Abstract
BLM and WRN are members of the RecQ family of DNA helicases, and in humans their loss is associated with syndromes characterized by genome instability and cancer predisposition. As the only RecQ DNA helicase in the yeast Saccharomyces cerevisiae, Sgs1 is known to safeguard genome integrity through its role in DNA recombination. Interestingly, WRN, BLM and Sgs1 are all known to be modified by the small ubiquitin-related modifier (SUMO), although the significance of this posttranslational modification remains elusive. Here, we demonstrate that Sgs1 is specifically sumoylated under the stress of DNA double strand breaks. The major SUMO attachment site in Sgs1 is lysine 621, which lies between the Top3 binding domain and the DNA helicase domain. Surprisingly, sumoylation of K621 was found to be uniquely required for Sgs1’s role in telomere–telomere recombination. In contrast, sumoylation was dispensable for Sgs1’s roles in DNA damage tolerance, supppression of direct repeat and rDNA recombination, and promotion of top3Δ slow growth. Our results demonstrate that although modification by SUMO is a conserved feature of RecQ family DNA helicases, the major sites of modification are located on different domains of the protein in different organisms. We suggest that sumoylation of different domains of RecQ DNA helicases from different organisms contributes to conserved roles in regulating telomeric recombination.
INTRODUCTION
Chromosomal DSBs, caused by replication fork disruption, environmental factors or endogenous nucleases are common yet potentially dangerous DNA lesions in all organisms. DNA DSBs are critical lesions that if unrepaired or misrepaired may be lethal for a cell or contribute to its malignant transformation. DSBs can be repaired either by homologous recombination or by non-homologous end joining pathways (1–6). In this regard, the budding yeast Saccharomyces cerevisiae has become the most intensely studied model system for DSB DNA repair.
RecQ proteins comprise a highly conserved family of 3′–5′ DNA helicases that includes the human BLM, WRN, RECQL4 and RECQ5 proteins, as well as Rqh1 from Schizosaccharomyces pombe and Sgs1 in S. cerevisiae (7–9). Werner’s, Bloom’s and Rothmund-Thomson’s genome instability syndromes are caused by mutation of the WRN, BLM and RECQL4 genes, respectively (10–12). RecQ DNA helicases have been implicated in several aspects of DNA metabolism (8), including a recently characterized role in the initial step of homologous recombination in S. cerevisiae (13,14). After a DSB is formed and recognized, Sae2 trims the ends to create a minimally resected intermediate. Sgs1 and Exo1 then rapidly process this intermediate to generate extensive tracts of single-strand DNA that serve as substrates for Rad51 in homologous recombination (13,14).
Posttranslational modification with the small ubiquitin-related modifier (SUMO), is a widespread mechanism for rapid and reversible changes in protein function. Sumoylation occurs by a process that is similar to ubiquitylation. An E1 activating protein (Aos1/Uba2) loads SUMO onto the E2 conjugating enzyme (Ubc9), which in turn transfers SUMO (Smt3 in budding yeast) onto specific lysine residues within target substrates (15). Sumoylation has recently been reported to regulate Rqh1 activity at telomeres in S. pombe (16). Moreover, WRN, BLM and Sgs1 were all previously shown to be sumoylated (17–19), although the exact role of this modification in homologous recombination is not completely understood.
In the absence of telomerase, immortalized mammalian cells and yeast may employ recombination-mediated pathways to maintain telomeres, termed alternative lengthening of telomeres (ALT), in mammalian cells (20–22). Telomerase-negative S. cerevisiae overcomes telomere crisis by utilizing one of two Rad52-dependent recombination-mediated pathways, termed Types I and II (23,24). Type I telomere lengthening requires Rad51, whereas Type II telomere lengthening requires Rad50 and the Sgs1/Top3 complex (25–29). Telomeric repeats in Type II survivors are amplified and often heterogeneous in length, whereas Type I survivors have amplified subtelomeric Y′-elements. Terminal telomeric repeats in human cells using ALT are long and heterogeneous, suggesting that a Type II-like mechanism is used in these pathways (20–22).
The association of Sgs1 with the Type II recombination pathway prompted us to hypothesize a conserved function of these RecQ helicases in recombination-mediated telomere lengthening. Furthermore, very recently, Rqh1 has been reported to control the fate of dysfunctional telomeres (16). In this study, we demonstrate that DSBs induced by ionizing radiation (IR) or chemicals, but not replication fork disruption or oxidative stress, promote Sgs1 sumoylation. The major SUMO attachment site in Sgs1 is lysine 621, which lies between the Top3 binding and DNA helicase domains (30,31). A conservative mutation at this residue reduces Type II telomere–telomere recombination, but does not alter the functions of Sgs1 in DSB repair and homologous recombination at other loci in the genome. This indicates that sumoylation of Sgs1 specifically facilitates telomere–telomere recombination.
MATERIALS AND METHODS
Yeast strains and plasmids
All the yeast operations were performed by standard methods (32). STY1525 (YPH499 SGS1-13Myc) was constructed by double crossing over the chromosomal SGS1 gene of YPH499 (24) with a 13Myc PCR fragment from pFA6a-13Myc-kanMX6 (33). STY1793 (YPH499 ubc9-1 SGS1-13Myc) was obtained by three backcrosses of MR966 ubc9-1 (34) with YPH499, ULP1 and ULP2 plasmids, which were kindly provided by Dr Mark Hochstrasser (35,36). pRS306-SGS1 was constructed as described below. Point mutations were introduced into pRS306-SGS1 using QuikChange site-directed mutagenesis (Stratagene). To generate chromosomal sgs1 mutants, pRS306-sgs1 mutants were linearized by AflII and transformed into SGS1 strains, and URA3 pop-out mutants were identified from 5-FOA-resistant colonies using PCR analysis. Both E3 deletion mutants were purchased from yeast deletion library (Invitrogen). The sgs1::HIS3 mutation was constructed by transforming these strains with an sgs1::HIS3 PCR fragment amplified from STY680 (sgs1::HIS3) (27) genomic DNA using an SGS1 upstream and downstream primer pairs. YPH499 exo1 strains were generated by transplacement of the YPH499 EXO1 locus with an exo1::kanMX6 fragment amplified from yeast deletion library (Invitrogen). STY1881 (YPH500 top3::URA3) was generated using a URA3 fragment that was PCR-amplified from pMPY-3xHA (37) using oligonucleotides with sequences homologous to the TOP3 upstream and downstream regions. This knockout fragment was then introduced into the indicated strains. For the inter-chromosomal recombination assay, MR966 and MR93-28c (34) were manipulated independently and mated to obtain diploid cells. The sgs1::URA3 mutation was generated by transforming strains with an sgs1::URA3 PCR fragment amplified using primers with sequences homologous to the SGS1 up- and downstream regions. pRS316-SGS1 was a backbone exchange construct of pRS314-SGS1 (27). A SacII-SacI fragment containing the C-terminal SGS1 and 13Myc regions was cloned into the SacII/SacI-digested pRS316-SGS1 to generate pRS316-SGS1-13Myc. pRS306-SGS1 for two-step gene replacement was cloned by ligation of an XhoI/BamHI fragment from pRS316-SGS1-13Myc into pRS306. pGEX-4T-SGS1 (410–713) was constructed by ligating the BamHI/PstI-Klenow-treated fragment containing the aa 410–713 of Sgs1 into the BamHI/SmaI-digested pGEX4T-1 (GE). All primer sequences for PCR and mutagenesis are available upon request.
Western blotting analysis and in vitro sumoylation assay
Early log phase cultures were grown to an OD600 of 0.5 and extracts were prepared for western blotting using an anti-Myc antibody. The signal was quantitated using ImageQuant software Version 5.2 (GE). The His6-tagged yeast SUMO E1, E2 and Smt3 expression plasmids were kindly provided by Drs Günter Blobel (38) and Ting-Fang Wang (39). Purification of these enzymes was performed as described (38). Recombinant GST-Sgs1 was expressed in the BL21 strain and purified according to the manufacturer (GE). For the in vitro sumoylation assay, 1 µg of SUMO, 2 μg of Aos1/Uba2, 2.5 μg of Ubc9 and 5 μg of substrate were incubated at 30°C for 2 h. Reactions were performed in 50 μl reaction buffer containing 50 mM HEPES (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 20 μg bovine serum albumin, 5 mM Tris, 0.1 mM DTT and 5 mM ATP.
Spot dilution assay
For the spot dilution assay, early log phase cultures were adjusted to an OD600 of 0.5 and serial 5-fold dilutions were made in sterial water. Five microliters were spotted onto YPD plates, followed by exposure to different temperatures, ionizing radiation (IR), or UV light. Alternatively, cells were spotted onto YPD plates containing various concentrations of methyl methanesulphonate (MMS), ethyl methanesulfonate (EMS), bleomycin or hydroxyurea (HU). Drug-containing plates were prepared 2 days before use.
Yeast telomere analysis
Liquid cultures were generated by inoculating spore colonies from the freshly dissected spores into 10 ml of liquid YEPD medium. Cultures were diluted repeatedly 1:10 000 into fresh medium and grown at 30°C for 48–72 h to reach the stationary phase. Solid-media experiments were performed by repeatedly streaking spore colonies from the dissection plates onto YEPD plates. To quantify the ratio of Type I versus Type II survivors, spore cells were serially restreaked onto YEPD medium as described (40) to obtain survivors on solid media. Genomic DNA was digested with a mixture of HaeIII, HinfI and MspI, and Southern blot analysis was performed using a telomere probe as previously described (40).
Competition assay for formation of survivors
The tlc1Δ and tlc1Δ sgs1-K621R cells were taken from freshly dissected spore colonies and mixed in a 50:50 ratio in YEPD medium. Following growth to stationary phase, cultures were diluted 1:10 000 every 2 days, for 12 days. Genomic DNA was extracted from cultures at the indicated time points and subjected to quantitative real-time PCR to determine the genomic background of SGS1 using specific primers. The tubulin gene was used as an internal control. The primer specific for wild-type SGS1 is GACTAACACTGGATTTCTCCCTTT and for sgs1-K621R is GACTAACACTGGATTTCTCCCTTC (the underline indicates the difference in the mutant). Control experiments showed that the primers did not cross-amplify non-specific products (data not shown). Data were expressed as a percentage of the sum of the signals amplified from SGS1 and sgs1-K621R in individual cultures. Other primer sequences for PCR are available upon request.
Assays for recombination frequency
Recombination between direct repeats was assayed in the indicated strains following transformation with pRS314-Lu (41). Fresh transformants were first patched onto YEPD plates for 24 h. Cells were then resuspended in water and diluted onto synthetic complete (SC) plates (with or without leucine) to score recombination frequency. To determine the inter-chromosomal recombination frequency between his1-1 and his1-7 heteroalleles, independent colonies of indicated strains were patched on YEPD plates for 24 h. Cells were then resuspended in water and diluted onto SC plates (with or without histidine) containing 0 or 0.01% of MMS to monitor viability and recombination frequency. It is worth noting that this assay could not distinguish between gene conversion and recombination.
rDNA recombination
The frequency of intra-chromosomal rDNA recombination was monitored by measuring the frequency of loss of the URA3 marker in strain STY1880 (rDNA::URA3 sgs1::HIS3). STY1880 was constructed from strain W979-3B (rDNA::URA3) as described above (42). Strains were then transformed with pRS314-based plasmids carrying either wild-type or mutant SGS1 genes. Five independent transformants of each genotype were grown to stationary phase in 2 ml of non-selective medium and then diluted appropriately and spread onto SC plates with or without 5-FOA.
RESULTS
Lysine 621 is the major sumoylation site of Sgs1
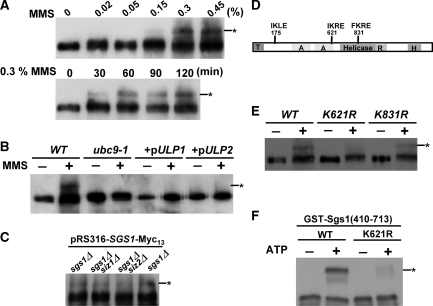
The finding that Sgs1 was sumoylated (19) prompted us to study its significance on the cellular functions of Sgs1. In order to optimize our ability to detect this modification, Sgs1 was chromosomally tagged with Myc13. We found that treatment of yeast cells with 0.3% MMS for 30 min induced a slower migrating form of Sgs1 as detected by immunoblot analysis (Figure 1A). The size of this mobility shift is consistent with the addition of one SUMO moiety (Smt3, ∼11 kDa). The fact that the slower band required wt SUMO E2 activity (UBC9) and was sensitive to overexpression of either de-sumoylase ULP1 or ULP2 (35,36) (Figure 1B) suggests that the upper band is the sumoylated form of Sgs1.
Figure 1.
The K621 residue of Sgs1 is critical for its sumoylation. (A) Sgs1 was chromosomally tagged with Myc13. Cells were grown to early log phase and subjected to various dosages and times of MMS treatment. Lysates were extracted and western blot analysis was performed using an anti-Myc antibody. (B) Wild-type and ubc9-1 strains overexpressing the indicated desumoylases were treated with or without 0.3% of MMS for 2 h and treated as above. (C) The sgs1Δ, sgs1Δ siz1Δ and sgs1Δ siz2Δ strains containing pRS316-SGS1-Myc13 were subjected to MMS treatment as described above. (D) Schematic representation of the position of SUMO consensus motifs and domains within Sgs1. T, Top3-interacting domain; A, acidic region; R, RecQ C-terminal homology region; H, Helicase and RNaseD C-terminal region. (E) Wild-type, sgs1-K621R and sgs1-K831R cells were subjected to MMS treatment as described above. Sumoylated Sgs1 is marked with an asterisk. (−) and (+) refers to treatment without or with MMS, respectively. (F) Recombinant Aos1/Uba2 (E1), Ubc9 (E2), Smt3 and purified GST-Sgs1(410–713) were incubated at 30°C for 2 h in the absence (−) or presence (+) of ATP as described in the ‘Materials and Methods’ section. Western blot analysis was then performed using an anti-GST antibody.
There are three major SUMO E3 ligases in budding yeast (Siz1, Siz2 and Mms21) (43), and a previous study showed that Mms21 was not required for the sumoylation of Sgs1 (19). Deletion of SIZ1 or SIZ2 did not significantly reduce the level of sumoylated Sgs1 (Figure 1C). This suggests that SIZ1 and SIZ2 are redundant in promoting the conjugation of SUMO onto Sgs1.
There are three SUMO consensus sites (ΨKxE) (44) within Sgs1 (aa 175, 621 and 831, Figure 1D). To identify the in vivo sumoylation site of Sgs1, each of these lysines was mutated to arginine. Sgs1 sumoylation was abolished by the K621R mutation, diminished by K831R and unaffected by K175R (Figure 1E; data not shown). This indicates that K621 is essential for sumoylation in vivo. To confirm that K621 is an authentic site for sumoylation, a fragment of Sgs1 containing this residue was purified from E. coli and used in an in vitro sumoylation assay. As shown in Figure 1F, the wild-type Sgs1 fragment was efficiently sumoylated, whereas protein containing the K621R mutation was not. These data indicate that K621 is the major sumoylation site of Sgs1.
Sumoylation of Sgs1 occurs in a lesion-dependent manner
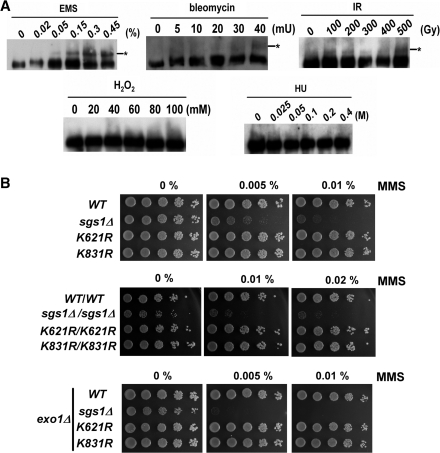
We next examined the conditions necessary for the induction of Sgs1 sumoylation. Modification of Sgs1 was rarely observed in wild-type cells grown under normal conditions, but a fraction of sumoylated protein appeared after culturing the cells in the presence of at least 0.3% MMS (Figure 1A). We, therefore, tested whether sublethal concentrations of the following DNA-damaging agents induced Sgs1 sumoylation: bleomycin and IR (which lead to DSB formation), hydroxyurea (which results in replication forks stalling) and H2O2 (which induces oxidative stress). In contrast to hydroxyurea- or H2O2-treated cells, Sgs1 sumoylation was observed because of treatment with bleomycin, IR or EMS (Figure 2A). These results indicate that Sgs1 sumoylation is elicited specifically under conditions that generate DSBs.
Figure 2.
Sumoylation of Sgs1 occurs in a lesion-dependent manner. (A) Cells were grown to early log phase and treated with either IR or the indicated concentrations of genotoxic agents for 2 h. Lysates were harvested and analyzed by western blot analysis as described in Figure 1. The efficiencies of damage levels were confirmed by Rad53 activation (bleomycin, IR, EMS and HU) or survival rate (H2O2) after treatment (data not shown). (B) Cells of the indicated genotype were grown to early log phase, serially diluted in 5-fold increments and spotted onto YPD plates containing the indicated levels of MMS. The plates were photographed after 2 days at 30°C.
We next asked whether Sgs1 sumoylation was important for growth in the absence of DNA damage. Relative growth rates were determined by an assay in which two types of cells are mixed and scored for their ability to proliferate. Wild-type Leu− cells were mixed at a 50:50 ratio with sgs1Δ, sgs1-K621R or sgs1-K831R Leu+ cells of the same mating type and allowed to grow without selection. Cultures were diluted every day, and after 7 days they were plated onto selective and nonselective media to measure the populations in the cultures. Data collected in this manner showed that sgs1Δ cells had reduced proliferative capacity. However, sgs1-K621R and sgs1-K831R cells gave results that were indistinguishable from those of the wild-type (Supplementary Figure S1). Thus, sumoylation of Sgs1 does not provide any growth advantage to yeast cells.
Because a major function of Sgs1 is to cope with the genotoxic insults, we tested whether sumoylation of Sgs1 contributes to DNA damage tolerance. Deletion of SGS1 confers sensitivity to DNA damage, a phenotype that is often attributed to defects in DNA repair (45). To determine if sgs1-K621R cells have similar repair defects, we tested their sensitivity to MMS, bleomycin and hydroxyurea by spotting serially diluted cells onto plates containing these drugs (Figure 2B, Supplementary Figure S2B). Alternatively, cells were treated with MMS before plating (Supplementary Figure S2A), or exposed to UV light immediately after plating (Supplementary Figure S2B). While sgs1Δ cells were MMS, bleomycin, hydroxyurea and UV sensitive, the sgs1-K621R and sgs1-K831R cells grew as well as wild-type cells under these treatments (Figure 2B; Supplementary Figure S2). Diploid sgs1Δ cells display heightened sensitivity to these agents relative to haploid strains (34). Therefore, we tested MMS sensitivity in homozygous diploid cells. Again, diploid mutant cells showed wt levels of DNA damage tolerance (Figure 2B). Further, Exo1 and Sgs1 have been found to collaborate in DNA DSB processing (13,14). We, therefore, tested whether deletion of EXO1 would amplify the contribution of Sgs1 sumoylation in DNA repair, by conducting similar experiments in the exo1 background. As shown in Figure 2B, exo1 sgs1-K621R and exo1 sgs1-K831R cells displayed the same MMS sensitivity as exo1 cells. Altogether, these data demonstrate that, unlike sgs1Δ cells, sgs1-K621R cells have wild- or near-wild-type abilities to repair exogenously generated DNA damage.
Sgs1 K621 sumoylation modulates telomere–telomere recombination
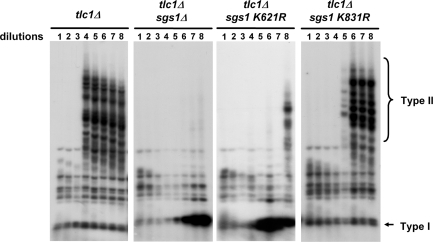
Previous studies have shown that Sgs1 deficiency eliminates telomere–telomere recombination in telomerase-deficient cells (25,27–29). We, therefore, analyzed whether the loss of Sgs1 sumoylation similarly influences this phenotype. Freshly generated tlc1Δ (telomerase deficient) sgs1 spore clones were repeatedly grown to saturation in liquid culture for analysis of telomeric recombination patterns by Southern blot. As shown in Figure 3, we analyzed recombinants from tlc1Δ, tlc1Δ sgs1Δ, tlc1Δ sgs1-K621R and tlc1Δ sgs1-K831R spores. Compared with the tlc1Δ and tlc1Δ sgs1-K831R strains, which displayed the Type II telomere–telomere recombination pattern after several dilutions, such a pattern was absent in the tlc1Δ sgs1Δ and tlc1Δ sgs1-K621R strains. Instead these cells produced the signal expected for Y′–Y′ (Type I) recombination. Moreover, the appearance of this Type I signal was significantly delayed relative to the onset of Type II recombinants. Thus, Sgs1 K621 sumoylation is functionally important for telomere–telomere recombination.
Figure 3.
Sgs1 K621 sumoylation promotes telomere–telomere recombination. Telomerase deficient (tlc1Δ) strains carrying the indicated SGS1 alleles were repeatedly diluted and grown to saturation in liquid culture. Following the indicated rounds of growth, cells were harvested and genomic DNA was isolated. DNA was then digested and Southern blotted with a telomeric probe as described in the ‘Materials and Methods’ section. Data shown here are representative of three or more experiments using independent spore clones. The increased intensity of the smallest band in panels 2 and 3 is due to Y′–Y′ amplification from Type I survivors.
Sgs1 K621 sumoylation promotes the generation of Type II survivors
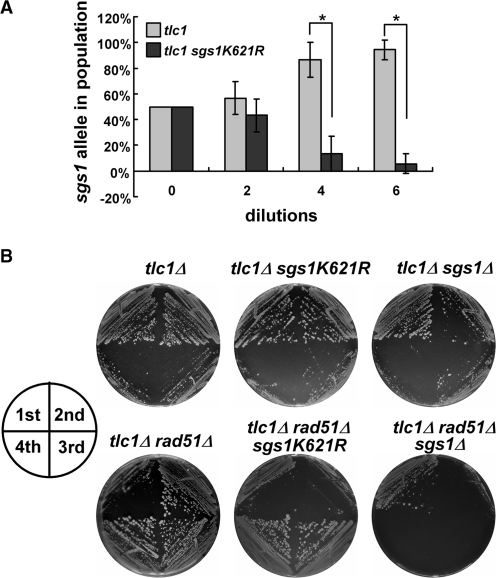
Since Sgs1 sumoylation is functionally important for telomeric recombination, we tested whether the modification stimulates Type II telomere–telomere recombination or whether it represses Type I Y′–Y′ amplification. Either mechanism would lead to the observed reduction in Type II recombinants in tlc1Δ sgs1-K621R cells (Figure 3). To distinguish between these two possibilities, a competition assay for survivor formation was performed. Cells from tlc1Δ and tlc1Δ sgs1-K621R spore colonies were mixed at a 50:50 ratio, and repeatedly diluted and grown to saturation. Quantitative PCR was used to show that, over time, the proliferation of the sgs1-K621R strain was compromised relative to the tlc1Δ single mutant (Figure 4A). The reduced survival of this strain indicates that telomeric healing is diminished and suggests that sumoylation promotes Type II events.
Figure 4.
Sgs1 K621 sumoylation promotes Type II telomere recombination. (A) A growth competition assay was performed on tlc1Δ and tlc1Δ sgs1-K621R strains to test the role of Sgs1 sumoylation in the absence of telomerase. After the indicated number of rounds of growth, genomic DNA was obtained and the fraction of cells carrying each allele was determined by real-time PCR. Plotted on the Y-axis is the average allele frequency from three independent experiments ± SDs. Significant differences (P < 0.001), as determined by the student’s t-test, are indicated by asterisks. (B) Freshly isolated spores of indicated strains were repeatedly streaked onto YPD plates and grown for 3 days. Growth from the first four streaks are shown.
Consistent with this interpretation, we performed senescence assays on solid media and isolated one hundred tlc1Δ sgs1-K621R survivors. DNA was then prepared from individual survivors and the telomere pattern was determined by Southern blot analysis. Significantly, all of the sgs1-K621R survivors (100 of 100) displayed the Type I pattern (data not shown). In contrast, tlc1Δ single mutants generate both Types I and II survivors (40). Taken together, these data indicate that Sgs1 K621 sumoylation stimulates Type II recombination.
To confirm this idea, we relied on the fact that Rad51 is required for Type I Y′–Y′ amplification and that tlc1Δ rad51Δ strains generate exclusively Type II survivors (24,26). If Sgs1 sumoylation is important for Type II telomere–telomere recombination, then sgs1-K621R should have an inhibitory effect on tlc1Δ rad51Δ survivors. The appropriate strains were generated from freshly dissected spores and examined by serial single-colony restreaking on solid media. As expected, tlc1Δ and tlc1Δ sgs1 strains showed a cellular senescence phenotype on the third restreak (Figure 4B), while the tlc1Δ rad51Δ sgs1Δ spores senesced at the first restreak and completely lost viability at the second restreak. These results indicate that the triple mutation prevents all three (telomerase, Types I and II) pathways for telomere maintenance and blocked survivor formation. While the tlc1Δ rad51Δ cells senesced at the first restreak and survivors developed at the second restreak, the tlc1Δ rad51Δ sgs1-K621R strain showed cellular senescence phenotype at both the first and second restreaks, and survivors developed at the third restreak (Figure 4B). This delayed survivor formation in the tlc1Δ rad51Δ background is consistent with the observation that the Sgs1-K621R protein is unable to stimulate Type II recombination (Figure 4A). Thus, the sgs1-K621R mutation reduces telomere–telomere recombination for telomeric maintenance and subsequently delays survivor formation in the tlc1Δ rad51Δ background.
Sumoylation of Sgs1 is dispensable for other homologous recombination pathways
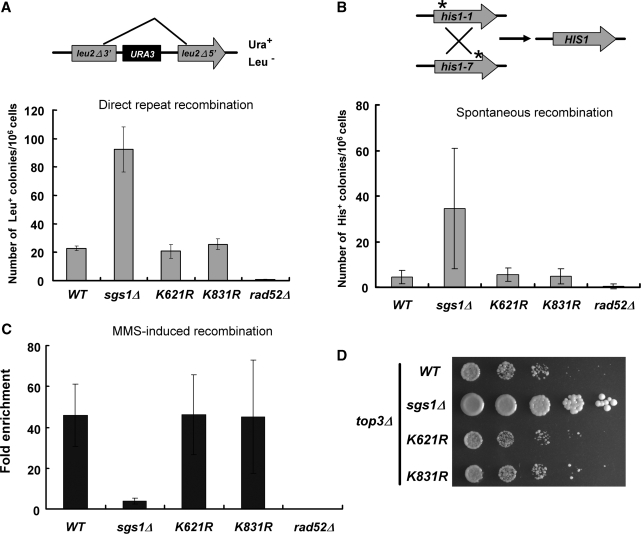
Sgs1 acts both early in the homologous recombination pathway (i.e. during 5′-end resection), and late (i.e. dissolution of hemi-catenated products) (13,14). To test whether Sgs1 sumoylation plays a role in homologous recombination, we examined the phenotype of the sumoylation-defective alleles in several recombination assays. To examine direct repeat recombination, we took advantage of a plasmid (41) containing a duplicated 600-bp internal fragment of LEU2, separated by intervening sequences (Figure 5A). If Sgs1 sumoylation is required for direct repeat recombination, the sgs1-K621R mutation would be expected to have phenotype like that of sgs1Δ cells. As shown in Figure 5A, both sgs1-K621R and sgs1-K831R mutants exhibited recombination frequencies similar to that of wild-type cells. And consistent with previous reports (46), a hyper-recombination phenotype was observed in sgs1Δ cells (Figure 5A). Based on these results, sumoylation of Sgs1 does not participate in the regulation of homologous recombination by direct repeats.
Figure 5.
Recombination phenotypes of SUMO-deficient Sgs1. (A) Recombination frequency of direct repeat was determined by the pRS314-Lu system (62). Recombination frequencies were calculated using the median method of Lea and Couison (63). (B) Schematic representation of inter-chromosomal recombination between his1-1 and his1-7 heteroalleles in diploid cells. Data shown here are the averages of four independent experiments (n = 12). Recombination-deficient rad52 mutant was served as a negative control. (C) MMS-induced inter-chromsomal recombination frequency was evaluated using the his1-1/his1-7 heteroallelic system described in (B). Data here are the fold enrichments upon 0.01% MMS treatment. (D) Slow growth phenotype was examined in strains combined with the top3 deletion. Exponentially growing cells were 5-fold serially diluted and spotted onto YPD plates.
Rad52 is a key player in most homologous recombination processes, including gene conversion, reciprocal exchange, single-strand annealing, telomere recombination and break-induced replication (47). Rad52 is also sumoylated, and mutation of its sumoylation sites has a minor effect on homologous recombination. However, inter-chromosomal recombination between heteroallelic markers is regulated by Rad52 sumoylation (48,49). We therefore tested whether Sgs1 sumoylation affected recombination between the his1-1 and his1-7 heteroalleles. However, as shown in Figures 5B and C, sgs1-K621R cells showed recombination frequencies that were similar to those of wild-type cells, including events induced by MMS. In contrast, the sgs1 null mutant showed severe defects in these assays (50).
It has been reported that the role of Sgs1 in DNA replication can be uncoupled from its role in homologous recombinational repair by mutating certain residues in the acidic region of Sgs1 (Figure 1D) (51). Since the K621 residue localizes to this region, we tested whether its sumoylation is required for Sgs1 to promote DNA replication. Deletion of SGS1 suppresses the mitotic cell cycle delay of a top3 mutant (42), and this implies that Sgs1 generates an intermediate that is normally resolved by Top3 in the S phase. However, as shown in Figure 5D, the slow growth of top3Δ cells was not suppressed by the sgs1-K621R mutation. Thus, sumoylation of Sgs1 does not influence the repair of DNA replication intermediates. Taken together, these assays demonstrate that sumoylation of Sgs1 is specifically required for telomere–telomere recombination, but is dispensable for other types of homologous recombination and replicative repair.
Sumoylation of Sgs1 is not involved in rDNA recombination
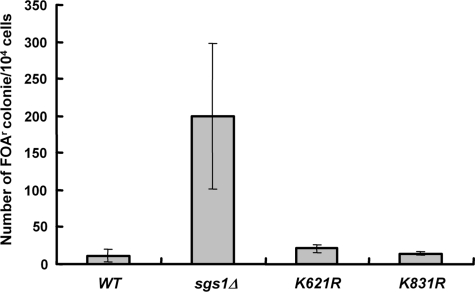
Since Sgs1 sumoylation is required for Type II telomere–telomere recombination but not other types of homologous recombination, we speculated that this might be due to its specialized telomeric heterochromatin. For example, sumoylation of Sgs1 might be required for the helicase to gain access to telomeric DNA that is maintained in the form of heterochromatin. To test the generality of this idea, we assayed recombination at the rDNA array because of its heterochromatic character (52,53). A URA3 marker was inserted in the rDNA locus of an sgs1Δ strain, and wild-type or mutant SGS1 plasmids were transformed into this strain. The frequency of rDNA recombination was determined by excision of the URA3 marker. As shown in Figure 6, no difference in the frequency of rDNA recombination was observed between the wild-type and sgs1-K621R strains. Based on these findings, we conclude that Sgs1 sumoylation does not play a general role in the regulation of homologous recombination, but specifically promotes Type II telomere–telomere recombination.
Figure 6.
SUMO-defective Sgs1 does not affect rDNA recombination. An sgs1 strain with the URA3 insertion in the rDNA locus was transformed with a CEN-based plasmid carrying wild-type SGS1 or sgs1 mutants. The rDNA recombination rate of each strain was calculated as described in Figure 5A. Data here are the averages of three independent experiments.
DISCUSSION
Several RecQ DNA helicases have been shown to be sumoylated (17–19). However, the exact function of this modification in homologous recombination is not well understood. Here, we have show that Sgs1 sumoylation is induced in response to DSBs, and that K621 is the major site of Sgs1 sumoylation. We were surprised to find that sumoylation is dispensable for most biological functions of Sgs1, including homologous recombination and promoting top3Δ slow growth. However, sumoylation of K621 is essential for promoting Type II telomere–telomere recombination. While it is not clear whether sumoylation is required for the human RecQ helicases to carry out homologous recombination, our data are consistent with the very recent findings that Rqh1 sumoylation is crucial for determining the pathway for repair of dysfunctional telomeres in S. pombe (16).
In budding yeast, Rad52, Smc5/6 and Sgs1 are all necessary for homologous recombination, and are all sumoylated. It has been suggested that concurrent sumoylation of several homologous recombination-related proteins may be required to orchestrate the efficient execution of DNA damage-induced homologous recombination (48). In that respect, it is important to note that many studies have found a link between specific subnuclear localization and protein modification by SUMO (18,38,54). Thus, upon DNA damage, multiple components of the homologous recombination machinery may necessarily co-localize at sites of sumoylation where some of them undergo SUMO modification to specifically alter their enzymatic activity. For example, Rad52 sumoylation, which is triggered by DSBs in an MRX-dependent manner, modifies Rad52 activity in inter-chromosomal recombination between heteroallelic markers (48). This mechanism appears to be conserved in human cells, where RecQ helicases and the recombination proteins RAD51, RPA and PML co-localize to PML bodies upon IR treatment (55), and sumoylation is required for the formation of the nuclear foci of these repair proteins (54,56). In our hands, however, Sgs1 sumoylation was not required for most of its roles in homologous recombination. One explanation for this result, although unattractive, is that Sgs1 sumoylation occurs non-specifically because of its close association with other recombination factors and an indiscriminate sumoylation machinery. However, it is more likely that the small fraction of Sgs1 molecules that is modified by SUMO in response to DSBs represents a pool of proteins whose novel functions are redundant with other recombination proteins or pathways. For example, in the absence of sumoylation, Sgs1-K621R may be activated for recombinational repair by back-up pathways or other posttranslational modifications. These modifications may be transient or constitute a pool size that is beyond our detection limit. If this second modification is essential for activating Sgs1-K621R, then its loss should result in synthetic interactions with sgs1-K621R. Thus, a screen for synthetic interactors with sgs1-K621R would be one approach to identify these other proteins or pathways.
If sumoylation does not contribute to the major roles of Sgs1 in homologous recombination, how does Sgs1 sumoylation specifically promote telomere–telomere recombination? A plausible explanation is that sumoylation is required to localize Sgs1 to telomeres for telomere–telomere recombination. As a mechanism for redistribution, sumoylation may provide better accessibility for Sgs1 to act on its telomeric substrates. In support of this idea, it has been shown that abolition of the Pli1 SUMO E3 ligase leads to defects in telomere clustering, silencing and length regulation in S. pombe (57). The ability of global sumoylation levels to influence telomeres in this way argues that sumoylation is an important player in telomere maintenance. Similarly, the SMC5/6 complex facilitates telomere–telomere recombination and elongation in ALT cells by promoting ALT-associated PML body formation via the sumoylation of multiple telomere binding proteins (58). And intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is also regulated by SUMO modification (18). All of these findings point to the possibility that sumoylation mediates the localization of multiple recombination proteins, including RecQ helicases, to telomeres to execute telomere–telomere recombination.
Sgs1 and its ortholog Rqh1 mediate the restart of stalled replication forks in both S. cerevisiae (59) and S. pombe (60). If telomeric recombination is initiated by the repair of stalled or collapsed replication forks, then it would be expected that a deficiency in Sgs1 would impact the frequency, and perhaps the mechanism, of telomere–telomere recombination. In addition, telomeric heterochromatin may present unique challenges to replication forks or their repair. For example, the telomere-specific phenotype observed here might be due to the fact that telomeric chromatin is actively repressed by Rap1 and other factors (61). Our data would seem to argue against this possibility since rDNA recombination was not modulated by Sgs1 sumoylation. However, the requirements for rDNA heterochromatin are not identical to those at the telomere (52,53). Thus, it remains a possibility that the strand transfer reactions that occur during telomere–telomere recombination must overcome obstacles not found in euchromatin or the rDNA. Our data suggests that Sgs1 sumoylation is involved in dealing with these challenges, perhaps by ensuring that the Sgs1 complex is targeted to these recombination sites.
Amino acid sequence alignment of the Sgs1, Rqh1 and BLM orthologs reveals a RecQ DNA helicase domain that is highly conserved, and an N-terminal domain of about 650 amino acids that shows little if any sequence conservation (Supplementary Figure S3). It is interesting to note that each of the sumoylation sites that have been identified in these three proteins are located in different domains of the protein (Supplementary Figure S3 and Figure 1D). This indicates that while sumoylation may be a conserved mechanism for modifying these RecQ helicases, there is no requirement that SUMO be conjugated to the same residue or even the same domain of the protein. Further studies will be needed to determine how sumoylation of the BLM orthologs affects each of their enzymatic activities and nuclear distribution. It is expected that such studies will shed light on whether the mechanism of SUMO modification is functionally conserved between these organisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Council (NRPGM-98-3112-B-002-039); National Health Research Institute of Taiwan (NHRI-EX98-9727BI to S.-C.T.); National Institutes of Health (GM071268 to S.J.B.). Funding for open access charge: National Health Research Institute of Taiwan (NHRI-Ex98-9727BI).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Andres Aguilera, Günter Blobel, Takemi Enomoto, Mark Hochstrasser, Rodney Rothstein and Ting-Fang Wang for providing plasmids and strains. We also thank Dr Tsai-Kun Li for his critical comments on the manuscript.
REFERENCES
- 1.Pastink A, Lohman PH. Repair and consequences of double-strand breaks in DNA. Mutat. Res. 1999;428:141–156. doi: 10.1016/s1383-5742(99)00042-3. [DOI] [PubMed] [Google Scholar]
- 2.Pastink A, Eeken JC, Lohman PH. Genomic integrity and the repair of double-strand DNA breaks. Mutat. Res. 2001;480–481:37–50. doi: 10.1016/s0027-5107(01)00167-1. [DOI] [PubMed] [Google Scholar]
- 3.van den Bosch M, Lohman PH, Pastink A. DNA double-strand break repair by homologous recombination. Biol. Chem. 2002;383:873–892. doi: 10.1515/BC.2002.095. [DOI] [PubMed] [Google Scholar]
- 4.Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22:5792–5812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- 5.West SC. Molecular views of recombination proteins and their control. Nat. Rev. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 6.Dudas A, Chovanec M. DNA double-strand break repair by homologous recombination. Mutat. Res. 2004;566:131–167. doi: 10.1016/j.mrrev.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Hickson ID. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 8.Hanada K, Hickson ID. Molecular genetics of RecQ helicase disorders. Cell Mol. Life Sci. 2007;64:2306–2322. doi: 10.1007/s00018-007-7121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khakhar RR, Cobb JA, Bjergbaek L, Hickson ID, Gasser SM. RecQ helicases: multiple roles in genome maintenance. Trends Cell Biol. 2003;13:493–501. doi: 10.1016/s0962-8924(03)00171-5. [DOI] [PubMed] [Google Scholar]
- 10.Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, et al. Positional cloning of the Werner’s syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 11.Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 12.Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 13.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz DC, Hochstrasser M. A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem. Sci. 2003;28:321–328. doi: 10.1016/S0968-0004(03)00113-0. [DOI] [PubMed] [Google Scholar]
- 16.Rog O, Miller KM, Ferreira MG, Cooper JP. Sumoylation of RecQ helicase controls the fate of dysfunctional telomeres. Mol. Cell. 2009;33:559–569. doi: 10.1016/j.molcel.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 17.Kawabe Y, Seki M, Seki T, Wang WS, Imamura O, Furuichi Y, Saitoh H, Enomoto T. Covalent modification of the Werner’s syndrome gene product with the ubiquitin-related protein, SUMO-1. J. Biol. Chem. 2000;275:20963–20966. doi: 10.1074/jbc.C000273200. [DOI] [PubMed] [Google Scholar]
- 18.Eladad S, Ye TZ, Hu P, Leversha M, Beresten S, Matunis MJ, Ellis NA. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum. Mol. Genet. 2005;14:1351–1365. doi: 10.1093/hmg/ddi145. [DOI] [PubMed] [Google Scholar]
- 19.Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- 20.Bryan TM, Marusic L, Bacchetti S, Namba M, Reddel RR. The telomere lengthening mechanism in telomerase-negative immortal human cells does not involve the telomerase RNA subunit. Hum. Mol. Genet. 1997;6:921–926. doi: 10.1093/hmg/6.6.921. [DOI] [PubMed] [Google Scholar]
- 21.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat. Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 22.Reddel RR, Bryan TM, Colgin LM, Perrem KT, Yeager TR. Alternative lengthening of telomeres in human cells. Radiat. Res. 2001;155:194–200. doi: 10.1667/0033-7587(2001)155[0194:alotih]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 23.Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 24.Teng SC, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen H, Sinclair DA. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc. Natl Acad. Sci. USA. 2001;98:3174–3179. doi: 10.1073/pnas.061579598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teng SC, Chang J, McCowan B, Zakian VA. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell. 2000;6:947–952. doi: 10.1016/s1097-2765(05)00094-8. [DOI] [PubMed] [Google Scholar]
- 27.Tsai HJ, Huang WH, Li TK, Tsai YL, Wu KJ, Tseng SF, Teng SC. Involvement of topoisomerase III in telomere-telomere recombination. J. Biol. Chem. 2006;281:13717–13723. doi: 10.1074/jbc.M600649200. [DOI] [PubMed] [Google Scholar]
- 28.Huang P, Pryde FE, Lester D, Maddison RL, Borts RH, Hickson ID, Louis EJ. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 2001;11:125–129. doi: 10.1016/s0960-9822(01)00021-5. [DOI] [PubMed] [Google Scholar]
- 29.Johnson FB, Marciniak RA, McVey M, Stewart SA, Hahn WC, Guarente L. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 2001;20:905–913. doi: 10.1093/emboj/20.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett RJ, Sharp JA, Wang JC. Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:9644–9650. doi: 10.1074/jbc.273.16.9644. [DOI] [PubMed] [Google Scholar]
- 31.Fricke WM, Kaliraman V, Brill SJ. Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J. Biol. Chem. 2001;276:8848–8855. doi: 10.1074/jbc.M009719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- 33.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 34.Maeda D, Seki M, Onoda F, Branzei D, Kawabe Y, Enomoto T. Ubc9 is required for damage-tolerance and damage-induced interchromosomal homologous recombination in S. cerevisiae. DNA Repair. 2004;3:335–341. doi: 10.1016/j.dnarep.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 35.Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 36.Li SJ, Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 2000;20:2367–2377. doi: 10.1128/mcb.20.7.2367-2377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- 38.Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl Acad. Sci. USA. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng CH, Lo YH, Liang SS, Ti SC, Lin FM, Yeh CH, Huang HY, Wang TF. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006;20:2067–2081. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai YL, Tseng SF, Chang SH, Lin CC, Teng SC. Involvement of replicative polymerases, Tel1p, Mec1p, Cdc13p, and the Ku complex in telomere-telomere recombination. Mol. Cell. Biol. 2002;22:5679–5687. doi: 10.1128/MCB.22.16.5679-5687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prado F, Aguilera A. Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes. Genetics. 1995;139:109–123. doi: 10.1093/genetics/139.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast Type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ulrich HD. The SUMO system: an overview. Methods Mol. Biol. 2009;497:3–16. doi: 10.1007/978-1-59745-566-4_1. [DOI] [PubMed] [Google Scholar]
- 44.Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett CB, Lewis LK, Karthikeyan G, Lobachev KS, Jin YH, Sterling JF, Snipe JR, Resnick MA. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 2001;29:426–434. doi: 10.1038/ng778. [DOI] [PubMed] [Google Scholar]
- 46.Watt PM, Hickson ID, Borts RH, Louis EJ. SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krogh BO, Symington LS. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 48.Ohuchi T, Seki M, Branzei D, Maeda D, Ui A, Ogiwara H, Tada S, Enomoto T. Rad52 sumoylation and its involvement in the efficient induction of homologous recombination. DNA Repair. 2008;7:879–889. doi: 10.1016/j.dnarep.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 49.Sacher M, Pfander B, Hoege C, Jentsch S. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat. Cell Biol. 2006;8:1284–1290. doi: 10.1038/ncb1488. [DOI] [PubMed] [Google Scholar]
- 50.Onoda F, Seki M, Miyajima A, Enomoto T. Involvement of SGS1 in DNA damage-induced heteroallelic recombination that requires RAD52 in Saccharomyces cerevisiae. Mol. Gen. Genet. 2001;264:702–708. doi: 10.1007/s004380000358. [DOI] [PubMed] [Google Scholar]
- 51.Bernstein KA, Shor E, Sunjevaric I, Fumasoni M, Burgess RC, Foiani M, Branzei D, Rothstein R. Sgs1 function in the repair of DNA replication intermediates is separable from its role in homologous recombinational repair. EMBO J. 2009;28:915–925. doi: 10.1038/emboj.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 53.Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 54.Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT, Ho CC, Chen YC, Lin TP, Fang HI, et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell. 2006;24:341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 55.Bischof O, Kim SH, Irving J, Beresten S, Ellis NA, Campisi J. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J. Cell Biol. 2001;153:367–380. doi: 10.1083/jcb.153.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol. Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xhemalce B, Riising EM, Baumann P, Dejean A, Arcangioli B, Seeler JS. Role of SUMO in the dynamics of telomere maintenance in fission yeast. Proc. Natl Acad. Sci. USA. 2007;104:893–898. doi: 10.1073/pnas.0605442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pebernard S, Schaffer L, Campbell D, Head SR, Boddy MN. Localization of Smc5/6 to centromeres and telomeres requires heterochromatin and SUMO, respectively. EMBO J. 2008;27:3011–3023. doi: 10.1038/emboj.2008.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003;22:4325–4336. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stewart E, Chapman CR, Al-Khodairy F, Carr AM, Enoch T. rqh1+, a fission yeast gene related to the Bloom’s and Werner’s syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lustig AJ, Kurtz S, Shore D. Involvement of the silencer and UAS binding protein Rap1 in regulation of telomere length. Science. 1990;250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- 62.Aguilera A. Genetic evidence for different RAD52-dependent intrachromosomal recombination pathways in Saccharomyces cerevisiae. Curr. Genet. 1995;27:298–305. doi: 10.1007/BF00352096. [DOI] [PubMed] [Google Scholar]
- 63.Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J. Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.