use of LC-MS/MS for the identification of plant and animal pathogens beyond those whose genomes have already been sequenced.
Supplementary Material
Acknowledgments
We thank Drs. Dilip Lakshman, USDA-ARS, Beltsville, MD, and Scott Gold and Steven Klosterman, University of Georgia, Athens, GA, for providing us with fungal isolates, Dr. Steve Stein, NIST Mass Spectrometry Data Center, Gaithersburg, MD for guidance. D.L.T. was supported by NIH P30 ES000267 and NIH 1 R01 CA126218-01. This work was funded in part by a National Research Initiative grant award to B.C. from USDA-CSREES (no. 2005-35605-15392).
Abbreviations
- MudPIT
multidimensional protein identification technology
- NR
nonredundant
- PDA
potato dextrose agar
- PDB
potato dextrose broth
5 References
- 1.Stokstad E. Plant pathologists gear up for battle with dread fungus. Science. 2004;306:1672–1673. doi: 10.1126/science.306.5702.1672. [DOI] [PubMed] [Google Scholar]
- 2.Lamour KH, Finley L, Snover-Clift KL, Stack JP, et al. Early detection of Asian soybean rust using PCR. Plant Health Prog. 2006 DOI: 10.1094/PHP-2006-0524-01-RS. [Google Scholar]
- 3.Padliya ND, Cooper B. Mass spectrometry-based proteomics for the detection of plant pathogens. Proteomics. 2006;6:4069–4075. doi: 10.1002/pmic.200600146. [DOI] [PubMed] [Google Scholar]
- 4.Dworzanski JP, Snyder AP. Classification and identification of bacteria using mass spectrometry-based proteomics. Expert Rev. Proteomics. 2005;2:863–878. doi: 10.1586/14789450.2.6.863. [DOI] [PubMed] [Google Scholar]
- 5.Fenselau C, Demirev PA. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom. Rev. 2001;20:157–171. doi: 10.1002/mas.10004. [DOI] [PubMed] [Google Scholar]
- 6.Cooper B, Eckert D, Andon NL, Yates JR, et al. Investigative proteomics: Identification of an unknown plant virus from infected plants using mass spectrometry. J. Am. Soc. Mass Spectrom. 2003;14:736–741. doi: 10.1016/S1044-0305(03)00125-9. [DOI] [PubMed] [Google Scholar]
- 7.Evans CR, Jorgenson JW. Multidimensional LC-LC and LC-CE for high-resolution separations of biological molecules. Anal. Bioanal. Chem. 2004;378:1952–1961. doi: 10.1007/s00216-004-2516-2. [DOI] [PubMed] [Google Scholar]
- 8.Hunt DF, Buko AM, Ballard JM, Shabanowitz J, et al. Sequence analysis of polypeptides by collision activated dissociation on a triple quadrupole mass spectrometer. Biomed. Mass Spectrom. 1981;8:397–408. doi: 10.1002/bms.1200080909. [DOI] [PubMed] [Google Scholar]
- 9.Hunt DF, Yates JR, III, Shabanowitz J, Winston S, et al. Protein sequencing by tandem mass spectrometry. Proc. Natl. Acad. Sci. USA. 1986;83:6233–6237. doi: 10.1073/pnas.83.17.6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Link AJ, Eng J, Schieltz DM, Carmack E, et al. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 11.Washburn MP, Wolters D, Yates JR., III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 12.Wolters DA, Washburn MP, Yates JR., III An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 13.Dworzanski JP, Deshpande SV, Chen R, Jabbour RE, et al. Mass spectrometry-based proteomics combined with bioinformatic tools for bacterial classification. J Proteome Res. 2006;5:76–87. doi: 10.1021/pr050294t. [DOI] [PubMed] [Google Scholar]
- 14.Dworzanski JP, Snyder AP, Chen R, Zhang H, et al. Identification of bacteria using tandem mass spectrometry combined with a proteome database and statistical scoring. Anal. Chem. 2004;76:2355–2366. doi: 10.1021/ac0349781. [DOI] [PubMed] [Google Scholar]
- 15.Verberkmoes NC, Hervey WJ, Shah M, Land M, et al. Evaluation of “shotgun” proteomics for identification of biological threat agents in complex environmental matrixes: Experimental simulations. Anal. Chem. 2005;77:923–932. doi: 10.1021/ac049127n. [DOI] [PubMed] [Google Scholar]
- 16.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 17.Cooper B, Garrett WM, Campbell KB. Shotgun identification of proteins from uredospores of the bean rust Uromyces appendiculatus. Proteomics. 2006;6:2477–2484. doi: 10.1002/pmic.200500630. [DOI] [PubMed] [Google Scholar]
- 18.Gatlin CL, Kleemann GR, Hays LG, Link AJ, et al. Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal. Biochem. 1998;263:93–101. doi: 10.1006/abio.1998.2809. [DOI] [PubMed] [Google Scholar]
- 19.Yates JR, III, McCormack AL, Link AJ, Schieltz D, et al. Future prospects for the analysis of complex biological systems using micro-column liquid chromatography-electrospray tandem mass spectrometry. Analyst. 1996;121:65R–76R. doi: 10.1039/an996210065r. [DOI] [PubMed] [Google Scholar]
- 20.Frank A, Pevzner P. PepNovo: De novo peptide sequencing via probabilistic network modeling. Anal. Chem. 2005;77:964–973. doi: 10.1021/ac048788h. [DOI] [PubMed] [Google Scholar]
- 21.Frank A, Tanner S, Bafna V, Pevzner P. Peptide sequence tags for fast database search in mass-spectrometry. J. Proteome. Res. 2005;4:1287–1295. doi: 10.1021/pr050011x. [DOI] [PubMed] [Google Scholar]
- 22.Feng J, Naimain DQ, Cooper B. A probability model for assessing proteins assembled from peptide sequences inferred from tandem mass spectrometry data. Anal. Chem. 2007;79:3901–3911. doi: 10.1021/ac070202e. [DOI] [PubMed] [Google Scholar]
- 23.Lee J, Cooper B. Alternative workflows for plant proteomic analysis. Mol. Biosyst. 2006;2:621–626. doi: 10.1039/b612326k. [DOI] [PubMed] [Google Scholar]
- 24.Basse CW. Dissecting defense-related and developmental transcriptional responses of maize during Ustilago maydis infection and subsequent tumor formation. Plant Physiol. 2005;138:1774–1784. doi: 10.1104/pp.105.061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otto CD, Kianian SF, Elias EM, Stack RW, et al. Genetic dissection of a major Fusarium head blight QTL in tetraploid wheat. Plant Mol. Biol. 2002;48:625–632. doi: 10.1023/a:1014821929830. [DOI] [PubMed] [Google Scholar]
- 26.Qutob D, Kamoun S, Gijzen M. Expression of a Phytophthora sojae necrosis-inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 2002;32:361–373. doi: 10.1046/j.1365-313x.2002.01439.x. [DOI] [PubMed] [Google Scholar]
- 27.Tyler BM, Tripathy S, Zhang X, Dehal P, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 28.Oard S, Rush MC, Oard JH. Characterization of antimicrobial peptides against a US strain of the rice pathogen Rhizoctonia solani. J. Appl. Microbiol. 2004;97:169–180. doi: 10.1111/j.1365-2672.2004.02291.x. [DOI] [PubMed] [Google Scholar]
- 29.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: The protein inference problem. Mol. Cell. Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Kapp EA, Schutz F, Connolly LM, Chakel JA, et al. An evaluation, comparison, and accurate benchmarking of several publicly available MS/MS search algorithms: Sensitivity and specificity analysis. Proteomics. 2005;5:3475–3490. doi: 10.1002/pmic.200500126. [DOI] [PubMed] [Google Scholar]
- 31.Peng J, Elias JE, Thoreen CC, Licklider LJ, et al. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: The yeast proteome. J. Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 32.Cargile BJ, Bundy JL, Stephenson JL., Jr. Potential for false positive identifications from large databases through tandem mass spectrometry. J. Proteome Res. 2004;3:1082–1085. doi: 10.1021/pr049946o. [DOI] [PubMed] [Google Scholar]
- 33.Qian WJ, Liu T, Monroe ME, Strittmatter EF, et al. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: The human proteome. J. Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 34.Geer LY, Markey SP, Kowalak JA, Wagner L, et al. Open mass spectrometry search algorithm. J. Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 35.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 36.Zhang N, Aebersold R, Schwikowski B. ProbID: A probabilistic algorithm to identify peptides through sequence database searching using tandem mass spectral data. Proteomics. 2002;2:1406–1412. doi: 10.1002/1615-9861(200210)2:10<1406::AID-PROT1406>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 37.Liska AJ, Shevchenko A. Expanding the organismal scope of proteomics: Cross-species protein identification by mass spectrometry and its implications. Proteomics. 2003;3:19–28. doi: 10.1002/pmic.200390004. [DOI] [PubMed] [Google Scholar]
- 38.Nesvizhskii AI, Roos FF, Grossmann J, Vogelzang M, et al. Dynamic spectrum quality assessment and iterative computational analysis of shotgun proteomic data: Toward more efficient identification of post-translational modifications, sequence polymorphisms, and novel peptides. Mol. Cell. Proteomics. 2006;5:652–670. doi: 10.1074/mcp.M500319-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Tabb DL, Saraf A, Yates JR., III GutenTag: High-throughput sequence tagging via an empirically derived fragmentation model. Anal. Chem. 2003;75:6415–6421. doi: 10.1021/ac0347462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bern M, Goldberg D, McDonald WH, Yates JR., III Automatic quality assessment of Peptide tandem mass spectra. Bioinformatics. 2004;20:I49–I54. doi: 10.1093/bioinformatics/bth947. [DOI] [PubMed] [Google Scholar]
- 41.Flikka K, Martens L, Vandekerckhove J, Gevaert K, et al. Improving the reliability and throughput of mass spectrometry-based proteomics by spectrum quality filtering. Proteomics. 2006;6:2086–2094. doi: 10.1002/pmic.200500309. [DOI] [PubMed] [Google Scholar]
- 42.Salmi J, Moulder R, Filen JJ, Nevalainen OS, et al. Quality classification of tandem mass spectrometry data. Bioinformatics. 2006;22:400–406. doi: 10.1093/bioinformatics/bti829. [DOI] [PubMed] [Google Scholar]
- 43.Xu M, Geer LY, Bryant SH, Roth JS, et al. Assessing data quality of peptide mass spectra obtained by quadrupole ion trap mass spectrometry. J. Proteome Res. 2005;4:300–305. doi: 10.1021/pr049844y. [DOI] [PubMed] [Google Scholar]
- 44.Venable JD, Yates JR., III Impact of ion trap tandem mass spectra variability on the identification of peptides. Anal. Chem. 2004;76:2928–2937. doi: 10.1021/ac0348219. [DOI] [PubMed] [Google Scholar]
- 45.Kilpatrick L, Mautner M, Neta P, Roth J, et al. 53rd Conference of the American Society for Mass Spectrometry; San Antonio, TX. 2005. [Google Scholar]
- 46.Yang F, Stenoien DL, Strittmatter EF, Wang J, et al. Phosphoproteome profiling of human skin fibroblast cells in response to low- and high-dose irradiation. J. Proteome Res. 2006;5:1252–1260. doi: 10.1021/pr060028v. [DOI] [PubMed] [Google Scholar]
- 47.Guldener U, Mannhaupt G, Munsterkotter M, Haase D, et al. FGDB: A comprehensive fungal genome resource on the plant pathogen Fusarium graminearum. Nucleic Acids Res. 2006;34:D456–D458. doi: 10.1093/nar/gkj026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Donnell K, Kistler HC, Tacke BK, Casper HH. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl. Acad. Sci. USA. 2000;97:7905–7910. doi: 10.1073/pnas.130193297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lam H, Deutsch EW, Eddes JS, Eng JK, et al. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics. 2007;7:655–667. doi: 10.1002/pmic.200600625. [DOI] [PubMed] [Google Scholar]
- 50.Craig R, Cortens JC, Fenyo D, Beavis RC. Using annotated peptide mass spectrum libraries for protein identification. J. Proteome Res. 2006;5:1843–1849. doi: 10.1021/pr0602085. [DOI] [PubMed] [Google Scholar]
- 51.Craig R, Cortens JP, Beavis RC. The use of proteotypic peptide libraries for protein identification. Rapid Commun. Mass Spectrom. 2005;19:1844–1850. doi: 10.1002/rcm.1992. [DOI] [PubMed] [Google Scholar]
- 52.Stein SE. Estimating probabilities of correct identification from results of mass spectral library searches. J. Am. Soc. Mass Spectrom. 1994;5:316–323. doi: 10.1016/1044-0305(94)85022-4. [DOI] [PubMed] [Google Scholar]
- 53.Stein SE, Scott DR. Optimization and testing of mass spectral library search algorithms for compound identification. J. Am. Soc. Mass Spectrom. 1994;5:859–866. doi: 10.1016/1044-0305(94)87009-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.