Abstract
The pineal complex of zebrafish consists of a pineal organ and a left-sided parapineal organ. Mutation of the floating head (flh) gene, which encodes a homeodomain protein, causes premature termination of pineal cell division without affecting specification or asymmetric placement of the parapineal. The from beyond (fby) mutation, a premature stop codon in the T-domain-containing protein Tbx2b, disrupts formation of the parapineal while leaving the pineal largely intact. However, flh is reported as being required for tbx2b transcription. To resolve the paradox that flhand tbx2b mutants have opposite phenotypes but have been placed in the same genetic pathway, we have examined transcriptional cross-regulation in single flh or fby mutants and genetic epistasis in double mutants. Careful analysis shows that flh is not required for tbx2b transcription and double mutants exhibit an additive phenotype. We conclude that Flh and Tbx2b regulate separate programs of pineal and parapineal development.
Keywords: homeodomain, t-box, epiphysis, epithalamus, dorsal diencephalon
Introduction
The epithalamus, or dorsal diencephalon, of the vertebrate brain includes the left and right habenular nuclei (habenulae) and the melatonin-secreting pineal complex. The habenulae receive afferent input from forebrain regions including the septum, globus pallidus, and lateral hypothalamus, and send efferent axons to the midbrain (Sutherland, 1982). The pineal complex includes the pineal organ, which is placed just to the left of the midline (Liang et al., 2000), and an accessory organ called the parapineal in lampreys and fish (Concha and Wilson, 2001). The parapineal organ is also asymmetrically placed in the brain, a phenomenon first described by Hill (1892). In zebrafish, the parapineal lies adjacent to the left habenula and dictates left-right (L-R) differences in size, neuropil and gene expression between the left and right habenulae (Concha et al., 2003; Gamse et al., 2003).
The zebrafish pineal complex anlage gives rise to both the pineal and parapineal organs. Lineage labeling at 22–24 hours post-fertilization (h) reveals that the anterior portion of the pineal complex anlage gives rise to both pineal and parapineal cells, while more posterior regions produce pineal cells only (Concha et al., 2003). Two transcription factors have been identified that regulate pineal or parapineal development: the homeodomain protein Floating head (Flh) and the T-box domain protein T-box 2b (Tbx2b, previously known as Tbx-c). Flh activity promotes the generation of pineal cells; in flhn1 mutants, neurogenesis initiates in the pineal complex but ends prematurely at 18 somites (s) stage (Masai et al., 1997). By contrast, Tbx2b is required for the specification and migration of parapineal cells; in fbyc144 mutants (a mutation in tbx2b), few or no parapineal cells develop and those that do fail to migrate to the left side of the brain (Snelson et al., 2008).
The regulatory relationship between flh and tbx2b is unclear. In the pineal complex anlage, flh expression begins at 80% epiboly (Masai et al., 1997), about 4 hr prior to tbx2b expression (Snelson et al., 2008). Expression of tbx2b and flh overlaps extensively within the pineal complex anlage at 24 h (Snelson et al., 2008). A previous publication (Cau and Wilson, 2003) suggested that Flh is required for expression of tbx2b. However, the phenotypes of flh and fby mutants are very different, arguing against the two genes acting in the same pathway. Asymmetrically-placed parapineal cells form in flh mutant larvae (Gamse et al., 2002, 2003), but not fby mutant larvae (Snelson et al., 2008). This observation prompted a re-evaluation of tbx2b and flh cross-regulation.
Examination of flh and tbx2b mRNA in fby and flh mutants reveals that they do not regulate one another's expression, and flh;fby double mutants exhibit an additive phenotype. Additionally, in flh mutants, specification and asymmetric migration of parapineal cells are indistinguishable from wild-type (WT) embryos. These results indicate that flh and tbx2b act independently of one another during pineal versus parapineal development.
Results
Parapineal Cell Division Ends Prior to Pineal Cell Division
To identify the period of development when pineal and parapineal cells are dividing, we performed BrdU pulse labeling at stages between 10 s and 30 hr and examined the fate of labeled cells at 4 days post-fertilization (d). To identify pineal and parapineal cells, embryos carrying the transgene Tg-(foxd3:gfp)fkg3 were used (Gilmour et al., 2002). Peak labeling of parapineal cells occurred when embryos received a half-hour pulse of BrdU at the 15 s stage (Fig. 1A,E,F and Table 1); BrdU pulses before (10 s) or after (18 s or later) labeled few or no parapineal cells (Fig. 1E,F and Table 1). Pineal cells also have maximal incorporation of BrdU at 15 s; interestingly, a second smaller peak of labeling occurs at 24 h (Fig. 1C,E,G and Table 1). In flh mutant larvae, pineal and parapineal BrdU labeling at the 10 s stage is similar to WT and increases at 15 s; however, almost no BrdU labeling of the pineal or parapineal occurs at or after 18 s (Fig. 1B,D,F,G and Table 1). The number of foxd3:gfp-positive cells in the pineal organ is reduced (Table 1), consistent with a premature end to pineal neurogenesis in flh mutants (Masai et al., 1997).
Fig. 1.
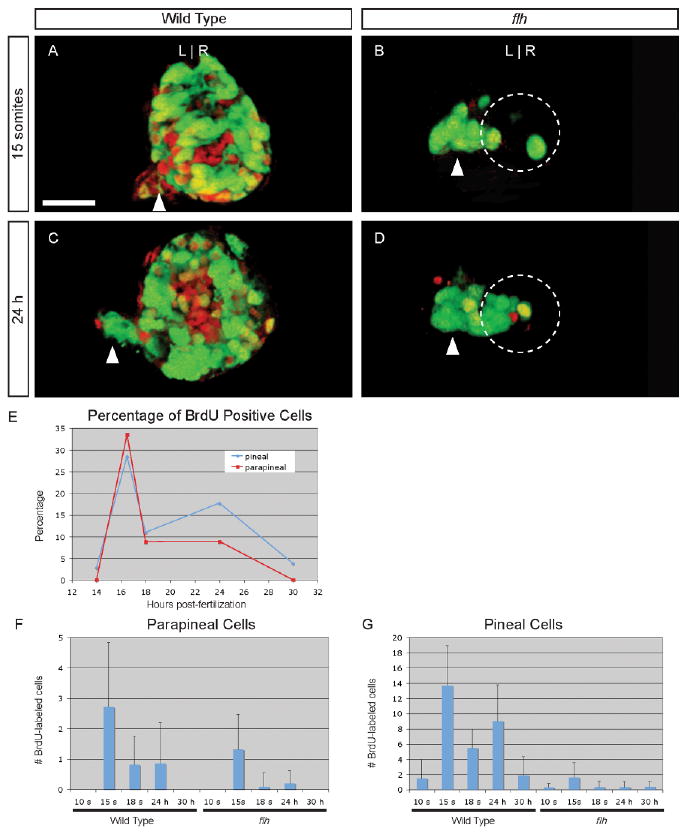
Cell division of parapineal precursors is largely complete by 18 s. A–D: Dorsal views of 4 d larvae, with BrdU (red) and foxd3:gfp (green) labeling in the epithalamus, following a BrdU pulse at (A, B) 15 s or (C, D) 24 hr stage in (A, C) WT or (B, D) flh mutant embryos. Dashed circles in B, D indicate the pineal cells. Scale bar = 25 μM. E: Line graph of the percentage of WT parapineal (red line) or pineal (blue line) cells that are labeled at 4 d, following a BrdU pulse at the indicated developmental stages. F: Bar graph showing the absolute number of BrdU-positive parapineal cells in WT or flh embryos. G: BrdU-positive pineal cells in WT or flh embryos.
TABLE 1. Fate of Pineal Complex Cells Labeled with BrdU Between 10 s and 30 hr.
| Genotype | Stagea | GFP+ pineal cells | BrdU+ pineal cells | GFP+ parapineal cells | BrdU+ parapineal cells | N |
|---|---|---|---|---|---|---|
| WT | 10 s | 47 +/− 6 | 1 +/− 3 | 8 +/− 2 | 0 | 11 |
| 15 s | 50 +/− 4 | 14 +/− 5 | 8 +/− 2 | 3 +/− 2 | 25 | |
| 18 s | 50 +/− 4 | 5 +/− 2b | 9 +/− 2 | 0.8 +/− 0.9b | 21 | |
| 24 h | 51 +/− 5 | 9 +/− 5b,c | 8 +/− 2 | 1 +/− 1b,d | 13 | |
| 30 h | 48 +/− 7 | 2 +/− 2 | 10 +/− 2 | 0 | 7 | |
| flh−/− | 10 s | 17 +/− 6 | 0.3 +/− 0.5e | 8 +/− 2 | 0b,e | 10 |
| 15 s | 16 +/− 5 | 2 +/− 2b | 11 +/− 2 | 1 +/− 1b | 25 | |
| 18 s | 15 +/− 4 | 0.3 +/− 0.8f | 7 +/− 2 | 0.1 +/− 0.4b,f | 21 | |
| 24 h | 14 +/− 5 | 0.3 +/− 0.7b | 8 +/− 2 | 0.2 +/− 0.4b | 15 | |
| 30 h | 15 +/− 6 | 0.4 +/− 0.7 | 10 +/− 3 | 0 | 17 |
Developmental stage when BrdU was first applied to embryos during a 0.5 hr pulse; all samples were fixed at 4 d for analysis.
Significantly different from WT 15 s by 2-tailed T-test, p < 0.03
Significantly different from WT 18 s by 2-tailed T-test, p < 0.01
Not significantly different from WT 18 s by 2-tailed T-test, p > 0.7
Not significantly different from WT 10 s by 2-tailed T-test, p > 0.17.
Significantly different from WT 18 s by 2-tailed T-test, p < 0.01
The Parapineal Organ in flh Mutants Develops Similarly to WT
In flh mutants, a single asymmetrically placed parapineal organ is always observed (Gamse et al., 2002) (although its placement is reversed in half of mutant larvae due to disrupted midline formation; Halpern et al., 1995). To confirm that the timing and migration of parapineal cells in flh mutants is similar to WT, time-lapse imaging was used in embryos carrying the transgene Tg(foxd3:gfp)fkg3 (Fig. 2). Similar to WT (Snelson et al., 2008; see Supplemental Movie 1, which can be viewed online), in flh mutants foxd3:gfp-positive parapineal cells are first visible at 31 h (Fig. 2A); cells continue to migrate unilaterally in a cluster between 32 and 46 h (Fig. 2B–P and Supplemental Movie 2).
Fig. 2.
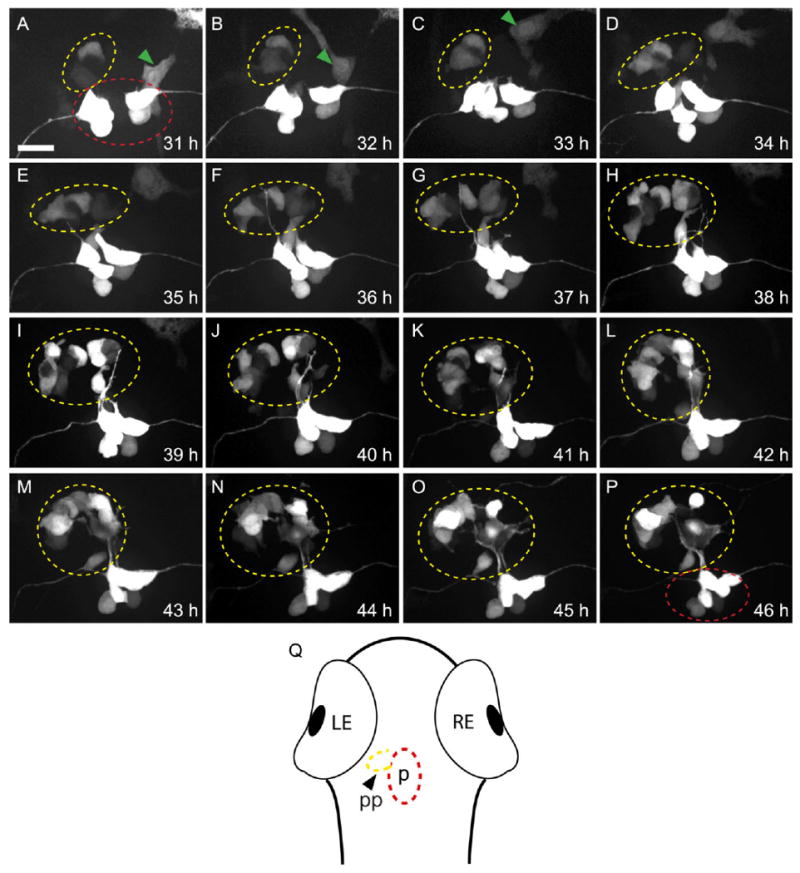
Parapineal cells migrate asymmetrically in flh mutants. All panels are dorsal views of foxd3:gfp labeling in the epithalamus of a single embryo at the times indicated. Parapineal cells are circled in yellow, pineal cells in red; a neural crest cell (which also expresses foxd3:gfp) in A–C is indicated by a green arrowhead. A: The first labeled parapineal cells are apparent near the midline at 31 hr. B–E: They migrate leftward. F–P: They are joined by more leftward-migrating parapineal cells during the subsequent 10 hr. Scale bar = 25 μm. Q: Diagram of a zebrafish larvae, showing the relative position of the pineal (p, red oval) and parapineal (pp, yellow oval) within the head.
To test whether the correct number of parapineal cells are specified in flh mutant larvae, we examined the expression of the gene growth factor independent-1 (gfi-1), which is expressed in parapineal cells from 48 h onward; no gfi-1 expression is seen in pineal cells (Dufourcq et al., 2004). In WT larvae, an average of 10 gfi-1 expressing cells are found on the left side of the brain in a tight cluster at 4 d (Fig. 3A and Table 2). Similarly, flh mutants form 10 gfi-1-expressing cells on average (Fig. 3B and Table 2).
Fig. 3.
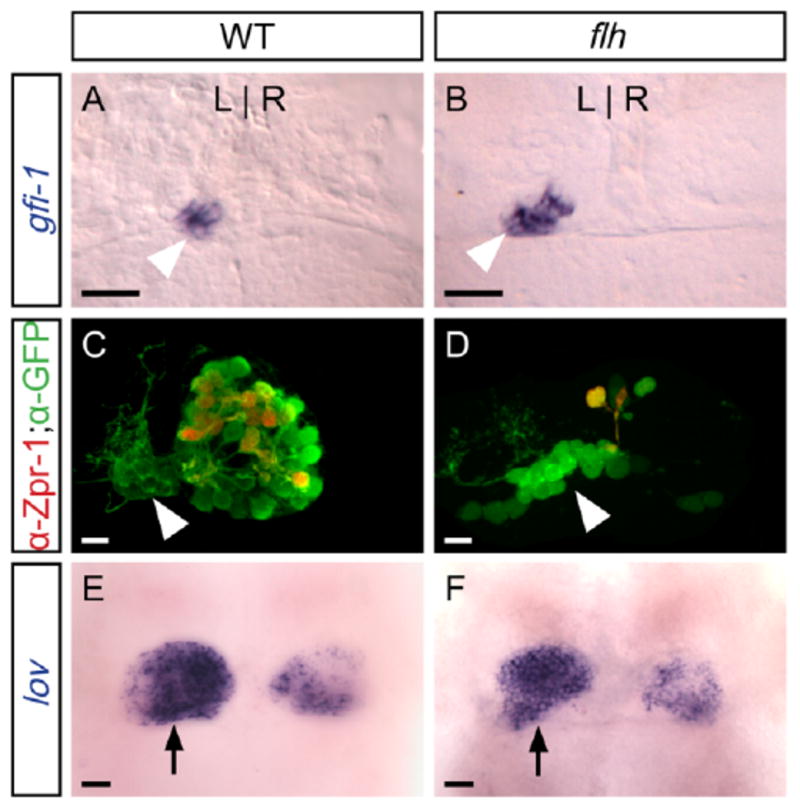
In flh mutants, parapineal specification is normal, but the pineal organ is reduced in size. All panels are dorsal views of the epithalamus of 4 d larvae. A: Expression of gfi-1 (blue), which is restricted to the parapineal organ (arrowhead), reveals ∼10 left-sided cells in WT and (B) flh larvae. C: In WT larvae, both the parapineal (arrowhead) and pineal express foxd3:gfp (green); many cells of the pineal organ are also labeled by the red-green cone marker Zpr-1 (Fret43) (red), but no parapineal cells are Zpr-1-labeled. D: In flh mutants, the pineal organ is drastically reduced in size, including fewer Zpr-1-labeled cells. As in WT, no Zpr-1-labeled cells are detected in the parapineal. E: At 4 d, lov expression is asymmetric in the habenular nuclei; the habenula adjacent to the parapineal (arrow) expresses lov extensively in WT and (F) flh mutants. Scale bars = 25 μm.
TABLE 2. Number of Labeled Cells Present in the Pineal Complex of Single and Double Mutants.
| Gene/protein (cell types labeled) | Genotype | Number of cellsa | N |
|---|---|---|---|
|
gfi-1 (parapineal cells) |
WT | 10 +/− 1 | 12 |
| flh | 10 +/− 1 | 14 | |
| fby;flh | 3 +/− 2b | 24 | |
| GFP in Tg(foxd3:gfp) (pineal and parapineal cells) |
WT | 52 +/− 10 | 5 |
| flh | 28 +/− 7c | 5 | |
| fby;flh | 24 +/− 10c | 7 | |
| Zpr-1 antibody (red-green double cone cells) |
WT | 30 +/− 5 | 5 |
| flh | 5 +/− 5c | 4 | |
| fby;flh | 7 +/− 1c | 4 |
Average number of cells labeled per larvae at 4 d, plus or minus one standard deviation.
Significantly different from WT and flh by 2-tailed T-test, p < 0.01.
Significantly different from WT by 2-tailed T-test, p < 0.03.
To confirm that parapineal cells in flh mutant larvae do not express a pineal-specific marker, expression of the Fret43 antigen was examined in the context of the transgene Tg(foxd3:gfp)fkg3 at 4 days. In WT larvae, Fret43 (labeled by the Zpr-1 antibody; Larison and Bremiller, 1990), is expressed on red-green double cone cells found throughout the pineal organ (Masai et al., 1997) but excluded from the parapineal organ (Fig. 3C and Table 2). As in WT, none of the parapineal cells in flh mutants express Zpr-1 (Fig. 3D and Table 2).
To assay the ability of the parapineal in flh mutants to dictate L-R differences in the habenulae, expression of the asymmetrically expressed gene leftover (lov) (Gamse et al., 2003) was examined at 4 days. In both WT and flh mutant larvae, expression of lov was found in more cells of the habenula adjacent to the parapineal than in the contralateral habenula (Fig. 3E,F).
In sum, all of the characteristics of the parapineal, including number of cells specified, asymmetric migration, and ability to direct habenular asymmetry, are comparable between flh mutants and WT.
Flh and Tbx2b Do Not Regulate One Another's Expression
To evaluate cross-regulation of flh and tbx2b, we examined expression of each gene in the pineal complex of tbx2b and flh mutants. In WT embryos, expression of tbx2b begins at the 6 s stage in bilateral groups of cells in the pineal complex anlage, which soon merge to form one medial domain. At 24 h, expression is robust (Dheen et al., 1999; Ruvinsky et al., 2000; Snelson et al., 2008; Fig. 4A,B). flh expression begins at 80% epiboly in the pineal complex anlage (Masai et al., 1997), and is found in a single medial domain at 6 s and 24 h stages (Fig. 4C,D). Double labeling reveals that expression of tbx2b overlaps with the flh expression domain at 6 s and 24 hr; flh expression extends further medially than tbx2b at 6 s (Fig. 4E,F). In flh mutants, tbx2b expression is still detected at both 6 s and 24 hr (Fig. 4G,H). The total number of tbx2b-expressing cells in the pineal complex anlage of flh mutants is reduced relative to wild type, consistent with the reduced size of the pineal organ that will develop. In fby mutants, the number of flh-expressing cells detected at 6 s and 24 h is similar to WT (Fig. 4I,J). The data demonstrate that tbx2b and flh expression is maintained in flh and fby mutants, respectively.
Fig. 4.
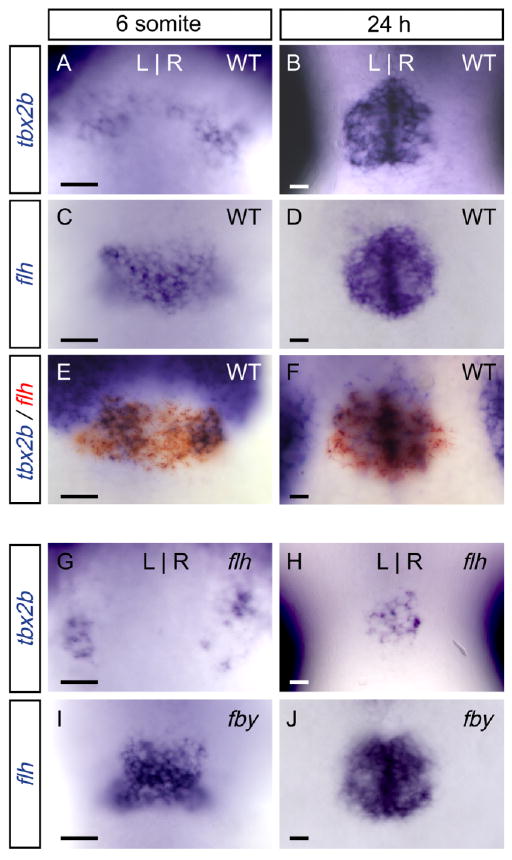
Flh and Tbx2b do not regulate one another's expression. All panels are dorsal views of the epithalamus at the stage indicated. (A,B) tbx2b and (C,D) flh are expressed in the pineal complex anlage at the 6-somite and 24-hr stages. E,F: Double labeling for tbx2b (blue) and flh (red) reveals that they overlap in expression at the 6-somite and 24-hr stages; flh is expressed further medially than tbx2b at the 6-somite stage. G: In flh mutants, expression of tbx2b initiates on time and (H) is maintained, albeit in a reduced number of cells. I,J: In fby mutants (a lesion in tbx2b), expression of flh is unaltered relative to WT. Scale bars = 25 μm.
flh;fby Double Mutants Have an Additive Phenotype
A sensitive test for genetic interaction is examining the phenotype of homozygous flh;fby double mutant embryos. In fby single mutants at 4 days, the pineal organ is similar in size to WT, but the gfi-1-expressing parapineal cells are reduced in number, remain near the midline, and fail to upregulate lov in the left habenula (Snelson et al., 2008; Fig. 5A,C,E).
Fig. 5.
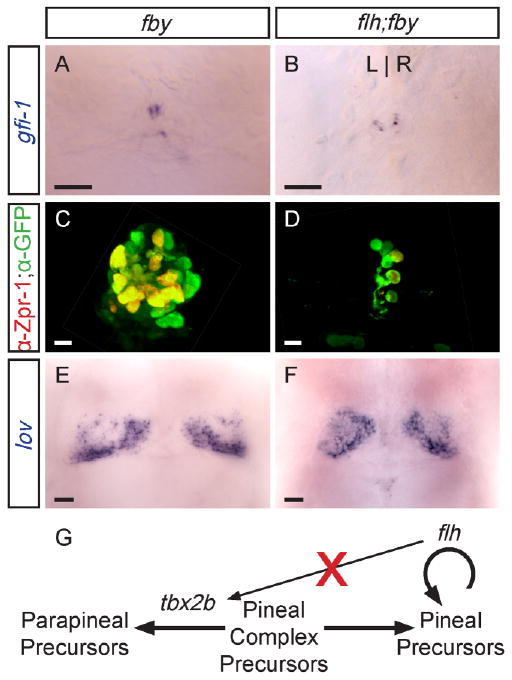
Double homozygous flh;fby mutants exhibit an additive phenotype. All panels are dorsal views of the epithalamus of 4 d larvae. A: In fby single mutants or (B) flh;fby double mutants, only a few disorganized parapineal cells are specified, and they fail to migrate to the left side of the brain. C: fby single mutants form a pineal organ. D: flh;fby double mutant larvae have both a reduced pineal, similar to flh single mutants (compare to Fig. 3D). E: In fby single mutants and (F) flh;fby double mutants, reduced lov expression is detected in the left habenula. Scale bars = 25 μm. G: Summary of tbx2b and flh activity in pineal complex development. The activity of tbx2b gives parapineal precursors their identity, while flh is needed for proliferation of pineal precursor cells. Expression of tbx2b does not require flh activity.
Similar to fby single mutants, in flh;fby double mutants the gfi-1-expressing parapineal cells are fewer in number than WT and do not migrate leftward (Fig. 5B). Additionally, lov expression in double mutants is not upregulated in the left habenulae, similar to fby single mutants (Fig. 5F). The number of GFP-expressing pineal cells in double mutants is reduced to a level similar to the number of pineal cells in flh single mutants. In addition, the number of Zpr-1-labeled cells in double mutants is ∼5-fold reduced compared to WT and indistinguishable from flh single mutants (Fig. 5D and Table 2).
These data indicate that flh;fby double mutants have an additive phenotype, exhibiting phenotypes in the pineal and parapineal organs characteristic of each single mutant.
Discussion
Previous work showed that Flh is a key regulator of pineal neurogenesis (Masai et al., 1997, Cau and Wilson, 2003) and suggested that Flh activates tbx2b transcription in pineal complex precursors (Cau and Wilson, 2003). The latter result implies that flh mutants should show defects in parapineal formation, which requires tbx2b function (Snelson et al., 2008). However, flh mutants develop a parapineal that is indistinguishable from WT larvae with regard to asymmetric migration and number of cells specified. Moreover, disruption of flh function does not eliminate the initiation or maintenance of tbx2b transcription in the pineal complex. Examination of homozygous double mutants reveals an additive phenotype and supports parallel rather than serial action of flh and tbx2b. These data argue strongly against tbx2b and flh acting in the same genetic pathway during the development of the pineal complex (Fig. 5G).
On the basis of mutational analyses and BrdU labeling, we propose the following model. Expression of flh and tbx2b before the 15 s stage defines a field of precursor cells that can divide to produce pineal or parapineal cells. An initial burst of cell division at the 15 s stage generates many new pineal complex cells; tbx2b activity assigns parapineal fate to some of these cells while the remainder develop with pineal fate. Subsequently, division of parapineal precursor cells drops off sharply, suggesting that tbx2b is largely dispensable for parapineal development after the 15 s stage. A method to conditionally inactivate tbx2b in the pineal complex will be necessary to test this hypothesis.
Beginning at 18 s, flh activity is required for continued production of cells in the pineal anlage (Masai et al., 1997, and this study). Almost all of the cells that divide after 18 s contribute to the pineal lineage. It is not clear why division of cells that give rise to the parapineal organ slows after the 15 s stage while division of pineal progenitors continues. One possibility is that separate groups of pineal and parapineal progenitor cells exist, and the latter group largely stops dividing after 15 s. At 6 s, the tbx2b-positive and tbx2b-negative cells within the flh-expressing domain may represent parapineal and pineal progenitors, respectively. A second explanation is that the same progenitor cells can give rise to either pineal or parapineal cells initially, but primarily produce pineal cells after 15 s. Careful lineage labeling of single cells in the pineal complex anlage will distinguish between these two scenarios.
A small number of pineal and parapineal cells still develop in flh;fby double mutant embryos. Presumably, there are other genes that contribute to proliferation and/or control specification of pineal and parapineal cells; however, these genes have yet to be isolated. Forward genetic screening in a sensitized background such as a tbx2b hypomorph should identify these genes.
The independent roles that tbx2b and flh play in the development of the pineal complex contrasts with the regulation of tbx2b expression by flh during notochord development. In flh mutants, tbx2b expression is absent in notochord precursor cells found in the chordoneural hinge (Dheen et al., 1999). In the notochord, flh appears to maintain tbx2b expression by blocking the action of another T-box transcription factor, spadetail/tbx16 (spt) (Dheen et al., 1999). By contrast, spt is not expressed in the pineal anlage (Ruvinsky et al., 1998), which may explain why flh and tbx2b activity are uncoupled in this tissue. Identification of other genes that are expressed in both the pineal complex anlage and notochord precursors, similar to flh and tbx2b, will illuminate the genetic networks involved in the patterning of these two tissues, and allow a comparison of evolutionary conservation and divergence between these networks.
Experimental Procedures
Zebrafish
Zebrafish were raised at 28.5°C on a 14/10 hr light/dark cycle and staged according to hours (h) or days (d) post-fertilization. The wild-type AB strain (Walker, 1999); the transgenic lines Tg(foxd3:GFP)fkg17 and Tg(foxd3:GF-P)fkg3 (Gilmour et al., 2002), and mutants carrying the spontaneous allele flhn1 (Halpern et al., 1995) and the ethylnitrosourea-induced null allele fbyc144 (Snelson et al., 2008) were used.
RNA In Situ Hybridization
Whole-mount RNA in situ hybridization was performed as described previously (Gamse et al., 2003), using reagents from Roche Applied Bioscience. RNA probes were labeled using fluorescein-UTP or digoxygenin UTP. To synthesize antisense RNA probes, znot (flh) (Talbot et al., 1995) and pBK-CMV-leftover (Gamse et al., 2003) were linearized with EcoRI and transcribed with T7 RNA polymerase; pCRII-tbx2b (Snelson et al., 2008) with BamHI and T7 RNA polymerase; and pBS-gfi1 (Dufourcq et al., 2004) with SacII and T3 RNA polymerase. Embryos were incubated at 70°C with probe and hybridization solution containing 50% formamide (or 65% for cyc). Hybridized probes were detected using alkaline phosphatase-conjugated antibodies and visualized by 4-nitro blue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) staining. All in situ data was collected on a Leica DM6000B microscope with a 20× or 40× objective.
BrdU Labeling
For pulse labeling with bromodeoxyuridine (BrdU, Roche), embryos were incubated with ice-cold 10 mM BrdU dissolved in embryo medium plus DMSO at 2% for 10–15 s embryos, 5% for 18-s-stage embryos, or 10% for 24-30-h embryos, over a period of 30 min. Embryos were then washed extensively with embryo medium and raised to 4 d, when they were fixed in 4% paraformaldehyde for 2 hours at room temperature.
Time-Lapse Imaging
For time-lapse imaging, WT or flh mutant embryos carrying the transgene (foxd3:GFP)fkg3 were mounted in a 0.8% solution of low melt agarose containing 0.003% PTU and anesthetized using 0.04% Tricaine. Images were collected on a Zeiss/Perkin Elmer spinning disk confocal microscope with a 40× oil-immersion objective every 15 min from 31 to 46 hr, and analyzed using Volocity software (Improvision).
Immunofluorescence
For whole-mount immunohistochemistry, 4 d larvae were fixed in 4% paraformaldehyde overnight (for anti-Zpr-1/anti-GFP) or 2 hr (for anti-BrdU/anti-GFP). Samples were permeabilized by treatment with 10 μg/ml Proteinase K (Roche Applied Bioscience) and refixed in 4% paraformaldehyde. For BrdU labeling, samples were additionally washed in deionized water following refixation, and incubated in 2 N HCl for 1 hr. All samples were blocked in PBS with 0.1% Triton X-100, 2% sheep serum, 1% DMSO, and 10% BSA (PB-STrS). For antibody labeling, rabbit anti-GFP (1:1,000, Torrey Pines Biolabs), mouse anti-Zpr-1 (1:250, Zebrafish International Resource Center, Eugene, OR), and mouse anti-BrdU antibody (1: 200, developed by Stephen Kaufman, obtained from the Developmental Studies Hybridoma Bank, University of Iowa) were used. Larvae were incubated overnight in primary antibody diluted in PBSTrS. Primary antibody was detected using goat-anti-rabbit or goat-anti-mouse secondary antibodies conjugated to the Alexa 568 or Alexa 488 fluorophores (1:350, Molecular Probes). Immunofluorescence data were collected on a Zeiss/Perkin Elmer spinning disk confocal microscope or a Zeiss LSM 510 Meta with a 40× oil-immersion objective and analyzed with Volocity software (Improvision).
Supplementary Material
Acknowledgments
We thank Erin Booton and Heidi Beck for expert fish care. We acknowledge the Developmental Studies Hybridoma Bank at the University of Iowa (developed under the auspices of the NICHD) for the anti-BrdU antibody, and the Vanderbilt Cell Imaging Shared Resource for microscopy support.
Grant sponsor: Whitehall Foundation; Grant number: 2005-08-79-APL.
Footnotes
The Supplementary Information referred to in this article can be viewed online.
References
- Cau E, Wilson SW. Ash1a and Neurogenin1 function downstream of Floating head to regulate epiphysial neurogenesis. Development. 2003;130:2455–2466. doi: 10.1242/dev.00452. [DOI] [PubMed] [Google Scholar]
- Concha ML, Wilson SW. Asymmetry in the epithalamus of vertebrates. J Anat. 2001;199:63–84. doi: 10.1046/j.1469-7580.2001.19910063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha ML, Russell C, Regan JC, Tawk M, Sidi S, Gilmour DT, Kapsimali M, Sumoy L, Goldstone K, Amaya E, Kimelman D, Nicolson T, Grunder S, Gomperts M, Clarke JD, Wilson SW. Local tissue interactions across the dorsal midline of the forebrain establish CNS laterality. Neuron. 2003;39:423–438. doi: 10.1016/s0896-6273(03)00437-9. [DOI] [PubMed] [Google Scholar]
- Dheen T, Sleptsova-Friedrich I, Xu Y, Clark M, Lehrach H, Gong Z, Korzh V. Zebrafish tbx-c functions during formation of midline structures. Development. 1999;126:2703–2713. doi: 10.1242/dev.126.12.2703. [DOI] [PubMed] [Google Scholar]
- Dufourcq P, Rastegar S, Strahle U, Blader P. Parapineal specific expression of gfi1 in the zebrafish epithalamus. Gene Expr Patterns. 2004;4:53–57. doi: 10.1016/s1567-133x(03)00148-0. [DOI] [PubMed] [Google Scholar]
- Gamse JT, Shen YC, Thisse C, Thisse B, Raymond PA, Halpern ME, Liang JO. Otx5 regulates genes that show circadian expression in the zebrafish pineal complex. Nat Genet. 2002;30:117–121. doi: 10.1038/ng793. [DOI] [PubMed] [Google Scholar]
- Gamse JT, Thisse C, Thisse B, Halpern ME. The parapineal mediates left-right asymmetry in the zebrafish diencephalon. Development. 2003;130:1059–1068. doi: 10.1242/dev.00270. [DOI] [PubMed] [Google Scholar]
- Gilmour DT, Maischein HM, Nusslein-Volhard C. Migration and function of a glial subtype in the vertebrate peripheral nervous system. Neuron. 2002;34:577–588. doi: 10.1016/s0896-6273(02)00683-9. [DOI] [PubMed] [Google Scholar]
- Halpern ME, Thisse C, Ho RK, Thisse B, Riggleman B, Trevarrow B, Weinberg ES, Postlethwait JH, Kimmel CB. Cell-autonomous shift from axial to paraxial mesodermal development in zebrafish floating head mutants. Development. 1995;121:4257–4264. doi: 10.1242/dev.121.12.4257. [DOI] [PubMed] [Google Scholar]
- Hill C. Development of the epiphysis in Coregonus Albus. J Morphol. 1892;5:503–510. [Google Scholar]
- Larison KD, Bremiller R. Early onset of phenotype and cell patterning in the embryonic zebrafish retina. Development. 1990;109:567–576. doi: 10.1242/dev.109.3.567. [DOI] [PubMed] [Google Scholar]
- Liang JO, Etheridge A, Hantsoo L, Rubinstein AL, Nowak SJ, Izpisua Belmonte JC, Halpern ME. Asymmetric nodal signaling in the zebrafish diencephalon positions the pineal organ. Development. 2000;127:5101–5112. doi: 10.1242/dev.127.23.5101. [DOI] [PubMed] [Google Scholar]
- Masai I, Heisenberg CP, Barth KA, Macdonald R, Adamek S, Wilson SW. floating head and masterblind regulate neuronal patterning in the roof of the forebrain. Neuron. 1997;18:43–57. doi: 10.1016/s0896-6273(01)80045-3. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Silver LM, Ho RK. Characterization of the zebrafish tbx16 gene and evolution of the vertebrate T-box family. Dev Genes Evol. 1998;208:94–99. doi: 10.1007/s004270050158. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Oates AC, Silver LM, Ho RK. The evolution of paired appendages in vertebrates: T-box genes in the zebrafish. Dev Genes Evol. 2000;210:82–91. doi: 10.1007/s004270050014. [DOI] [PubMed] [Google Scholar]
- Snelson CD, Santhakumar K, Halpern ME, Gamse JT. Tbx2b is required for the development of the parapineal organ. Development. 2008;135:1693–1702. doi: 10.1242/dev.016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland RJ. The dorsal diencephalic conduction system: a review of the anatomy and functions of the habenular complex. Neurosci Biobehav Rev. 1982;6:1–13. doi: 10.1016/0149-7634(82)90003-3. [DOI] [PubMed] [Google Scholar]
- Talbot WS, Trevarrow B, Halpern ME, Melby AE, Farr G, Postlethwait JH, Jowett T, Kimmel CB, Kimelman D. A homeobox gene essential for zebrafish notochord development. Nature. 1995;378:150–157. doi: 10.1038/378150a0. [DOI] [PubMed] [Google Scholar]
- Walker C. Methods in Cell Biology. London: Elsevier; 1999. Haploid screens and gamma-ray mutagenesis; pp. 43–70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


