Abstract
An ambitious plan to collect, curate, and make accessible information on genetic variations affecting human health is beginning to be realized.
It has been 60 years since the first variation causing inherited disease was defined at the protein level. Currently, at least one such mutation is known to have occurred in 3000 of the 20,000 recognized human genes. In the next few years, the number of genes in which disease-causing mutations are recognized will increase dramatically. Despite good intentions, efforts to develop and build databases have failed to keep up with this pace.
Thus, clinicians and diagnostic laboratories must waste their time trawling through many publications and databases to determine whether a mutation found in a patient has been previously characterized. Availability of previous characterizations of all mutations and their effects would allow them to base their diagnoses and prognoses on evidence rather than guesswork and conjecture. For inherited diseases, rapid access to curated information on all mutations in all genes from all populations is needed. Note that those who gain most by the availability of up-to-date gene variant data are usually downloading information only and are failing to add their findings to further improve the quality of the data collected. Changing this attitude and collecting all data seem to be mammoth tasks, but they are essential.
The Human Variome Project (www.human-variomeproject.org/), initiated in 2006 (1–3) is the global community’s effort to collect, curate, and make accessible information on all genetic variations affecting human health. The specific objectives are to encourage the development and adoption of standards; define, reach consensus on, and implement ethical requirements (including informed consent forms and approaches for protecting patient confidentiality); develop systems for automated data submission; develop community education and communication programs; enable participation by developing countries; support curation processes; promote evidence-based genetic medicine; and create usable systems for contribution, curation, search, and retrieval.
The Human Variome Project is complementary to the massive resequencing projects that contribute on a daily basis to variation databases such as dbSNP and the increasing information from genomewide association studies. These large data sets provide excellent population data on all variations compatible with life and also variations associated with common diseases. But ascertainment of mutations through observation of rare phenotypes provides a quality of information that will not otherwise be captured.
Proofs of Principle
Recent progress indicates that the collection of mutations in all genes worldwide is possible. For example, the International Society for Gastrointestinal HereditaryTumours (InSiGHT; www.insight-group.org), since February 2007, has embarked on developing a pipeline to collect legacy and new data, to place all data from disparate databases on the Leiden Open (source) Variation Database (4) (LOVD; www.lovd.nl) with freely available software and to develop submissions to National Center for Biotechnology Information (NCBI, U.S. National Institutes of Health); European Bioinformatics Institute (EBI); and University of California at Santa Cruz (UCSC) Genome Browser. InSiGHT has begun a pilot project for the collection of all mutations from all countries in four mismatch repair genes that are altered in colon cancer patients. Strategies need to be developed and appropriate software created and put in place in the next 3 years that will enable the seamless, effortless, and low-cost collection of data from laboratories, clinics, and hospital records and their delivery to appropriate databases.
One project that has just begun represents a creative approach to funding for supporting data curation. This cost varies depending on the extent of work involved and the number of mutations per gene, but estimates range from $1000 to $200,000 per year. The Adopt-a-Gene Program through the Human Variome Project is encouraging industry and patient support groups to sponsor the curation of specific genes. The first of these partnerships is already under way, with CMO Global Services supporting the Familial Hemiplegic Migraine Variation Database (http://lovd.nl/FHM).
When a variation is found in a patient, the clinician or diagnostician has to decide whether it is causing the disease. Numerous algorithms have been developed that predict pathogenicity on the basis of such features as evolutionary conservation, frequency, nature of missense change, and protein structural changes. Ideally, an algorithm should incorporate all of these features to give a probability of pathogenicity. International efforts to create such an algorithm are ongoing. Below, we describe efforts to create the next generation of databases for one group of disorders.
Neurological Disorders
The brain has the largest number of expressed transcripts of all organ systems, and this is reflected in the very large number of neurogenetic disorders. Over 30% of Mendelian diseases have neurological manifestations (5). When the molecular lesion is singular and easily detected, such as the triplet repeat expansion in Huntington disease, molecular testing has revolutionized clinical practice. Accurate diagnosis and genetic counseling can now be given.
However, the situation is more complicated in most neurogenetic diseases. Neurogenetics is full of examples of genetic heterogeneity (e.g., Charcot-Marie-Tooth disease); allelic disorders (e.g., hemiplegic migraine and episodic ataxia); variable expressivity (e.g., synucleinopathies); incomplete penetrance (e.g., dopa-responsive dystonia); anticipation (e.g., spastic paraparesis); pheno-copies (e.g., familial epilepsies); and imprinting (e.g., Angelman and Prader-Willi syndromes), all of which are biological phenomena that entangle genotype-phenotype relationships. To further complicate matters, mitochondrial mutations add their particular inheritance mechanism and heterogeneous phenotypic expression [e.g., Leigh syndrome and Neuropathy, Ataxia, and Retinitis Pigmentosa (NARP)].
Neurological phenotypes are extremely varied and often complex and can evolve over time in a given patient. Two individuals with the same genetic and pathologic process may show different phenotypes, whereas the same phenotype (e.g., lack of fine motor coordination) may be manifested in totally different pathogenic events and disorders.
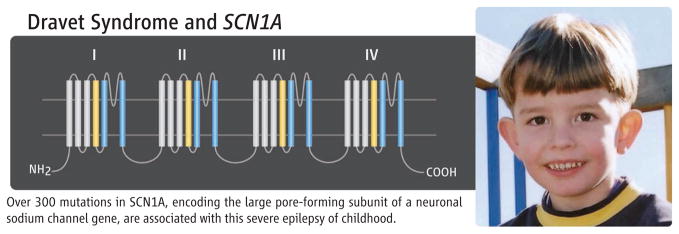
A particularly good example in which a complete catalog of variation would be invaluable is in a severe epilepsy, Dravet syndrome (see figure on page 861). It begins at 6 months of life with febrile seizures, followed by later intellectual decline and a somber prognosis. Some evidence suggests that early aggressive treatment may improve outcome, making early diagnosis imperative. The disease is associated in ~80% of cases with variation in the neuronal sodium channel subunit gene SCN1A; a large gene with 26 exons. Half the cases have mutations predicting truncation of the protein, but half are missense. However, there is a milder epilepsy syndrome [generalized epilepsy with febrile seizures + (GEFS+)] that begins at the same time (with febrile seizures) for which treatment is often not required, the outcome is good, and missense mutations in SCN1A are found in about 10% of cases. Our knowledge base is insufficient to predict the phenotype associated with particular missense mutations in SCN1A.
Figure.
Over 300 mutations in SCN1A, encoding the large pore-forming subunit of a neuronal sodium channel gene, are associated with this severe epilepsy of childhood.
A number of public neurological databases already exist, such as the Alzheimer Disease and Frontotemporal Dementia Mutation Database (www.molgen.ua.ac.be/ADMutations) and the Inherited Neuropathies Database (www.molgen.ua.ac.be/CMTMutations). However, they generally contain limited data on the phenotype and on the evidence of pathogenicity. Phenotype-based databases, on the other hand, such as the London Neurogenetics Database (6) or GeneReviews (www.geneclinics.org/) are detailed in disease description but usually not in the interpretation of specific mutations and variations in a particular gene.
Among the challenges to meet for neurogenetic databases is that of finding appropriate forums and funding policies that allow the confluence of “genotypers” and “phenotypers,” i.e., multidisciplinary teams that include molecular geneticists, cell physiologists, and biochemists, as well as clinicians (adult and pediatric neurologists, clinical neurophysiologists, and neurosurgeons).
What organizations are best suited to call for convocation of such multidisciplinary teams? International networks for neurological disorders such as spinocerebellar ataxias, Charcot-Marie-Tooth disease, or spastic paraparesis could facilitate collection of genetic and clinical information from basic science and clinical research groups. However, the information generated by these consortia often remains available only to members, with important legal and ethical questions emerging from the use of these large patient data sets and sample collections. Additional limitations of disease-centered consortia are their dependence on financial support for a specific time-frame, marked by funding or mission views.
Alternative forums might be neurogenetics societies; however, the current landscape of neurogenetics meetings still needs to evolve to face the multidisciplinary effort involved. There are neurogenetics study groups within some neurological societies, such as the Spanish Society for Neurology (SEN), the American Academy of Neurology (AAN), or the European Federation of Neurological Societies (EFNS). However, these panels are generally small, run by neurologists with an interest in genetics, and with very limited participation of geneticists and basic researchers. Associations for human genetics generally lack specific neurogenetics committees, plus clinical neurologists are absent or minimally represented in these genetic forums. (One exception is the German Society for Neurogenetics.)
An example of the difficulties can be found in Spain, where nine large networking national centers for research have recently been created by the Spanish Health Institute Carlos III (CIBERs), and are responsible for coordinating translational biomedical research in specific areas. Among the CIBERs, the Center for Research on Rare Diseases (CIBERER) has established a neurogenetics committee, but virtually no clinical neurologists, neurophysiologists, or neurosurgeons belong to the CIBERER. Another CIBER on neurodegenerative disorders (CIBERNED) includes basic neuroscientists, pathologists, and clinical neurologists, but only one or two clinical diagnostic geneticists. Any initiative aimed at organizing the data on genetic neurological disorders in Spain would have to bring these groups together. There is currently an initiative to launch a Neurogenetics Association in Spain.
A strategy for the development of neurological locus-specific databases could start with international, multidisciplinary, disease-centered networks. These would be organized into a multidisciplinary neurogenetics society or network with representatives of each disease consortium and/or locus-specific database. They would integrate the information, propose common guidelines, discuss common coding issues, and facilitate navigation from one database to another. The role of the Human Variome Project will be to foster such a strategy, to get disease-centered networks involved, and to promote or host coordination forums.
Supplementary Material
Footnotes
References and Notes
- 1.Editorial. Nat Genet. 2007;39:423. [Google Scholar]
- 2.Ring HZ, Kwok PY, Cotton RG. Pharmacogenomics. 2006;7:969. doi: 10.2217/14622416.7.7.969. [DOI] [PubMed] [Google Scholar]
- 3.Cotton RG, et al. Nat Genet. 2007;39:433. doi: 10.1038/ng2024. [DOI] [PubMed] [Google Scholar]
- 4.Fokkema IF, den Dunnen JT, Taschner PE. Hum Mutat. 2005;26:63. doi: 10.1002/humu.20201. [DOI] [PubMed] [Google Scholar]
- 5.Costa T, Scriver CR, Childs B. Am J Med Genet. 1985;21:231. doi: 10.1002/ajmg.1320210205. [DOI] [PubMed] [Google Scholar]
- 6.Baraitser M, Winter RM. London Dysmorphology Database, London Neurogenetics Database & Dysmorphology Photo Library on CD-ROM. Oxford Univ. Press; Oxford: 2001. [Google Scholar]
- 7.We thank I. Scheffer and the patient’s family for the photograph.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.