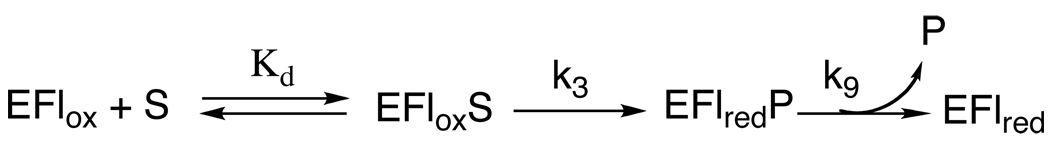Abstract
In mammalian cells, the flavoprotein spermine oxidase (SMO) catalyzes the oxidation of spermine to spermidine and 3-aminopropanal. Mechanistic studies have been carried out with the recombinant human enzyme. The initial velocity pattern when the ratio between the concentrations of spermine and oxygen is kept constant establishes the steady-state kinetic pattern as ping-pong. Reduction of SMO by spermine in the absence of oxygen is biphasic. The rate constant for the rapid phase varies with the substrate concentration, with a limiting value (k3) of 49 s−1 and an apparent Kd value of 48 µM at pH 8.3. The rate constant for the slow step is independent of the spermine concentration, with a value of 5.5 s−1, comparable to the kcat value of 6.6 s−1. The kinetics of the oxidative half-reaction depend on the aging time after spermine and enzyme are mixed in a double mixing experiment. At an aging time of 6 s the reaction is monophasic with a second order rate constant of 4.2 mM−1 s−1. At an aging time of 0.3 s the reaction is biphasic with two second order constants equal to 4.0 and 40 mM−1 s−1. Neither is equal to the kcat/KO2 value of 13 mM−1s−1. These results establish the existence of more than one pathway for the reaction of the reduced flavin intermediate with oxygen. The kcat/KM value for spermine exhibits a bell-shaped pH-profile, with an average pKa value of 8.3. This profile is consistent with the active form of spermine having three charged nitrogens. The pH profile for k3 shows a pKa value of 7.4 for a group that must be unprotonated. The pKi-pH profiles for the competitive inhibitors N,N’-dibenzylbutane-1,4-diamine and spermidine show that the fully protonated forms of the inhibitors and the unprotonated form of an amino acid residue with a pKa of about 7.4 in the active site are preferred for binding.
The polyamines spermine, spermidine, and putrescine are important in cell proliferation, differentiation, and survival (1). Because of the absolute requirement of these compounds for cell growth, the polyamine metabolic pathway has been extensively studied. Polyamine catabolism is mediated by the activity of three enzymes (2). Spermidine/spermine N1-acetyltransferase acetylates spermine and spermidine to produce the N-acetylated compounds (3). These can be exported from the cell or oxidized by the peroxisomal enzyme polyamine oxidase (PAO) to yield spermidine or putrescine, hydrogen peroxide, and 3-acetamidopropanol (4). Alternatively, the cytosolic enzyme spermine oxidase (SMO) can catalyze the oxidation of spermine directly to spermidine (Scheme 1), bypassing the necessity for acetylation (5, 6). The relative contribution of these two enzymes to polyamine catabolism has not been established. While PAO is constitutive, SMO is induced by polyamine analogs (7). Indeed, it has been suggested (8) that SMO is responsible for the antitumor effects of polyamine analogs.
Scheme 1.
Human SMO has a strong preference for spermine as a substrate (6), while PAO prefers N1-acetylspermidine and N1-acetylspermine (9). In contrast, plant polyamine oxidases preferentially oxidize spermine rather than N1-acetylspermine. In addition, the plant enzymes oxidize the endo CN bond of the substrate, while the mammalian enzymes oxidize the exo bond (10). The yeast enzyme Fms1 is also classified as a spermine oxidase, but the kcat/KM values for spermine, N1-acetylspermidine and N1-acetylspermine are comparable (11). The structural bases for these differences in specificity and reactivity are not known.
Both SMO and PAO are flavoproteins and thus members of the family of flavin amine oxidases. Structurally, most flavin amine oxidases can be classified as members of the monoamine oxidase (MAO) family that includes MAO A and B, lysine-specific demethylase (LSD1), and L-amino acid oxidase, or as members of the D-amino acid oxidase (DAAO) family that includes DAAO, sarcosine oxidase, and glycine oxidase, among others (12). The three-dimensional structures of maize PAO and yeast Fms1 have been determined (13, 14), establishing that they have the same overall structure as MAO (15) and that the mammalian SMO and PAO also belong to the MAO structural family. While amine oxidation by members of the DAAO family is more generally accepted to involve direct hydride transfer (16–20), the mechanism of amine oxidation by MAO has been a matter of controversy (12, 21–23).
Because of its relatively recent discovery (5, 7), the mechanism of SMO has not been established. We describe here analysis of the kinetic mechanism of human SMO using both steady-state and rapid reaction kinetic methods and analysis of the effects of pH on the steady-state and reductive half-reaction kinetics. The results provide insight into the mechanism of amine oxidation by SMO and the basis for substrate specificity.
MATERIALS AND METHODS
Materials
Spermine and spermidine trihydrochloride were purchased from Acros Organics (Geel, Belgium). 1,12-Diaminododecane was from Sigma-Aldrich (Milwaukee, WI). N,N’-Dibenzyl-1,4-diaminobutane (DBDB) was from Prime Organics (Woburn, MA). The pET28b(+) vector was from Novagen (Madison, WI). The vector pJ10:G04331 encoding human spermine oxidase was obtained from DNA 2.0 (Menlo Park, CA). Plasmid pGro7 was from Fisher Scientific. The nickel-nitrilotriacetic acid (Ni-NTA) agarose resin was purchased from Invitrogen (Carlsbad, CA).
Expression and Purification of Human Spermine Oxidase
The DNA encoding the human SMO in a pDrive vector with codons optimized for expression in E. coli was subcloned into pET28b(+) vector using the NdeI and EcoRI sites at the 5’ and 3’ ends, respectively, for expression of the His-tagged enzyme. The plasmid was transformed into BL21(DE3) E. coli together with plasmid pGro7 to assist protein folding during expression. After induction with 0.15 mM isopropyl-β-d-thioglucanopyranoside, the cells were grown overnight at 18 °C in LB broth containing kanamycin and chloramphenicol. The cells were harvested by centrifugation at 5000g for 30 min at 4 °C. The cell paste was resuspended in 50 mM HEPES (pH 8.0), 10% glycerol, 2 µM pepstatin, 2 µM leupeptin, 0.1 mM NaCl, 100 µg/mL phenylmethanesulfonyl fluoride, and 100 µg/mL lysozyme, and lysed by sonication. After the lysate was centrifuged at 22400g, solid ammonium sulfate was added to the supernatant to reach 35% saturation. After centrifugation at 22400g for 30 min at 4 °C, the supernatant was brought to 50% ammonium sulfate saturation. The pellet was resuspended in 50 mM HEPES (pH 8.0), 10% glycerol, 0.1 mM NaCl, 2 µM pepstatin, and 2 µM leupeptin, and loaded onto a 10 mL Ni-NTA column equilibrated with this buffer. The protein was eluted with a linear gradient from 0 to 90 mM imidazole in 20 column volumes of the same buffer. Fractions containing SMO were pooled and concentrated by the addition of solid ammonium sulfate to 50% saturation. The pellet resulting from the final precipitation was resuspended in 50 mM HEPES (pH 8.0), 10% glycerol, and dialyzed against three changes of the same buffer. The resulting protein sample was centrifuged at 22400g for 30 min at 4 °C to remove precipitated protein and stored at −80 °C.
Extinction Coefficient of Human Spermine Oxidase
An equal volume of 10 M urea was added to enzyme in 50 mM HEPES, 10% glycerol, pH 8.0. Any precipitated protein was removed by centrifugation for 10 min at 14000g, and the visible absorbance spectrum of the supernatant was recorded. The change in absorbance between the enzyme-bound FAD and the free FAD after denaturation gave an extinction coefficient of 11.7 mM−1 cm−1 at 458 nm for the flavin in SMO based on the extinction coefficient of FAD of 11.3 mM−1cm−1 (24). This value was used to determine the enzyme concentration.
Assays
Enzyme activity was determined in a buffer containing 10% glycerol by following oxygen consumption with a Yellow Springs Instrument Model 5300 biological oxygen monitor. All assays were carried out at 25 °C. The buffers were 200 mM Tris-HCl from pH 7.0 to 8.75 and 200 mM CHES from pH 9.0 to 9.35. For assays not run in air-saturated buffer, the appropriate mixture of O2/N2 was bubbled into the assay mixture for 10 min prior to starting the reaction.
Rapid Reaction Kinetics
Rapid reaction kinetic measurements were performed using an Applied Photophysics SX-20 stopped-flow spectrophotometer. The instrument was made anaerobic by filling the system with a solution of 30 nM glucose oxidase and 1 mM glucose in anaerobic buffer the night before the experiment was carried out. Anaerobic conditions were obtained by applying several cycles of vacuum and oxygen-scrubbed argon to enzyme solutions, while substrate solutions were bubbled with argon. Glucose and glucose oxidase at final concentrations of 5 mM and 36 nM, respectively, were added to all solutions to maintain anaerobic conditions. For pH-profiles the buffers were 200 mM Tris-HCl and 200 mM CHES from pH 7.0 to 8.75 and 9.0 to 9.4, respectively. For the sequential mixing stopped-flow experiment used to study the reaction of the reduced enzyme with oxygen, 60 µM enzyme was first mixed anaerobically with 60 µM spermine. After aging times of 6 and 0.3 s, this was mixed with the oxygenated buffer equilibrated with different oxygen concentrations. All experiments were conducted at 25 °C.
Data Analysis
The kinetic data were analyzed using the program KaleidaGraph (Synergy Software, Reading, PA). To determine kcat/KM and KM values, initial rate data were fitted to the Michaelis-Menten equation. Equation 1 was used to analyze data for inhibition by competitive inhibitors and obtain the Ki values. Equation 2 was used to analyze the pH-dependence of kinetic parameters that decrease at both low and high pH. Equation 3 was used to analyze the pH-dependence of kinetic parameters that decrease at low pH. In these equations, Y is the experimental value of the kinetic parameter of interest, K1 and K2 are the dissociation constants for the ionizable groups, and C is the pH-independent value of the parameter. To determine the kinetic parameters for the reduction of SMO by spermine, stopped-flow traces were fitted to eq 4, which describes a biphasic exponential decay, where λ1 and λ2 are the first-order rate constants for each phase, A1 and A2 are the absorbances of each species at time t, and A∞ is the final absorbance. The resulting pseudo-first rate constants were analyzed using eq 5, where k3 is the rate constant for flavin reduction and Kd is the apparent dissociation constant for the substrate. The rate constant for flavin oxidation was obtained by fitting the change in absorbance of the flavin with time to eqs 4 and 6 at the delay times of 0.3 and 6 s, respectively. The value of k7 in Scheme 4 was calculated using Microsoft Excel 2004.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
Scheme 4.
RESULTS
Kinetic Mechanism of Human Spermine Oxidase
Flavoprotein oxidases can exhibit either a parallel or an intersecting line pattern in double reciprocal plots when both substrates are varied. The former results when flavin reduction is effectively irreversible, even though these enzymes do not follow the standard ping-pong kinetic mechanism (25). For a bisubstrate enzyme reaction, the proper steady-state kinetic equation can readily be identified using the fixed ratio approach (26). In this method, the ratio between the concentrations of both substrates is kept constant and the initial rate is determined as a function of the concentration of one of the substrates. Curvature in a double reciprocal plot suggests a sequential pattern, since there will be a term containing the square of the concentrations of one of the substrates in the rate equation (eq 7 with a = [O2]/[spermine]). A linear plot will arise if there is no term in the rate equation containing the concentrations of both substrates, as in a ping-pong mechanism (eq 8). For SMO a double reciprocal plot of initial rate vs. the spermine concentration at a fixed ratio of spermine to oxygen is linear (Figure 1). A fit of these data to the Michaelis-Menten gives a kcat value of 6.6 ± 0.2 s−1. The individual kcat/KM values for spermine and oxygen were determined in separate analyses by varying each at a fixed concentration of the other. The resulting values are given in Table 1. In addition, the kcat/KM value for N1-acetylspermine was determined at a constant concentration of oxygen. The value establishes that human SMO prefers spermine to N1-acetylspermine as a substrate by about 50-fold at pH 8.3 (Table 1). We could not detect any activity with spermidine as substrate.
| (7) |
| (8) |
Figure 1.
Double reciprocal plot of the initial rate vs. the spermine concentration at a fixed ratio of spermine to oxygen concentrations of two at pH 8.3, 25°C.
Table 1.
Steady-State Kinetic Parameters for Human Spermine Oxidasea
| Kinetic parameter | Value |
|---|---|
| kcat (spermine) (s−1)b | 6.6 ± 0.2 |
| kcat/Kspermine (mM−1 s−1)c | 37 ± 7 |
| Kspermine (µM) | 190 ± 32 |
| kcat/KO2 (spermine) (mM−1 s−1)d | 12.6 ± 1.6 |
| KO2 (spermine) (µM) | 565 ± 66 |
| kcat (N1-acetylspermine) (s−1)c | 0.40 ± 0.06 |
| kcat/KN-acetylspermine (mM−1 s−1)c | 0.8 ± 0.2 |
| KN-acetylspermine (µM)c | 492 ± 97 |
Conditions: pH 8.3 and 25 °C.
Determined by varying the concentrations of both oxygen and spermine.
Determined at 250 µM oxygen.
Determined at 500 µM spermine.
pH-Dependence of kcat/Kspermine
To gain insight into the basis for substrate specificity and the roles of amino acid residues in binding and catalysis, the effect of pH on the kcat/KM value for spermine was determined. A plot of the data (Figure 2) shows a bell-shaped dependence with a maximum at pH 8.3. This pH profile with limiting slopes of unity on either side of the pH maximum indicates the importance of two ionizable groups in the free enzyme or free substrate. The data can be fitted to eq 2 to extract the pKa values. However, the two pKa values are too close together to resolve, so that only the average of the two pKa values (8.3 ± 0.03) can be determined with confidence (Table 2).
Figure 2.
kcat/KM-pH profile for spermine oxidase with spermine as substrate. The line is from a fit of the data to eq 2.
Table 2.
pKa values for Spermine Oxidase
| Kinetic parameter | pK1 | pK2 |
|---|---|---|
| kcat/Kspermine | 8.3 ± 0.03 | 8.3 ± 0.03 |
| Ki (spermidine) | 7.9 ± 0.03 | 7.9 ± 0.03 |
| Ki (DBDB) | 7.3 ± 0.2 | 8.6 ± 0.1 |
| k3 (spermine) | 7.4 ± 0.02 | — |
pH Dependence of pKi Profiles
To corroborate the required protonation states of individual nitrogens in the substrate and aid in further distinguishing between groups that are responsible for binding or catalysis, several substrate analogs (Scheme 2) were examined as inhibitors. Spermidine and N,N’-dibenzyl-1,4-diaminobutane (DBDB) are inhibitors for SMO, while concentrations of 1, 12-diaminododecane up to 2 mM do not result in any detectable inhibition even at concentrations of spermine well below the KM value. Both spermidine and DBDB exhibit competitive inhibition vs. spermine (data not shown) with Ki values at pH 8.3 of 0.55 ± 0.02 mM and 0.32 ± 0.08 mM, respectively. The effects of pH on these Ki values were determined. As shown in Figure 3 both pH-pKi profiles are bell-shaped, so that the data were fit to eq 2 to extract the pKa values (Table 2). For spermidine this gives pKa values too close together to resolve accurately (27); consequently, only the average of the two pKa values can be estimated reliably with this inhibitor. For DBDB the two pKa values obtained by fitting the data to eq 2 are further apart and can be determined.
Scheme 2.
Figure 3.
pKi-pH profiles for spermidine (circles) and DBDB (triangles) as inhibitors of spermine oxidase. The lines are from fits to eq 2.
Reductive Half-Reaction
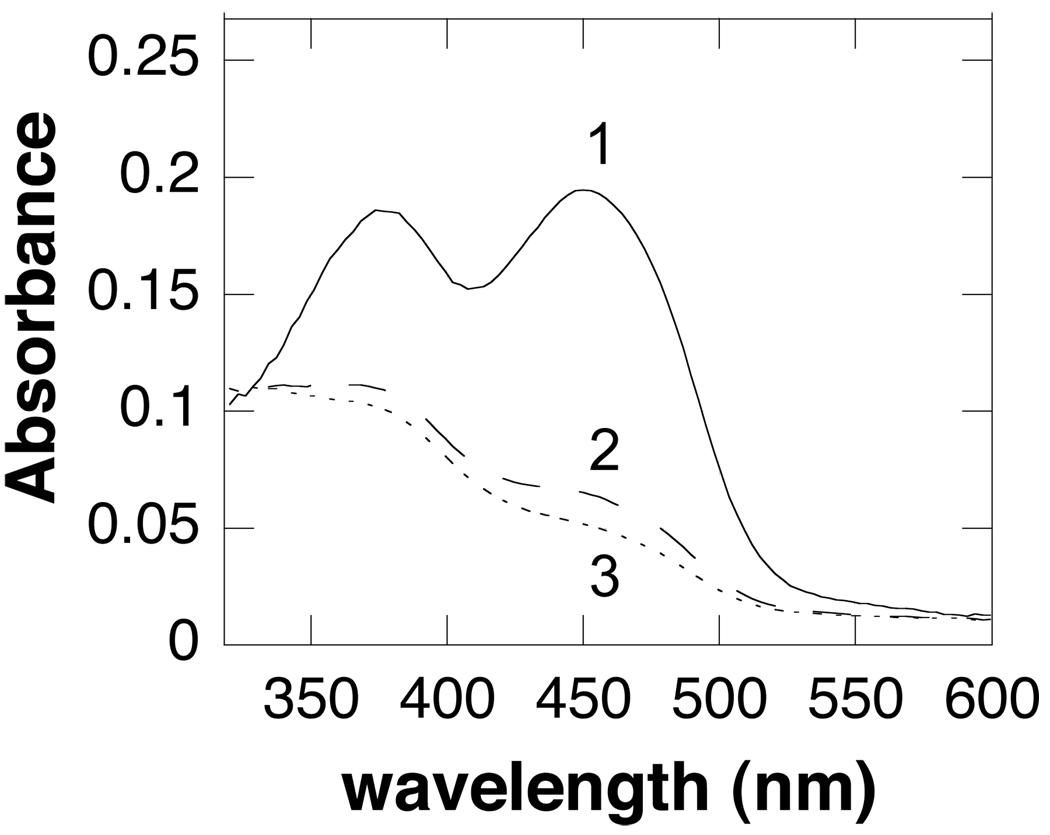
Stopped-flow spectroscopy was used to determine the kinetics of flavin reduction. When SMO and spermine are mixed rapidly in the absence of oxygen, the spectrum of oxidized flavin rapidly changes to the one of reduced flavin, followed by a slower phase with a much smaller absorbance change (Figure 4). Only the first-order rate constant for the fast phase is substrate-dependent. The observed rate constants at different spermine concentrations for this phase can be fit to eq 5. At pH 8.3, this gives values of 48 ± 8.2 µM for the apparent Kd value for spermine and 49 ± 1.3 s−1 for k3, the rate constant for flavin reduction at saturating concentrations of spermine. The rate constant for the slower phase is independent of the spermine concentration, with a value of 5.5 ± 0.3 s−1 at pH 8.3. These results are consistent with the mechanism in Scheme 3. The first phase is due to binding of spermine with no significant absorbance change, followed by amine oxidation and flavin reduction. The slower second phase can be attributed to dissociation of the oxidized amine from the enzyme. The spectrum of the intermediate does not show any evidence of the charge-transfer absorbance band often seen with flavoprotein oxidases; no such band has been reported for the related enzymes PAO (28, 29) or MAO (30).
Figure 4.
Absorbance spectra of flavin intermediates observed in the reductive half reaction of spermine oxidase (16.8 µM) by spermine at pH 8.3: 1, spectrum of spermine oxidase before reaction; 2, spectrum at the end of the first phase; 3, final spectrum.
Scheme 3.
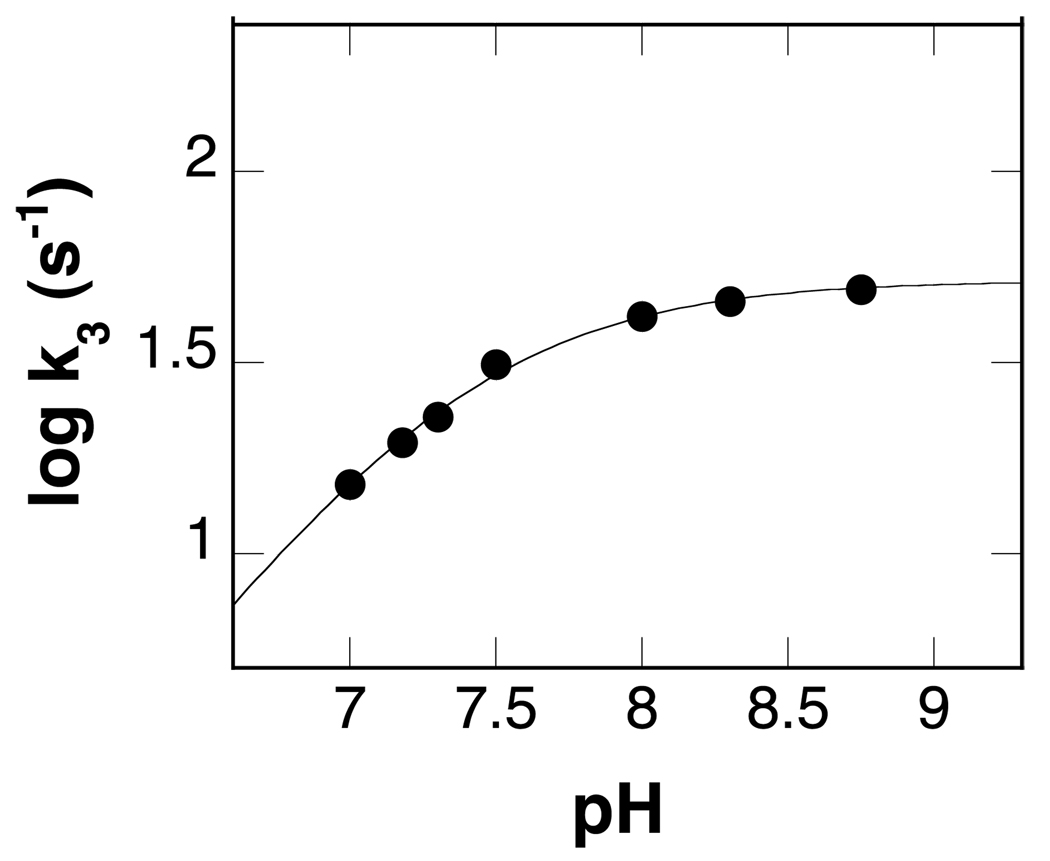
The effect of pH on the value of k3 was determined. As shown in Figure 5, the value of k3 decreases at low pH, but is independent of pH at high pH. These data were fit to eq 3 to obtain a pKa value of 7.4 ± 0.02 for the group that must be unprotonated for reduction.
Figure 5.
pH dependence of the rate constant for reduction of spermine oxidase by spermine at 25°C. The line is from a fit of the data to eq 3.
Oxidative Half-Reaction
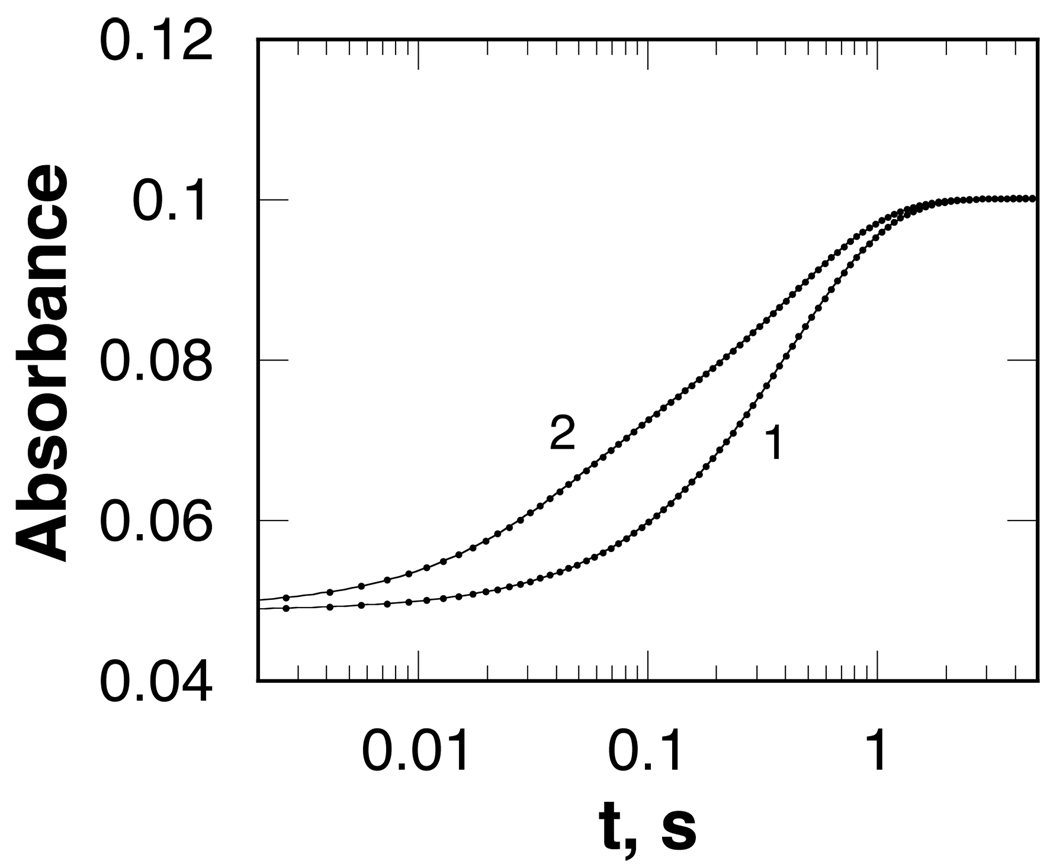
Stopped-flow spectrophotometry was also used to determine the kinetics of the reaction of the reduced enzyme with oxygen. This was done in a double-mixing experiment. SMO was first mixed with spermine in the absence of oxygen to form the reduced complex. After allowing the enzyme to be reduced, it was then mixed with buffer equilibrated with different concentrations of oxygen. When SMO and spermine are allowed to react for 6 s before mixing with oxygen, the increase in absorbance due to flavin oxidation is monophasic (Figure 6). The observed first order rate constant for the reaction varies directly with the concentration of oxygen (Figure 7), giving a second-order rate constant for the reaction of the reduced enzyme with oxygen of 4.2 ± 0.03 mM−1s−1. This kinetic behavior is consistent with an irreversible second-order reaction between the reduced enzyme and oxygen, with no evidence for an oxygen-enzyme complex. In contrast, when the aging time is shortened to 0.3 s, the absorbance increase is faster and biphasic (Figure 6). The rate constants for both phases vary directly with the concentration of oxygen (Figure 7), again consistent with irreversible bimolecular reactions. For the fast phase the second-order rate constant is 40 ± 5 mM−1s−1, while for the slow phase it is 4.0 ± 0.04 mM−1s−1. The latter value is essentially identical to the rate constant when the delay time is 6 s.
Figure 6.
Absorbance changes during the reaction of reduced spermine oxidase with 607 µM oxygen at pH 8.3 after first allowing the enzyme to react with spermine for 6 s (1) or 0.3 s (2). For clarity, only every fifth point is shown. The lines are from fits of the data to eq 4 or 6.
Figure 7.
The dependence of the rate constants for flavin oxidation on the oxygen concentration at pH 8.3, 25°C, when enzyme and spermine are first allowed to react for 6 s (A) or 0.3 s (B).
DISCUSSION
The results of the steady-state and rapid-reaction kinetic analyses of human SMO described here are consistent with the kinetic mechanism of Scheme 4 and the values for the individual rate constants given in Table 3. The ping-pong kinetic pattern established by the data in Figure 1 establishes that reduction is effectively irreversible (25). The Kd value, the first-order rate constant for flavin reduction (k3), and the constant for product release from the reduced EFLredP complex (k9) can all be obtained from the stopped-flow analyses of the reductive half-reaction in the absence of oxygen. The value of k9 is comparable to kcat, so that this step could be along the catalytic pathway for SMO. The values of k5 and k11 are from the stopped-flow analyses of the oxidative half-reaction.
Table 3.
Intrinsic Kinetic Parameters for Spermine Oxidase with Spermine as Substrate at pH 8.3
| Kinetic Parameter | Value |
|---|---|
| Kd (µM) | 48 ± 8 |
| k3 (s−1) | 49 ± 1 |
| k5 (mM−1 s−1) | 40 ± 5 |
| k7 (s−1) | 5.9 ± 0.6 |
| k9 (s−1) | 5.5 ± 0.3 |
| k11 (mM−1 s−1) | 4.0 ± 0.04 |
The oxidative reaction cannot be described as a simple reaction of only one of the SMO intermediates with oxygen since the absorbance change during oxidation is either monophasic or biphasic depending on the aging time. At an aging time of 6 s, all of the oxidized product would have dissociated from the reduced enzyme, so that oxygen is reacting with the free reduced enzyme. At an aging time of 0.3 s, a significant fraction of the enzyme still has the oxidized product bond, resulting in a biphasic reaction in which the rate constants for both phases depend on the oxygen concentration. The rapid phase can be attributed to the reaction of the reduced enzyme-product complex with oxygen. The excellent agreement of the rate constant for the slow phase at the shorter aging time with the rate constant for the reaction at the longer aging time supports the conclusion that the slow phase is due to the reaction of the free reduced enzyme. An increase in the rate constant for oxidation when product is bound to a reduced flavoprotein oxidase has been observed with other flavin amine oxidases (31).
Mechanisms including only one path for reaction of the reduced flavin intermediate with oxygen result in the rate constant for the oxidative reaction being equal to the kcat/KO2 value (32). However, the values of the rate constants for both phases in the present case are significantly different from the kcat/KO2 value of 12.6 ± 1.6 mM−1s−1. When flavin oxidation occurs by both pathways in Scheme 4, the kcat/KO2 value is not a simple function of the individual rate constants. The kinetic equation for this mechanism is eq 9. This is given in double-reciprocal form to more readily illustrate the oxygen dependency. The only rate constant in Scheme 4 that was not measured directly is k7, the rate constant for dissociation of the product from the oxidized enzyme. To obtain the value of k7, the initial rate of spermine oxidation was calculated using eq 9 and the values of the other kinetic parameters in Table 3. The differences between the observed and calculated rates at the concentrations of oxygen in the experiment shown in Figure 8 are minimized when k7 is 5.94 s−1.2 Fitting these calculated rates to the Michaelis-Menten equation yields a kcat/KO2 value of 11.5 ± 1.5 mM−1s−1 and a kcat value of 7.6 ± 0.8 s−1. The agreement of these values with the experimentally determined values (Table 1) establishes that the branched mechanism of Scheme 4 provides a better model for the kinetics than simpler models involving only the upper or lower pathway for the oxidative half-reaction. The possibility of such a bifurcated mechanism for the oxidative half-reaction has been suggested previously for flavoprotein oxidases (33), but the ability of the calculated rates to be fit by the Michaelis-Menten equation establishes that such behavior would be difficult to detect in steady-state kinetic assays.
| (9) |
Figure 8.
Comparison of the observed rates of spermine oxidation by spermine oxidase as a function of oxygen concentration (circles) with values calculated from the mechanism of Scheme 4 and the kinetic parameters in Table 3 (triangles), at pH 8.3, 25°C. The line is from a fit of the calculated data to the Michaelis-Menten equation.
The kcat/KM-pH profile for spermine is most consistent with the active form of the substrate having three of the four nitrogens protonated. The concentrations of the mono-, di- and triprotonated forms of spermine are maximal at pH 10.5, 9.5, and 8.4 (28, 34–37). The expected pH distribution of the triprotonated form agrees well with the pH optimum in the kcat/KM-pH profile for spermine of 8.3. The data do not allow us to identify the unprotonated nitrogen, but it is most likely to be the secondary nitrogen at the site of oxidation. Earlier studies with mouse PAO have established that the reacting nitrogen must be neutral for amine oxidation (28), and a neutral nitrogen would be required for hydride transfer to the flavin (12). The need for this nitrogen to be neutral provides a ready explanation for the pKa of 7.4 seen in the k3 pH profile. If the fully protonated form of spermine can bind but not react, its pKa when bound to the enzyme will be seen in this profile. The alternate possibility that this pKa is due to an amino acid residue that must be unprotonated cannot be ruled out, especially in light of the results with inhibitors (vide infra).
The ability of the incorrectly protonated form of spermine to bind the enzyme is supported by the pKi-pH profiles for spermidine and DBDB. The mono- and diprotonated forms of spermidine have their maximum concentrations at pH 10.4 and 9.1, too high for this profile to be due only to the protonation state of spermidine. However, the triprotonated form of this inhibitor has a pKa value of 8.3 ± 0.1, primarily due to the secondary nitrogen (28, 35–38). This suggests that the average value of 7.9 obtained from the pKi-pH profile is due to binding of the triprotonated form of this inhibitor to a form of the enzyme in which an active site residue is unprotonated. The average value of the pKas of 7.9 and the pKa value of 8.3 for the triprotonated form of spermine yield a pKa value of 7.5 for the group on the enzyme. While pKa values have not been reported for DBDB, they can be estimated. Beginning with the pKa of N-ethylbenzene of 9.7 (39), adjusting for the effects of another protonatable nitrogen four carbons away (40) allows estimation of the macroscopic pKa values for the diprotonated and monoprotonated forms of DBDB as 8.5 and 9.8. The pKa for the diprotonated form of DBDB is in good agreement with the pKa of 8.6 in the pKi-pH profile for this inhibitor, consistent with the doubly charged form being the active inhibitor. This suggests that the pKa of 7.3 for a group that must be unprotonated can be assigned to an amino acid residue on the protein. This value is in good agreement with the value of 7.5 calculated from the pH profile for spermidine. Thus, the acidic pKi in the pKi-pH profiles for both spermidine and DBDB is most likely due to an amino acid residue in the active site of SMO that must be unprotonated for binding of either inhibitor, while the basic limb can be attributed to the need for all of the nitrogens in the inhibitors to be protonated. Based on the sequence of SMO and the structures of maize PAO and Fms1, likely candidates for the amino acid residue are His82 and His212.
The requirements for productive binding of spermine and for binding of inhibitors are clearly different. This suggests that these two inhibitors bind to the protein in a slightly different fashion than does spermine. In the case of DBDB it is certainly possible that the inhibitor binds such that neither nitrogen is positioned where the reactive nitrogen of spermine would be located in the active site. Spermidine could bind with a primary nitrogen in place of the reactive secondary nitrogen of spermine. In both cases the out of register binding would be assisted by an unprotonated amino acid residue. Structures have been described of Fms1, a yeast spermine oxidase, with spermidine and 1, 8-diaminooctane bound (pdb codes 3cn8 and 3bi5). These show that both inhibitors bind differently from spermine (14), in that neither has an atom near the N5 position of the flavin, the site to which hydride transfer would occur.
SMO differs from mammalian PAOs in the relative preference for spermine versus N-acetylspermine. The kcat-KM pH profiles for the two enzymes suggest that the two enzymes discriminate between these two substrates based on their protonation states. PAO requires a form of the substrate with only one protonated nitrogen (28), while SMO requires the triprotonated form of spermine. N-Acetylspermine would then be a poor substrate for SMO because acetylation of N1 prevents it from being protonated. Conversely, acetylation increases the amount of substrate with only a single positive charge, the required form for PAO. The different pH optima are also consistent with the different locations of the enzymes in the cell. The pH of the peroxisome where PAO is located is 8–8.5 (41, 42), consistent with its higher pH optimum (28) compared to the cytosolic SMO.
In conclusion, the complete kinetic mechanism of human SMO has been determined using both steady-state and rapid reaction kinetic methods, and the individual rate constants have been measured. With spermine as substrate, product release occurs from both the EFlredP and EFloxP complexes. In addition, the triprotonated form of spermine is the active form of the substrate, whereas the inhibitors spermine and DBDB bind most tightly when fully protonated.
Abbreviations
- SMO
spermine oxidase
- DBDB
N,N’-dibenzyl-1,4-diaminobutane
- PAO
polyamine oxidase
- Ni-NTA
nickel-nitrilotriacetic acid
- MAO
liver monoamine oxidase
- LSD1
lysine-specific demethylase
- DAAO
D-amino acid oxidase
Footnotes
This work was supported in part by NIH grant GM058698.
The best way of estimating the confidence interval for the value of k7 is not obvious. Varying the value by ± 0.1 from a value of 5.94 results in a visibly poorer fit to the data. However, the precision of the individual rates is only 5–10%, and the values of k5 and k9 have precisions of ~10%. Thus, a reasonable estimate for the precision of k7 is 10%.
REFERENCES
- 1.Gerner EW, Meyskens FL., Jr Polyamines and cancer: old molecules, new understanding. Nat. Rev. Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- 2.Seiler N. Catabolism of polyamines. Amino Acids. 2004;26:217–233. doi: 10.1007/s00726-004-0070-z. [DOI] [PubMed] [Google Scholar]
- 3.Casero R, Jr, Pegg A. Spermidine/spermine N1-acetyltransferase--the turning point in polyamine metabolism. FASEB J. 1993;7:653–661. [PubMed] [Google Scholar]
- 4.Seiler N, Peter M, Yu KFT, Alan AB. Polyamine oxidase, properties and functions. Prog. Brain Res. 1995;106:333–344. doi: 10.1016/s0079-6123(08)61229-7. [DOI] [PubMed] [Google Scholar]
- 5.Vujcic S, Diegelman P, Bacchi CJ, Kramer DL, Porter CW. Identification and characterization of a novel flavin-containing spermine oxidase of mammalian cell origin. Biochem. J. 2002;367:665–675. doi: 10.1042/BJ20020720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Murray-Stewart T, Devereux W, Hacker A, Frydman B, Woster PM, Casero RA., Jr Properties of purified recombinant human polyamine oxidase, PAOh1/SMO. Biochem. Biophys. Res. Commun. 2003;304:605–611. doi: 10.1016/s0006-291x(03)00636-3. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Devereux W, Woster PM, Stewart TM, Hacker A, Casero RA., Jr Cloning and characterization of a human polyamine oxidase that is inducible by polyamine analogue exposure. Cancer Res. 2001;61:5370–5373. [PubMed] [Google Scholar]
- 8.Pledgie A, Huang Y, Hacker A, Zhang Z, Woster PM, Davidson NE, Casero RA., Jr Spermine oxidase SMO(PAOh1), not N1-acetylpolyamine oxidase PAO, is the primary source of cytotoxic H2O2 in polyamine analogue-treated human breast cancer cell lines. J. Biol. Chem. 2005;280:39843–39851. doi: 10.1074/jbc.M508177200. [DOI] [PubMed] [Google Scholar]
- 9.Wu T, Yankovskaya V, McIntire WS. Cloning, sequencing, and heterologous expression of the murine peroxisomal flavoprotein, N1-acetylated polyamine oxidase. J. Biol. Chem. 2003;278:20514–20525. doi: 10.1074/jbc.M302149200. [DOI] [PubMed] [Google Scholar]
- 10.Sebela M, Radová A, Angelini R, Tavladoraki P, Frébort I, Pec P. FAD-containing polyamine oxidases: a timely challenge for researchers in biochemistry and physiology of plants. Plant Sci. 2001;160:197–207. doi: 10.1016/s0168-9452(00)00380-0. [DOI] [PubMed] [Google Scholar]
- 11.Landry J, Sternglanz R. Yeast Fms1 is a FAD-utilizing polyamine oxidase. Biochem. Biophys. Res. Commun. 2003;303:771–776. doi: 10.1016/s0006-291x(03)00416-9. [DOI] [PubMed] [Google Scholar]
- 12.Fitzpatrick PF. Oxidation of amines by flavoproteins. Arch. Biochem. Biophys. 2010 doi: 10.1016/j.abb.2009.07.019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binda C, Coda A, Angelini R, Federico R, Ascenzi P, Mattevi A. A 30 Å long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase. Structure. 1999;7:265–276. doi: 10.1016/s0969-2126(99)80037-9. [DOI] [PubMed] [Google Scholar]
- 14.Huang Q, Liu Q, Hao Q. Crystal structures of Fms1 and its complex with spermine reveal substrate specificity. J. Mol. Biol. 2005;348:951–959. doi: 10.1016/j.jmb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 15.Binda C, Mattevi A, Edmondson DE. Structure-function relationships in flavoenzyme dependent amine oxidations. A comparison of polyamine oxidase and monoamine oxidase. J. Biol. Chem. 2002;277:23973–23976. doi: 10.1074/jbc.R200005200. [DOI] [PubMed] [Google Scholar]
- 16.Denu JM, Fitzpatrick PF. Intrinsic primary, secondary, and solvent kinetic isotope effects on the reductive half-reaction of D-amino acid oxidase: Evidence against a concerted mechanism. Biochemistry. 1994;33:4001–4007. doi: 10.1021/bi00179a029. [DOI] [PubMed] [Google Scholar]
- 17.Mattevi A, Vanoni MA, Todone F, Rizzi M, Teplyakov A, Coda A, Bolognesi M, Curti B. Crystal structure of D-amino acid oxidase: A case of active site mirror-image convergent evolution with flavocytochrome b2. Proc. Natl. Acad. Sci. USA. 1996;93:7496–7501. doi: 10.1073/pnas.93.15.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurtz KA, Rishavy MA, Cleland WW, Fitzpatrick PF. Nitrogen isotope effects as probes of the mechanism of D-amino acid oxidase. J. Am. Chem. Soc. 2000;122:12896–12897. [Google Scholar]
- 19.Ralph EC, Fitzpatrick PF. pH and kinetic isotope effects on sarcosine oxidation by N-methyltryptophan oxidase. Biochemistry. 2005;44:3074–3081. doi: 10.1021/bi047716h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ralph EC, Hirschi JS, Anderson MA, Cleland WW, Singleton DA, Fitzpatrick PF. Insights into the mechanism of flavoprotein-catalyzed amine oxidation from nitrogen isotope effects on the reaction of N-methyltryptophan oxidase. Biochemistry. 2007;46:7655–7664. doi: 10.1021/bi700482h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scrutton NS. Chemical aspects of amine oxidation by flavoprotein enzymes. Nat. Prod. Rep. 2004;21:722–730. doi: 10.1039/b306788m. [DOI] [PubMed] [Google Scholar]
- 22.Ralph EC, Anderson MA, Cleland WW, Fitzpatrick PF. Mechanistic studies of the flavoenzyme tryptophan 2-monooxygenase: Deuterium and 15N kinetic isotope effects on alanine oxidation by an L-amino acid oxidase. Biochemistry. 2006;45:15844–15852. doi: 10.1021/bi061894o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edmondson DE, Binda C, Mattevi A. Structural insights into the mechanism of amine oxidation by monoamine oxidases A and B. Arch. Biochem. Biophys. 2007;464:269–276. doi: 10.1016/j.abb.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitby LG. New method for preparing flavin-adenine dinucleotide. Biochem. J. 1953;54:437–442. doi: 10.1042/bj0540437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer G, Massey V. Mechanisms of flavoprotein catalysis. In: Singer TP, editor. Biological oxidation. New York: John Wiley and Sons; 1968. pp. 263–300. [Google Scholar]
- 26.Rudolph FB, Fromm HJ. Plotting methods for analyzing enzyme rate data. Methods Enzymol. 1979;63:138–159. doi: 10.1016/0076-6879(79)63009-4. [DOI] [PubMed] [Google Scholar]
- 27.Cleland WW. Enzyme kinetics as a tool for determination of enzyme mechanisms. In: Bernasconi CF, editor. Investigation of Rates and Mechanism. 4th Ed. Part I. Vol. 6. New York: John Wiley & Sons; 1986. pp. 791–870. [Google Scholar]
- 28.Henderson Pozzi M, Gawandi V, Fitzpatrick PF. pH Dependence of a Mammalian Polyamine Oxidase: Insights into Substrate Specificity and the Role of Lysine 315. Biochemistry. 2009;48:1508–1516. doi: 10.1021/bi802227m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Royo M, Fitzpatrick PF. Mechanistic studies of mouse polyamine oxidase with N1,N12-bisethylspermine as a substrate. Biochemistry. 2005;44:7079–7084. doi: 10.1021/bi050347k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller JR, Edmondson DE, Grissom CB. Mechanistic probes of monoamine oxidase B catalysis: Rapid-scan stopped flow and magnetic field independence of the reductive half-reaction. J.Am.Chem.Soc. 1995;117:7830–7831. [Google Scholar]
- 31.Pollegioni L, Blodig W, Ghisla S. On the mechanism of D-amino acid oxidase. Structure/linear free energy correlations and deuterium kinetic isotope effects using substituted phenylgylcines. J.Biol.Chem. 1997;272:4924–4934. doi: 10.1074/jbc.272.8.4924. [DOI] [PubMed] [Google Scholar]
- 32.Fitzpatrick PF, Massey V. The kinetic mechanism of D-amino acid oxidase with D-α-aminobutyrate as substrate: Effect of enzyme concentration on the kinetics. J. Biol. Chem. 1982;257:12916–12923. [PubMed] [Google Scholar]
- 33.Porter DJT, Bright HJ. Flavoprotein oxidase mechanisms. In: Singer TP, editor. Flavins and Flavoproteins. Amsterdam: Elsevier Scientific Publishing Company; 1976. pp. 225–237. [Google Scholar]
- 34.Frassineti C, Ghelli S, Gans P, Sabatini A, Moruzzi MS, Vacca A. Nuclear magnetic resonance as a tool for determining protonation constants of natural polyprotic bases in solution. Anal. Biochem. 1995;231:374–382. doi: 10.1006/abio.1995.9984. [DOI] [PubMed] [Google Scholar]
- 35.Lomozik L, Gasowska A, Bolewski L. Copper (II) ions as a factor interfering in the interaction between bioligands in systems with adenosine and polyamines. J. Inorg. Biochem. 1996;63:191–206. [Google Scholar]
- 36.Bencini A, Bianchi A, Garcia-Espana E, Micheloni M, Remirez JA. Proton coordination by polyamine compounds in aqueous solution. Coord. Chem. Rev. 1999;188:97–156. [Google Scholar]
- 37.da Silva JA, Felcman J, Lopes CC, Lopes RSC, Villar JDF. Study of the protonation/deprotonation sequence of two polyamines: bis-[(2S)-2-pyrrolidinylmethyl] ethylenediamine and spermidine by 1H and 13C nuclear magnetic resonance. Spectrosc. Lett. 2002;35:643–661. [Google Scholar]
- 38.Kimberly M, Goldstein JH. Determination of pKa values and total proton distribution pattern of spermidine by carbon-13 nuclear magnetic resonance titrations. Anal. Chem. 1981;53:789–793. [Google Scholar]
- 39.Hall HK. Correlation of the Base Strengths of Amines. J. Am. Chem. Soc. 1957;79:5441–5444. [Google Scholar]
- 40.Aikens D, Bunce S, Onasch F, Parker R, 3rd, Hurwitz C, Clemans S. The interactions between nucleic acids and polyamines. II. Protonation constants and 13C-NMR chemical shift assignments of spermidine, spermine, and homologs. Biophys. Chem. 1983;17:67–74. doi: 10.1016/0301-4622(83)87015-x. [DOI] [PubMed] [Google Scholar]
- 41.van Roermund CWT, de Jong M, IJist L, van Marie J, Dansen TB, Wanders RJA, Waterham HR. The peroxisomal lumen in Saccharomyces cerevisiae is alkaline. J. Cell Sci. 2004;117:4231–4237. doi: 10.1242/jcs.01305. [DOI] [PubMed] [Google Scholar]
- 42.Dansen TB, Wirtz KWA, Wanders RJA, Pap EHW. Peroxisomes in human fibroblasts have a basic pH. Nat. Cell Biol. 1999;2:51–53. doi: 10.1038/71375. [DOI] [PubMed] [Google Scholar]