Abstract
Most of the opioids used in clinical practice exert their effects through μ opioid receptors. Yet, subtle but important pharmacological differences have been observed among the mu opioids. Their potency, effectiveness, and adverse effects can vary unpredictably among patients. These clinical differences among the mu opioids strongly argue against a single receptor mediating their actions. The cloning of the μ opioid receptor has greatly enhanced our understanding of the complexity of this system and has provided possible mechanisms to explain these observations. A single mu opioid receptor gene has been identified, but we now know that it generates a multitude of different mu opioid receptor subtypes through a mechanism commonly used to enhance protein diversity, alternative splicing. Early studies identified a number of splice variants involving the tip of the C-terminus. This region of the receptor is far away from the binding pocket, explaining why these variants still exhibit the same selectivity for mu opioids. However, the differences in structure at the C-terminus influence the activation patterns of the μ opioids. In addition, a second series of variants have been isolated that involve alternative splicing at the N-terminus. Together, these sets of mu opioid receptor splice variants may help explain the clinical variability of the mu drugs among patients and provide insights into why it is so important to individualize therapy for every patient in pain.
Historical Perspective
Pain is a difficult sensation to quantify, in part due to its subjectivity and variability among individuals. However, even for a single individual, the perception of pain from the same nociceptive input can vary widely depending on the setting in which it occurs. Pain is an integrated perception with subconscious components that differ from moment to moment and is unique to each individual, raising the possibility of a sensory filtering system capable of modulating pain perception. Indeed, evidence now suggests that opioids act through the activation of this system.
In the early 1970s,1 Mayer and Liebeskind found that electrical stimulation of specific regions of rat brains, such as the periventricular gray, the central gray, and parts of the midbrain, produced profound analgesia, a response that could be blocked by the opioid antagonist, naloxone. The sensitivity of the response to naloxone implied that electrical stimulation was releasing endogenous opioid-like materials within the brain, compounds subsequently identified as enkephalins and endorphins.2,3 Opioids activate these filtering systems by mimicking the actions of the endogenous opioids. What truly sets the opioids apart from other analgesics is their unique ability to diminish pain perception without altering more objective sensations, such as light touch, temperature or position, providing insights into the characteristics and power of this sensory filtration system.
Three families of endogenous opioids (enkephalins, dynorphins and β-endorphin) have been identified, as have three major families of opioid receptors (respectively: μ, δ, and κ1), a family of evolutionally conserved receptors present in most vertebrates.4 Of these receptors, the μ opioid receptors are particularly important clinically, since most of the opioids used in patients target these receptors, including the drug morphine.
Behavioral Observations Supporting the Existence of μ Opioid Receptor Subtypes
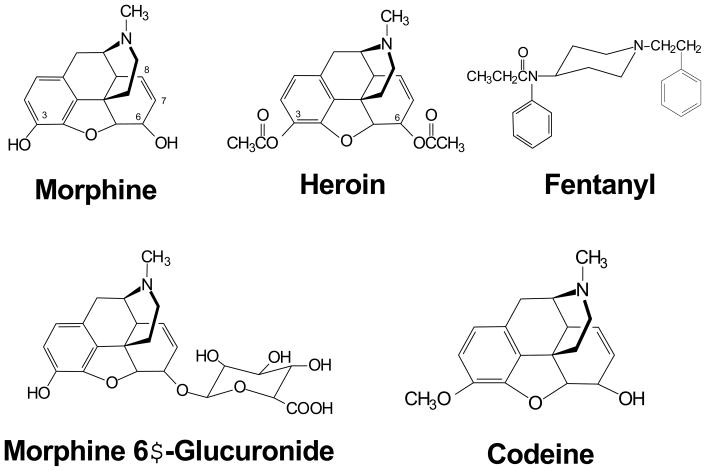
μ opioids are defined by their selectivity for the μ opioid receptor, MOR-1. Compounds that are highly selective for μ opioid receptors include morphine (which is metabolized in the liver to morphine-3-glucuronide an inactive and potentially toxic byproduct, and morphine-6β-glucuronide, which is a potent analgesic in its own right),5,6 codeine, fentanyl, heroin, methadone, oxymorphone and oxycodone. For years, many pharmacologists considered these agents interchangeable, consistent with similar structures across many of the opioids and their similar selectivity for μ receptors (Figure 1). However, clinicians have long been aware of subtle, but important pharmacological differences among them. Clinically, their potency and effectiveness can vary unpredictably among patients, with some patients achieving excellent relief with one agent, while other patients requiring a different drug. Side effects such as nausea and vomiting also differ among individuals, wherein one μ drug is intolerable, but another μ analgesic is suitable. Thus, the most effective drug for one patient may not be the best one for another individual. Finally, the presence of incomplete cross-tolerance among the μ opioids is another example of differences among the drugs. Patients highly tolerant to one μ opioid may regain their sensitivity when switched to another μ opioid, an observation which has led to the practice of opioid rotation.7 In opioid rotation, patients no longer able to tolerate further escalation of a drug due to side effects, is switched to a different opioid, often leading to the restoration of analgesic sensitivity at doses of the second drug far lower than might have been anticipated.
Figure 1.
Structures of Common μOpioids.
Animal models illustrate the importance of genetic backgrounds in these subtle differences. For example, a fixed dose of morphine yields widely varying analgesic responses in different strains of mice. In one study, a fixed dose of morphine produced analgesia in 90% of BALB-c mice, but in none of the CXBK strain, with other strains showing intermediate responses. However, it was not simply a matter of one strain being more, or less, sensitive to all of the μ drugs. Although the CXBK mice were insensitive to morphine, they responded normally to heroin, methadone, fentanyl and morphine-6β-glucuronide,8 clearly differentiating the responses to these drugs on a genetic basis, very much like the clinical observations of varying patient responses to different opioid drugs.9,10 These preclinical studies support the hallmark of opioid therapy—the need to individualize treatment for each individual.
The complexity of μ opioid pharmacology is also indicated by analgesic studies exploring different combinations of opioids administered together.11 Despite the fact that the drugs in these studies were all classified as μ-selective, select combinations displayed profound synergy, implying that perhaps, their mechanistic effects were unique. This was particularly evident with combinations of methadone and either morphine, morphine-6β-glucuronide, codeine, or the active metabolite of heroin, 6-acetylmorphine.
It is difficult to reconcile all of these observations with a single receptor. Indeed, pharmacological studies led to the suggestion of the existence of multiple μ opioid receptors more than 25 years ago.12 Much of this evidence has been derived from studies examining several unusual μ antagonists in the laboratory. In molecular studies, these μ antagonists differentially block one subpopulation of μ opioid receptor binding sites.13,14 In vivo studies of these same antagonists have clearly dissociated morphine analgesia from respiratory depression, the inhibition of GI transit, and many aspects of physical dependence.15 Based upon these classical studies, two μ opioid receptor subtypes were proposed.12 However, it was not until the μ receptor was cloned that we began to appreciate its true complexity.
Alternative Splicing of the C-terminus of theμ Opioid Receptor
Several groups first cloned the μ opioid receptor a little more than 15-years ago. The cloned receptor, MOR-1, is derived from a single gene, raising the question of how to reconcile a single gene with multiple receptors. We now know that the single μ opioid receptor gene generates dozens of protein subtypes through a prevalent mechanism used to enhance protein diversity, termed alternative splicing.
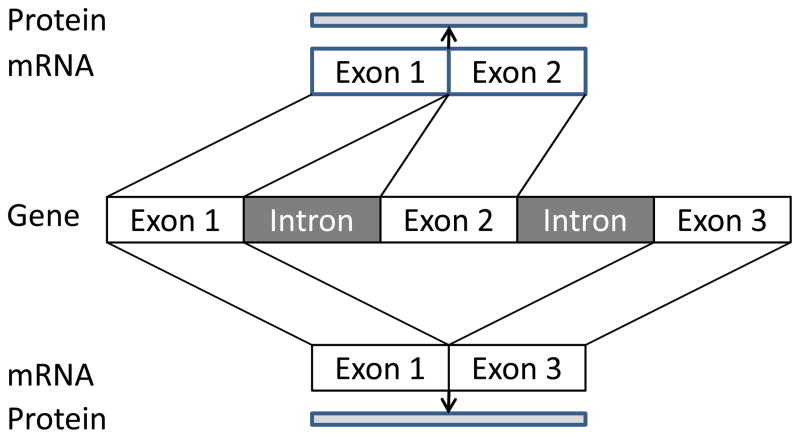
Although the human genome contains only approximately 20,000–30,000 genes,16,17 these genes are responsible for generating many-fold more proteins, thereby greatly enhancing the diversity and complexity of organisms. Genes comprise sequences that are contained within mRNA, termed exons, and sequences that are not, termed introns. By changing the composition of the exons, many different proteins can be assembled. The process of combining the exons together is termed splicing, while including different sets of exons within an mRNA is alternative splicing (Figure 2). It has been estimated that at least 60% of genes produce at least one alternative mRNA.18 The process of alternative splicing involves a macromolecular machine composed of proteins and RNAs—the spliceosome that removes introns and joins exons together to form mRNAs, which are then read by the ribosome to generate proteins.19,20 Alternative splicing is under a number of controls, making it an important mechanism ultimately leading to differences among individuals in many areas, including cancer, asthma and other diseases.18
Figure 2.
Alternative mRNA Splicing.
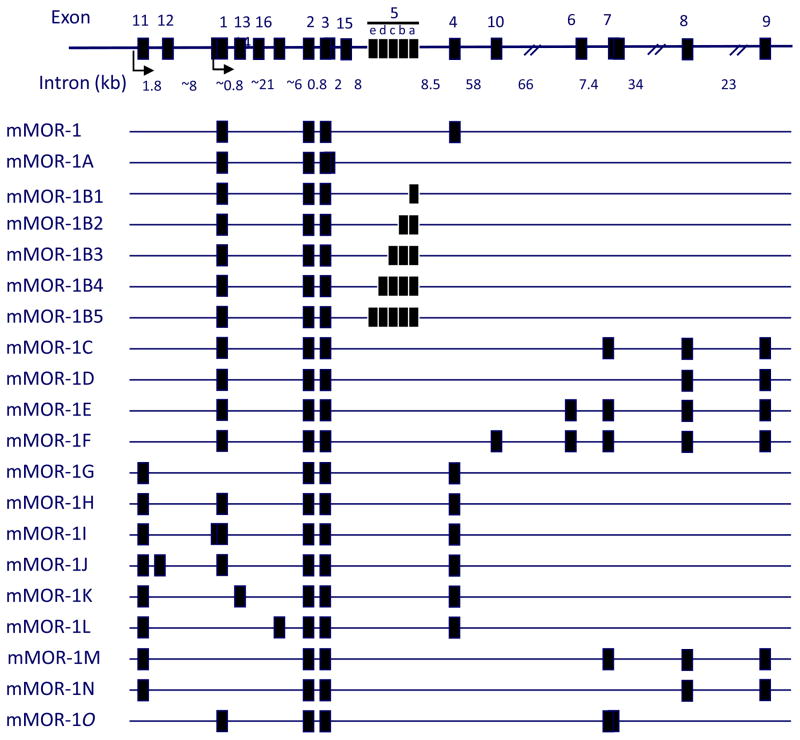
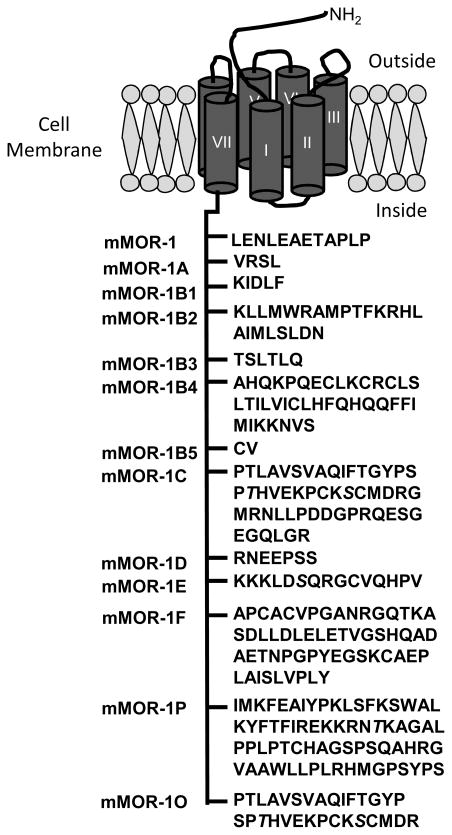
The μ opioid receptor MOR-1 is comprised of a number of exons that combine to form a wide range of splice variants (Figure 3); MOR-1 itself contains four exons. Like other G-protein coupled receptors, MOR-1 is comprised of seven transmembrane (TM) domains with an extracellular N-terminus and an intracellular C-terminus (Figure 4). The first three exons are responsible for encoding the N-terminus and all seven TM domains, while exon-4 encodes only the last 12 amino acids in the intracellular C-terminus. The major series of μ opioid receptor variants in mice involves replacing exon-4 with a set of alternative exons,21–23 a splicing pattern that is relatively conserved between humans24,25–27 mice21–23 and rats.28,29 The binding pocket for all of the full-length μ opioid receptor variants is identical, since the pocket is comprised of the seven TM domains encoded by exons-1, -2 and -3. Their differences are limited to only the tip of the C-terminus, far from the binding pocket, explaining why the variants all retain their high affinity and selectivity for μ opioids.27 However, the differences in the amino acid sequence at the C-terminus do appear to be important. Although their binding affinities are similar, the activation patterns of the various μ opioids for the full-length receptor differ, possibly explaining their subtle pharmacological differences.
Figure 3. Alternative mRNA Splicing of MOR-1 Leads to a Combinatorial Diversity of μ Opioid Receptor Proteins.
MOR-1 receptor splice variants are depicted.
Figure 4.
Domains of the MOR-1 Protein and Alternative Splice Variants of the C-Terminus.
Immunohistochemical imaging studies reveal that the MOR-1 variants are widely distributed throughout the nervous system, including within regions of the brain and spinal cord involved in pain processing. Yet, confocal microscopy of cells labeled for two different variants, MOR-1 and MOR-1C, in the dorsal horn of the spinal cord reveal that cells typically generate one variant and not the another, raising interesting possibilities regarding the mechanisms and control of splicing.9 Staining studies also demonstrate that MOR-1 receptors are present in the periphery, labeling dorsal horn neurons and peripheral nerves, including some innervating the skin and blood vessels. Evidence suggests that these peripheral receptors can be targeted to help relieve pain.
Beyond varying regional distributions,30 the receptor variants also likely serve different functions. Although the binding pockets of the full-length variants are identical, their overall pharmacological functions are defined by both the transduction systems through which they couple and by the neurons, and thus the pathways, in which they are found. Although we often envisage the receptor in the membrane associated with a G-protein, it is, in fact, a component of a large molecular complex of numerous additional proteins. The composition of the intracellular receptor complex is defined by many factors, including the amino acid sequences located at the tip of the intracellular C-terminus. These complexes, called the μ opioid receptorsome, are integral to transducing the signal initiated at the μ opioid receptor. While much of the emphasis has been placed upon the G-proteins and identifying which specific ones associate with the receptor variants, many other proteins influence the activation of the G-protein, and thus, the effects of opioids. Studies have implicated a number of different proteins in the signal transduction process, but they have not been systematically examined for specificity to individual variants. Indeed, the full complement of proteins within a single opioid receptor complex of a receptor has not yet been fully documented. Still, there is evidence that the μ opioid receptor variants do differ functionally.
One feature of an opioid is its ability to stimulate the opioid receptor, which biochemically is associated with its ability to activate G-proteins. This can be measured by examining the incorporation of a radiolabeled GTP analog. Using this approach, it has been clearly shown that different variants can be activated to differing extents by each opioid. In other words, each opioid has a distinct activation profile of specific receptor variants. While this can easily be demonstrated with variants singly expressed in a cell, characterization of the variants expressed in the brain—where all the variants coexist, may be more difficult, raising the interesting question of how the actions of a drug should be interpreted.
The affinity of an individual opioid has little variation between receptor subtypes. Thus, when administered in vivo, it would be expected that the opioid would bind to all the variants equally well; however, the ability of the drug to activate each variant is different. Ultimately, the outcome of administering an opioid to an individual is the result of the sum of the effects of binding of the drug simultaneously to the multiple receptor splice variants. Thus, differing composite activation profiles of the drugs are likely to explain the nuances and subtle differences in responses to opioids among patients.31
Knockout mice
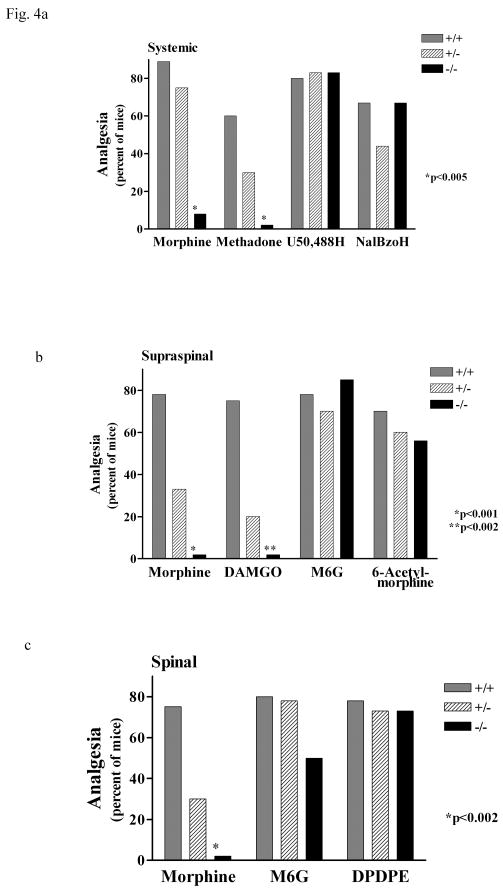
Establishing the functional significance of a cloned gene can be most elegantly explored using animals that have portions of that gene deleted. Indeed, the importance of the full-length MOR-1 variants has clearly been demonstrated in studies with animals that have a disrupted MOR-1 gene.32–34 Removal of exon-1 eliminates all of the full-length variants, since exon-1 is a necessary component within all of them. Therefore, it was not surprising to see that administering morphine to these knockout models was without effect. Yet, one exon-1 knockout mouse presented with particularly interesting characteristics. Like the others, morphine actions were totally lost, as were those of methadone, when given systemically (Figure 5a; Table 1). Similarly, the analgesic actions of morphine and a μ peptide, DAMGO ([D-Ala2,MePhe4,Gly(ol)5]-enkephalin) given centrally were also lost in the exon-1 knockout mouse (Figure 5b), while both morphine-6-glucuronide and 6-acetylmorphine, the active metabolite of heroin, retained their analgesic activity.34 The lack of effects of the deletion of exon-1 on the two κ drugs: U50,488H and naloxone benzoylhydrazone (NalBzoH), as well as the δ-selective peptide: DPDPE ([D-Pen2,D-Pen5]enkephalin), were important controls. While these observations implied that morphine-6-glucuronide and heroin have different mechanisms of action than morphine, additional experiments with selective antagonists and antisense approaches clearly showed that they were still acting through μ opioid receptors. Furthermore, this analgesia persisted in triple knockout mice also lacking δ and κ receptors, as well as exon-1 of MOR-1, indicating that this residual analgesia was not the result of cross activating δ and κ opioid receptors.
Figure 5. Opioid Analgesia in a MOR-1 Exon-1 Knockout Mouse34.
Analgesic action of opioids in wild-type, heterozygous or homozygous mice. (a) Groups of mice (n = 8) received subcutaneous injections of 13 mg/kg morphine, 2 mg/kg methadone, 5 mg/kg of the κ1 drug U50,488H, or 60 mg/kg of the κ3 drug NalBzoH (naloxone benzoylhydrazone), and were tested for analgesia 30 minutes later. (b) Groups of mice (n = 9) received intracerebroventricular injections of 0.7 μg morphine, 6 ng of the μ peptide DAMGO (([D-Ala2,MePhe4,Gly(ol)5]-enkephalin), 12.5 ng M6G (morphine-6β-glucuronide) or 1.2 μg 6-acetylmorphine, and were tested for analgesia 15 minutes later. (c) Groups of mice (n = 8) received intrathecal injections of 0.8 μg morphine, 12.5 ng M6G or 500 ng of the δ peptide DPDPE ([D-Pen2,D-Pen5]enkephalin) and were tested for analgesia 15 minutes later. Analgesia is expressed as the percentage of mice responding.[Permission needed.]
Table 1.
Effect of Exon-specific Knockouts of MOR-1 on μ Opioid Analgesia
| Exon-1 (mg/kg, s.c.) | Exon-11 (mg/kg, s.c.) | |||||
|---|---|---|---|---|---|---|
| Drug | WT | KO | Shift | WT | KO | Shift |
| Morphine | 5.0 | >100 | >20 | 1.6 | 2.6 | 1.6 |
| M6G | 3.8 | 8.5 | 2.3 | 0.92 | 19.3 | 21 |
| Heroin | 1.2 | 4.0 | 2.7 | 0.58 | 3.2 | 5.5 |
| Fentanyl | 0.024 | 0.230 | 9.7 | |||
The exon-1 knockout mice also continued to make mRNA transcripts of the MOR-1 gene, as shown by RT-PCR and immunohistochemistry—which revealed the continued expression of epitopes associated with some of the splice variants. The immunostaining of the exon-4 epitope was markedly diminished, but not completely abolished. Staining with the exon 7/8/9 epitope was similar to that seen in wild type mice. These data raised the question of whether or not the persistent analgesia seen in these animals might result from a MOR-1 variant lacking exon-1. While the presence of variants in which exon-4 was replaced with combinations of other exons had been well established, no N-terminal splicing had been observed. However, our laboratory explored this possibility in depth and identified a second promoter associated with a new exon, exon-11, which was located more than 20 kb upstream of exon-1 and its promoter. Additional cloning studies then isolated a series of exon-11-associated variants of MOR-1 with splicing at the N-terminus of the receptor, in both mice and humans, including clones that did not contain exon-1.27,35
Like the full-length C-terminus variants, these exon-11-associated variants displayed a regional distribution of mRNA distinct from MOR-1 itself and most of the other variants. Three of the exon-11 variants generate the same receptor at the protein level as MOR-1, while a different three, MOR-1G, MOR-1M and MOR-1N, predict proteins lacking exon-1 and possessing the exon-11 peptide epitope. Western blots confirm their presence within the brain, and immunohistochemistry images show unique distributions. However, these three variants predict only six TM regions, not the seven normally associated with G-protein coupled receptors. A full understanding of how these truncated receptors act at the molecular level has not been elucidated, although there are some evidence suggesting that they may act through dimerization with full-length variants. Several of the other exon-11-associated variants predict small (<10 kDa) peptides, however these have not been identified within the brain, raising questions regarding their endogenous expression and/or relevance.
Potentially, the variants lacking exon-1 could be responsible for the residual analgesia from heroin and morphine-6β-glucuronide. To assess this possibility, we generated another knockout mouse in which exon-11 was eliminated, while leaving exon-1 intact.36 These animals responded normally to morphine and methadone, with ED50 values that were not statistically different from wild-type animals (Table 1).34,36 However, the potency of heroin, morphine-6β-glucuronide and fentanyl was markedly reduced, implying that exon-11-associated variants lacking exon-1 are important in the analgesic actions of heroin and other opioids.
Conclusions
Clinicians have been at the forefront of opioid pharmacology, recognizing subtleties in drug actions that would not be possible using animal models. Extensive experience has clearly documented the importance of individualizing opioid therapy. Variable responses among patients for individual drugs and for multiple drugs within a single patient preclude “rules” in opioid use. Although many questions remain, recent molecular studies suggest intriguing possible explanations regarding why this variability among a single class of drug exists, such as the existence of multiple μ opioid receptor subtypes.
A single μ opioid receptor gene has been identified, however this does not preclude the existence of many distinct μ opioid receptors. Alternative splicing is an important mechanism for generating protein diversity and it has been well documented regarding the μ opioid receptor gene in mice, rats and humans. Two general classes of splice variants have been identified. The major class of μ opioid receptor splice variants includes the full-length seven TM domain and involves exchanging exon-4 for combinations of alternative exons. These are interesting in several respects. Their binding pockets are identical since they are formed from a portion of the protein that is completely conserved among the variants. Yet, C-terminal differences among the variants appear to impact the proteins comprising the receptor complex and thus their transduction systems. Furthermore, while many of the variants are present in the same regions of the nervous system, they are often expressed by different cells suggesting further potential functional differences at the level of the neuronal circuitry. Opioids likely bind to all of these seven TM variants equally well, but they differ in their activation profiles, perhaps explaining their subtle, but real, pharmacological differences. From the perspective of pharmacologists, this offers an interesting new approach towards drug design, since it may be possible to engineer novel drugs based upon these relative activation profiles and not simply based on affinity.
The second set of splice variants involves the N-terminus that also show C-terminus splicing similar to the first group, as well. These variants include a set of three six-TM variants that appear to be important in heroin and morphine-6β-glucuronide analgesia. The pharmacological relevance of these truncated variants raise intriguing questions regarding G-protein coupled receptors in general.
Research into μ opioid receptor pharmacology has made enormous strides in recent years, but the more we learn, the more complex it becomes. One can only assume that this will continue. These findings support the clinical paradigm of the individualization of therapy for every patient in pain and provide a mechanistic understanding of why this approach is needed. The existence of multiple μ opioid receptor splice variants also provides some insight into a potential explanation for the variability among drugs in one individual and among patients. Genetic backgrounds are important in animal models and it is reasonable to assume that genetics are an important factor impacting patients, as well.
Identifying the genetic difference(s) will be a major goal in the coming years. Single polynucleotide polymorphisms (SNPs) are the most common genetic alteration, and a number of these have been identified within the μ opioid receptor gene. One, involving a single nucleotide change at position 118 (i.e. A118G) has been suggested to influence opioid function by clinical studies, but the effects of this SNP in laboratory studies have been somewhat controversial. In addition, many other SNPs exist that have not yet been fully evaluated. Most investigations have focused upon MOR-1 itself. Extending these studies to the splice variants may prove quite interesting. More extensive mutations are possible and this possibility also needs to be explored. How these mutations may impact the sensitivity of the patient to a drug may not be simple. We often believe that the genetic changes will alter affinity or activation profiles, and this is certainly possible. However, mutations in either exons or introns alike may also impact relative splicing patterns, and thus, influence absolute or relative levels of the variants. Genetic alterations may alter trafficking of the mRNA or the receptor within the cell, leading to functional changes. Finally, genetic variability may reside “downstream” from the receptor itself; in much the same way as cutting any link in a chain will give the same results. This may help explain the association of opioid sensitivity to other neurotransmitter systems. The future promises further interesting developments in the field of opioid pharmacology.
Acknowledgments
The work described in this article was funded by the National Institute on Drug Abuse through grants (DA02615, DA07242 and DA06241) and a Senior Scientist Award (DA00220) to GWP. The author gratefully acknowledges MK Medical Communications, LLC and the editorial contributions of Rebecca Bachmann, PhD.”
Footnotes
Disclosure: Dr. Pasternak reports no conflicts of interest related to this paper.
References
- 1.Mayer DJ, Liebeskind JC. Pain reduction by focal electrical stimulation of the brain: an anatomical and behavioral analysis. Brain Res. 1974;68(1):73–93. doi: 10.1016/0006-8993(74)90534-4. [DOI] [PubMed] [Google Scholar]
- 2.Lord JA, Waterfield AA, Hughes J, et al. Endogenous opioid peptides: multiple agonists and receptors. Nature. 1977;267(5611):495–499. doi: 10.1038/267495a0. [DOI] [PubMed] [Google Scholar]
- 3.Martin WR, Eades CG, Thompson JA, et al. The effects of morphine- and nalorphine- like drugs in the nondependent and morphine-dependent chronic spinal dog. J Pharmacol Exp Ther. 1976;197(3):517–532. [PubMed] [Google Scholar]
- 4.Dreborg S, Sundstrom G, Larsson TA, et al. Evolution of vertebrate opioid receptors. Proc Natl Acad Sci U S A. 2008;105(40):15487–15492. doi: 10.1073/pnas.0805590105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasternak GW, Bodnar RJ, Clark JA, et al. Morphine-6-glucuronide, a potent mu agonist. Life Sci. 1987;41(26):2845–2849. doi: 10.1016/0024-3205(87)90431-0. [DOI] [PubMed] [Google Scholar]
- 6.Paul D, Standifer KM, Inturrisi CE, et al. Pharmacological characterization of morphine-6 beta-glucuronide, a very potent morphine metabolite. J Pharmacol Exp Ther. 1989;251(2):477–483. [PubMed] [Google Scholar]
- 7.Pasternak GW. The pharmacology of mu analgesics: from patients to genes. Neuroscientist. 2001;7(3):220–231. doi: 10.1177/107385840100700307. [DOI] [PubMed] [Google Scholar]
- 8.Rossi GC, Brown GP, Leventhal L, et al. Novel receptor mechanisms for heroin and morphine-6 beta-glucuronide analgesia. Neurosci Lett. 1996;216(1):1–4. doi: 10.1016/0304-3940(96)12976-1. [DOI] [PubMed] [Google Scholar]
- 9.Pasternak GW. Incomplete cross tolerance and multiple mu opioid peptide receptors. Trends Pharmacol Sci. 2001;22(2):67–70. doi: 10.1016/s0165-6147(00)01616-3. [DOI] [PubMed] [Google Scholar]
- 10.Pasternak GW. Molecular biology of opioid analgesia. J Pain Symptom Manage. 2005;29(5 Suppl):S2–S9. doi: 10.1016/j.jpainsymman.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Bolan EA, Tallarida RJ, Pasternak GW. Synergy between mu opioid ligands: evidence for functional interactions among mu opioid receptor subtypes. J Pharmacol Exp Ther. 2002;303(2):557–562. doi: 10.1124/jpet.102.035881. [DOI] [PubMed] [Google Scholar]
- 12.Wolozin BL, Pasternak GW. Classification of multiple morphine and enkephalin binding sites in the central nervous system. Proc Natl Acad Sci U S A. 1981;78(10):6181–6185. doi: 10.1073/pnas.78.10.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson HA, Pasternak GW, Snyder SH. Differentiation of opiate agonist and antagonist receptor binding by protein modifying reagents. Nature. 1975;253(5491):448–450. doi: 10.1038/253448a0. [DOI] [PubMed] [Google Scholar]
- 14.Elliott J, Smart D, Lambert DG, et al. Characterisation of mu-opioid receptors on SH-SY5Y cells using naloxonazine and beta-funaltrexamine. Eur J Pharmacol. 1994;268(3):447–450. doi: 10.1016/0922-4106(94)90072-8. [DOI] [PubMed] [Google Scholar]
- 15.Ling GS, Paul D, Simantov R, et al. Differential development of acute tolerance to analgesia, respiratory depression, gastrointestinal transit and hormone release in a morphine infusion model. Life Sci. 1989;45(18):1627–1636. doi: 10.1016/0024-3205(89)90272-5. [DOI] [PubMed] [Google Scholar]
- 16.Human Genome Project. The Science Behind the Human Genome Project. 2008 [Google Scholar]
- 17.Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 18.Passetti F, Ferreira CG, Costa FF. The impact of microRNAs and alternative splicing in pharmacogenomics. Pharmacogenomics J. 2009;9(1):1–13. doi: 10.1038/tpj.2008.14. [DOI] [PubMed] [Google Scholar]
- 19.Fox-Walsh KL, Hertel KJ. Splice-site pairing is an intrinsically high fidelity process. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0813128106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.House AE, Lynch KW. Regulation of alternative splicing: more than just the ABCs. J Biol Chem. 2008;283(3):1217–1221. doi: 10.1074/jbc.R700031200. [DOI] [PubMed] [Google Scholar]
- 21.Pan YX, Xu J, Bolan E, et al. Isolation and expression of a novel alternatively spliced mu opioid receptor isoform, MOR-1F. FEBS Lett. 2000;466(2–3):337–340. doi: 10.1016/s0014-5793(00)01095-4. [DOI] [PubMed] [Google Scholar]
- 22.Pan YX, Xu J, Bolan E, et al. Identification and characterization of three new alternatively spliced mu-opioid receptor isoforms. Mol Pharmacol. 1999;56(2):396–403. doi: 10.1124/mol.56.2.396. [DOI] [PubMed] [Google Scholar]
- 23.Pan YX, Xu J, Bolan E, et al. Identification of four novel exon 5 splice variants of the mouse mu-opioid receptor gene: functional consequences of C-terminal splicing. Mol Pharmacol. 2005;68(3):866–875. doi: 10.1124/mol.105.011858. [DOI] [PubMed] [Google Scholar]
- 24.Shabalina SA, Zaykin DV, Gris P, et al. Expansion of the Human {micro}-Opioid Receptor Gene Architecture: Novel Functional Variants. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan L, Xu J, Yu R, et al. Identification and characterization of six new alternatively spliced variants of the human mu opioid receptor gene, Oprm. Neuroscience. 2005;133(1):209–220. doi: 10.1016/j.neuroscience.2004.12.033. [DOI] [PubMed] [Google Scholar]
- 26.Pan YX, Xu J, Mahurter L, et al. Identification and characterization of two new human mu opioid receptor splice variants, hMOR-1O and hMOR-1X. Biochem Biophys Res Commun. 2003;301(4):1057–1061. doi: 10.1016/s0006-291x(03)00089-5. [DOI] [PubMed] [Google Scholar]
- 27.Xu J, Xu M, Hurd YL, et al. Isolation and characterization of new exon 11-associated N-terminal splice variants of the human mu opioid receptor gene, OPRM1. J Neurochem. 2009;108(4):962–972. doi: 10.1111/j.1471-4159.2008.05833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasternak DA, Pan L, Xu J, et al. Identification of three new alternatively spliced variants of the rat mu opioid receptor gene: dissociation of affinity and efficacy. J Neurochem. 2004;91(4):881–890. doi: 10.1111/j.1471-4159.2004.02767.x. [DOI] [PubMed] [Google Scholar]
- 29.Zimprich A, Bacher B, Hollt V. Cloning and expression of an isoform of the rmu-opioid receptor (rmuOR1B) Reg Peptides. 1994;54(1):347–348. [Google Scholar]
- 30.Abbadie C, Pan YX, Pasternak GW. Differential distribution in rat brain of mu opioid receptor carboxy terminal splice variants MOR-1C-like and MOR-1-like immunoreactivity: evidence for region-specific processing. J Comp Neurol. 2000;419(2):244–256. doi: 10.1002/(sici)1096-9861(20000403)419:2<244::aid-cne8>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 31.Bolan EA, Pan YX, Pasternak GW. Functional analysis of MOR-1 splice variants of the mouse mu opioid receptor gene Oprm. Synapse. 2004;51(1):11–18. doi: 10.1002/syn.10277. [DOI] [PubMed] [Google Scholar]
- 32.Matthes HW, Maldonado R, Simonin F, et al. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the mu-opioid-receptor gene. Nature. 1996;383(6603):819–823. doi: 10.1038/383819a0. [DOI] [PubMed] [Google Scholar]
- 33.Sora I, Takahashi N, Funada M, et al. Opiate receptor knockout mice define mu receptor roles in endogenous nociceptive responses and morphine-induced analgesia. Proc Natl Acad Sci U S A. 1997;94(4):1544–1549. doi: 10.1073/pnas.94.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuller AG, King MA, Zhang J, et al. Retention of heroin and morphine-6 beta-glucuronide analgesia in a new line of mice lacking exon 1 of MOR-1. Nat Neurosci. 1999;2(2):151–156. doi: 10.1038/5706. [DOI] [PubMed] [Google Scholar]
- 35.Pan YX, Xu J, Mahurter L, et al. Generation of the mu opioid receptor (MOR-1) protein by three new splice variants of the Oprm gene. Proc Natl Acad Sci U S A. 2001;98(24):14084–14089. doi: 10.1073/pnas.241296098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan YX, Xu J, Xu M, et al. Involvement of exon 11-associated variants of the mu opioid receptor MOR-1 in heroin, but not morphine, actions. Proc Natl Acad Sci U S A. 2009;106(12):4917–4922. doi: 10.1073/pnas.0811586106. [DOI] [PMC free article] [PubMed] [Google Scholar]