Abstract
In shotgun proteomics, tandem mass spectra of peptides are typically identified through database search algorithms such as Sequest. We have developed DirecTag, an open-source algorithm to infer partial sequence tags directly from observed fragment ions. This algorithm is unique in its implementation of three separate scoring systems to evaluate each tag on the basis of peak intensity, m/z fidelity, and complementarity. In data sets from several types of mass spectrometers, DirecTag reproducibly exceeded the accuracy and speed of InsPecT and GutenTag, two previously published algorithms for this purpose. The source code and binaries for DirecTag are available from http://fenchurch.mc.vanderbilt.edu.
Keywords: sequence tagging, bioinformatics, de novo, multi-platform, peptide identification
Introduction
Proteomic database search algorithms have evolved considerably since their introduction in 1994. In Sequest, Yates et al. applied a cross correlation scoring algorithm borrowed from signal processing.1 A host of successors has contributed a diverse collection of scoring metrics.2–8 More recently, though, the field has emphasized the statistics of evaluating match scores rather than the way in which these scores are produced. In their 2003 article, Fenyo and Beavis described the utility of expectation values for recognizing outstanding scores.5 As journals have tightened their requirements for publication, the field has developed a consensus on the importance of false discovery rate estimation.9 This new emphasis has made it possible to evaluate database search algorithms on a level playing field.6,10
Automated inference of sequences from tandem mass spectra has posed a significant challenge. In 1990, Bartels proposed transforming the tandem mass (MS/MS) spectrum to a “sequence graph” in order to simplify reading sequences from MS/MS,11 and his technique has been the basis for most de novo algorithms to infer full peptide sequences. This approach requires that the software be trained on data sets from each mass spectrometer type to learn which ions can be expected. Taylor and Johnson created the LuteFisk software implementing this strategy in 1997.12 In 1999, Dancik et al. introduced “Sherenga,” a tool that ascribed greater importance to peaks accompanied by the neutral losses that are characteristic of fragment ion series.13 Most recent de novo algorithms have followed the Bartels paradigm in which the software first constructs an abstraction of the spectrum and then interprets the abstraction.14–16
Sequence tagging comprises a middle path between database search and de novo strategies. Software infers a partial sequence from a tandem mass spectrum, and then this partial sequence is evaluated against protein database sequences to interpret the remainder of the spectrum.17 Mann and Wilm introduced the sequence tagging approach to peptide identification in 1994,18 the same year as Sequest, but strategies to infer tags through software did not appear until later. Tabb et al. offered GutenTag as a software tool to automate peptide identification through sequence tagging, using a strategy that scored inferred sequences directly against the observed spectrum rather than an abstraction derived from it.19 Frank et al. generated tag sequences with PepNovo, their de novo sequence inference tool, demonstrating this software’s high accuracy.14 Tanner et al. created InsPecT for sequence tagging, inferring partial sequences by a technique similar to PepNovo.20 These tools are beginning to mature, but they have yet to gain significant traction outside of the laboratories that developed them.
In this manuscript, we evaluate DirecTag, new software for sequence tagging. DirecTag seeks to apply statistical models to the problem of tag inference. It evaluates multiple chemical properties of each tag, generating p-values from these tests that can be combined to generate a single score for the tag. Because the properties evaluated by DirecTag are common across multiple instrument types, the software does not require training for each instrument. We compare the accuracy and computational efficiency achieved by this tool to that of GutenTag and InsPecT.
Experimental Section
Data Sources
This evaluation of sequence tagging algorithms employs several publicly available data sets; URLs for the corresponding sites are given in the Supporting Information. Thermo RAW files (San Jose, CA) were converted to mzXML format by the LibMSR conversion tool, available from http://fenchurch.mc.vanderbilt.edu, with settings that exported only tandem mass spectra, writing m/z and intensity values with 32-bit precision. Other instrument data were transcoded as described below. All sequence databases included a set of 77 contaminant proteins such as proteases, keratins, and immunoglobulin constant chains. All databases contained sequences in both forward and reversed orientations for identification error rate assessment. Specific databases are described below. MyriMatch6 and Sequest1 identified the tandem mass spectra against these sequences, with mass accuracies as shown in Table 1 (full configurations are available in Supporting Information). All searches allowed for the possibility of oxidation of methionine, formation of pyroglutamic acid from N-terminal glutamines, and carboxamidomethylation of cysteines. Unless otherwise stated, candidate peptides were required to result from trypsin cleavages on both ends to be matched to peptides. Throughout this manuscript, database search score thresholds were derived to yield a 2% False Discovery Rate. Peptides passing these thresholds were treated as legitimate identifications for evaluating tag accuracy.
Table 1.
Key Configuration Parameters for MyriMatch, Sequest, DirecTag, and InsPecTa
| precursor/fragment analyzer | LTQ/LTQ | FTMS/LTQ | Orbi/LTQ | Orbi/Orbi | Quad/TOF | TOF/TOF |
|---|---|---|---|---|---|---|
| Precursor m/z tolerance | 1.25 | 0.1 | 0.1 | 0.1 | 0.25 | 0.25 |
| Fragment m/z tolerance | 0.5 | 0.5 | 0.5 | 0.1 | 0.25 | 0.25 |
| Precursor mass type | avg | mono | mono | mono | mono | mono |
| Precursor isotope adjust | none | ± neutron | ± neutron | ± neutron | ± neutron | ± neutron |
| InsPecT mode | ESI-ION-TRAP | FT-Hybrid | FT-Hybrid | QTOF | QTOF | QTOF |
These settings reflect the different mass accuracies of the various data sets employed in this manuscript. InsPecT did not have a mode ideal for the Orbi/Orbi data, and so its QTOF mode was employed for this set. MyriMatch was allowed to adjust precursor mass by one neutron in isotopically resolved MS scans; Sequest did not perform these adjustments. Exhaustive configuration details are available in Supporting Information.
Subscore Evaluation Data Sets
To evaluate the three scoring metrics implemented in DirecTag, we made use of data sets collected on the Thermo Fisher LTQ, the AB 4700 TOF/TOF, and the Thermo Fisher LTQ Orbitrap. The LTQ data represented an RPLC analysis of immunoisolated human gastric vesicles at Vanderbilt University Medical Center.21 Spectra from “klc_498_KA_062805b_2_HKATPase” were identified against the IPI Human database, version 3.37.
The TOF/TOF platform was exemplified by the “Aurum” set from the University of Michigan.22 In this set, an Applied Biosystems 4700 TOF/TOF mass spectrometer analyzed MALDI spots for 250 trypsin-digested human proteins. The files were downloaded as a series of MGF files. These were concatenated and converted via LibMSR to a single mzXML file containing 10 097 tandem mass spectra. The IPI Human v3.37 database was employed to identify these spectra as well.
The third data set featured tandem mass spectra of high resolution and mass accuracy. The RPLC separation of the second MudPIT fraction for a Rhodopseudomonas palustris lysate at Oak Ridge National Laboratory was analyzed in the Thermo Fisher LTQ Orbitrap (methods were similar to those in VerBerkmoes et al.23). For this data set, both MS and MS/MS scans were collected in the Orbitrap (yielding 4465 MS/MS scans). Spectra were identified against the R. palustris database available from ORNL. The 7500 resolution used for the MS/MS scans was the lowest resolution mode available on the instrument in order to maximize the number of scans.
Algorithm Reproducibility Data Sets
We employed data sets with multiple replicates to test the reproducibility of tag inference. The NCI Mouse Models of Human Cancers Consortium tested different means of increasing dynamic range in identifying serum proteins.24 In one test, the group employed a MARS column to deplete the serum of the 12 most abundant proteins. At Fred Hutchinson Cancer Research Center, researchers digested these samples and produced 10 replicate RPLC runs on a Thermo Fisher LTQ linear ion trap (San Jose, CA). The files averaged 19 378 MS/MS scans. MyriMatch and Sequest identified peptides using the IPI Human v3.37 database. DTAs were generated for Sequest via “ScanSifter,” a tool under development at Vanderbilt University Medical Center. OUT files were merged into pepXML via SQTer, available at http://fenchurch.mc.vanderbilt.edu.
The NCI Clinical Proteomic Technology Assessment for Cancer program has generated a sample yeast lysate for evaluating instrument reproducibility. This lysate was reduced, alkylated, and digested at National Institute of Standards and Technology. Vanderbilt produced eight RPLC analyses of this sample on an LTQ Orbitrap. The runs were grouped in two sets of triplicates and one pair of runs, where sets of replicates differed in date and minor instrument configuration details. Files averaged 13 187 MS/MS scans. The numbers of confidently identified spectra and peptides were consistent from run to run in searches against the Saccharomyces Genome Database orf_trans_all.fasta file, downloaded in April of 2007.
Multiplatform Algorithm Performance Data Sets
The Institute for Systems Biology employs a mix of 18 standard proteins to test its instruments.25 The reduced and digested protein mixture was evaluated in replicate RPLC runs on a Thermo Fisher LTQ, an LTQ FT, an LTQ Orbitrap, an Applied Biosystems QSTAR Pulsar I QqTOF, and an Applied Biosystems 4800 TOF/TOF. Protein Pilot 2.01 generated peak lists from QSTAR WIFF files for Mix 2 and wrote them to MGF files. These, in turn, were transcoded to mzXML files by LibMSR. For the AB 4800, DTA files were downloaded and then converted to mzXML by LibMSR. A semitryptic search of a database containing known proteins for this mixture plus the sequences for Haemophilus influenzae as decoy sequences was employed to identify a pool of approximately 40 proteins (including contaminants and impurities) from these mixtures.
DirecTag Algorithm
The “DirecTag” software was created in C++. Its multithreaded design can exploit multiple CPUs and multicore CPUs, and its use of the Message Passing Interface library enables it to distribute operations over a cluster of computers. DirecTag accepts spectra provided in the mz-Data, mzXML, and MGF file formats via use of the “LibMSR” library. Source code for the algorithm is available from the Tabb group Web site: http://fenchurch.mc.vanderbilt.edu.
Setup and Preprocessing
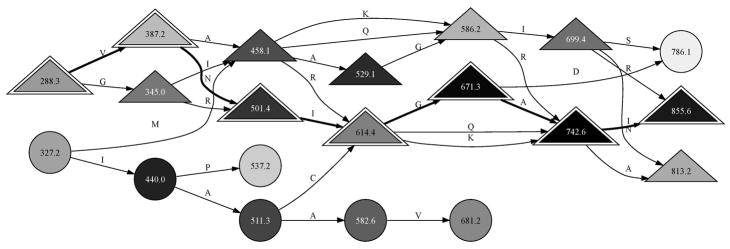
To begin preprocessing, DirecTag consolidates isotopic fragment ion clusters in the spectrum. In brief, the software estimates the proportion of ions at each mass expected to incorporate one or two 13C atoms, and it reapportions the intensity observed in these isotopic peaks to the monoisotope.26 DirecTag seeks out complementary pairs of fragment ions and stores a pointer to the complementary peak for each fragment ion, if present. The software retains a user-configured number of peaks for each spectrum, filtering out those of low intensity, and it stores the intensity rank of each retained peak. In this report, spectra were always reduced to their top 100 peaks. The software next seeks out pairs of peaks that are separated by amino acid masses. The spectrum can then be evaluated as a graph, with peaks represented by nodes and amino acid gaps between peaks represented by edges (see Figure 1).
Figure 1.
A spectrum may be viewed as a graph for tag inference. This graph represents the MS/MS observed for the peptide DAGTIAGLNVLR. Each major peak in the spectrum is shown as a node, with the most intense peaks given the darkest shading. When peaks are separated by the mass of an amino acid, they are joined by an edge labeled with the amino acid symbol. Complemented peaks are denoted by triangles. The 3-letter sequence tags for this spectrum are sequences of four connected nodes. DirecTag was configured to retain the top 100 peaks for each spectrum, yielding graphs with far more possible tags than shown here. The true sequence of this peptide can be reconstructed from a path of ions stretching from 288.3 to 855.6 m/z (indicated by outlined triangles). It is reversed with respect to m/z order because the fragments are y ions.
Tag Enumeration
When a set of peaks is joined by consecutive edges in this graph, the set constitutes a tag. In this manuscript, we focus on tags of three residues (three edges that connect four peaks). Tag length is dictated by the realities of sequence tag-based identification. Two-residue tags can be generated more quickly than three-residue tags, but their ability to screen out erroneous sequences in identification is limited; there are only 324 different two-residue tags if Leu is indiscernible from Ile and Gln is indiscernible from Lys. Longer sequence tags can be generated at a slower speed, but the percentage of identifiable spectra that yield valid four-residue tags is lower than the percentage of identifiable spectra that will yield valid three-residue tags (data not shown). This phenomenon can be explained by the fact that DirecTag relies upon a consecutive series of fragment ions to infer tags. Tags of three amino acids pose an ideal balance between tag selectivity and tag generation sensitivity. DirecTag recursively enumerates all possible tags of four peaks/three gaps from the graph, and each tag is subjected to scoring. Limiting each spectrum to a particular number of peaks prevents the number of potential tags from becoming too large.
DirecTag can infer sequences from series of +1 fragment ions or from +2 fragments. It only seeks +2 tags from spectra that result from the fragmentation of peptide ions charged +3 or higher. The gaps for any tag must correspond to only +1 amino acid masses or only +2 amino acid masses, not a mixture of charges.
Intensity Subscore
DirecTag scores tags on the basis of their peak intensities. Tags that contain intense peaks are more likely to be correct than those that contain average peaks. Rather than work with intensity directly, the software evaluates peaks by their intensity ranks. For each tag, DirecTag computes a rank sum. For example, if a tag includes the fourth, 10th, 17th, and 50th most intense peaks, the tag rank sum is 81.
Like all subscores in DirecTag, this metric is converted to a p-value. During run setup, the software has computed the probability of every possible rank sum for a spectrum containing N peaks when T peaks are chosen at random. When a particular rank sum is observed, the software can look up the probability that a lower or equal rank sum would result. The distribution of these rank sums is identical to the distribution of the U statistic in the Wilcoxon–Mann–Whitney test. In essence, the intensity subscore is evaluating whether the four selected peaks come from the same intensity distribution as the remaining peaks.
The use of intensity rank sums makes more efficient use of intensity information than an intensity classification system such as was used in MyriMatch.6 This application of intensity rank bears similarity to OMSSA 2.0.27 The intensity scoring in DirecTag reflects Bern’s observation that the significance of fragment ions may be judged more accurately by their intensity ranks than by their relative intensities.28
m/z Fidelity Subscore
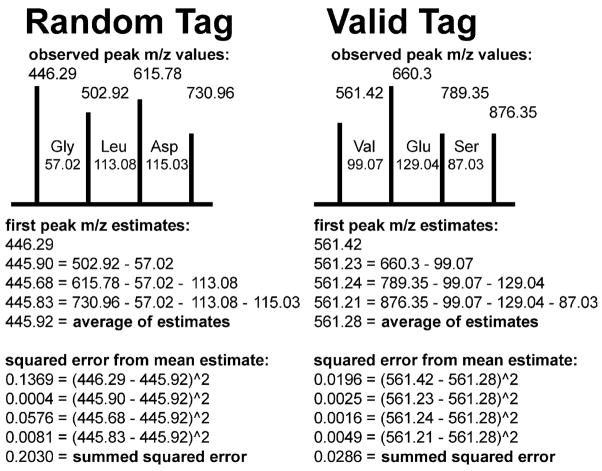
For a set of four peaks to be evaluated as a putative tag, each peak must be separated from the next by the mass of an amino acid within some tolerance (typically 0.5 m/z for an ion trap tandem mass spectrum). If these four peaks are a valid tag sequence, the error in the observed peak m/z values is likely to be smaller than if these four peaks are not related to each other. In database search, the positions of peaks can be computed precisely from the full sequence of the peptide, but in sequence tagging, only the distances between peaks are known exactly.
DirecTag evaluates each of the four peaks in the tag to yield a different estimate of the position of the lowest m/z peak in the tag (see Figure 2). The m/z observed for the first peak, of course, can be used for this purpose without manipulation. The algorithm subtracts the m/z of the appropriate amino acids from the subsequent peak m/z values to compute the other first peak m/z estimates. The mean of these m/z estimates is the optimal location for the first peak to minimize m/z position error for the entire tag.
Figure 2.
m/z fidelity for a tag can be characterized through SSE. DirecTag evaluates the consistency of fragment ion m/z values for each tag. The gap defined by each pair of fragment ions in a valid tag will be close to an amino acid mass. In a random collection of peaks, though, the gap masses will have random differences from amino acid masses. This computation yields a summed squared error (SSE) to reflect the fidelity of the peak m/z spacings.
DirecTag computes the summed squared error (SSE) of the first peak m/z estimates for each tag. When all four peaks represent C-terminal y ions, the SSE value will be very low. If the four peaks do not bear this relationship to each other, the SSE value will be higher. By use of a random simulation at startup, DirecTag can evaluate the probability that an equal or lower SSE value would occur by random chance (see Figure 2). This p-value is the m/z Fidelity subscore.
Complementarity Subscore
Peaks that match to complementary ions within the spectrum are more trustworthy than other peaks. DirecTag assesses the number and concordance of complementary ions for each tag. If a spectrum contains 100 peaks, 20 of which match to complementary ions, the probability that three of four peaks chosen at random would match to complements is 2.3%, a value which can be computed through application of the hypergeometric distribution. The probability for matching 0, 1, 2, 3, or 4 peaks is the first element of the complementary score.
Because multiple tags may have the same number of complementary ions, DirecTag employs a secondary metric to differentiate tags from each other. The sum of a fragment ion mass and its complement ion mass estimates the mass of the peptide. The software sums each fragment/complement m/z pair for the tag and computes the SSE of these sums. A low SSE indicates that the m/z values for complementary pairs are in accord with each other.
In the complementarity subscore, tags for each spectrum are sorted first by the number of complements that they contain and then by the concordance of their m/z sums. For example, a tag with two complementary ions receives a p-value that sums over these elements:
the probability of a tag matching four complementary ions by random chance
the probability of a tag matching three complementary ions by random chance
the probability that a tag with two complementary ions would yield an equal or lower SSE by random chance
Tag Score Reporting
Tags are evaluated on three different axes: intensity, m/z fidelity, and complementarity. Each of these subscores results in a p-value. The p-value represents the probability that a better score would have resulted for a random collection of four peaks. DirecTag employs Fisher’s Method for combining these p-values.29 In brief, the method rolls multiple p-values together to generate a test statistic that follows a chi-square distribution. The joint p-value is the probability that an equal or lower test statistic would be observed in this distribution. In spectra where no complementary peak pairs are found, the complementarity subscore is excluded from the overall score computation.
The joint p-values for all tags from a spectrum are computed, and the tags are ranked by these p-values. Some spectra produce large numbers of tags and others yield only a few. As a result, the best result from a spectrum producing 10 000 tags is likely to have a lower joint p-value by random chance than the best tag from a spectrum producing only ten tags. For reporting, DirecTag multiplies the p-value for tags by the number of tags generated for each spectrum. This yields the “expectation value” metric, an approach that has been demonstrated to be useful in the context of database search.5,7
DirecTag stores the sequences it infers into a tab-delimited file. As HUPO PSI standardization efforts continue to progress, DirecTag will implement a standardized XML-based reporting format. DirecTag does not record all tags, just those that rank highly by score. The number of tags retained by the software is configurable by the user.
To compare DirecTag to other algorithms, the output of those algorithms must be in comparable format. We modified the source code of InsPecT 20070613 to enable it to output the same format. The output format of GutenTag v1.02 was similar enough that our evaluation tools could read either type of file. Initial efforts to include PepNovo in this comparison were forestalled when we found that the current release performed less accurately and consistently than earlier versions. As a result, this comparison was limited to GutenTag and InsPecT, the two primary peer-reviewed algorithms designed explicitly for tag inference.
TagValidate
TagValidate is software written in the C++ language to evaluate the inferences supplied in a tag file. It can limit tags to those that are at a lower rank than a specified value, or it can apply score thresholds to the tags. The software is designed to read raw spectral identifications in SQT or pepXML format, selecting those that achieve a user-specified false discovery rate (2%, in this case) in a way similar to DBValidate.6 Using this information, TagValidate can discern which tags are valid (matching both the sequence of the confident identification and the masses that would flank the sequence). Its outputs were then evaluated in R scripts to produce the figures shown here.
Results and Discussion
We evaluated DirecTag through three sets of tests. In the first test, we compared the subscores of DirecTag to determine their relative effectiveness for generating correct tag sequences in data sets of different mass accuracies. The second examination pitted DirecTag against InsPecT and GutenTag in evaluating replicate data from Thermo LTQ and OrbiTrap instruments. The final test examined the performance of DirecTag in replicate defined mixture data from various instrument platforms. These tests establish DirecTag as a high-throughput algorithm suitable for implementation in a variety of instrumental workflows.
Subscore Evaluation
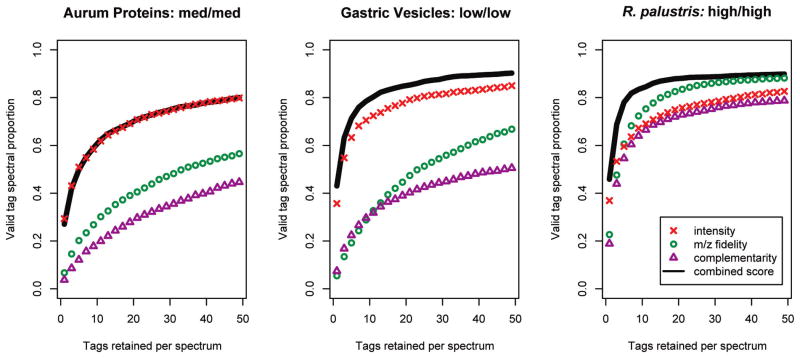
If the subscores differ considerably in their discriminatory power, assigning an equal weight to each in Fisher’s Method may reduce the discrimination of DirecTag overall. We configured DirecTag seven different ways for three data sets containing low, medium, and high mass-accuracy tandem mass spectra. DirecTag was configured to appropriate tolerances for each instrument platform (see Table 1). The first three runs tested each subscore independently. The next three runs tested pairs of subscores, and the final run employed all three subscores. In each case, DirecTag retained the best 50 tags for each spectrum.
The peptide identifications from a MyriMatch6 database search for these three files were used to evaluate the inferred sequence tags. The included identifications had an estimated false discovery rate of 2%; the error rate for these peptides is estimated to be one error in 50 identifications. To be marked “valid,” sequence tags were required to match both the peptide sequences and flanking masses. The N-terminal flanking mass was based on the precursor mass minus the highest y ion mass and so was permitted to differ from the database sequence by up to 2.5 Da. The C-terminal mass was based solely on the lowest y ion mass and was permitted to differ by up to 1.0 Da. In each case, the proportion of identified spectra for which the algorithm inferred at least one valid tag was used as the metric of success (see Figure 3). If an algorithm generated valid tags for a larger number of spectra, the curve was positioned higher on the graph. Better algorithms sort valid tags to lower ranks, yielding a faster rise to maximum curve height.
Figure 3.
Three subscores contribute to DirecTag scoring accuracy. Tests on data sets of three different mass accuracies revealed different discrimination in DirecTag’s three subscores. The spectra used in these tests had been successfully identified by database search. The curves reflect the fraction of these spectra for which a valid tag was generated by DirecTag. When more tags were retained for each spectrum, DirecTag succeeded on a larger proportion of spectra. The best discrimination, however, pushes valid tags to the best ranks, making it possible to retain fewer tags per spectrum. Combining the three subscores performed better than any single subscore in almost all cases. The mass accuracy of precursor and fragment ions is shown after the sample name for each panel.
In two of the data sets, the intensity subscore was dominant. The gastric vesicle (LTQ) and Aurum (TOF/TOF) data both demonstrated that computing intensity rank sums for tags was a very effective way to score them. In the LTQ sample, the subscores for m/z fidelity and complementarity contributed to scoring when all three subscores were combined, but the overall scoring discrimination did not improve for the TOF/TOF files when m/z fidelity and complementarity were taken into account. The high-resolution tandem mass spectra from the LTQ Orbitrap showed a different balance among the three subscores; each contributes nearly equally to DirecTag performance. When the subscores were considered in pairs instead (data not shown), the combination of intensity and m/z fidelity subscores was generally the best combination. By relying on multiple metrics for each tag, DirecTag is able to achieve high performance on diverse data sets.
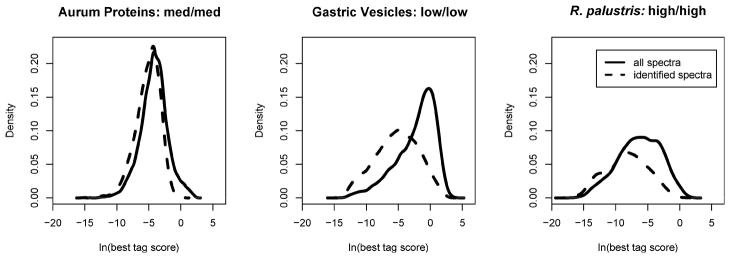
Having examined tagging performance for identified spectra, we broadened the analysis to evaluate scores for all spectra, whether identified or not (see Figure 4). TagValidate reported the best tag score for each spectrum. The density plots in this figure show the resulting distributions for each data set. Spectra that were identified by MyriMatch were associated with lower expectation values. In the case of LTQ data for gastric vesicles, the many unidentified spectra shifted the distribution of scores to higher expectation values. We plan to explore the use of DirecTag for quality assessment of spectra in a subsequent paper.
Figure 4.
The best tag scores for identifiable spectra are better than those of other spectra. These three density plots examine the best tag scores observed for each spectrum. Each score is expressed as an expectation value, so the distributions evaluate the natural logarithms of the scores. “Identified spectra” are those for which the MyriMatch score exceeded the highest-scoring reversed peptide in the set. The differences between these distributions suggest that tag scores may be used to quality filter spectra prior to identification.
In this analysis DirecTag inferred valid tags for more than 80% of the identified peptides. The use of all three subscores was at least equivalent to the use of one or two; for all remaining experiments, all three subscores were used together. The next test compared DirecTag to other tools for inferring partial sequences from spectra.
Comparison of Sequence Tagging Algorithms
We evaluated three tag inference algorithms to place DirecTag in context. GutenTag is Java software from the Yates Laboratory19 that employs an ad-hoc scorer to separate valid tags from random ones. InsPecT20 is a tool from the Pevzner Laboratory (written in C and C++) that builds a Prefix Residue Mass (PRM) graph as a first step. DirecTag is like GutenTag in its direct approach to scoring tags against spectral m/z and intensity values, but its scoring system is considerably different, and it is implemented in C++ rather than Java. InsPecT was run in “Tags-Only” mode to forestall database search, but GutenTag always conducts this process. To reduce the performance penalty of this lookup process, the algorithm was provided a FASTA database containing a single sequence for this step.
The replicate data sets enabled measurement of performance variability. The “Serum” data set included 10 replicate RPLC separations of depleted human serum from a Thermo LTQ linear ion trap. The “Yeast” data set included eight replicate RPLC separations of Saccharomyces cerevisiae lysate on a Thermo Orbitrap. We used these replicates to compare the time required for each algorithm to run on a single processor and the number of identified spectra for which a valid tag was produced.
The algorithms differed considerably in their run-times on a Dell Optiplex 745 with an Intel Core 2 Duo 6400 processor and 1 GB of RAM. The average time required to process a raw file from the serum and yeast samples with GutenTag was 3324 and 712 s, respectively. This algorithm, the only one to require DTA files, was the slowest in the test. In single processor operation, DirecTag and InsPecT were comparable, requiring 487 or 631 s for each serum file, respectively. The yeast samples took longer per file on DirecTag (570 s) but less time per file on InsPecT (471 s). When DirecTag was allowed to use both cores of the processor in multithreaded operation, its times on both files diminished (312 s for each serum file and 299 for each yeast file). Tag inference can be conducted rapidly enough that data analysis can proceed more quickly than data acquisition.
Sequence tag inference differs from database search in that multiple tags are typically retained for each spectrum. If at least one of them corresponds to the correct sequence, reconciling the tag list to a sequence database can identify the spectrum. Judging the correct number of tags to retain, however, is dependent on the accuracy of the tag inference engine. We evaluated the proportion of identified spectra for which at least one valid tag had been generated as the number of tags retained for each spectrum scaled from 1 to 50. Because both data sets featured multiple replicates, we present the interquartile range of this proportion; a broad range of performance indicates variation in the accuracy produced by an algorithm, while a narrow range is associated with consistent accuracy among multiple replicates.
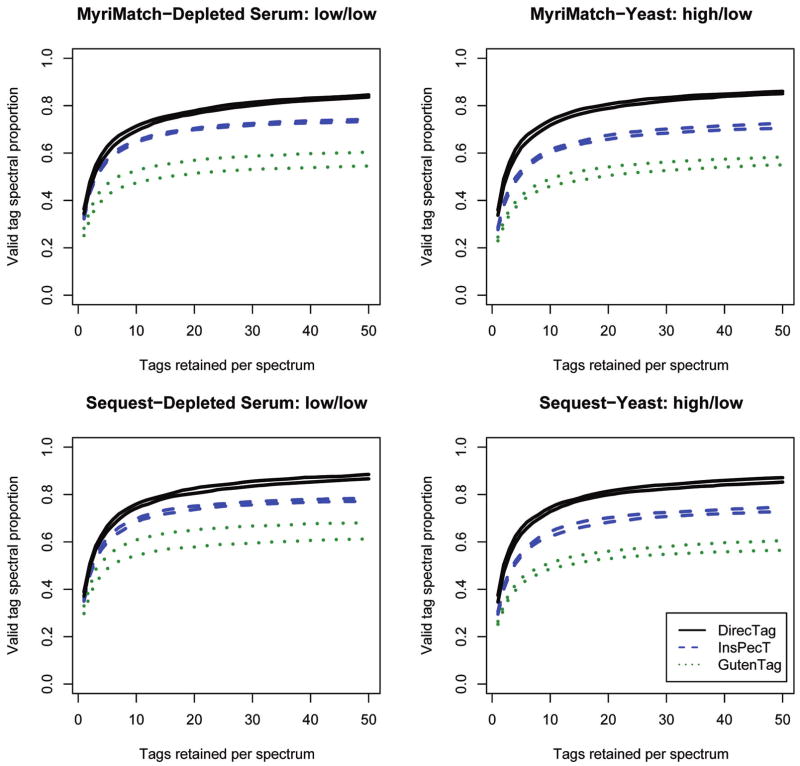
The serum data set from the Thermo LTQ separates the algorithms into two classes (see Figure 5, left column). To ensure that the algorithm generating peptide identifications was not an important factor, the tagging algorithms were compared to identifications from both MyriMatch (top row) and Sequest (bottom row). GutenTag falls behind the other algorithms in accuracy and is more variable, in part because it is limited to only spectra from doubly charged precursors. InsPecT and DirecTag are more directly comparable to each other; each is applicable to peptides of multiple charge states. InsPecT derives tags from a PRM abstraction; however, DirecTag works directly from the peaks observed in the spectrum to generate potential tags. InsPecT employs a training-dependent log-odds ratio scoring technique for its tags, while DirecTag generates p-values under a random model that requires no training. If these two algorithms are permitted to retain only one tag per spectrum, their accuracy is very similar, but as more tags are retained for each spectrum, DirecTag consistently generates valid tags for a larger proportion of identifiable spectra than does InsPecT.
Figure 5.
Tagging accuracy for DirecTag, InsPecT, and GutenTag. Replicate RPLC separations of serum and yeast were analyzed on a Thermo LTQ or LTQ Orbitrap, respectively. In “high/low,” the first word describes the precursor mass accuracy and the second word describes the fragment ion mass accuracy. Identifications were generated from these sets by MyriMatch and Sequest, and the spectra were processed through three sequence tag generation tools. These graphs show the fraction of confidently identified spectra that produce valid tags from each of the three tools. An ideal sequence tagger is one that can produce valid tags for a large proportion of identifiable spectra (i.e., have the highest curve) and then rank those valid tags at the top of the list (i.e., produce a curve that rises more quickly). The two traces for each algorithm describe the interquartile performance across the replicates; the higher shows the 75%ile performance, and the lower shows the 25%ile performance.
The yeast data set (see Figure 5, right column) differed from the serum by having highly mass-accurate precursors measured in the Orbitrap. For DirecTag, this may have translated to more accurately assessing complementary pairs of fragment ions. The problems of inferring sequences, however, were unchanged since the fragment ion m/z values were only as accurate as the LTQ mass analyzer could achieve. This data set yielded the same ranking of the three algorithms, though the separation between DirecTag and InsPecT was increased.
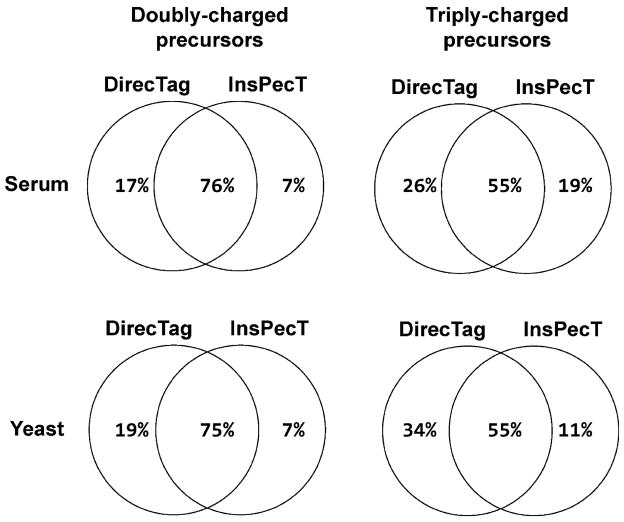
To explore the relative performance of these two algorithms further, we constructed a Venn diagram comparing the identified spectra for which DirecTag and InsPecT produced valid tags (see Figure 6). Only tags ranked in the top 20 positions for each spectrum were considered. The diagrams look very similar for serum and yeast data sets, but the overlap in performance for doubly charged precursors was greater than that for triply charged precursors. The disparities on triply charged precursors probably reflect algorithmic differences. For DirecTag, all peaks for a particular tag must be singly charged, or all must be doubly charged. InsPecT can mix evidence from both singly charged and doubly charged fragment ions in a single tag.
Figure 6.
Overlapping performance for DirecTag and InsPecT. These Venn diagrams describe the overlap in spectra with valid tags. The precursor ion charge was a better predictor of overlap than the instrument that produced the spectra. These two algorithms were more similar in spectra from doubly charged precursors than in spectra from triply charged precursors.
DirecTag Performance across Instrument Platforms
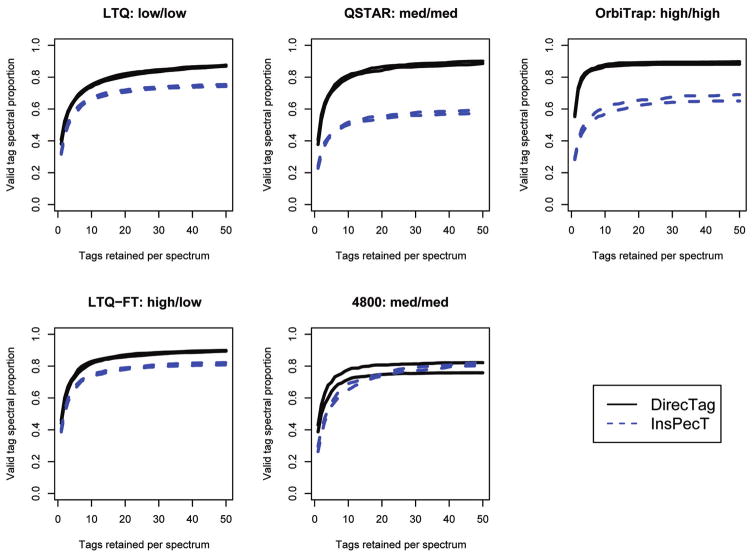
We compared DirecTag to InsPecT across data from five different instrument platforms at the Institute for Systems Biology. These instruments ranged from unit mass accuracy (such as the LTQ) to the higher accuracy of TOF analyzers to the ppm mass accuracy of the Orbitrap and FTMS. Each data set featured 10 replicates, making it possible to gauge the reproducibility of tag inference as in the complex serum and yeast data sets.
The diversity of these sets put some constraints on the comparison. Because GutenTag is designed solely for doubly charged peptides analyzed in ion traps, we excluded it from this comparison. InsPecT did not feature a mode for processing tandem mass spectra collected in the Orbitrap cell or MS/MS from TOF/TOF analyzers, and so these data were processed in the software’s “QTOF” mode. The configuration used for each data set is shown in Table 1.
DirecTag consistently displayed better accuracy for all instruments than did InsPecT (see Figure 7). The significant difference in the Orbitrap MS/MS spectra is probably attributable to InsPecT’s lack of a profile for this data type; while DirecTag could be configured to expect fragment ions to be separated by gaps that were within 0.1 m/z of amino acid masses, InsPecT did not offer a configuration that would require such stringent mass accuracy.
Figure 7.
DirecTag and InsPecT compared on multiple instruments. These curves compare tagging performance for a defined mixture on a variety of instruments. In each case, the distance between the two curves for each instrument reflects the interquartile consistency of tag inference. Mass accuracy for each instrument is summarized as high, medium, or low, with precursor mass accuracy described first and fragment ion mass accuracy described second. In each case, the tag inference accuracy achieved by DirecTag was superior to that observed for InsPecT.
Working with data from such diverse instruments also provided lessons in data conversion. The data from the Applied Biosystems QSTAR Pulsar I, for example, yielded fewer identifications when mzWiff was used to generate mzXML files than when Protein Pilot 2.01 was employed to generate MGF files instead.
Conclusion
The scorers in DirecTag propelled it to performance beyond that of InsPecT. In the past, de novo sequence inference has largely been advanced through machine learning approaches. It is clear, though, that statistical scorers may be adapted from database search identification algorithms to good effect. DirecTag’s emphasis on scoring tag sequences directly against observed fragment ions resulted in a scoring algorithm of high discrimination.
Building this sequence inference engine is the first step of several. In future work, we intend to put DirecTag to work in identifying peptides that may be missed through database search. Sequence tagging is well-positioned to assist in quality assessment of tandem mass spectra, identifying mutated peptides, and mapping post-translational modifications to peptides. These efforts will build upon the tag inference strategy described here.
Supplementary Material
Acknowledgments
D.L.T., Z.-Q.M., and M.C.C. were supported by NIH Grants R01 CA126218, U24 CA126479, P30 ES000267, and by an American Cancer Society Institutional Research Grant (no. IRG-58-009-48) through the Sartain-Lanier Family Foundation and VICC Discovery Grant Programs. The cross-platform data sets were collected under P50 GM076547 by D.B.M. at the Institute for Systems Biology. Yeast replicates were collected by A.-J.L.H. under U24 CA126479. D.L.T. thanks Sean L. Seymour of Applied Biosystems for WIFF to MGF conversion in this data set. GutenTag was used under license from The Scripps Research Institute. The authors gratefully acknowledge public datasets provided by Jim Goldenring (VUMC), Nathan VerBerkmoes (ORNL), Phil Andrews (U-Michigan), and Mandy Paulovich (FHCRC).
Footnotes
Supporting Information Available: URLs for the publicly available data sets, DirectTag, MyriMatch and Sequest configurations. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Yates JR, III, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67(8):1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 2.Sadygov RG, Yates JR., III A hypergeometric probability model for protein identification and validation using tandem mass spectral data and protein sequence databases. Anal Chem. 2003;75(15):3792–3798. doi: 10.1021/ac034157w. [DOI] [PubMed] [Google Scholar]
- 3.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J Proteome Res. 2004;3(5):958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 4.Zhang N, Aebersold R, Schwikowski B. ProbID: a probabilistic algorithm to identify peptides through sequence database searching using tandem mass spectral data. Proteomics. 2002;2(10):1406–1412. doi: 10.1002/1615-9861(200210)2:10<1406::AID-PROT1406>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 5.Fenyo D, Beavis RC. A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Anal Chem. 2003;75(4):768–774. doi: 10.1021/ac0258709. [DOI] [PubMed] [Google Scholar]
- 6.Tabb DL, Fernando CG, Chambers MC. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res. 2007;6(2):654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20(9):1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 8.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 9.Tabb DL. What’s driving false discovery rates. J Proteome Res. 2008;7(1):45–46. doi: 10.1021/pr700728t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balgley BM, Laudeman T, Yang L, Song T, Lee CS. Comparative evaluation of tandem MS search algorithms using a target-decoy search strategy. Mol Cell Proteomics. 2007;6(9):1599–1608. doi: 10.1074/mcp.M600469-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Bartels C. Fast algorithm for peptide sequencing by mass spectroscopy. Biomed Environ Mass Spectrom. 1990;19(6):363–368. doi: 10.1002/bms.1200190607. [DOI] [PubMed] [Google Scholar]
- 12.Taylor JA, Johnson RS. Sequence database searches via de novo peptide sequencing by tandem mass spectrometry. Rapid Commun Mass Spectrom. 1997;11(9):1067–1075. doi: 10.1002/(SICI)1097-0231(19970615)11:9<1067::AID-RCM953>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 13.Dancik V, Addona TA, Clauser KR, Vath JE, Pevzner PA. De novo peptide sequencing via tandem mass spectrometry. J Comput Biol. 1999;6(3–4):327–342. doi: 10.1089/106652799318300. [DOI] [PubMed] [Google Scholar]
- 14.Frank A, Pevzner P. PepNovo: de novo peptide sequencing via probabilistic network modeling. Anal Chem. 2005;77(4):964–973. doi: 10.1021/ac048788h. [DOI] [PubMed] [Google Scholar]
- 15.DiMaggio PA, Jr, Floudas CA. De novo peptide identification via tandem mass spectrometry and integer linear optimization. Anal Chem. 2007;79(4):1433–1446. doi: 10.1021/ac0618425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer B, Roth V, Roos F, Grossmann J, Baginsky S, Widmayer P, Gruissem W, Buhmann JM. NovoHMM: a hidden Markov model for de novo peptide sequencing. Anal Chem. 2005;77(22):7265–7273. doi: 10.1021/ac0508853. [DOI] [PubMed] [Google Scholar]
- 17.Sunyaev S, Liska AJ, Golod A, Shevchenko A, Shevchenko A. MultiTag: multiple error-tolerant sequence tag search for the sequence-similarity identification of proteins by mass spectrometry. Anal Chem. 2003;75(6):1307–1315. doi: 10.1021/ac026199a. [DOI] [PubMed] [Google Scholar]
- 18.Mann M, Wilm M. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal Chem. 1994;66(24):4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 19.Tabb DL, Saraf A, Yates JR., III GutenTag: high-throughput sequence tagging via an empirically derived fragmentation model. Anal Chem. 2003;75(23):6415–6421. doi: 10.1021/ac0347462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal Chem. 2005;77(14):4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- 21.Lapierre LA, Avant KM, Caldwell CM, Ham AJ, Hill S, Williams JA, Smolka AJ, Goldenring JR. Characterization of immunoisolated human gastric parietal cells tubulovesicles: identification of regulators of apical recycling. Am J Physiol: Gastrointest Liver Physiol. 2007;292(5):G1249–1262. doi: 10.1152/ajpgi.00505.2006. [DOI] [PubMed] [Google Scholar]
- 22.Falkner JA, Kachman M, Veine DM, Walker A, Strahler JR, Andrews PC. Validated MALDI-TOF/TOF mass spectra for protein standards. J Am Soc Mass Spectrom. 2007;18(5):850–855. doi: 10.1016/j.jasms.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 23.VerBerkmoes NC, Shah MB, Lankford PK, Pelletier DA, Strader MB, Tabb DL, McDonald WH, Barton JW, Hurst GB, Hauser L, Davison BH, Beatty JT, Harwood CS, Tabita FR, Hettich RL, Larimer FW. Determination and comparison of the baseline proteomes of the versatile microbe Rhodopseudomonas palustris under its major metabolic states. J Proteome Res. 2006;5(2):287–298. doi: 10.1021/pr0503230. [DOI] [PubMed] [Google Scholar]
- 24.Whiteaker JR, Zhang H, Eng JK, Fang R, Piening BD, Feng LC, Lorentzen TD, Schoenherr RM, Keane JF, Holzman T, Fitzgibbon M, Lin C, Zhang H, Cooke K, Liu T, Camp DG, II, Anderson L, Watts J, Smith RD, McIntosh MW, Paulovich AG. Head-to-head comparison of serum fractionation techniques. J Proteome Res. 2007;6(2):828–836. doi: 10.1021/pr0604920. [DOI] [PubMed] [Google Scholar]
- 25.Klimek J, Eddes JS, Hohmann L, Jackson J, Peterson A, Letarte S, Gafken PR, Katz JE, Mallick P, Lee H, Schmidt A, Ossola R, Eng JK, Aebersold R, Martin DB. The standard protein mix database: a diverse data set to assist in the production of improved Peptide and protein identification software tools. J Proteome Res. 2008;7(1):96–103. doi: 10.1021/pr070244j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senko MW, Beu SC, McLafferty FW. Automated assignment of charge states from resolved isotopic peaks for multiply charged ions. J Am Soc Mass Spectrom. 1995;6(1):52–56. doi: 10.1016/1044-0305(94)00091-D. [DOI] [PubMed] [Google Scholar]
- 27.Geer LY, BDL, Kowalak JA, Chi A, Xu M, Shabanowitz J, Markey SP, Hunt DF, Bryant SH. Reducing false positive rates in MS/MS sequence searching and incorporating intensity into match based statistics. Proceedings of the 54th American Society of Mass Spectrometry Conference. 2006 [Google Scholar]
- 28.Bern M, Goldberg D, McDonald WH, Yates JR., III Automatic quality assessment of peptide tandem mass spectra. Bioinformatics. 2004;20(Suppl 1):i49–154. doi: 10.1093/bioinformatics/bth947. [DOI] [PubMed] [Google Scholar]
- 29.Fisher RA. Combining independent tests of significance. Am Stat. 1948;2(5):30. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.