Abstract
Borrelia burgdorferi, an agent of Lyme disease, encodes the β3-chain integrin ligand P66. P66 is expressed by B. burgdorferi in the mammal, in laboratory media, and as the bacteria are acquired or transmitted by the tick, but is not expressed by the bacterium in unfed ticks. Attempts to reveal factors influencing expression revealed that P66 was expressed in all in vitro conditions investigated. Candidate regulators identified in a search of the B. burgdorferi genome for homologs to other bacterial transcription factors were cloned and introduced into E. coli carrying a p66 promoter-signal sequence-phoA (alkaline phosphatase, or AP) fusion. Three candidate transcription factors—two that decreased AP activity (Hbb and BB0527), and one that increased AP activity (BBA23)—were identified. BBA23 and BB0527 did not bind to the p66 promoter at physiologically relevant concentrations. In contrast, several promoter fragments, including p66, were bound by Hbb (BB0232), with slightly different affinities. Consistent with results from other laboratories, Hbb appears to recognize multiple DNA sequences. Changes in the expression of p66 and bb0232 in the tick at various points with respect to feeding on mice, along with the results of the reporter experiment in the surrogate host E. coli, are consistent with Hbb/BB0232 being involved in regulating p66 expression.
INTRODUCTION
Lyme disease is the most prevalent arthropod-borne disease in the USA (1,2). The causative agent, Borrelia burgdorferi, migrates between vertebrate hosts and the tick vector. The relatively small genome size of B. burgdorferi belies the complex regulation one would expect to be necessary for living in these different environments. Only three sigma (σ) factors have been identified in B. burgdorferi, relatively few when compared with the larger numbers found in other organisms. Borrelia burgdorferi encodes rpoD, the sigma 70 (σ70) subunit, in addition to rpoS (σ38) and rpoN (σ54) homologs. Typically, RpoD homologs are involved with housekeeping functions, whereas RpoS controls gene expression in stationary phase or the stress response, and RpoN is often involved in responses to nitrogen limitation or other stresses. Borrelia burgdorferi differs significantly from the classical patterns, in that RpoS, RpoN and the response regulator Rrp2 participate in a regulatory pathway that is critical to how B. burgdorferi responds to certain environmental changes (3–10).
A number of laboratories have investigated in vitro culture methods thought to model the differences between the unfed tick, the feeding tick and the mammal. The pH of the unfed tick midgut is slightly alkaline (11), and the temperature is that of the tick’s surroundings. However, as the tick begins to take its blood meal, the pH in the midgut drops to ∼6.8 (11), and the temperature is near the surface temperature of the mammal, between 34°C and 37°C (11,12). Temperature, growth phase or cell density, pH, oxygen concentration, the addition of blood to the medium, migration between the vertebrate and tick environments, and time and tissue localization within a mammal have been shown to affect the expression of many genes in B. burgdorferi (11,13–26). Other genes affected by environmental cues were identified using dialysis membrane chambers (DMCs) implanted in rats (27,28), which at least partially allow for adaptation to the mammalian host by B. burgdorferi.
P66 is a B. burgdorferi surface-exposed outer membrane protein and a ligand for the β3-chain integrins (29–32). P66 is commonly recognized by Lyme disease patient sera (33,34), demonstrating that the protein is produced when the bacteria are in a mammalian host. Mapping of the transcriptional start site revealed that p66 expression is apparently under the control of a σ70-dependent promoter (29). This is corroborated by the fact that its expression was not changed in the rpoS or rpoN deletion mutants used for microarray experiments (6,8,35).
Indirect immunofluorescence studies showed that P66 protein levels do, however, vary at the different stages in the life cycle of the tick (36). As compared with flagellin expression, P66 is not produced by B. burgdorferi in the midguts of unfed ticks, but as the ticks take their blood meal, P66 is expressed by a majority of B. burgdorferi (36). At 7 days post-repletion, P66 is still present in the majority of bacterial cells, but by 16 days post-repletion, P66 is produced by only a fraction of the B. burgdorferi cells, indicating that the production of P66 is tied to the presence of the mammalian blood meal.
In similar studies, OspC undergoes a sharp peak of production coincident with transmission from the tick to the mammal (37). OspA is produced in the midgut of the unfed tick, but expression decreases as the ticks reach repletion, and increases again in the days following repletion (37). These patterns of expression led us to believe that differential regulation of P66 protein and/or p66 mRNA occurs under different conditions, especially since p66 and flaB appear to be transcribed from σ70 dependent promoters. Because proteases that might degrade P66 are not known to be present in either the tick midgut or in B. burgdorferi, we searched for potential regulators of p66 at the transcriptional level as the first step to understanding how expression and production of this integrin ligand are regulated.
MATERIALS AND METHODS
Infection, feeding and collection of ticks
Ixodes scapularis larvae were infected with B. burgdorferi strain B31 by artificial infection (38). Infected larvae and control, uninfected larvae were fed separately on Rocky Mountain Laboratories (RML) mice, an outbred strain of Swiss‐Webster mice, and allowed to molt to nymphs. Nymphs were held in the unfed state, or were fed on naïve mice and collected when replete. At unfed, replete and 9 days post-repletion time points, midguts of 15, 10 and 10 ticks, respectively, were dissected and stored in water at −80°C until RNA extraction. Animal experiments were performed under protocols approved by the RML Animal Care and Use Committee, prepared according to National Institutes of Health guidelines. The RML Animal Facility is accredited by the International Association for Assessment and Accreditation of Laboratory Animal Care.
Bacterial strains and culture conditions
Escherichia coli strains JM109 and BLR were used for cloning of genes; CC118 and CC118(DE3)pLysS were used for Alkaline Phosphatase (AP) assays, and BL21(DE3)pLysS and Rosetta-gami2(DE3)pLysS were used for protein expression and purification. Proteins expressed in and purified from the Rosetta-gami2(DE3)pLysS strain were used for electrophoretic mobility shift assays (EMSAs). Strain genotypes are provided in Table 1.
Table 1.
Escherichia coli strain genotypes
 |
Borrelia burgdorferi strains N40 (clone D10E9) and B31 (uncloned) were used for all pH, temperature, growth phase, mammalian components and tick cell culture medium experiments. Borrelia burgdorferi were grown in either BSK-II (39) or MKP (40) medium. For some in vitro tests of how the manipulations of culture conditions affect p66 expression, BSK-H (Sigma-Aldrich, St. Louis, MO) was also used. Tick cell culture medium (41) was used for some experiments.
Borrelia were inoculated from frozen stocks (passage 4 for N40 or passage 5 B31) into fresh medium, and grown to late exponential phase (∼1 × 108 bacteria/ml). Bacteria were then subcultured 1 : 200 into fresh medium and grown again to late exponential phase (unless otherwise specified). Bacterial cell densities were determined by dark-field microscopy. Cultures were then divided so that 7 ml of culture was harvested for SDS‐PAGE and immunoblot analysis, and 3 ml of culture was harvested for RNA analysis. The bacteria were pelleted, washed in 1 ml PBS (phosphate-buffered saline) plus 0.2% BSA (bovine serum albumin), then again in 200 µl PBS. The pellets were resuspended in Trizol (Invitrogen, Carlsbad, CA) for subsequent RNA extraction, or in HEPES-buffered saline plus protease inhibitors [25 mM HEPES pH 7.8, 150 mM NaCl, 1 mM EDTA, 0.1 TIU/ml aprotinin, 1 mM benzamidine, 10 µM pepstatin A and 1 mM PMSF (Sigma-Aldrich Co., St. Louis, MO)] for analysis of protein expression and stored at −70°C. In total, 5 × 107 B. burgdorferi were loaded per lane and SDS‐PAGE and immunoblotting were performed according to standard protocols (42,43) (data not shown).
Analysis of mRNA levels
RNA from ticks or cultured B. burgdorferi samples was purified using the Trizol fractionation method according to the manufacturer’s instructions, then treated with RNase-free DNase and repurified using the RNeasy kit (Qiagen, Valencia, CA). First-strand synthesis was performed using Superscript II RT (Gibco, Carlsbad, CA) and gene-specific reverse primers listed in Table 2. Each cDNA sample was then amplified by either conventional polymerase chain reaction (PCR) or quantitative PCR using gene-specific primers as listed in Table 2. All primers were checked against the NCBI database for potential homology to and amplification from the other genomes that had been sequenced, and were tested for sensitivity using known quantities of DNA purified from cultured B. burgdorferi. Conventional RT‐PCR products were analyzed by electrophoresis through 10% acrylamide gels in Tris‐borate‐EDTA buffer. Quantitative RT‐PCR (qRT‐PCR) samples were amplified by use of the SYBR Green master mix (Qiagen, Valencia, CA) and either the ABI Prism 7700 sequence detection system (PE Biosystems/Applied Biosystems Inc., Foster City, CA) or the Stratagene MxPro 3000P system (Stratagene, La Jolla, CA). Melt point analysis was performed to assess homogeneity of the amplified product. All samples were amplified in parallel with standard curves using known quantities of purified genomic DNA from B. burgdorferi using the same primer sets in the same 96-well plates. Copy numbers of the target sequence cDNAs in the tick samples were determined from the Ct values of triplicate samples using the standard curves from the same reaction plate. Each mRNA preparation was analyzed by performing at least two cDNA synthesis reactions and subsequent qRT‐PCR reactions. The data shown for the tick experiments are from one of two separate cohorts of ticks (biological replicates) that showed the same trends, although the absolute numbers of mRNA copies differed between the cohorts.
Table 2.
Primers used for qRT‐PCR
| Target | Forward primer, 5′–3′ | Reverse primer, 5′–3′ | Limit of detection, copy number of genomes |
|---|---|---|---|
| flaA (bb0668) | aaagtcacacagttcaaaagagc | gattcttcaggtttttcactctc | <15 |
| flaB (bb0147) | aacggcacatattcagatgcagacagagg | aagacgcttgagaccctgaaagtgatgc | <15 |
| p66-MM (bb0603) | atttaaaagcacttactatggattcc | tcgttttgatcaagtagattttttattgg | 150 |
| p66-CC (bb0603) | tgaacaaagttcaacaagcacaaag | agcacttccaatagcagcattattt | <15 |
| ospA (bba15) | tgaaggcgtaaaagctgacaaa | ttctgttgatgacttgtctttggaa | 150 |
| ospC (bbb19) | tgttaaagggcctaatcttacagaaataa | taccaatagctttggtagcaagttcat | 150 |
| hbb (bb0232) | aagaagaccaaaggttactaagtc | atgatctaggaccttaacatactcc | 15 |
Primers were designed and tested by PCR, then by conventional RT–PCR prior to use in qRT–PCR.
Plasmid construction and cloning of candidate transcription factors
The pET30a (Novagen, Gibbstown, NJ) plasmid was modified by the insertion of a His6-HA-HA tag in place of the His6 and S-tags present in the original vector as previously described (44). Each candidate regulator gene was cloned in this vector after PCR amplification from B. burgdorferi strain N40 genomic DNA using primers listed in Table 3. Candidate clones were screened for inserts by PCR using pET multiple cloning site-specific primers flanking the cloning site (Novagen, Gibbstown, NJ). Six candidates for each gene cloned were sequenced (Tufts University Core Facility, Boston, MA), and a clone with no PCR-generated errors was chosen for protein production (for sequencing primers also, see Table 3).
Table 3.
Oligonucleotides used for cloning and sequencing
| Target, purpose | Genome coordinates | Forward primer, 5′–3′ | Reverse primer, 5′–3′ |
|---|---|---|---|
| bba23 cl,seqa | 15303–15886 | tagcggatccaaaatgtttattgagaaaatattacaaagc | tacgctcgagtcatggttgggtttgggc |
| bb0225 cl,seqa | 229242–230266 | tagcggatccatagccccaatggtaaacattacagacg | tacgctcgagtcattcctaaaagtttggttaagtatacaggc |
| bb0232 cl,seqa,b | 237956–237620 | tagcggatccatgtctttttcaagaagaccaaagg | tacgctcgagtcataaccggcatttaacctttgatacc |
| bb0345 cl,seqa | 354572–353303 | tagcggatccttgactcttgaaatggtagctgagg | tacgctcgagtcagcaattataactcttgttttaccacttcc |
| bb0355 cl,seqa | 365570–365181 | tagcggatcctatccaatgcatggagtaggtacg | tacgctcgagtcaatcttaaagaatcaaacacagcaaatcc |
| bb0462 cl,seqa,b | 484254–484820 | tagcggatccttggagcaagtgaagttctggagg | tacgctcgagtcaacctctcctaactccatctgg |
| bb0468 cl,seqa | 488226–489096 | tagcggatccttgaatgtaagagatttgtcttttaagc | tacgctcgagtcatgaacttaatttaacatactttgcaacc |
| bb0527 cl,seqa | 537538–538491 | tagcggatcctcagaattgataattgatattggaaataccagc | atcgctcgagtcacgtagcaaggaaaattatgtagaatcaaacg |
| bb0647 cl,seqa,b | 686504–685854 | tagcggatccgacaacataatagacgtacattcc | tacgctcgagtcatgtcaatttcttctatgtttttagg |
| His6-HA-HA Tc cloning | n/a | tatgcaccaccaccaccaccacaccggttatccttacgacgtacctg actacgcagcaggatacccatacgacgtcccagactacgctggtac | |
| His6-HA-HA Bc cloning | n/a | cagcgtagtctgggacgtcgtatgggtatcctgctgcgtagtca ggtacgtcgtaaggataaccggtgtggtggtggtggtggtgca | |
| bb0225seqd | 229722,229294 | ttattgtacatgcaagg | ttgtaatcactagataaaggcc |
| bb0345seqd | 353840,354168,353867 | aaattgccgagcagc | 1 atctccctctttaagagaaacg 2 ttcccttaacatcaaactgcc |
| bb0468seqd | 488607, 488724 | atggcggagttaatctagg | attgcaaccttattcaccg |
| bb0527seqd | 538038,538042 | 1 agaattgataattgatattggaaatacc 2 attccccattagcactcc | ttggagtgctaatggg |
| p66 promoter cloning | 626808–627175 | agaggatcctggacctttacaaacacaaatggca | agaggatcctttaatgcgtctgctgcaaata |
Restriction sites are underlined. n/a: not applicable.
acl,seq denotes that the oligonucleotides were used for both cloning and sequencing.
bThese primers were described in a previous study (44) but are included here for the sake of convenience.
cThese oligonucleotides were annealed to form the His6-HA-HA tag inserted in the cloning vector pET30a, and do not correspond to sequences in the B. burgdorferi genome.
dThese oligonucleotides were used for internal sequencing of candidate clones.
Production, purification, and analysis of recombinant candidate regulators
BL21(DE3)pLysS or Rosetta-gami2(DE3)pLysS E. coli strains containing the recombinant plasmids were grown in LB (Luria‐Burtani) medium plus 30 µg/ml choloramphenicol and 50 µg/ml kanamycin overnight at 30°C. Five milliliters of overnight culture was added to 500 ml 2XYT (Yeast Extract Tryptone) medium containing 0.2% dextrose, chloramphenicol and kanamycin and grown to mid-exponential phase (OD600 0.4–0.6) at 30°C. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to cultures to a final concentration of 1 mM for induction of protein production, after which cultures were aerated for 1–2 h at 250 rpm and 30°C. Cells were harvested, chilled on ice for 30 min, and centrifuged at 4230×g for 10 min at 4°C. Pellets were resuspended in 20 ml HEPES-buffered saline, and centrifuged at 4230×g for 10 min at 4°C. The pellets were stored at −70°C. Pellets were lysed in a French pressure cell, in the presence of protease inhibitors (0.1 TIU/ml aprotinin, 1 mM benzamidine, 10 µM pepstatin A and 1 mM PMSF, Sigma-Aldrich Co., St Louis, MO). Lysed cells were centrifuged at 26 890×g for 30 min at 4°C. The supernatant was decanted and centrifuged at 38 720×g for 20 min at 4°C. His-bind kit columns (Novagen/EMD Biosciences, Gibbstown, NJ) were used to purify the recombinant His-6 tagged proteins from the supernatants. The purification was performed as per the manufacturer’s instructions, except that protease inhibitors (0.1 TIU/ml aprotinin, 1 mM benzamidine and 10 µM pepstatin A) were added to the binding and elution buffers, and 10% glycerol was added to the elution buffer. Protein purity was assessed by SDS‐PAGE (42) (data not shown). Protein concentrations were determined for each sample using both the Bradford (BioRad, Hercules, CA) and BCA (Pierce, Rockford, IL) reagents. The tags were not cleaved from the recombinant proteins to allow detection throughout the experiments.
Alkaline phosphatase reporter activity assays
A fragment encoding the P66 secretion signal (amino acids 1–19) plus an additional 476 base pairs upstream of the start codon (primer sequences are in Table 2) were cloned in pAWLP2 (a gift of A. Wright, Tufts University School of Medicine) to generate a translational fusion to alkaline phosphatase codon 14. pAWLP2 is derived from pBR322 and carries the alkaline phosphatase gene from which the first 13 codons of the gene have been deleted, which replaces much of the bla gene. The plasmid confers resistance to tetracycline. The p66‐phoA construct was sequenced to ensure that the p66 promoter and secretion signal were intact and in frame. The pET30 derivatives in which the candidate regulators were cloned confer resistance to kanamycin, so all experiments were performed in the presence of both antibiotics, which resulted in maintenance of both replicons despite their having the same origins of replication. Both plasmids were introduced by electroporation into E. coli CC118, which is phoA−. Expression of the candidate regulator genes was induced by addition of IPTG to 0.1 mM and incubation for 1 h at 30°C. AP activity was then determined as described previously (45), using p-nitrophenolphosphate as a colorimetric substrate; color was measured at OD405. Activity was calculated based on the OD600 of the bacterial suspensions and the incubation time with the substrate. To allow inclusion of all data obtained in multiple experiments for every clone, the units in each sample were normalized to the vector control, which was set at one. The means and standard deviations of the normalized values were calculated, and statistical analyses were performed using the Student’s two-tailed t-test.
Electrophoretic mobility shift assays (EMSA)
DNA containing the regions upstream of the p66, flaB, ospA and ospC genes, and containing the oriC region of the linear chromosome in B. burgdorferi (46) were prepared by PCR utilizing the primers listed in Table 4, and the p66 promoter fragment then purified by agarose gel electrophoresis for most experiments. DNA was labeled with γ32P-ATP (Perkin-Elmer, Waltham, MA) and T4 polynucleotide kinase (New England Biolabs, Ipswich, MA). Purified Hbb (BB0232), BB0527 and BBA23 at various concentrations (indicated in figures/legends) were mixed with the radiolabeled DNA at concentrations indicated in figure legends in a buffer containing 20 mM HEPES, pH 7.6, 50 mM NaCl, 5 mM DTT, 5% glycerol, 50 µg/ml BSA and 1 mM MgCl2. For competition assays, salmon sperm DNA was added to each reaction for a final concentration of 1 µg per reaction. BSA served as a control protein in initial experiments, and was added as part of the EMSA buffer in subsequent experiments. The mixtures were incubated at 33°C for 30 min before loading on to 5% polyacrylamide nondenaturing gels. After electrophoresis, gels were dried and exposed to Maximum Sensitivity (MS) X-ray film (Kodak, Rochester, NY) with an intensifier screen.
Table 4.
Primers used for amplification of DNAs for EMSAs
| Target | Genome coordinates | Forward primer, 5′–3′ | Reverse primer, 5′–3′ | Fragment length (bp) |
|---|---|---|---|---|
| oriC | 457923–458387 | aaacccattcaacagtgctttattc (46) | caatgcactccaaatatcattcata (46) | 465 |
| flaBp | 149013–148493 | ttgtctgtcgcctcttgtggcttcc (91) | aagtggaaggtgaacttaataccttgg (91) | 522 |
| p66p | 626808–627175 | tggacctttacaaacacaaatggc | tcctttaatgcgtctgctgc | 558 |
| ospAp | 9018–9457 (lp54) | atcctgaatttacgctttttgatacc | aacattttgcttacatgctattaaggc | 439 |
| ospCp | 16561–17016 (cp26) | tagtaaggtattacttttgtataaacgcc | aacagactcatcagcagaatttgc | 455 |
RESULTS
Expression of p66 in different laboratory culture conditions
We first attempted to mimic the conditions of the unfed tick midgut versus the fed tick midgut in vitro with regard to p66 expression by growing B. burgdorferi in BSK and MKP media under conditions in which we varied the temperature, starting pH and growth phase at which the cultures were harvested. Medium to which the mammalian-derived gelatin, serum and BSA had not been added was also tested. As previously reported, no significant differences in P66 were seen at the protein level, while at the mRNA level a maximum decrease of only 10-fold was observed in p66 mRNA levels in bacteria grown in medium devoid of the mammalian components serum, gelatin and BSA, as compared with levels seen in bacteria grown in standard medium (36). We also tested tick cell culture medium with similar results. Although a 10-fold decrease may be significant in terms of expression of some genes, in repeated attempts, the fold changes at the mRNA level differed with the batch of base medium used, so no definitive in vitro culture conditions that reliably mimic the midgut of the unfed tick were identified with regard to P66 production. Moreover, the bacteria do not survive for more than a few days in medium devoid of the mammalian-derived components, and in fact do not replicate more than once, so changes in the mRNA level that are seen may not be reflected at the protein level if the protein turnover rate is slow. These results suggest, however, that P66 levels may be regulated at both the transcriptional and post-transcriptional levels. We therefore determined whether p66 mRNA levels vary in the tick with feeding status.
Assessment of p66 mRNA levels in the tick midgut
To determine whether changes at the protein level were reflected at the mRNA level, we examined six sets of tick samples: unfed, replete (‘drop-off’), or 9 days post-repletion samples from ticks that were either uninfected or infected with B. burgdorferi. The time course differed slightly from the samples used in the immunofluorescence studies (36), but still included the conditions we were interested in, namely the absence or presence of the mammalian blood meal. qRT‐PCR analysis of these samples examined expression of p66, flaA, ospA and ospC. Flagellar expression is used in B. burgdorferi as a control and a marker for spirochete presence, as flagellar operons are thought to be constitutively expressed in B. burgdorferi. Attempts to perform qRT‐PCR using primers against the B. burgdorferi 16S rRNA proved unsuccessful due to high levels of background, even in the uninfected tick midgut samples, presumably due to the presence of other bacteria. OspA and OspC have long been considered hallmarks of differential expression during the tick-mammal cycle (47), and we examined their expression as controls.
For qRT‐PCR, expression within the tick midguts was compared with standard curves of known concentrations of serial dilutions of B. burgdorferi genomic DNA. In this way, we were able to calculate limits of detection for each set of primers used (Table 1). We also used the BLAST program (http://www.ncbi.nlm.nih.gov/blast/) (48) to ensure that there were no obvious sequences related to the primers we designed in other organisms.
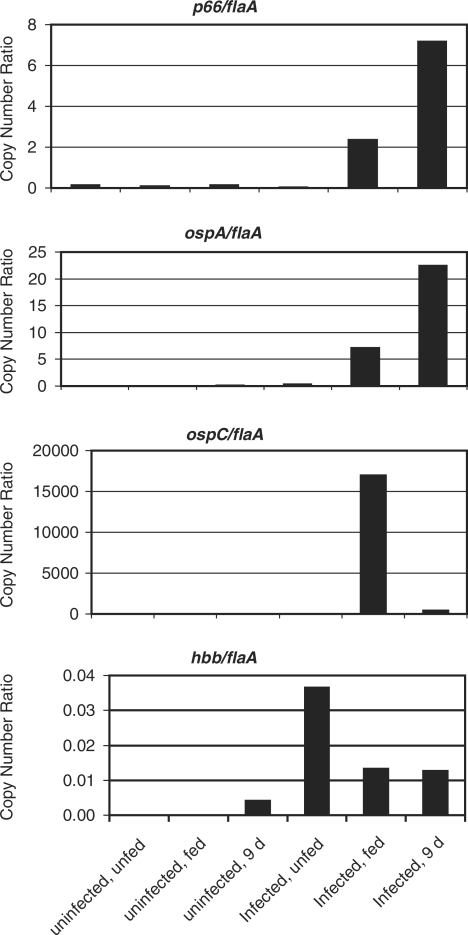
There is a clear increase in the expression of all genes, including flaA, in both the replete and the 9 days post-repletion samples. This likely reflects both an increase in the number of B. burgdorferi cells and transcriptional activity of the bacteria, so data for each gene are shown normalized to the flaA transcript (Figure 1). By immunofluorescence intensity, flagellar expression appears consistent in all infected tick samples, and was not seen in uninfected tick midguts (36). By qRT‐PCR, p66 (bb0603) is not detectable above background in the infected, unfed ticks, indicating that P66 protein is not simply being degraded in the tick midgut (Figure 1). p66 is expressed to higher levels at repletion, and its expression is highest at 9 days. P66 protein expression was not detected in unfed tick midguts, was highest in the midguts of ticks at repletion, and was still highly expressed at 7 days post repletion (36). However, the expression of P66 at 16 days post-repletion was significantly decreased in the immunofluorescence studies. While the time points analyzed in the two experiments differ, p66 expression still increases dramatically in the presence of the mammalian blood meal by both immunofluorescence and qRT‐PCR [this study, (36)]. At the mRNA level, the increase in p66 expression is greater than that of flaA expression following the tick blood meal.
Figure 1.
Quantification of mRNA levels of select B. burgdorferi genes in ticks. Midguts were removed from ticks that were either uninfected or infected with B. burgdorferi, and unfed, fed to repletion (‘fed’), or 9 days post-repletion. The cDNA prepared from total RNA was used as the template for quantitative PCR. Ct values were determined for each cDNA sample generated in the presence and absence of RT. Copy numbers were determined against standard curves of genomic DNA. The flaA and flaB primer sets generated similar standard curves, so data shown are normalized to flaA. The p66 results are shown for primer set p66-CC (Table 1). Results shown are from one cohort of ticks. A second cohort gave rise to higher ‘background’ in the uninfected controls, and somewhat different absolute transcript numbers, but the expression trends were similar. Note the very different quantities of the transcripts for each gene normalized to flaA.
The qRT‐PCR data for ospA show that little transcript is detected in the unfed, infected tick, more is detected in the replete tick, and still more is found in the post-repletion tick midgut (Figure 1). These are the only results that are somewhat inconsistent with what was seen at the protein level (37,47). OspA by immunofluorescence is expressed to high levels in unfed tick midguts, and to slightly lower levels at repletion and 13 days post-repletion. However, the ospA qRT‐PCR results may indicate transcriptional changes occurring that are not directly reflected at the protein level. The relatively quiescent bacteria in the unfed tick midgut may not turn over OspA protein rapidly, and again our results are consistent with complex regulation of gene expression versus protein production for at least some B. burgdorferi genes. For ospC, the qRT‐PCR results are most directly consistent with the immunofluorescence data (Figure 1) (37,47). By qRT‐PCR, there is a sharp peak in ospC expression in the infected, replete ticks. Expression is highest at repletion, lower at 9 days post-repletion, and lowest in unfed tick midguts. Based on the immunofluorescence studies, OspC is expressed to high levels briefly before repletion during tick feeding, but not at the other time points (37,47).
The qRT‐PCR results are consistent with the immunofluorescence data in that transcription of p66 increases dramatically when the blood meal is taken. The increases in transcript levels of all genes are likely to reflect increases in bacterial numbers and metabolic activity as well as gene-specific increases in expression levels, but the increases in bacterial numbers and metabolic activity are most likely reflected by the changes in the flaA mRNA levels. Although the expression of flaA and p66 both increased in the fed versus unfed tick environments, the fold change for p66 was much greater than that for flaA (Figure 1), suggesting the possibility that p66 expression may be repressed in the unfed tick or activated in the fed tick. In addition, it is clear that p66 production patterns at the protein level are distinct from those of flaA, ospA and ospC, despite the similarity of the p66 and ospA mRNA expression patterns. These results suggest that there are additional specific factors encoded in the B. burgdorferi genome that may bind to and regulate the expression of the p66 promoter, and possibly, expression of the gene at the post-transcriptional level.
Identification of candidate transcriptional regulators
The published B. burgdorferi strain B31 genome was surveyed for candidate regulators based on several parameters. First, we looked for orthologs of known transcriptional regulators in other bacteria, including the five main families of transcription factors (araC/xylS, argR, lacI, lysR and ompR families). We also looked for homologs of proteins outside the five major transcription factor families that have been identified as participating in transcriptional regulation. Finally, we looked for proteins predicted to contain DNA-binding domains such as helix-turn-helix (HTH) or helix-loop-helix motifs. We identified 14 potential transcription factors in the B. burgdorferi genome and further evaluated nine of these candidates (Table 5), only one of which, Hbb, had been characterized at the start of this investigation. Although we were able to amplify the remaining five genes by PCR, and were able to obtain clones of some these candidates that were stable in E. coli, some are not in themselves likely to affect gene expression, but to cooperate with other proteins in a more complex network. Examples include proteins with GGDEF and EAL domains, which would potentially modulate cyclic di-GMP levels, and Rrp2, which is part of the RpoN-RpoS regulatory cascade that does not affect p66 expression (49). We therefore focused on candidates that were predicted to have domains that might interact with DNA directly. Several of the candidate genes, bb0225, bb0345, bb0355 and bb0468, have not previously been studied in B. burgdorferi, although all are predicted to encode proteins with motifs of possible interest regarding regulation of gene expression, e.g. homology to known transcription factors in other bacteria, and HTH or other putative DNA-binding motifs (Table 4).
Table 5.
Candidate regulators of p66 expression
| Genome locus | Description/possible roles | Putative functions |
|---|---|---|
| BBA23 | MarR-type Putative Transcription Factor | MarR-type transcriptional regulator family. Located on plasmid lp54-A in B31 |
| BB0225 | TIM-barrel protein | Putative transcriptional regulator, function unknown, conserved hypothetical |
| BB0232 | Hbb (HU-like protein) | Histone-like, potential roles in DNA replication, transcriptional regulation (46,53) |
| BB0345 | HTH-3 | Conserved hypothetical protein, family includes phage transcriptional control, bacterial methylase, plasmid copy control |
| BB0355 | Transcription Factor (CarD-like) | Transcriptional regulator homolog (92) |
| BB0462 | EbfC | Transcriptional regulator homolog (44,54) |
| BB0468 | Putative Transcription Factor | Homologous to Lac I family transcription factors in Brucella spp. |
| BB0527 | Transcriptional Activator (Baf family) | Putative transcriptional activator, putative B. pertussis toxin operon regulator |
| BB0647 | Fur family | Transcription factor activity, Borrelia oxidative stress regulator, BosR (13,44,50,52) |
Three candidates that we identified, BB0232 (Hbb), BB0462 (EbfC) and BB0647 (Fur/BosR) have been identified by other laboratories as having site-specific DNA binding activity (13,46,50–54), and were examined in a previous study of the regulation of oppA promoters (44). Hbb was originally identified on the basis of complementation of E. coli mutants defective in λ phage packaging, and is a homolog of the HU/IHF family of DNA binding proteins in E. coli (53). Hbb was later shown to bind specifically to a site in the dnaA-dnaN intergenic region in the origin of replication of the B. burgdorferi chromosome (46). Hbb also binds to the upstream regions of hbb, ospC and gacA (46), but Hbb did not interact specifically with any oppA promoter (44). EbfC (BB0462) was previously identified as a regulator of specific erp loci (54) and interacts with the oppA5 promoter (44). Fur/BosR (BB0647) (13,50,52,55) has been studied by several groups, and although there is some controversy regarding its activities, they are consistent with its similarity to Fur and Per proteins found in other organisms. Fur/BosR interacts with the oppA4 promoter as well as its own promoter and those of bb0153 (sodA), bb0646 (a predicted hydrolase) and bb0690 (napA) (44,50,52,56).
BB0527 is homologous to Baf (Bvg accessory factor), which helps to activate the Bvg two-component system of Bordetella pertussis (57). In that system, BvgS is a transmembrane sensor kinase, and BvgA the positive regulator, of virulence gene expression in response to certain environmental signals in B. pertussis (58). Baf augments BvgAS effects in E. coli, and is essential for B. pertussis viability (57), suggesting additional possible roles for the protein.
BBA23 in B. burgdorferi is a MarR homolog, and is the only one of the candidate transcription factors identified to date that is located on one of the plasmids in strain B31 (59,60). In E. coli, MarR encodes a repressor of the marRAB operon, which controls the response of the bacterial cell to multiple antibiotics and to other environmental stresses (61). MarR may bind as oligomers both at the −35 and −10 region of the mar operator, and at the marR ribosome-binding site (61). Binding of this protein to the operator appears to be influenced by a wide variety of signals, including antibiotics and reactive oxygen species.
Functional characterization of candidate regulators
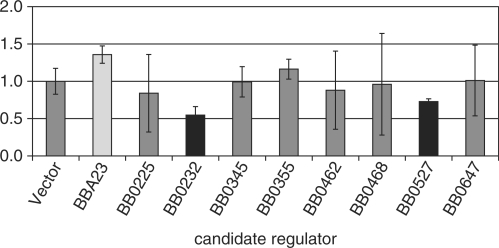
Nine candidate regulators expressed from an E. coli pET expression vector were introduced into phoA− E. coli carrying a second plasmid containing the p66 promoter plus secretion signal fused to the alkaline phosphatase (AP) reporter gene devoid of promoter and signal sequence. This fusion was generated to account for the possibility that some regulatory factors might bind to sites overlapping the translational start site, so the fragment cloned included the 5′ end of the structural gene. In addition, to account for the possibility that some factors might bind to sites considerably 5′ to the −35 element of the promoter, the fragment cloned was 556 bp in length. The AP assays were performed after growth of the dually transformed strains in liquid culture. At least three independent experiments were performed for each candidate regulator. Controls in all experiments included the p66 promoter fusion alone, with and without the pET vector with no B. burgdorferi DNA; both of these control strains expressed similar low levels of AP activity that we termed ‘basal’. Three of the nine candidates tested reproducibly affected production of AP activity (P-value of <0.01 by the Students’ two-tailed t-test in comparison to the control; Figure 2). These candidates were BB0232 (Hbb), which resulted in decreased AP expression from the p66 promoter, BB0527 (Baf homolog), which also resulted in decreased AP activity, and BBA23 (MarR homolog), which resulted in an increase in AP activity. The decrease in p66-phoA expression is distinct from the effects of Hbb on the expression of the oppA1-oppA5 promoters in a similar reporter system, in which, if Hbb expression affected reporter activity at all, an increase was observed (44). The other candidates did not significantly affect the basal AP activity, although all nine candidate regulators were expressed after induction as analyzed by immunoblot (data not shown). Therefore, no further characterization of these six candidate regulators was performed as part of this study, although some were shown to affect expression of particular oppA promoters (44).
Figure 2.
Activity of a p66 promoter-signal sequence-phoA fusion reporter in the presence of candidate regulators. phoA‐ E. coli carrying the p66 promoter-signal sequence-phoA fusion, were transformed with plasmids encoding nine candidate B. burgdorferi regulatory proteins or the vector only control, and were tested for AP activity. Shown are the means ± standard deviations of three to five replicate experiments. To incorporate all data from independent experiments, activities in the presence of the candidate regulators were normalized to that of the vector only control. Statistically significant differences were observed for BBA23 (light bar, increased activity), and for BB0232, and BB0527 (black bars, decreased activity) with a P-value of less than 0.01 based on a Students’ two-tailed t-test.
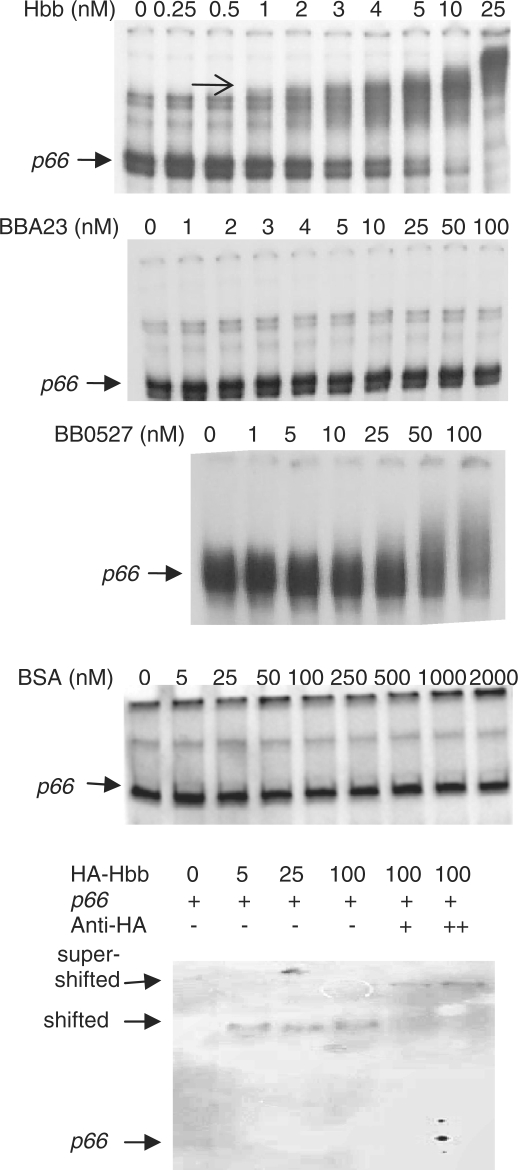
To determine whether Hbb, BB0527 and BBA23 interact directly with the p66 promoter region, we performed electrophoretic mobility shift assays (EMSAs, or gel shift assays) with purified, recombinant preparations of the three proteins identified in the reporter screen described above. Hbb bound to the p66 promoter at 1 nM, while BBA23 and showed no binding, and BB0527 only very inefficient binding, although this was reproducible (Figure 3). Therefore, within the E. coli cell, additional factors may contribute to the interaction of BB0527 and BBA23 with the p66 promoter, or their effects may be indirect. While BB0527 may interact with the p66 promoter, it appears that the interaction is of very low affinity and therefore may not be physiologically relevant. However, Hbb binding occurs at much higher affinity, and Hbb binding to the p66 promoter fragment may occur to multiple sites of different affinities, as several distinct species appear with increasing concentrations of Hbb. The shifted bands are due to Hbb-specific interaction with the DNA fragments, as ‘supershift’ assays in which antibody to the HA tag fused to the Hbb resulted in a further decrease in migration of the promoter fragment (Figure 3). The p66 promoter fragment was not shifted in the presence of BSA (Figure 3). Because Hbb does bind directly to the p66 promoter, this interaction was characterized further.
Figure 3.
Interaction of purified Hbb, BBA23 and BB0527 with the p66 promoter region. Various concentrations of purified recombinant proteins were incubated with 100 pM radiolabeled DNA for 30 min, and the reactions were run on nondenaturing 5% polyacrylamide gels. The gels were dried and exposed to film overnight at −70°C and were visualized by autoradiography. Hbb starts to shift the p66 promoter fragment at 1 nM (arrow), while BB0527 requires much higher concentrations, and BBA23 did not appear to interact with this DNA fragment. The p66 promoter fragment is homogeneous when analyzed by agarose gel electrophoresis but generally heterogeneous by acrylamide gel electrophoresis, even after gel purification.
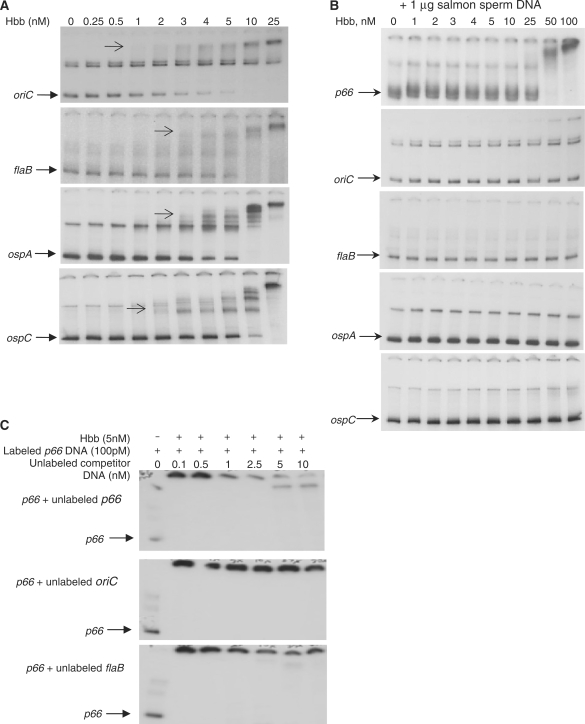
To assess the specificity of the interaction of Hbb with the p66 promoter region, we also examined the promoter regions of flaB, ospA and ospC. In addition, the origin of replication (oriC) of the linear chromosome of B. burgdorferi was tested because it is known to interact with Hbb (46). Hbb bound to all of these DNA fragments at nanomolar concentrations, although concentrations differed slightly between the different fragments (Figure 4A). In the presence of excess salmon sperm DNA, the binding by Hbb to each of the DNAs is disrupted at lower Hbb concentrations, but at high Hbb concentrations, only the p66 and oriC fragments are still bound by Hbb (Figure 4B). These results suggest that the p66 promoter and oriC fragments bind Hbb with higher affinity than do the flaB, ospA, or ospC promoter fragments. Finally, Hbb binding to the p66 promoter was competed by unlabeled p66 promoter DNA, but not by flaB or oriC DNA (Figure 4C).
Figure 4.
Specificity of interaction of purified Hbb with B. burgdorferi DNA fragments. (A) Varying concentrations of Hbb were incubated with 100 pM radiolabeled DNA fragments, and the samples fractionated on non-denaturing gels, as described for Figure 3. Arrows indicate the first shifted band apparent for each DNA fragment, all of which appear at 1–2 nM Hbb. (B) One microgram salmon sperm DNA was added to each binding reaction as a competitor. (C) Hbb was held constant, and unlabeled DNA fragments were added at the concentrations shown.
Together, our results suggest that Hbb binds to the p66 promoter with an affinity similar to that reported for the oriC fragment (46). In their work with the oriC region, Kobryn et al. (46) demonstrated specificity sufficient to identify a binding site using DNase 1, KMnO4 and hydroxyl radical footprinting. In the p66 promoter fragment used in this study, there is a site similar to that identified by Kobryn et al. and refined by Mouw and Rice (46,62) (Figure 5). A 41 bp fragment (shown in blue in Figure 5A) containing this site was bound by Hbb, as was a shorter p66 promoter fragment in which this site was truncated, but both were bound by Hbb only at >2 nM (Figure 5B and data not shown), suggesting that binding with maximal affinity occurs only when an extended p66 upstream region is present.
Figure 5.
The p66 promoter region and Hbb-binding sites. (A) The p66 upstream region is shown 5′–3′ with the −35 and −10 highlighted in red, the transcriptional start site highlighted in red (29), the putative ribosome binding site in green, the translational start site in green, and the candidate Hbb binding site tested by EMSA highlighted in blue. Mismatches to a perfect palindrome within this site are highlighted in pink, the base at the axis of symmetry is in orange. The DNA sequences bound by Hbb and their affinities are from (46,62). Their binding site studies were refined with double stranded oligonucleotides (designated oligo 1, oligo 2 and oligo 3 in the alignment, shown without flanking 10 bp fragments) designed to test the roles of symmetry and IHF consensus sequences (62). The p66 bases that are exact matches to the sequences bound by Hbb with affinities of <30 nM are indicated by black asterisks. (B) Hbb binds to the 41 bp region shown in blue in (A). The DNA fragment was formed by annealing two oligonucleotides, and tested for binding to Hbb by EMSA as described in Figures 3–5, with the exception that the samples were run on a higher percentage acrylamide gel.
Quantitative RT‐PCR analysis of hbb mRNA levels in tick midguts
To determine whether the expression of hbb reflects the expression of p66 by B. burgdorferi in tick midguts, we performed qRT‐PCR on the tick midgut samples used for the experiments shown in Figure 1. The pattern of hbb expression in infected ticks seen by qRT‐PCR generally reflects that seen in the cases of p66, flaA and ospA in that the highest absolute transcript levels are seen post-repletion, and the lowest expression occurs when the ticks are unfed. The major difference between hbb and the other genes analyzed is that the hbb transcript levels are much lower, and that transcript is detected in the ticks that were not infected with B. burgdorferi at 9 days post repletion. This is most likely an artifact of the extremely low signals of both the hbb transcripts in all samples combined with the low flaA transcript backgrounds in the non-infected ticks, so that any signal for hbb would appear elevated due to the low flaA signal in the non-infected ticks. In addition, this analysis is likely to be complicated by amplification of hu/ihf homologs of tick bacterial endosymbionts, which may replicate in response to the blood meal. However, even in the ticks infected with B. burgdorferi, the copy numbers are very low for this gene, since all of the values fell at the lower end of the standard curve. Notably, in contrast to the data presented for p66, ospA and ospC, the hbb/flaA ratio decreases in the infected fed tick and the 9 days post repletion tick samples in comparison to the unfed tick (Figure 1). This decrease is apparent even in the 9 days post repletion sample when, potentially, bacterial endosymbionts with imperfect matches in the primer hybridization sites might be contributing to the overall signal despite the competition from the B. burgdorferi RNA. This, together with the AP assay results, is consistent with a model in which Hbb participates in repression of p66 transcription by altering the DNA structure upstream of the promoter, as the fold increase in hbb mRNA is considerably less than that for p66.
DISCUSSION
Borrelia burgdorferi is intimately tied to both the tick vector and the vertebrate (usually mammalian) host. The bacteria reside in the tick midgut in apparent quiescence, then replicate and change gene and protein expression as the tick takes its blood meal. Several B. burgdorferi proteins and genes are produced or expressed very differently between the two environments. Changes in temperature, pH, cell density and other as-yet unidentified signals are thought to contribute to these changes. The paradigm for this change in gene expression is the induction of OspC in conjunction with decreased OspA expression while the tick is feeding (47). Changes in the expression of many other genes and gene products by B. burgdorferi in preparation for transit between tick and mammal have subsequently been observed by multiple investigators (6,12,35–37,47,63–73). Because digestion of the blood meal occurs intracellularly (74,75), and B. burgdorferi is extracellular, degradation of surface-exposed bacterial proteins by tick lytic enzymes is thought unlikely. qRT–PCR analysis of B. burgdorferi in tick midguts or salivary glands suggests transcriptional regulation of gene expression by B. burgdorferi during the tick life cycle (76–80). Post-transcriptional regulation, e.g. by small RNAs (81), or regulation of protein half-lives are other possible mechanisms by which B. burgdorferi could be altering its protein complement within the tick.
Transcriptional regulation of gene expression has been demonstrated for a number of B. burgdorferi genes. Primer extension was performed on p66 RNA, and a putative σ70-dependent promoter was identified (34). In accordance with this work, microarray studies of rpoN and rpoS mutants of B. burgdorferi do not indicate significant roles for these alternative sigma factors in p66 expression (6,8,35). We therefore hypothesized that σ70-dependent expression of p66 is modulated by accessory factors that result in the differences observed in fed versus unfed tick, especially since p66, ospA and the flagellar genes flaA and flaB are all thought to be σ70-dependent (29,82), but show different patterns of expression. In fact, the repression of ospA in the mammalian host is RpoS-dependent (3). These observations led us to search for potential factors in B. burgdorferi that affect p66 expression.
Of the candidate regulators identified, only Hbb (BB0232) appeared to interact with the p66 promoter in settings consistent with physiologic relevance, although additional factors may influence p66 mRNA and protein expression in B. burgdorferi during the tick-mammal infectious cycle. Hbb is a homolog of the architectural DNA-binding proteins, including the H-NS, IHF, FIS and HU-like proteins (46,53). Proteins in this family are known to play a variety of roles, including roles in DNA replication (46), promoting complex assembly by inducing severe bends in DNA (83), and transcriptional regulation by virtue of DNA bending or stabilizing sequence- and temperature-dependent secondary structures (84). HU- and IHF-like proteins, to which Hbb is more closely similar, tend to be involved in the activation of transcription due to formation of an open nucleoprotein complex, especially in circular DNA (83,85,86). Conversely, H-NS proteins tend to be involved in repression by virtue of DNA compaction (85,86). Of particular interest, H-NS in Yersinia pseudotuberculosis and Y. enterocolitica represses expression of the inv gene, which encodes the integrin ligand invasin. This repression is countered by competition for overlapping binding sites upstream of the −35 sequence by RovA, a marR family member. These results parallel those reported here, in which Hbb repressed the activity of a p66 promoter fusion to the reporter alkaline phosphatase, while BBA23, another MarR family member, increased reporter activity (87,88). It will be interesting to determine the effects of deletion of hbb (bb0232) and bba23 on p66 during the tick-mouse infectious cycle, but those experiments will require the generation of the mutants in an infectious strain background, which remains technically challenging in B. burgdorferi. It is also likely that, since B. burgdorferi appears to encode a very limited number of proteins involved in the regulation of gene expression, deletion of each gene will have pleiotropic effects. Further studies of the regulation of expression of hbb itself will also be required, as contradictory reports are in the literature. Expression of hbb was increased in both the rpoN and rpoS mutants in the earlier of two studies (6), but these results were contradicted by the findings of a different group (8), which used a different array, a different background strain for generating the mutants, and different culture conditions. Unfortunately, it remains difficult to perform global analyses of gene expression in B. burgdorferi in situ within vertebrate hosts and ticks, especially since available data suggest variations with time and tissue environment (37,47,89).
Our results are consistent with Hbb being involved in the reduction of p66 expression. Because Hbb causes bends in the DNA to which it binds, full assembly of the RNA polymerase complex may not occur adjacent to a significant bend. Upstream (5′) of the −35 site of the p66 promoter there is a candidate Hbb binding site consistent with the requirements outlined by Mouw and Rice (62) (Figure 5A). Hbb does bind to this fragment (Figure 5B), but additional work is required to assess specificity and stoichiometry. One model consistent with our data is that Hbb is expressed at low levels, and that hbb transcription is not increased at a rate sufficient to keep up with bacterial replication as the tick feeds. Hbb would then be diluted as the bacteria divide, allowing access of RNA polymerase to promoters, such as that of p66, that were previously rendered inaccessible by Hbb. In fact, although hbb expression at the mRNA level does increase in the tick after the blood meal, this increase does not keep pace with the increases in either p66 or flaA expression. The p66/flaA ratio is 0.033 in the unfed tick, increases to 2.36 in the fed tick and further increases to 7.18 in the tick 9 days post repletion. In contrast, the hbb/flaA ratio is 0.037 in the unfed tick, and decreases to 0.013 in the fed tick and remains at that level in the tick at 9 days. As the bacteria slow or stop replication, the intracellular Hbb concentration may then increase to levels to allow interaction with sites such as the p66 promoter, and resumption of repression of transcription from this promoter. In preliminary studies using anti-Hbb generated in the laboratory of Dr Scott Samuels (90), Hbb in laboratory-cultivated B. burgdorferi is present at less than 750 copies of the protein molecule per cell (data not shown), which is low considering that Hbb forms a dimer (62) and that it is thought to have roles in DNA replication (46) and transcription. Our model, however, requires validation through additional experimentation. Hbb may also serve as a type of scaffold, upon which additional factors could build to influence gene expression. It is also possible that BBA23 competes with Hbb binding, and results in increased gene expression, but all of these possibilities require future experimentation. Future studies will also be required to define the Hbb binding site in the p66 promoter. Ultimately, the most important experiment would be to mutate the gene in infectious B. burgdorferi and determine whether the bacteria are able to transcribe genes appropriately in the tick, in the mammal, and in transitions between these two hosts.
FUNDING
National Institutes of Health (grant R01 AI51407 and R01 AI059505 to J.C.); National Institutes of Health training (grant T32-AI-07422 and F31 AI52495, partial); Division of Intramural Research, National Institutes of Allergy and Infectious Diseases, National Institutes of Health (to T.G.S. and P.F.P.). Funding for open access charge: National Institutes of Health grant. We are grateful for thesupport of Anne Kane, MD and the Tufts-NEMC Center for Gastroenterology Research on Absorptive and Secretory Processes funded by a grant from National Institutes of Diabetes and Digestive and Kidney Diseases (P30DK39428).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The E. coli strain CC118 E. coli was a kind gift from Andrew Wright, Ph.D. Tufts University. We thank Carla Cugini for design and testing of primer sets for p66, ospA and ospC, Scott Samuels and Sharyl Fyffe for anti-Hbb antiserum, Scott Samuels for critical review of the manuscript, and Brendan O’Malley and David Lazinski for sharing unpublished information that helped with the construction of the HISHAB and HISHAT oligonucleotides.
REFERENCES
- 1.CDC. Lyme disease ‐ United States, 2003-2005. MMWR Morb. Mortal. Wkly Rep. 2007;56:573–576. [PubMed] [Google Scholar]
- 2.Shapiro ED, Gerber MA. Lyme disease. Clin. Infect. Dis. 2000;31:533–542. doi: 10.1086/313982. [DOI] [PubMed] [Google Scholar]
- 3.Caimano MJ, Eggers CH, Gonzalez CA, Radolf JD. Alternate sigma factor RpoS is required for the in vivo-specific repression of Borrelia burgdorferi plasmid lp54-borne ospA and lp6.6 genes. J. Bacteriol. 2005;187:7845–7852. doi: 10.1128/JB.187.22.7845-7852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caimano MJ, Eggers CH, Hazlett KR, Radolf JD. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect. Immun. 2004;72:6433–6445. doi: 10.1128/IAI.72.11.6433-6445.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elias AF, Bono JL, Carroll JA, Stewart P, Tilly K, Rosa P. Altered stationary-phase response in a Borrelia burgdorferi rpoS mutant. J. Bacteriol. 2000;182:2909–2918. doi: 10.1128/jb.182.10.2909-2918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher MA, Grimm D, Henion AK, Elias AF, Stewart PE, Rosa PA, Gherardini FC. Borrelia burgdorferi sigma54 is required for mammalian infection and vector transmission but not for tick colonization. Proc. Natl Acad. Sci. USA. 2005;102:5162–5167. doi: 10.1073/pnas.0408536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hubner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl Acad. Sci. USA. 2001;98:12724–12729. doi: 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ouyang Z, Blevins JS, Norgard MV. Transcriptional interplay among the regulators Rrp2, RpoN and RpoS in Borrelia burgdorferi. Microbiology (Reading, England) 2008;154:2641–2658. doi: 10.1099/mic.0.2008/019992-0. [DOI] [PubMed] [Google Scholar]
- 9.Smith AH, Blevins JS, Bachlani GN, Yang XF, Norgard MV. Evidence that RpoS (sigmaS) in Borrelia burgdorferi is controlled directly by RpoN (sigma54/sigmaN) J. Bacteriol. 2007;189:2139–2144. doi: 10.1128/JB.01653-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang XF, Alani SM, Norgard MV. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc. Natl Acad. Sci. USA. 2003;100:11001–11006. doi: 10.1073/pnas.1834315100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X, Goldberg MS, Popova TG, Schoeler GB, Wikel SK, Hagman KE, Norgard MV. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol. Microbiol. 2000;37:1470–1479. doi: 10.1046/j.1365-2958.2000.02104.x. [DOI] [PubMed] [Google Scholar]
- 12.Schwan TG, Piesman J. Vector interactions and molecular adaptations of lyme disease and relapsing fever spirochetes associated with transmission by ticks. Emerging Infect. Dis. 2002;8:115–121. doi: 10.3201/eid0802.010198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boylan JA, Posey JE, Gherardini FC. Borrelia oxidative stress response regulator, BosR: a distinctive Zn-dependent transcriptional activator. Proc. Natl Acad. Sci. USA. 2003;100:11684–11689. doi: 10.1073/pnas.2032956100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carroll JA, Garon CF, Schwan TG. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect. Immun. 1999;67:3181–3187. doi: 10.1128/iai.67.7.3181-3187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang FT, Nelson FK, Fikrig E. Molecular adaptation of Borrelia burgdorferi in the murine host. J. Exp. Med. 2002;196:275–280. doi: 10.1084/jem.20020770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang FT, Yan J, Mbow ML, Sviat SL, Gilmore RD, Mamula M, Fikrig E. Borrelia burgdorferi changes its surface antigenic expression in response to host immune responses. Infect. Immun. 2004;72:5759–5767. doi: 10.1128/IAI.72.10.5759-5767.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margolis N, Rosa PA. Regulation of expression of major outer surface proteins in Borrelia burgdorferi. Infect. Immun. 1993;61:2207–2210. doi: 10.1128/iai.61.5.2207-2210.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDowell JV, Sung SY, Price G, Marconi RT. Demonstration of the genetic stability and temporal expression of select members of the lyme disease spirochete OspF protein family during infection in mice. Infect. Immun. 2001;69:4831–4838. doi: 10.1128/IAI.69.8.4831-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montgomery RR, Malawista SE, Feen KJ, Bockenstedt LK. Direct demonstration of antigenic substitution of Borrelia burgdorferi ex vivo: exploration of the paradox of the early immune response to outer surface proteins A and C in Lyme disease. J. Exp. Med. 1996;183:261–269. doi: 10.1084/jem.183.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, Barbour A, Jasinskas A, Benach J, Katona L, Radolf J, et al. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 2003;71:1689–1705. doi: 10.1128/IAI.71.4.1689-1705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Posey JE, Gherardini FC. Lack of a role for iron in the Lyme disease pathogen. Science. 2000;288:1651–1653. doi: 10.1126/science.288.5471.1651. [DOI] [PubMed] [Google Scholar]
- 22.Ramamoorthy R, Philipp MT. Differential expression of Borrelia burgdorferi proteins during growth in vitro. Infect. Immun. 1998;66:5119–5124. doi: 10.1128/iai.66.11.5119-5124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramamoorthy R, Scholl-Meeker D. Borrelia burgdorferi proteins whose expression is similarly affected by culture temperature and pH. Infect. Immun. 2001;69:2739–2742. doi: 10.1128/IAI.69.4.2739-2742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwan TG, Burgdorfer W, Garon CF. Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect. Immun. 1988;56:1831–1836. doi: 10.1128/iai.56.8.1831-1836.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevenson B, Schwan TG, Rosa PA. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 1995;63:4535–4539. doi: 10.1128/iai.63.11.4535-4539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tokarz R, Anderton JM, Katona LI, Benach JL. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect. Immun. 2004;72:5419–5432. doi: 10.1128/IAI.72.9.5419-5432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brooks CS, Hefty PS, Jolliff SE, Akins DR. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 2003;71:3371–3383. doi: 10.1128/IAI.71.6.3371-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Revel AT, Talaat AM, Norgard MV. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl Acad. Sci. USA. 2002;99:1562–1567. doi: 10.1073/pnas.032667699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bunikis J, Noppa L, Bergstrom S. Molecular analysis of a 66-kDa protein associated with the outer membrane of Lyme disease Borrelia. FEMS Microbiol. Lett. 1995;131:139–145. doi: 10.1111/j.1574-6968.1995.tb07768.x. [DOI] [PubMed] [Google Scholar]
- 30.Coburn J, Chege W, Magoun L, Bodary SC, Leong JM. Characterization of a candidate Borrelia burgdorferi beta3-chain integrin ligand identified using a phage display library. Mol. Microbiol. 1999;34:926–940. doi: 10.1046/j.1365-2958.1999.01654.x. [DOI] [PubMed] [Google Scholar]
- 31.Coburn J, Cugini C. Targeted mutation of the outer membrane protein P66 disrupts attachment of the Lyme disease agent, Borrelia burgdorferi, to integrin alphavbeta3. Proc. Natl Acad. Sci. USA. 2003;100:7301–7306. doi: 10.1073/pnas.1131117100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Probert WS, Allsup KM, LeFebvre RB. Identification and characterization of a surface-exposed, 66-kilodalton protein from Borrelia burgdorferi. Infect. Immun. 1995;63:1933–1939. doi: 10.1128/iai.63.5.1933-1939.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bunikis J, Luke CJ, Bunikiene E, Bergstrom S, Barbour AG. A surface-exposed region of a novel outer membrane protein (P66) of Borrelia spp. is variable in size and sequence. J. Bacteriol. 1998;180:1618–1623. doi: 10.1128/jb.180.7.1618-1623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bunikis J, Olsen B, Westman G, Bergstroom S. Variable serum immunoglobulin responses against different Borrelia burgdorferi sensu lato species in a population at risk for and patients with Lyme disease. J. Clin. Microbiol. 1995;33:1473–1478. doi: 10.1128/jcm.33.6.1473-1478.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, Schwartz I, Radolf JD. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol. Microbiol. 2007;65:1193–1217. doi: 10.1111/j.1365-2958.2007.05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cugini C, Medrano M, Schwan TG, Coburn J. Regulation of expression of the Borrelia burgdorferi beta(3)-chain integrin ligand, P66, in ticks and in culture. Infect. Immun. 2003;71:1001–1007. doi: 10.1128/IAI.71.2.1001-1007.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwan TG, Piesman J. Temporal changes in outer surface proteins A and C of the lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J. Clin. Microbiol. 2000;38:382–388. doi: 10.1128/jcm.38.1.382-388.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Policastro PF, Schwan TG. Experimental infection of Ixodes scapularis larvae (Acari: Ixodidae) by immersion in low passage cultures of Borrelia burgdorferi. J. Med. Entomol. 2003;40:364–370. doi: 10.1603/0022-2585-40.3.364. [DOI] [PubMed] [Google Scholar]
- 39.Barbour AG, Burgdorfer W, Hayes SF, Peter O, Aeschlimann A. Isolation of a cultivatible spirochete from Ixodes ricinus ticks of Switzerland. Curr. Microbiol. 1983;8:123–126. [Google Scholar]
- 40.Preac-Mursic V, Wilske B, Schierz G. European Borrelia burgdorferi isolated from humans and ticks culture conditions and antibiotic susceptibility. Zentralbl. Bakteriol. Mikrobiol. Hyg. [A] 1986;263:112–118. doi: 10.1016/s0176-6724(86)80110-9. [DOI] [PubMed] [Google Scholar]
- 41.Munderloh UG, Liu Y, Wang M, Chen C, Kurtti TJ. Establishment, maintenance and description of cell lines from the tick Ixodes scapularis. J. Parasitol. 1994;80:533–543. [PubMed] [Google Scholar]
- 42.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 43.Towbin H, Staeheli T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl Acad. Sci. USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Medrano MS, Ding Y, Wang XG, Lu P, Coburn J, Hu LT. Regulators of expression of the oligopeptide permease A proteins of Borrelia burgdorferi. J. Bacteriol. 2007;189:2653–2659. doi: 10.1128/JB.01760-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffman CS, Wright A. Fusions of secreted proteins to alkaline phosphatase: an approach for studying protein secretion. Proc. Natl Acad. Sci. USA. 1985;82:5107–5111. doi: 10.1073/pnas.82.15.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobryn K, Naigamwalla DZ, Chaconas G. Site-specific DNA binding and bending by the Borrelia burgdorferi Hbb protein. Mol. Microbiol. 2000;37:145–155. doi: 10.1046/j.1365-2958.2000.01981.x. [DOI] [PubMed] [Google Scholar]
- 47.Schwan TG, Piesman J, Golde WT, Dolan MC, Rosa PA. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl Acad. Sci. USA. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blevins JS, Xu H, He M, Norgard MV, Reitzer L, Yang XF. Rrp2, a {sigma}54-dependent transcriptional activator of Borrelia burgdorferi, activates rpoS in an enhancer-independent manner. J. Bacteriol. 2009;191:2902–2905. doi: 10.1128/JB.01721-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katona LI, Tokarz R, Kuhlow CJ, Benach J, Benach JL. The fur homologue in Borrelia burgdorferi. J. Bacteriol. 2004;186:6443–6456. doi: 10.1128/JB.186.19.6443-6456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hyde JA, Seshu J, Skare JT. Transcriptional profiling of Borrelia burgdorferi containing a unique bosR allele identifies a putative oxidative stress regulon. Microbiology (Reading, England) 2006;152:2599–2609. doi: 10.1099/mic.0.28996-0. [DOI] [PubMed] [Google Scholar]
- 52.Seshu J, Boylan JA, Hyde JA, Swingle KL, Gherardini FC, Skare JT. A conservative amino acid change alters the function of BosR, the redox regulator of Borrelia burgdorferi. Mol. Microbiol. 2004;54:1352–1363. doi: 10.1111/j.1365-2958.2004.04352.x. [DOI] [PubMed] [Google Scholar]
- 53.Tilly K, Fuhrman J, Campbell J, Samuels DS. Isolation of Borrelia burgdorferi genes encoding homologues of DNA-binding protein HU and ribosomal protein S20. Microbiology (Reading, England) 1996;142(Pt 9):2471–2479. doi: 10.1099/00221287-142-9-2471. [DOI] [PubMed] [Google Scholar]
- 54.Babb K, Bykowski T, Riley SP, Miller MC, Demoll E, Stevenson B. Borrelia burgdorferi EbfC, a novel, chromosomally encoded protein, binds specific DNA sequences adjacent to erp loci on the spirochete's; resident cp32 prophages. J. Bacteriol. 2006;188:4331–4339. doi: 10.1128/JB.00005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seshu J, Boylan JA, Gherardini FC, Skare JT. Dissolved oxygen levels alter gene expression and antigen profiles in Borrelia burgdorferi. Infect. Immun. 2004;72:1580–1586. doi: 10.1128/IAI.72.3.1580-1586.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boylan SA, Redfield AR, Brody MS, Price CW. Stress-induced activation of the sigma B transcription factor of Bacillus subtilis. J. Bacteriol. 1993;175:7931–7937. doi: 10.1128/jb.175.24.7931-7937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wood GE, Friedman RL. The Bvg accessory factor (Baf) enhances pertussis toxin expression in Escherichia coli and is essential for Bordetella pertussis viability. FEMS Microbiol. Lett. 2000;193:25–30. doi: 10.1111/j.1574-6968.2000.tb09397.x. [DOI] [PubMed] [Google Scholar]
- 58.DeShazer D, Wood GE, Friedman RL. Identification of a Bordetella pertussis regulatory factor required for transcription of the pertussis toxin operon in Escherichia coli. J. Bacteriol. 1995;177:3801–3807. doi: 10.1128/jb.177.13.3801-3807.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, et al. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 60.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 61.Alekshun MN, Kim YS, Levy SB. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol. Microbiol. 2000;35:1394–1404. doi: 10.1046/j.1365-2958.2000.01802.x. [DOI] [PubMed] [Google Scholar]
- 62.Mouw KW, Rice PA. Shaping the Borrelia burgdorferi genome: crystal structure and binding properties of the DNA-bending protein Hbb. Mol. Microbiol. 2007;63:1319–1330. doi: 10.1111/j.1365-2958.2007.05586.x. [DOI] [PubMed] [Google Scholar]
- 63.Bykowski T, Woodman ME, Cooley AE, Brissette CA, Brade V, Wallich R, Kraiczy P, Stevenson B. Coordinated expression of Borrelia burgdorferi complement regulator-acquiring surface proteins during the Lyme disease spirochete's; mammal-tick infection cycle. Infect. Immun. 2007;75:4227–4236. doi: 10.1128/IAI.00604-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grimm D, Eggers CH, Caimano MJ, Tilly K, Stewart PE, Elias AF, Radolf JD, Rosa PA. Experimental assessment of the roles of linear plasmids lp25 and lp28–1 of Borrelia burgdorferi throughout the infectious cycle. Infect. Immun. 2004;72:5938–5946. doi: 10.1128/IAI.72.10.5938-5946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grimm D, Tilly K, Byram R, Stewart PE, Krum JG, Bueschel DM, Schwan TG, Policastro PF, Elias AF, Rosa PA. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc. Natl Acad. Sci. USA. 2004;101:3142–3147. doi: 10.1073/pnas.0306845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hodzic E, Feng S, Freet KJ, Borjesson DL, Barthold SW. Borrelia burgdorferi population kinetics and selected gene expression at the host-vector interface. Infect. Immun. 2002;70:3382–3388. doi: 10.1128/IAI.70.7.3382-3388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jewett MW, Byram R, Bestor A, Tilly K, Lawrence K, Burtnick MN, Gherardini F, Rosa PA. Genetic basis for retention of a critical virulence plasmid of Borrelia burgdorferi. Mol. Microbiol. 2007;66:975–990. doi: 10.1111/j.1365-2958.2007.05969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jewett MW, Lawrence K, Bestor AC, Tilly K, Grimm D, Shaw P, VanRaden M, Gherardini F, Rosa PA. The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol. Microbiol. 2007;64:1358–1374. doi: 10.1111/j.1365-2958.2007.05746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miller JC, von Lackum K, Babb K, McAlister JD, Stevenson B. Temporal analysis of Borrelia burgdorferi Erp protein expression throughout the mammal-tick infectious cycle. Infect. Immun. 2003;71:6943–6952. doi: 10.1128/IAI.71.12.6943-6952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramamoorthi N, Narasimhan S, Pal U, Bao F, Yang XF, Fish D, Anguita J, Norgard MV, Kantor FS, Anderson JF, et al. The Lyme disease agent exploits a tick protein to infect the mammalian host. Nature. 2005;436:573–577. doi: 10.1038/nature03812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.von Lackum K, Miller JC, Bykowski T, Riley SP, Woodman ME, Brade V, Kraiczy P, Stevenson B, Wallich R. Borrelia burgdorferi regulates expression of complement regulator-acquiring surface protein 1 during the mammal-tick infection cycle. Infect. Immun. 2005;73:7398–7405. doi: 10.1128/IAI.73.11.7398-7405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.von Lackum K, Ollison KM, Bykowski T, Nowalk AJ, Hughes JL, Carroll JA, Zuckert WR, Stevenson B. Regulated synthesis of the Borrelia burgdorferi inner-membrane lipoprotein IpLA7 (P22, P22-A) during the Lyme disease spirochaete's; mammal-tick infectious cycle. Microbiology (Reading, England) 2007;153:1361–1371. doi: 10.1099/mic.0.2006/003350-0. [DOI] [PubMed] [Google Scholar]
- 73.Yang XF, Pal U, Alani SM, Fikrig E, Norgard MV. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J. Exp. Med. 2004;199:641–648. doi: 10.1084/jem.20031960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ribeiro JM. The midgut hemolysin of Ixodes dammini (Acari:Ixodidae) J. Parasitol. 1988;74:532–537. [PubMed] [Google Scholar]
- 75.Sojka D, Hajdusek O, Dvorak J, Sajid M, Franta Z, Schneider EL, Craik CS, Vancova M, Buresova V, Bogyo M, et al. IrAE: an asparaginyl endopeptidase (legumain) in the gut of the hard tick Ixodes ricinus. Int. J. Parasitol. 2007;37:713–724. doi: 10.1016/j.ijpara.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fingerle V, Goettner G, Gern L, Wilske B, Schulte-Spechtel U. Complementation of a Borrelia afzelii OspC mutant highlights the crucial role of OspC for dissemination of Borrelia afzelii in Ixodes ricinus. Int. J. Med. Microbiol. 2007;297:97–107. doi: 10.1016/j.ijmm.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 77.Fingerle V, Rauser S, Hammer B, Kahl O, Heimerl C, Schulte-Spechtel U, Gern L, Wilske B. Dynamics of dissemination and outer surface protein expression of different European Borrelia burgdorferi sensu lato strains in artificially infected Ixodes ricinus nymphs. J. Clin. Microbiol. 2002;40:1456–1463. doi: 10.1128/JCM.40.4.1456-1463.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gilmore RD, Jr, Piesman J. Inhibition of Borrelia burgdorferi migration from the midgut to the salivary glands following feeding by ticks on OspC-immunized mice. Infect. Immun. 2000;68:411–414. doi: 10.1128/iai.68.1.411-414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Piesman J, Schneider BS, Zeidner NS. Use of quantitative PCR to measure density of Borrelia burgdorferi in the midgut and salivary glands of feeding tick vectors. J. Clin. Microbiol. 2001;39:4145–4148. doi: 10.1128/JCM.39.11.4145-4148.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Piesman J, Zeidner NS, Schneider BS. Dynamic changes in Borrelia burgdorferi populations in Ixodes scapularis (Acari: Ixodidae) during transmission: studies at the mRNA level. Vector Borne Zoonotic Dis. 2003;3:125–132. doi: 10.1089/153036603768395825. [DOI] [PubMed] [Google Scholar]
- 81.Lybecker MC, Samuels DS. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol. Microbiol. 2007;64:1075–1089. doi: 10.1111/j.1365-2958.2007.05716.x. [DOI] [PubMed] [Google Scholar]
- 82.Ge Y, Old IG, Girons IS, Charon NW. The flgK motility operon of Borrelia burgdorferi is initiated by a sigma 70-like promoter. Microbiology (Reading, England) 1997;143(Pt 5):1681–1690. doi: 10.1099/00221287-143-5-1681. [DOI] [PubMed] [Google Scholar]
- 83.Swinger KK, Lemberg KM, Zhang Y, Rice PA. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003;22:3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Falconi M, Colonna B, Prosseda G, Micheli G, Gualerzi CO. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 1998;17:7033–7043. doi: 10.1093/emboj/17.23.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dorman CJ, Deighan P. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 2003;13:179–184. doi: 10.1016/s0959-437x(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 86.Dorman CJ, Hinton JC, Free A. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 1999;7:124–128. doi: 10.1016/s0966-842x(99)01455-9. [DOI] [PubMed] [Google Scholar]
- 87.Ellison DW, Miller VL. H-NS represses inv transcription in Yersinia enterocolitica through competition with RovA and interaction with YmoA. J. Bacteriol. 2006;188:5101–5112. doi: 10.1128/JB.00862-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heroven AK, Nagel G, Tran HJ, Parr S, Dersch P. RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol. Microbiol. 2004;53:871–888. doi: 10.1111/j.1365-2958.2004.04162.x. [DOI] [PubMed] [Google Scholar]
- 89.Hodzic E, Feng S, Freet KJ, Barthold SW. Borrelia burgdorferi population dynamics and prototype gene expression during infection of immunocompetent and immunodeficient mice. Infect. Immun. 2003;71:5042–5055. doi: 10.1128/IAI.71.9.5042-5055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Knight SW, Kimmel BJ, Eggers CH, Samuels DS. Disruption of the Borrelia burgdorferi gac gene, encoding the naturally synthesized GyrA C-terminal domain. J. Bacteriol. 2000;182:2048–2051. doi: 10.1128/jb.182.7.2048-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bono JL, Elias AF, Kupko JJ, 3rd, Stevenson B, Tilly K, Rosa P. Efficient targeted mutagenesis in Borrelia burgdorferi. J. Bacteriol. 2000;182:2445–2452. doi: 10.1128/jb.182.9.2445-2452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang XF, Goldberg MS, He M, Xu H, Blevins JS, Norgard MV. Differential Expression of a Putative CarD-Like Transcriptional Regulator, LtpA, in Borrelia burgdorferi. Infect. Immun. 2008;76:4439–4444. doi: 10.1128/IAI.00740-08. [DOI] [PMC free article] [PubMed] [Google Scholar]