Abstract
The (H3-H4)2 histone tetramer forms the central core of nucleosomes and, as such, plays a prominent role in assembly, disassembly and positioning of nucleosomes. Despite its fundamental role in chromatin, the tetramer has received little structural investigation. Here, through the use of pulsed electron-electron double resonance spectroscopy coupled with site-directed spin labelling, we survey the structure of the tetramer in solution. We find that tetramer is structurally more heterogeneous on its own than when sequestered in the octamer or nucleosome. In particular, while the central region including the H3-H3′ interface retains a structure similar to that observed in nucleosomes, other regions such as the H3 αN helix display increased structural heterogeneity. Flexibility of the H3 αN helix in the free tetramer also illustrates the potential for post-translational modifications to alter the structure of this region and mediate interactions with histone chaperones. The approach described here promises to prove a powerful system for investigating the structure of additional assemblies of histones with other important factors in chromatin assembly/fluidity.
INTRODUCTION
In addition to packaging eukaryotic genomes within the confines of nuclei, chromatin provides a means of regulating access to the underlying genetic material. The fundamental subunit of chromatin is the nucleosome that consists of 147 bp of DNA wrapped in 1.6 superhelical turns around an octet of four core histones (1). The core histones are arranged as a central (H3-H4)2-tetramer that is capped on either side by an H2A-H2B dimer. This organization mirrors chromatin assembly, where the tetramer is deposited onto the DNA template first (known as the tetrasome), forming a high-affinity binding platform onto which the H2A-H2B dimers can load (2).
Nucleosomes are not static entities and can undergo a number of dynamic transitions. Among the best characterized of these is the dissociation of the outer turns of DNA (3). It has been possible to observe that DNA remains wrapped for periods of the order 250 ms before dissociating for ∼25 ms. The histone composition of chromatin is not fixed; Instead, the histones within a subpopulation of chromatin exchange rapidly (4–6). A range of factors have been implicated in the destabilization of histones within nucleosomes including the underlying DNA sequence (7), the binding of transcription factors (8,9), the action of remodelling enzymes (10–13) and polymerases (14).
Rearrangement of the histone composition within nucleosomes results in the generation of non-canonical chromatin particles of which a range have been reported including, hexasomes (14–16), tetrasomes (15,17), di-tetrasomes (18) and others (19–21). The interaction between H3 and H4 is central to many of these structures. Current insight as to how H3 and H4 interact with each other is largely derived from crystallographic structures of nucleosomes and octamers. There is evidence that, in the absence of histones H2A and H2B, the arrangement of H3 and H4 can differ from that observed in the crystal structures. For example, a chiral transition within the tetramer has been proposed to enable wrapping of DNA with a right-handed pitch (22,23). Structural studies of H3 and H4 in association with the histone chaperone Asf1 indicate a complex with a single H3-H4 dimer (24,25). There is also evidence that dimers of H3 and H4 associate with the chaperones CAF-1 and HIRA (26). These observations highlight the need for structural studies of non-canonical chromatin particles.
The tetramer particle has received little structural investigation. Crystallographic studies have met with only partial success (27), possibly due to structural heterogeneity within the tetramer. Nuclear magnetic resonance has been used to investigate the structure of the H2AZ-H2B dimers in solution (28) complexed with a histone chaperone peptide; however, the histone tetramer is typically too large for structure determination by this method. A previous study has examined the tetramer using deuterium exchange/mass spectrometry (29). The study of deuterium exchange has the potential to provide information on the dynamic nature of the tetramer, but in this case the technique was used to compare the wild-type tetramer with one in which histone H3 had been replaced by CENP-A. Förster resonance energy transfer (FRET) has been proved to be fruitful in investigating the dynamics of the DNA component of nucleosome particles (30), but due to the dimeric nature of the histone core, attaching a single acceptor and donor fluorophore is difficult to achieve, thus FRET has not been able to relay structural information from the underlying histone component of nucleosomes.
In this current study, we have employed site-directed spin labelling (SDSL) (31) and pulsed electron-electron double resonance (PELDOR) spectroscopy on both the core histone octamer [(H3-H4)2(H2A-H2B)2] and tetramer (H3-H4)2. PELDOR is a powerful tool for measuring long-range distances between unpaired electrons, in the range 2–8 nm (32). It has been utilized to investigate proteins (33–37), nucleic acids (38–42) and protein–nucleic acid complexes (43). PELDOR can be applied to chemically identical spin label pairs, thus this method is ideally suited to protein systems that are homo-dimeric.
We show that distance distributions measured from the octamer correlate well with a published high-resolution crystal structure and contrast this with distance distributions measured from the tetramer. While a region surrounding the histone fold dimerization interface retains a structure comparible with that observed in the octamer, other regions, most notably the H3 αN helix, deviate from this. The flexibility observed in this region is likely to influence functional interactions.
MATERIALS AND METHODS
Protein purification and sample preparation
Site directed mutagenesis was used to introduce cysteine residues at strategic locations on Xenopus laevis histone H3C110A and wild-type H4, aided by the crystal structure of the histone octamer (1TZY.pdb) using standard cloning procedures. Recombinant Xenopus histones were purified from Escherichia coli [strain BL21(DE3) pLysS, Stratagene] as described previously (1), dialysed into H2O and stored as lyophilates. Spin labelling of the histone octamer was carried out as described in Ward et al. (40). Briefly, unfolded histones, H2A, H2B, H3 C110A and H4, carrying the desired mutations for spin labelling, were mixed in equimolar ratios and refolded by dialysis into 2 M sodium chloride, 10 mM Tris–HCl pH 7.5, 1 mM EDTA and 5 mM β-mercaptoethanol (refolding buffer). Refolded histones were purified from misfolded aggregates by gel filtration chromatography, Superdex S200 (GE Healthcare), in refolding buffer without β-mercaptoethanol (Supplementary Figure S5), which additionally served to remove excess reducing agent prior to spin labelling with MTSL. Before chromatography, 20 mM dithiothrietol was added to the sample to fully reduce cysteine residues. Spin labelling was carried-out overnight at 4°C with a 10-fold excess of MTSL and a protein concentration of 20–100 µM octamer (variation within this range did not affect the efficiency of labelling as monitored by MALDI-TOF mass spectrometry; data not shown). Unreacted MTSL was removed by dialysis into 2 × 1 L of refolding buffer without β-mercaptoethanol. Spin labelled octamer was prepared for PELDOR by buffer exchanging into refolding buffer without β-mercaptoethanol made with deuterated water (Sigma-Aldrich), using an Amicon Ultra-4 spin concentrator with a molecular weight cut-off of 10 000 (Millipore). An equimolar excess of H2A-H2B dimers, prepared in deuterated refolding buffer as for the octamer, was added to octamer samples as this had previously been shown to suppress octamer dissociation (40). Finally, the sample was diluted 1:1 with D8-glycerol and stored at −20°C until distance extraction by PELDOR. Spin concentration of the samples was typically between 100 and 200 µM. Spin labelling of the histone tetramer was essentially the same for the octamer except that H2A and H2B were omitted from the refolding reaction. None of the mutations used in this study inhibited the binding of H2A-H2B dimers, as judged by size exclusion chromatography (for a typical chromatogram of refolded octamer and tetramer see Supplementary Figure S5). The presence of free tetramer within octamer preparations does introduce some extra problem in data interpretation; the fact that all octamer data are compared with pure tetramer preparations resolves this problem. Far UV circular dichroism of wild-type H4 and H4E63C tetramers (both tetramers containing the H3C110A mutation) was carried out in 10 mM Tris–HCl pH 7.5, 0.9 M sodium fluoride and 1 mM EDTA. Spectra were recorded in a 0.02-cm path length quartz cuvette using a protein concentration of 0.4 mg/ml or 19 µM.
PELDOR experiment
PELDOR experiments were executed using a Bruker ELEXSYS E580 spectrometer operating at X-band with a dielectric ring resonator and a Bruker 400 U second microwave source unit. All measurements were made at 50 K with an overcoupled resonator giving a Q factor of ∼100. The video bandwidth was set to 20 MHz. The four-pulse, dead-time free, PELDOR sequence was used, with the pump pulse frequency positioned at the centre of the nitroxide spectrum; the frequency of the observer pulses was increased by 80 MHz. The observer sequence used a 32-ns π-pulse; the pump π-pulse was typically 16 ns. The experiment repetition time was 4 ms, and the number of scans used was sufficient to obtain a suitable signal (typically >400 scans) with 50 shots at each time point. Total experiment time varied according to the distance under investigation. Good signal to noise data on short distances (3 nm) could be obtained in <7 h, while longer distances (≥5 nm) required at least 16 h of averaging time.
PELDOR data analysis
In brief, the experimentally obtained time domain trace is processed so as to remove any unwanted intermolecular couplings, which is called the background decay. Tikhonov regularization (44,45) is then used to simulate time trace data that give rise to distance distributions, P(r), of different peak width depending on the regularization factor, α. The α-term used was judged by reference to a calculated L-curve. The L-curve is a plot of the α-term against quality of fit, measured by mean square deviation between the experimental data and simulation. The most appropriate α-term to be used is at the inflection of the L-curve, since this provides the best compromise between smoothness (artefact suppression) and fit to the experimental data. PELDOR data were analysed using the DeerAnalysis 2006 software package (46). The dipolar coupling evolution data were corrected for background echo decay using a homogeneous 3D spin distribution. The starting time for the background fit was optimized to give the best fit Pake pattern in the fourier transformed data and the lowest root mean square deviation background fit.
Spin label dynamics (generation of synthetic distributions)
Coordinates were taken from PDB code 1TZY and mutated within PyMol (47) to replace required amino acid positions with cysteine. Parameter and topology files for MTSSL were created using PRODG (48). Coordinates for the MTSSL spin label were generated and minimized using the program Ghemical (49), and then melded with the protein structures by common atom superposition within PyMol. Molecular dynamics, using XPLOR-NIH (50), was carried out on residues within 1.2 nm of the selected site, as ascertained using PyMol, in order to generate distributions for each spin label attached to a cysteine mutant site. Backbone C, N and O atoms were restrained by harmonic function to initial positions and the unrestrained atoms were allowed to move under molecular dynamics at a temperature of 500°K. Structures were taken at regular intervals and the distance between pairs of spin label nitroxide nitrogen atoms were calculated, binned in 1-Å groups to give synthetic distance distributions.
RESULTS
Selection of sites for introduction of the spin probe
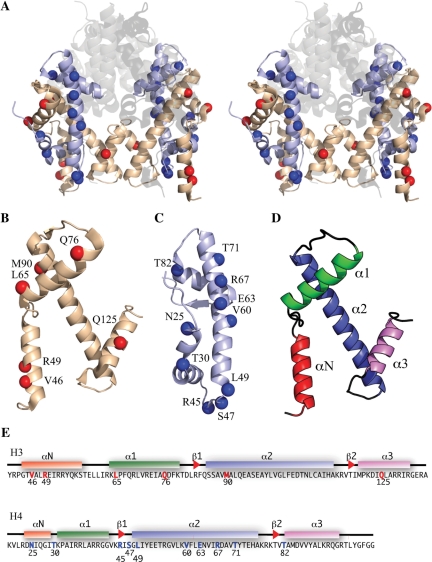
In order to investigate the structure of the histone tetramer by using SDSL coupled with PELDOR, it was first important to select labelling sites that minimally perturb the overall protein complex. Solvent-accessible positions, which were not part of any known interface with the H2A-H2B heterodimers, were chosen based on the octamer crystal structure PDB code 1TZY (Figure 1A). In order to be sensitive to the differences between the octamer and tetramer, sites were also selected to be on, or close to, each of the secondary-structural elements of the histone folds (Figure 1B–E; and Supplementary Table S1). Since PELDOR can measure distances of between 2 and 8 nm, the distances between the α-carbons of the selected mutants were measured to ensure that the spin labels would be in this range (Supplementary Table S1). SDSL was carried out by first mutating the selected positions to cysteines and then reacting them with (1-Oxyl-2,2,5,5-tetramethyl-pyrroline-3-methyl) methanethiosulphonate to form the disulphide product. This new side chain is referred to as R1 (Supplementary Figure S1) (51).
Figure 1.
Known structure and spin labelling positions. (A) Stereo cartoon from the crystal structure (1.9 Å) of the histone octamer (PDB code 1TZY): H2A, H2B represented by transparent/grey colour; H3 by wheat; H4 by pale blue. Spin label positions indicated by blue (H4) or red (H3) spheres. (B) H3 labelling positions (red) on monomer. (C) H4-labelling positions (blue) monomer. (D) Histone fold (colour coded). (E) H3 and H4 amino acid sequences with labelling sites coloured red and blue, respectively. Helices indicated by coloured boxes and β-sheets by red arrowheads.
The histone octamer complex adopts a largely homogeneous structure in aqueous solution
Electron paramagnetic resonance can be used to extract nanometre-range distances between two unpaired electrons, spins A and B, by reporting the dipole–dipole coupling between them. The coupling is proportional to the cubed root of the spin–spin distance. To measure the dipolar coupling, the two allowed transitions of both spins A and B must be excited. This can be achieved either by using a single or a double frequency pulse experiment. The PELDOR experiment is a double frequency method that is capable of exciting spins A and B selectively based on their angular relationship to the external magnetic field (52). The analysis protocol used to obtain the distance distribution is contained within the program DeerAnalysis2006. This package is well documented (46). The experimentally obtained time domain trace is processed to remove any unwanted intermolecular couplings that make contributions to the background decay. Tikhonov regularization is then used to obtain an appropriate distance distribution. In order to be confident that distance distributions are accurate, PELDOR data should describe at least one full oscillation and decay to baseline. In general, such data describes a dominant distribution following regularization and can be interpreted with confidence. However, in the cases where the oscillation is not well defined, caution is required as mobile or severely heterogeneous structures can give rise to misleading distance distributions.
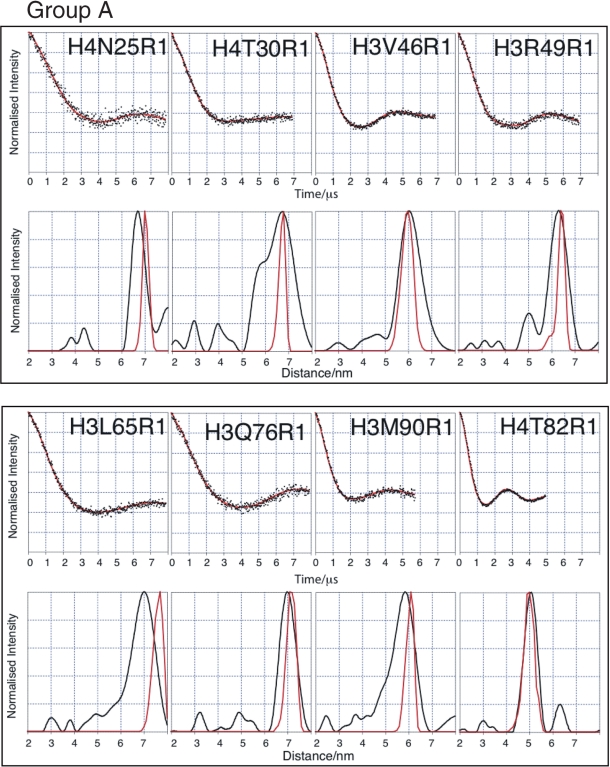
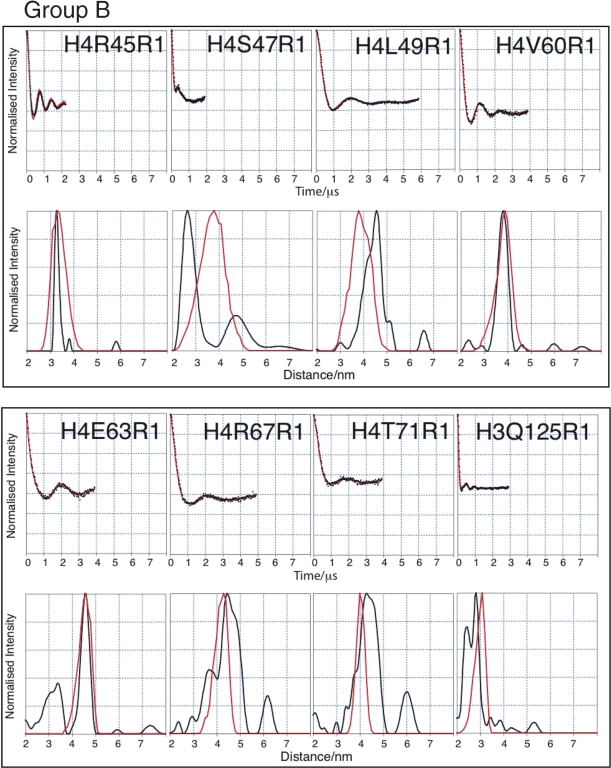
The histone octamer has been crystallized to high resolution (1.9 Å) in a high-ionic strength environment (53), suggesting that under such conditions the histone octamer is a stable, structurally homogeneous complex. It should be noted that the core structure of the crystalized octamer is almost identical to those seen in crystal structures of the nucleosome. Thus to aid our interpretation of the tetramer structure, we first carried out SDSL and PELDOR on the octamer. In order to simplify the description of the octamer PELDOR data, the spin labelled mutants have been divided into two groups depending on whether their Cα–Cα distance is greater (Group A) or less than (Group B) 4 nm based on the coordinates from the crystal structure (Figures 2 and 3). It is notable that some of the labelling sites in Group A, namely H4N25R1, H4T30R1, H3V46R1 and H3R49R1, give some of the longest distances so far measured from a biological macromolecule using PELDOR (54). It can be seen that the corrected dipolar evolution data for Group A gives oscillations between 2.5 (e.g. H4T82) and 7 µs (e.g. H3Q76). The clearly discernible dipolar oscillation obtained for each mutant, except for H4T30, results in discrete distance distributions. The broad and complex distance distribution data for H4T30 suggest that this position is not as well defined as the crystal structure might suggest. The data for Group B show much shorter time period oscillations, between 0.38 (e.g. H4S47) and 2 µs (e.g. H4L49), resulting in much shorter distances for these mutants as one would expect. The observation of tight distance distributions for the majority of labelling sites indicates that the histone octamer forms a well-defined structure under the experimental conditions used.
Figure 2.
PELDOR data from the histone octamer for Group A. Results derived from label positions >4 nm apart based on predicted Cα-Cα distance. For each mutant two graphs of data are shown. Top graphs: Background corrected echo oscillations (black), Tikhonov fit (red). Bottom graphs: Tikhonov regularization-derived distance distributions (black), simulated distributions (red).
Figure 3.
PELDOR data from the histone octamer for Group B. Results derived from label positions <4 nm apart based on predicted Cα-Cα distance. For each mutant two graphs of data are shown. Top graphs: Background corrected echo oscillations (black), Tikhonov fit (red). Bottom graphs: Tikhonov regularization-derived distance distributions (black), simulated distributions (red).
Close agreement between PELDOR-derived and simulated distance distributions
In order to evaluate the PELDOR data further, a protocol for the simplistic molecular modelling of the spin label distributions has been used. First, the residue R1 (Supplementary Figure S1) was incorporated at the site of interest, using coordinates from the octamer crystal structure. Molecular dynamics was carried out on residues within 1.2 nm of the selected site. Backbone C, N and O atoms were restrained and a simplified force-field was used (no electrostatics or attractive van der Waals potential). For each spin-pair position, 100 structures were extracted from the molecular dynamics trajectory. From the 100 structures, all possible spin label nitrogen–nitrogen distances were measured, binned and used to simulate distance distributions (Figures 2 and 3).
It can be seen that, in almost all cases, the modal distances derived from the PELDOR data agree very well, with those obtained from the dynamics simulations (Table 1). Out of the 16 distances measured on the octamer, only three exhibit serious discrepancies (H3L65, H4S47 and H4L49). The quality of the experimental data and the close agreement with the crystal structure, of the majority of derived distances, gives us a high degree of confidence in our analysis while at the same time suggesting that the three exceptional distance measurements are indicative of a real (although possibly minor) structural difference between the structures under PELDOR conditions and those under the conditions of crystallization.
Table 1.
Distance data obtained from PELDOR and dynamic simulation for Groups A and B
| Probe position | Distance (EPR) (nm) | Distance (Dyn) (nm) | ΔDistance (Dyn − EPR) |
|---|---|---|---|
| Group A | |||
| H3V46 | 6.0 | 6.0 | 0.0 |
| H3R49 | 6.3 | 6.4 | 0.1 |
| H3L65 | 7.0 | 7.7 | 0.7 |
| H3Q76 | 7.0 | 7.1 | 0.1 |
| H3M90 | 5.9 | 6.1 | 0.2 |
| H4N25 | 6.7 | 7.0 | 0.3 |
| H4T30 | 6.7 | 6.8 | 0.1 |
| H4T82 | 5.1 | 5.0 | −0.1 |
| Group B | |||
| H3Q125 | 2.8 | 3.1 | 0.3 |
| H4R45 | 3.3 | 3.3 | 0.0 |
| H4S47 | 2.6 | 3.8 | 1.2 |
| H4L49 | 4.6 | 3.8 | −0.8 |
| H4V60 | 3.8 | 3.9 | 0.1 |
| H4E63 | 4.6 | 4.6 | 0.0 |
| H4R67 | 4.5 | 4.3 | −0.2 |
| H4T71 | 4.3 | 4.0 | −0.3 |
The PELDOR data provide two pieces of information concerning the spin labels: the modal distances, which are generally robust, and the distance distributions, which are more susceptible to artefacts and inaccuracies caused by noise, data truncation and the choice of processing parameters. When comparing simulated with experimental distance distributions, one would expect the simulated to be narrower, since no account is taken of backbone, or indeed global, protein dynamics. The PELDOR distance distribution is a convolution of different effects that determine the position of the spin labels; these include both global and local structural heterogeneity of the protein and spin label. Comparison of the simulations to the experimental distance distributions should shed some light on the underlying protein structure and dynamics. In the majority of cases, the simulations give similar distribution widths to the experimental data. The similarity is surprising in the light of the fact that no protein backbone dynamics was allowed for. Interestingly, positions H4R45 and H4S47 have much narrower PELDOR-derived distributions than the simulations, implying that the spin label itself is more restrained than the simple dynamics predicts. In highly restricted environments, the output from the PELDOR experiment can be affected by the rigid alignment of the spin labels. Alignment can be tested by moving the observe pulse position and observing changes in the calculated distance distribution. H4R45 and H4S47 spin-pairs were tested in this manner and no anomalous orientation effects were observed (data not shown). Overall, these observations confirm our impression that the octamer, under the conditions used, is largely stable and undynamic, adopting the same conformation in aqueous solution as it does in a crystal lattice.
Comparison between the octamer and tetramer
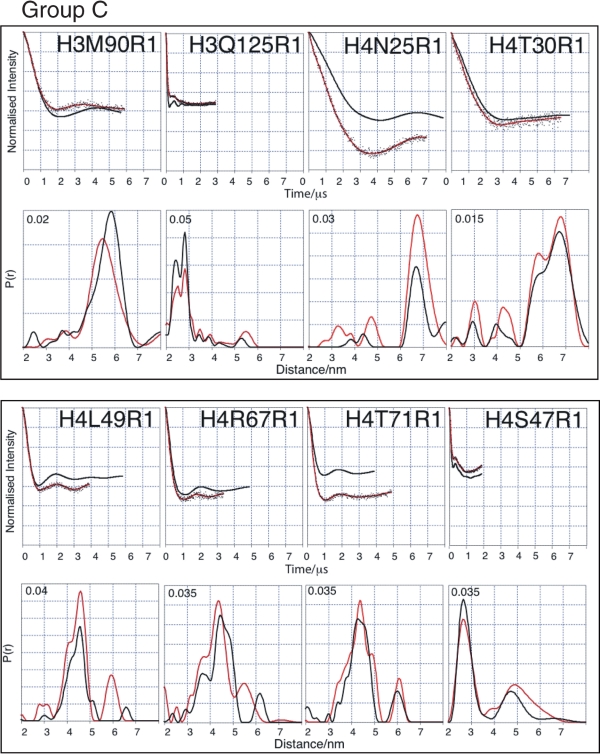
Having established the validity of the PELDOR approach using the histone octamer, we next sought to apply the same approach to study the H3-H4 tetramer for which there is far less structural information available. At first inspection, the data can be classified into two groups. Measurements made from labelling sites that show only minor differences, in comparison between the octamer and tetramer, are collected as Group C (H3M90, H3Q125, H4N25, H4T30, H4S47, H4L49, H4R67 and H4T71), and illustrated in Figure 4 and Table 2. In these cases, the echo oscillation for both tetramer and octamer describe similar shapes and phase persistence.
Figure 4.
Comparison of tetramer and octamer PELDOR data in Group C (minor differences between octamer and tetramer measurements). For each mutant two graphs of data are shown. Top graphs: Tikhonov-derived fit to octamer PELDOR data (black), background-corrected echo oscillation from tetramer (black dots) and Tikhonov-derived fit to tetramer (red). Bottom graphs: Tikhonov regularization-derived distance distributions octamer (black), tetramer (red).
Table 2.
Comparison of tetramer and octamer modal distances (nanometre) obtained from PELDOR data (where multiple peaks are present, and the modal distance is taken from the most intense peak)
| Probe positions | Tetramer (nm) | Octamer (nm) | ΔDistance (octamer − tetramer) |
|---|---|---|---|
| Group C | |||
| H3M90a | 5.5 | 5.9 | 0.4 |
| H3Q125 | 2.8 | 2.8 | 0.0 |
| H4N25 | 6.8 | 6.7 | −0.1 |
| H4T30 | 6.8 | 6.7 | −0.1 |
| H4S47 | 2.7 | 2.6 | −0.1 |
| H4L49 | 4.6 | 4.6 | 0.0 |
| H4R67 | 4.4 | 4.5 | 0.1 |
| H4T71 | 4.4 | 4.3 | −0.1 |
| Group D | |||
| H4R45 | 3.4 | 3.3 | −0.1 |
| H4V60 | 3.7 | 3.8 | 0.1 |
| H4E63 | 2.3 | 4.6 | 2.3 |
| H4T82 | 4.8 | 5.1 | 0.3 |
| H3V46 | 5.5 | 6.0 | 0.5 |
| H3R49 | 6.4 | 6.3 | −0.1 |
| H3L65 | 7.0 | 7.0 | 0.0 |
| H3Q76 | 7.0 | 7.0 | 0.0 |
aThe apparently large Δdistance is due to a change in the shape of the distribution rather than a gross movement in the distribution position.
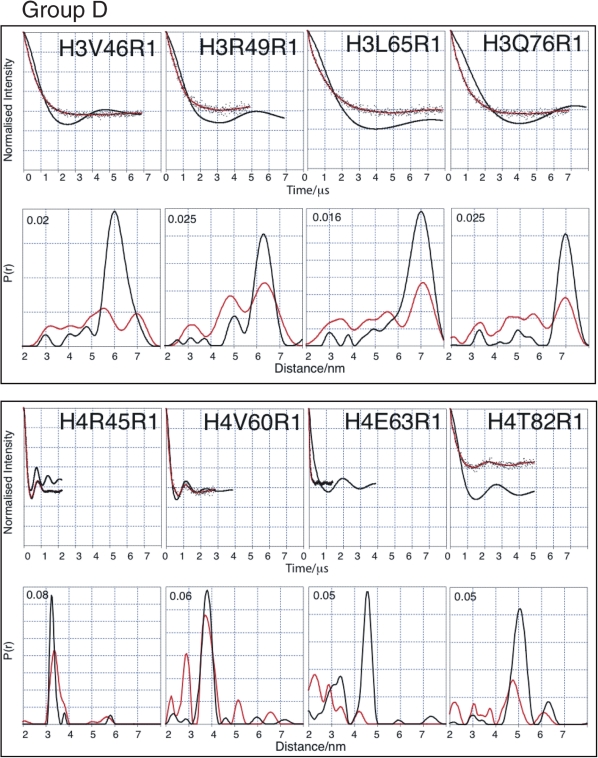
Measurements that differ markedly between octamer and tetramer are collected in Group D (H3V46, H3R49, H3L65, H3Q76, H4R45, H4V60, H4E63 and H4T82) (Figure 5 and Table 2). The echo intensity measured from positions H3V46, H3R49, H3L65 and H3Q76 drop faster than seen for the octamer and their echo oscillations are less well defined, which results in additional short-range distance distributions and generalized broadening. The echoes measured from positions H4R45 and H4T82 show dampened oscillations and consequently relatively broadened distance distributions.
Figure 5.
Comparison of tetramer and octamer PELDOR data in Group D (major differences between octamer and tetramer measurements). For each mutant two graphs of data are shown. Top graphs: Tikhonov-derived fit to octamer PELDOR data (black), background-corrected echo oscillation from tetramer (black dots) and Tikhonov-derived fit to tetramer (red). Bottom graphs: Tikhonov regularization-derived distance distributions octamer (black), tetramer (red).
Data derived from positions H4V60 and H4E63 are of special note. H4E63 shows by far the biggest difference in modal distance between the octamer and tetramer. The modal distance difference is 2.3 nm with virtually no distance distribution remaining at the original octamer distance of 4.6 nm. Position H4V60 is situated one helical turn away from E63 and, although still showing the same modal distance in the tetramer as in the octamer (3.8 nm), exhibits a new and significant shorter distance at 2.9 nm. To rule out the possibility that it is the E63C mutation itself causing a dramatic change in the tetramer structure, which is then restored upon addition of H2A/H2B, we carried out circular dichroism on the H4E63C mutant tetramer and compared it with that of the H4 wild-type tetramer (both tetramers containing the H3C110A mutation). No discernable difference was observed (Supplementary Figure S2); thus we conclude that there is no major secondary structural effect of the E63C mutation, however, we cannot rule out local disturbances in secondary structure of this region. Consistent with this, other studies have used H4E63C for the attachment of fluorophores (55), without observing major perturbations to nucleosome structure.
Taken together, these observations suggest that in the regions covered by labelling sites falling within Group D, the structure of the tetramer is less well ordered than in the octamer. Given that the PELDOR data are taken from the frozen state, it may be reasonable to infer that the observed disordered regions would be dynamic in non-frozen solution.
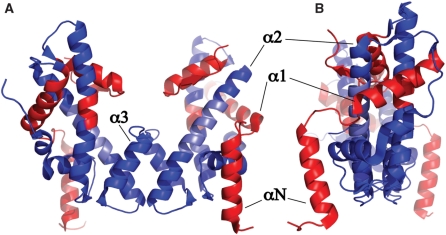
If the regions defined by Groups C and D are mapped onto the structure of the tetramer (taken from the octamer crystal structure), it can be seen that the core of the molecule, centred on the H3-H3′ dimerization interface, largely retains the structure present in the octamer. The regions that differ, either in terms of observed modal distance, or structural heterogeneity (inferred dynamics), are situated largely at the periphery of the structure; these include the αN and α1 helices of H3 and the α3 helix of H4 (Figure 6).
Figure 6.
Cartoon view of H3/H4 histone tetramer coloured as per histone core octamer structure. Very similar regions (blue), very dissimilar or heterogeneous regions (red), region with lack of reporting data (light blue) are depicted. The helices of histone H3 are labelled for clarity. (A) front view showing H3-H3′ interface at centre. (B) side view.
DISCUSSION
PELDOR has been shown to be a powerful tool for obtaining long-distance measurements from the histone core octamer. Only small deviations were observed between the PELDOR-derived and crystal structure-predicted distances from the octamer. This serves as a validation of the application of PELDOR to study chromatin, and confirms that the octamer structure is maintained under different experimental conditions. Using exactly the same positions for spin labelling, we compared the arrangement of histones H3 and H4 in the octamer to that in the histone tetramer for which very little structural data of any kind exist. Careful analysis of the tetramer data reveals a variety of differences including modal distance changes, distance distribution broadening and, in some cases, multiple distance distributions indicative of structural heterogeneity. The altered conformation of the isolated histone tetramer is anticipated to influence its interaction with binding partners during the course of chromatin metabolism.
The effect of H2A-H2B dimers on the stability of the tetramer
One of the clearest conclusions that can be drawn from this study is that for positions H3M90, H3Q125, H4N25, H4T30, H4S47, H4L49, H4R67 and H4T71, the presence or absence of the H2A-H2B dimers has little effect. The underlying secondary structure probed by these positions is intact and the region covered by these sites appears to adopt a very similar global structure in both the tetramer and octamer (regions coloured blue in Figure 6).
In contrast, removal of the H2A and H2B histones from the octamer results in significant changes to the structure at other locations. Of these locations showing changes, the centre of H4 α2 region is reported on by H4E63 and H4V60. At H4V60 a very significant, shorter distance is detected in the tetramer although the original distance distribution seen from the octamer still persists. At the nearby H4E63, a more dramatic change occurs involving the loss of the major octamer measurement and the appearance of several shorter distance distributions. As there are only minor changes at positions H4R67 and H4T71, which are further along the H4 α2 helix, these data suggest that this helix develops a discontinuity at, or close to, H4E63, which also affects H4V60 to a lesser extent.
On first analysis, this relatively serious distortion within the major helix of the H4 histone fold, upon removal of the H2A/H2B dimers, seems unlikely due to the fact that this region has no direct contacts with either H2A or H2B. Upon closer examination, it would seem possible that this effect might be transmitted through the α2 helix of H3 since it makes hydrophobic contacts with both the H2A C-terminus (H3V101 to H2AV107) and the H4 α2 helix (H3V101 to H4F61). The interaction between the H3 α2 and the H4 α2 helixes has been observed before in a study (29) which used deuterium exchange mass spectrometry to investigate the structural impact of CENP-A, a centromere-specific H3 variant, on the tetramer. This study used the rates at which backbone amide protons exchanged with deuterium, from deuterated water, as a measure of relative solvent accessibility and secondary structure stability. When the data were compared between wild-type H3 and CENP-A, it was found that the most significant change was an increase in the stability of the H3 α2 helix. In addition, it was found that CENP-A stabilized the H4 α2 helix more than the wild-type H3, and the region that was most affected included the residues V60 and E63, two of the positions that show significant change on going from octamer to tetramer.
Another region that displays significant changes on removal of the H2A-H2B dimers is the region probed by labels at positions H3V46, H3R49, H3L65 and H3Q76. These regions in the tetramer show significant structural heterogeneity. Because of the nature of the measurements, it is not possible to determine whether this heterogeneity is a reflection of large-scale dynamics or the existence of meta-stable states. H3V46 and H3R49 are situated on the αN helix of H3, with H3L65 and H3Q76 being situated on the adjoining H3 α1 helix. It is interesting to note that in going from the octamer to the tetramer, the C-terminal region of H2A is removed from making a significant number of contacts with both the H3 αN helix (53) and the H4 α1-α2 loop (L1). The structural heterogeneity reported on by H3V46 and H3R49 may propagate to H4R45, which also displays a relatively broadened distribution in the tetramer. The octamer crystal structure shows that the H4R45 side chain position is constrained by the H3 αN helix. Movement or disorganization of the H3 αN helix could cause the observed increase in modal distance and distribution monitored by the H4R45 position.
The picture that emerges from this interpretation is one in which the H3 αN helix becomes severely destabilized by the loss of the H2A, possibly leading to the generation of meta-stable states in some of which the αN helix is relocated to fill the void left by the H2A C-terminal extension, creating a more compact structure with shorter spin–spin distances. The destabilization of this region may be accompanied by some level of structural disorder giving rise to an extended random coil N-terminal tail. Movement of the H3 αN helix would be expected to influence the structure of the adjoining H3 α1 helix and that is indeed what is observed in the data from H3L65 and H3Q76.
Biological implications of structural heterogeneity within the H3 αN helix of soluble tetramer
It is known that within the cell soluble histones are frequently found to be associated with histone chaperones, which are a broad family of proteins that are both structurally and functionally diverse (56,57). One such chaperone, conserved from yeast to man, is Asf1. Asf1 has recently been shown, through biochemical and structural means, to disrupt the histone tetramer by binding to the H3-H3′ interface (24,25,58), forming an Asf1-H3-H4 heterotrimer. This raises the issue whether the histone tetramer is stable under physiological ionic conditions. Our own PELDOR measurements carried out in low salt conditions (Supplementary Figure S3) confirm that the tetramer is a stable entity under these conditions, which is in agreement with previous observations (59,60).
Our finding that several regions of the tetramer can undergo structural rearrangements in comparison with their organization in the octamer provides a new basis for interpreting the interaction between histones and their chaperones. For example, the recent crystal structures of the small subunit of the highly conserved chromatin assembly factor-1 (CAF-1) complex (p55 in flies and RbAp46 in humans) (61,62) show that histone binding is mediated, at least in part, via the α1 helix of H4. In the crystal structure of the histone octamer, the H4 α1 helix is obscured by the H3 αN helix, thus instability of the αN helix in the soluble tetramer could allow access to the α1 helix of H4 by RbAp46/p55 (Supplementary Figure S4).
Additionally, structural heterogeneity within the H3 αN helix may have a role in presenting acetylation sites within the soluble histone tetramer to histone acetyl transferases before incorporation into chromatin. It is known that the acetyl transferase Rtt109 acetylates sites within the unstructured tail of H3 (lysine 9) and within the αN helix (lysine 56) (63–65). This has raised the question of how Rtt109 accommodates both α-helix and random coil structure in its active site (66,67). The instability that we see in the H3 αN helix leads to a loss of secondary structure. Thus, it is quite possible that H3 in its soluble state presents random-coil structure to Rtt109 for acetylation of both lysine 9 and 56.
The αN helix of H3 has been implicated previously in nucleosome dynamics through the action of chromatin remodellers and post-translational modification (68–70). We suggest that this helix, and additionally the α1-helix of H3, may also function in soluble histone interactions. Such interactions may include the binding to histone chaperones and the increase in accessibility of acetylation sites that have previously been thought to reside on the globular core.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biotechnology and Biological Sciences Research Council (BB/E022286/1); Engineering and Physical Sciences Research Council (EP/F0390341). Funding for open access charge: Biotechnology and Biological Sciences Research Council and The Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank David Keeble (Dundee) and Graham Smith (St Andrews) for discussion and Sharon Kelly (Glasgow) for circular dichroism measurements. A.B. would like to acknowledge receipt of a Wellcome Trust studentship.
REFERENCES
- 1.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 2.Jorcano JL, Ruiz-Carrillo A. H3.H4 tetramer directs DNA and core histone octamer assembly in the nucleosome core particle. Biochemistry. 1979;18:768–774. doi: 10.1021/bi00572a005. [DOI] [PubMed] [Google Scholar]
- 3.Polach KJ, Widom J. Mechanism of protein access to specific DNA-sequences in chromatin – a dynamic equilibrium-model for gene-regulation. J. Mol. Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 4.Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiriet C, Hayes JJ. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 2005;19:677–682. doi: 10.1101/gad.1265205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 7.Segal E, Fondufe-Mittendorf Y, Chen LY, Thastrom A, Field Y, Moore IK, Wang JPZ, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.OwenHughes T, Workman JL. Remodeling the chromatin structure of a nucleosome array by transcription factor-targeted trans-displacement of histones. EMBO J. 1996;15:4702–4712. [PMC free article] [PubMed] [Google Scholar]
- 9.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorch Y, Zhang M, Kornberg RD. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- 11.Bruno M, Flaus A, Stockdale C, Rencurel C, Ferreira H, Owen-Hughes T. Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol. Cell. 2003;12:1599–1606. doi: 10.1016/s1097-2765(03)00499-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Badis GC, Esther T, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 15.Binastein M, Simpson RT. Specific folding and contraction of DNA by histones H-3 and H-4. Cell. 1977;11:609–618. doi: 10.1016/0092-8674(77)90078-2. [DOI] [PubMed] [Google Scholar]
- 16.Baer BW, Rhodes D. Eukaryotic RNA polymerase-II binds to nucleosome cores from transcribed genes. Nature. 1983;301:482–488. doi: 10.1038/301482a0. [DOI] [PubMed] [Google Scholar]
- 17.Ruiz-Carrillo A, Jorcano JL. An octamer of core histones in solution: central role of the H3-H4 tetramer in the self-assembly. Biochemistry. 1979;18:760–768. doi: 10.1021/bi00572a004. [DOI] [PubMed] [Google Scholar]
- 18.Flaus A, Luger K, Tan S, Richmond TJ. Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals. Proc. Natl Acad. Sci. USA. 1996;93:1370–1375. doi: 10.1073/pnas.93.4.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. Plos Biol. 2007;5:1798–1809. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 21.Engeholm M, de Jager M, Flaus A, Brenk R, van Noort J, Owen-Hughes T. Nucleosomes can invade DNA territories occupied by their neighbors. Nat. Struct. Mol. Biol. 2009;16:151–158. doi: 10.1038/nsmb.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamiche A, Carot V, Alilat M, De Lucia F, O’Donohue MF, Revet B, Prunell A. Interaction of the histone (H3-H4)2 tetramer of the nucleosome with positively supercoiled DNA minicircles: potential flipping of the protein from a left- to a right-handed superhelical form. Proc. Natl Acad. Sci. USA. 1996;93:7588–7593. doi: 10.1073/pnas.93.15.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bancaud A, Wagner G, Conde ESN, Lavelle C, Wong H, Mozziconacci J, Barbi M, Sivolob A, Le Cam E, Mouawad L, et al. Nucleosome chiral transition under positive torsional stress in single chromatin fibers. Mol. Cell. 2007;27:135–147. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 24.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 26.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 27.Lattman E, Burlingame R, Hatch C, Moudrianakis EN. Crystallization of the tetramer of histones H3 and H4. Science. 1982;216:1016–1018. doi: 10.1126/science.7079748. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Z, Feng H, Hansen DF, Kato H, Luk E, Freedberg DI, Kay LE, Wu C, Bai Y. NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B. Nat. Struct. Mol. Biol. 2008;15:868–869. doi: 10.1038/nsmb.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 30.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 31.Altenbach C, Marti T, Khorana HG, Hubbell WL. Transmembrane protein structure: spin labeling of bacteriorhodopsin mutants. Science. 1990;248:1088–1092. doi: 10.1126/science.2160734. [DOI] [PubMed] [Google Scholar]
- 32.Schiemann O, Prisner TF. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Q. Rev. Biophys. 2007;40:1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 33.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc. Natl Acad. Sci. USA. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanson SM, Van Eps N, Francis DJ, Altenbach C, Vishnivetskiy SA, Arshavsky VY, Klug CS, Hubbell WL, Gurevich VV. Structure and function of the visual arrestin oligomer. EMBO J. 2007;26:1726–1736. doi: 10.1038/sj.emboj.7601614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Z, DeSensi SC, Stein RA, Brandon S, Song L, Cobb CE, Hustedt EJ, Beth AH. Structure of the cytoplasmic domain of erythrocyte band 3 hereditary spherocytosis variant P327R: band 3 Tuscaloosa. Biochemistry. 2007;46:10248–10257. doi: 10.1021/bi700948p. [DOI] [PubMed] [Google Scholar]
- 36.Smirnova I, Kasho V, Choe JY, Altenbach C, Hubbell WL, Kaback HR. Sugar binding induces an outward facing conformation of LacY. Proc. Natl Acad. Sci. USA. 2007;104:16504–16509. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Z, DeSensi SC, Stein RA, Brandon S, Dixit M, McArdle EJ, Warren EM, Kroh HK, Song L, Cobb CE, et al. Solution structure of the cytoplasmic domain of erythrocyte membrane band 3 determined by site-directed spin labeling. Biochemistry. 2005;44:15115–15128. doi: 10.1021/bi050931t. [DOI] [PubMed] [Google Scholar]
- 38.Schiemann O, Cekan P, Margraf D, Prisner TF, Sigurdsson ST. Relative orientation of rigid nitroxides by PELDOR: beyond distance measurements in nucleic acids. Angew. Chem. Int. Ed. Engl. 2009;48:3292–3295. doi: 10.1002/anie.200805152. [DOI] [PubMed] [Google Scholar]
- 39.Schiemann O, Piton N, Mu Y, Stock G, Engels JW, Prisner TF. A PELDOR-based nanometer distance ruler for oligonucleotides. J. Am. Chem. Soc. 2004;126:5722–5729. doi: 10.1021/ja0393877. [DOI] [PubMed] [Google Scholar]
- 40.Ward R, Keeble DJ, El-Mkami H, Norman DG. Distance determination in heterogeneous DNA model systems by pulsed EPR. ChemBiochem. 2007;8:1957–1964. doi: 10.1002/cbic.200700245. [DOI] [PubMed] [Google Scholar]
- 41.Cai Q, Kusnetzow AK, Hubbell WL, Haworth IS, Gacho GP, Van Eps N, Hideg K, Chambers EJ, Qin PZ. Site-directed spin labeling measurements of nanometer distances in nucleic acids using a sequence-independent nitroxide probe. Nucleic Acids Res. 2006;34:4722–4730. doi: 10.1093/nar/gkl546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin PZ, Butcher SE, Feigon J, Hubbell WL. Quantitative analysis of the isolated GAAA tetraloop/receptor interaction in solution: a site-directed spin labeling study. Biochemistry. 2001;40:6929–6936. doi: 10.1021/bi010294g. [DOI] [PubMed] [Google Scholar]
- 43.Sen KI, Logan TM, Fajer PG. Protein dynamics and monomer-monomer interactions in AntR activation by electron paramagnetic resonance and double electron-electron resonance. Biochemistry. 2007;46:11639–11649. doi: 10.1021/bi700859p. [DOI] [PubMed] [Google Scholar]
- 44.Jeschke G, Koch A, Jonas U, Godt A. Direct conversion of EPR dipolar time evolution data to distance distributions. J. Magn. Reson. 2002;155:72–82. doi: 10.1006/jmre.2001.2498. [DOI] [PubMed] [Google Scholar]
- 45.Chiang YW, Borbat PP, Freed JH. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J. Magn. Reson. 2005;172:279–295. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 46.Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, Jung H. DeerAnalysis2006 – a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 2006;30:473–498. [Google Scholar]
- 47.DeLano WL. CA, USA: Palo Alto; 2002. DeLano Scientific. [Google Scholar]
- 48.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 49.Hassinen T, Perakyla M. New energy terms for reduced protein models implemented in an off-lattice force field. J. Comput. Chem. 2001;22:1229–1242. [Google Scholar]
- 50.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 51.McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 52.Jeschke G. Distance measurements in the nanometer range by pulse EPR. Chemphyschem. 2002;3:927–932. doi: 10.1002/1439-7641(20021115)3:11<927::AID-CPHC927>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 53.Wood CM, Nicholson JM, Lambert SJ, Chantalat L, Reynolds CD, Baldwin JP. High-resolution structure of the native histone octamer. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2005;61:541–545. doi: 10.1107/S1744309105013813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ward R, Bowman A, El-Mkami H, Owen-Hughes T, Norman DG. Long distance PELDOR measurements on the histone core particle. J. Am. Chem. Soc. 2009;131:1348–1349. doi: 10.1021/ja807918f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muthurajan UM, Bao Y, Forsberg LJ, Edayathumangalam RS, Dyer PN, White CL, Luger K. Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. EMBO J. 2004;23:260–271. doi: 10.1038/sj.emboj.7600046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nat. Struct. Mol. Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 57.Park YJ, Luger K. Histone chaperones in nucleosome eviction and histone exchange. Curr. Opin. Struct. Biol. 2008;18:282–289. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.English CM, Maluf NK, Tripet B, Churchill MEA, Tyler JK. ASF1 binds to a heterodimer of histories H3 and H4: a two-step mechanism for the assembly of the H3-H4 heterotetramer on DNA. Biochemistry. 2005;44:13673–13682. doi: 10.1021/bi051333h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baxevanis AD, Godfrey JE, Moudrianakis EN. Associative behavior of the histone (H3-H4)2 tetramer: dependence on ionic environment. Biochemistry. 1991;30:8817–8823. doi: 10.1021/bi00100a013. [DOI] [PubMed] [Google Scholar]
- 60.Banks DD, Gloss LM. Folding mechanism of the (H3-H4)2 histone tetramer of the core nucleosome. Protein Sci. 2004;13:1304–1316. doi: 10.1110/ps.03535504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song JJ, Garlick JD, Kingston RE. Structural basis of histone H4 recognition by p55. Genes Dev. 2008;22:1313–1318. doi: 10.1101/gad.1653308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han J, Zhou H, Li Z, Xu RM, Zhang Z. The Rtt109-Vps75 histone acetyltransferase complex acetylates non-nucleosomal histone H3. J. Biol. Chem. 2007;282:14158–14164. doi: 10.1074/jbc.M700611200. [DOI] [PubMed] [Google Scholar]
- 64.Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol. Cell Biol. 2008;28:4342–4353. doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang Y, Holbert MA, Wurtele H, Meeth K, Rocha W, Gharib M, Jiang E, Thibault P, Verreault A, Cole PA, et al. Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP. Nat. Struct. Mol. Biol. 2008;15:998. doi: 10.1038/nsmb.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stavropoulos P, Nagy V, Blobel G, Hoelz A. Molecular basis for the autoregulation of the protein acetyl transferase Rtt109. Proc. Natl Acad. Sci. USA. 2008;105:12236–12241. doi: 10.1073/pnas.0805813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ferreira H, Somers J, Webster R, Flaus A, Owen-Hughes T. Histone tails and the H3 alphaN helix regulate nucleosome mobility and stability. Mol. Cell Biol. 2007;27:4037–4048. doi: 10.1128/MCB.02229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Somers J, Owen-Hughes T. Mutations to the histone H3 alpha N region selectively alter the outcome of ATP-dependent nucleosome-remodelling reactions. Nucleic Acids Res. 2009;37:2504–2513. doi: 10.1093/nar/gkp114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neumann H, Hancock S, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol. Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.