Abstract
In many eukaryotic mRNAs one or more short ‘upstream’ open reading frames, uORFs, precede the initiator of the main coding sequence. Upstream ORFs are functionally diverse as illustrated by their variety of features in polyamine pathway biosynthetic mRNAs. Their propensity to act as sensors for regulatory circuits and to amplify the signals likely explains their occurrence in most polyamine pathway mRNAs. The uORF-mediated polyamine responsive autoregulatory circuits found in polyamine pathway mRNAs exemplify the translationally regulated dynamic interface between components of the proteome and metabolism.
INTRODUCTION
Eukaryotic mRNAs are considered to be overwhelmingly monocistronic even though many, an under-appreciated proportion (1), have a short upstream coding sequence (uORF) 5′ of the main coding sequence (main ORF). With the predominant mode of eukaryotic translation initiation stemming from 5′ cap dependent scanning, sensing of the uORF initiation codons is an important issue since reinitiation following termination occurs in only special circumstances (2). The identity of specific nucleotides flanking translation initiation codons, the Kozak consensus, influence the efficiency of initiation (3). Setting the relative efficiency of initiation at a uORF, when present, and the main coding sequence has a big influence on the efficiency of expression. With a poor initiation context for a uORF, a substantial proportion of the scanning complexes will continue past it (‘leaky scanning’), and initiate at the beginning of the main ORF, assuming it has an efficient initiating context. In a hypothetic series with increasingly greater efficiency of uORF initiation, there is a disproportionate repressive effect on initiation at the main ORF. This can be illustrated with a graph where the efficiency of initiation at the uORF and main ORF are arbitrarily set to be the same (Figure 1A and B). For uORF initiation, utilization rises linearly to 100% with increasing efficiency whereas for the downstream main ORF initiation utilization rises and falls in a parabolic curve with a maximal utilization of 25%. For example, as overall efficiency increases from 20 to 80%, utilization of the uORF initiator increases from 20 to 80% but utilization of the main ORF initiator stays the same at 16%, a 4-fold relative repression of main ORF initiation. When initiation of uORF translation leads to a physical barrier, for instance ribosome stalling, repression of main ORF initiation is, of course, much greater. Particular nascent peptide sequences, while still within the ribosome, can lead to ribosome stalling. Upstream ORFs which exhibit such sequence dependent stalling are in contrast to those uORFs whose coding sequence identity is immaterial—the degree of influence of this category is wholly dependent on the efficiency of their translation initiation. Occasionally, uORFs have regulatory functions. Either, or both uORF initiation and stalling, efficiencies may be affected by alteration in the concentration of small molecules, such as an amino acid or polyamines, and so a subset of uORFs serve a regulatory role. Genes encoding polyamine biosynthetic pathway components are a rich source of regulatory uORFs and are reviewed here. Diverse features including non-AUG start codons, and multiple uORFs, are also found elsewhere, but are especially striking in polyamine pathway genes.
Figure 1.
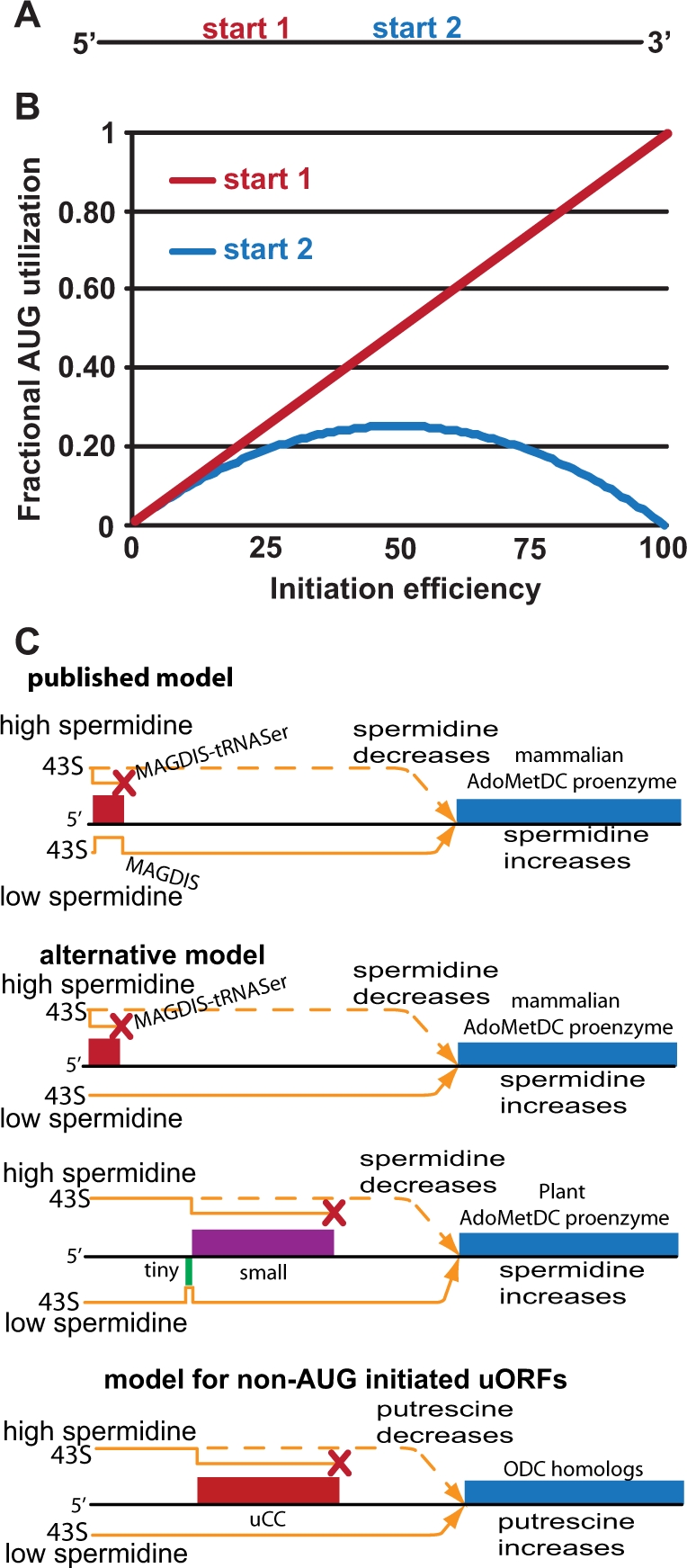
Schematic models for the effect of upstream initiation on initiation downstream. (A) Schematic representation of mRNA with an upstream and a downstream initiation sites. (B) Relationship between increasing initiation efficiency and fractional utilization of each initiation codon. (C) Schematic models for the role of uORFs in regulating the expression of various mRNAs encoding proteins in the polyamine biosynthetic pathway.
Polyamines are small organic polycations. Their linearly distributed positive charges facilitate binding to nucleic acids but polyamines are involved in many other interactions and functions. Among these are the essential post-translational modification, hypusination, of translation elongation factor “e1F5A” (4) and inward rectification of potassium channels (5). In bovine lymphocytes and rat liver, most polyamine content was found to be bound to RNA (6). The requirement of polyamines for cell proliferation and especially their elevated levels in cancerous cells, is a particular focus of interest (7).
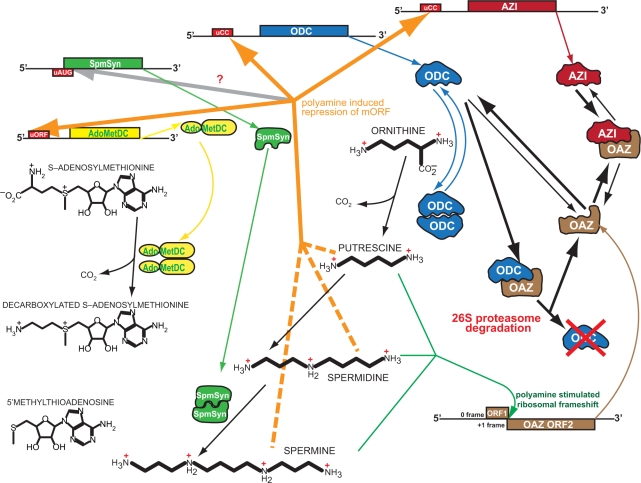
The initial step in polyamine biosynthesis is the production of putrescine from ornithine by the highly regulated enzyme ornithine decarboxylase (8). ODC protein level is regulated by an inhibitory protein termed antizyme, and antizyme itself is regulated by an ODC homolog termed antizyme inhibitor. Spermidine is formed from putrescine and spermine from spermidine by the symmetrical addition of aminopropyl groups transferred from decarboxylated S-adenosylmethionine (dcAdoMet) by spermidine synthase and spermine synthase, respectively. The dcAdoMet is produced from AdoMet by the activity of AdoMet decarboxylase (AdoMetDC) (Figure 2). Both spermidine and spermine can be acetylated by spermidine/spermine acetyltransferase (SSAT) leading to eventual catabolism and/or export from the cell. We discuss here the prominent role of uORFs in polyamine-responsive regulation of the polyamine pathway genes and reveal the presence of previously unsuspected uORFs in the mRNAs of spermine synthase and SSAT mRNAs. That polyamines differentially affect translation of eukaryotic mRNAs has been shown in many studies (9). Given the importance of polyamine concentrations, it is not unexpected that the synthesis of polyamine pathway components is exceptionally responsive to polyamine levels.
Figure 2.
Cartoon representation of the biosynthesis of polyamines in vertebrate cells and the accompanying translational regulation through upstream open reading frames. The mRNA structure for the catabolic enzyme SSAT which features in vertebrates an uORF that overlaps the main coding sequence and in many cases a discrete uORF further 5′, is not shown. ODC, ornithine decarboxylase; OAZ, ornithine decarboxylase antizyme; AZI, antizyme inhibitor; AdoMetDC, S-adenosylmethionine decarboxylase; SpmSym, spermine synthase.
AdoMetDC mRNA: Leaky scanning and ribosome stalling, one uORF in mammals, two in plants
S-Adenosylmethionine decarboxylase (AdoMetDC) catalyzes a rate limiting step in the synthesis of spermidine and spermine by committing S-adenosylmethionine to a role in polyamine synthesis (10). Early evidence for translational regulation of mammalian AdoMetDC suggested that this is due to an uORF encoding the hexapeptide sequence MAGDIS that starts only 14 nucleotides downstream of the 5′ cap (11). The uORF is so small and close to the 5′ cap that ribosomes terminating translation of the ORF would occlude its start codon thereby preventing loading of the 43S preinitiation complex of ribosome small subunit and initiation factors onto the mRNA. Interestingly, ribosomes translating the uORF stall just before termination, in response to specific features of the nascent MAGDIS peptide sequence, and so have such an occluding effect. This in turn prevents ribosome access to the downstream AdoMetDC initiation codon as there is no internal ribosome entry site or shunting and its initiation is 5′ cap and scanning dependent. There is evidence for polyamine dependent regulation of the length of the ribosome stalling at the end of the uORF presumably due to polyamine effects on the nascent peptide-ribosome interaction (12); perhaps involving the internal wall of the ribosomal peptide exit tunnel. However, polyamines also influence ribosome pausing near the termination codon on non-inhibitory variants of the uORF peptide (11). Small molecule regulation via effects on termination stalling, are also known in the Escherichia coli tryptophanase operon—in this case by tryptophan itself (13). Counterpart arginine regulated ribosome stalling in decoding the N. crassa arg-2 uORF is not termination linked (14) and at low arginine concentrations most ribosomes ignore the uORF due to leaky scanning (15). With AdoMetDC mRNA it has been claimed that there is no polyamine regulation of uORF translation initiation (11). However, in our view the data presented do not prove this point, and an additional model, is presented in Figure 1C. In this model the uORF is not translated by a proportion of the ribosomes, which then scan downstream and initiate at the AdoMetDC ORF. The proportion of ribosomes that do not initiate at the uORF initiation codon diminishes with elevating spermidine concentration. Since the start codon of the uORF is so close to the 5′ cap it is possible that polyamines influence sensing of the uORF initiation codon by the loading 43S preinitiation complex of small ribosomal subunit and initiation factors. Thus when polyamine levels are low, the 43S preinitiation complex ignores the uORF and initiates at the AdoMetDC ORF causing synthesis of AdoMetDC and increasing polyamine levels. When polyamine levels rise, the 43S preinitiation complex recognizes the uORF AUG more efficiently, and the ribosome stalls before termination of the uORF, blocking access to the downstream AdoMetDC ORF so the short-lived AdoMetDC activity is rapidly decreased and polyamine levels fall.
The plant AdoMetDC mRNA is also translationally regulated in response to polyamines. There are two conserved, overlapping uORFs in the mRNA 5′ leader: the 5′ ‘tiny’ uORF and the 3′ ‘small uORF’ and in most terrestrial plant species, the two uORFs overlap by just one nucleotide (16). The tiny uORF encodes 2–3 amino acids and the small uORF encodes 48–55 amino acids. Overexpression of the AdoMetDC mRNA in transgenic plants did not lead to any change in AdoMetDC activity or polyamine levels, probably due to a polyamine responsive homeostatic mechanism that involves the tiny uORF/small uORF pair (17). However, abrogation of the uORF-mediated translational regulation of the overexpressed AdoMetDC mRNA led to disrupted polyamine homeostasis and abnormal plant growth and development (17,18). The small uORF is sequence-dependent and the sequence is well conserved in land plants. Its function is to inhibit downstream translation (17,19). In contrast, the tiny uORF is not inhibitory and its function appears to occlude the small uORF AUG initiation codon. If the tiny uORF is recognized by the scanning 43S preinitiation complex, then the inhibitory small uORF is not translated and the ribosome will efficiently reinitiate translation at the downstream AdoMetDC proenzyme ORF. A model for polyamine-responsive translational regulation of the plant AdoMetDC mRNA stipulates that under conditions of low polyamine concentration, the scanning 43S preinitiation complex recognizes the tiny uORF AUG and after translation termination, reinitiates efficiently at the downstream AdoMetDC ORF, thereby increasing AdoMetDC activity and polyamine levels rise. When polyamine levels are excessive, the scanning 43S preinitiation complex ignores the weaker tiny uORF AUG and recognizes the stronger small uORF AUG, resulting in translation of the small uORF and sequence-dependent translational repression so that the downstream AdoMetDC ORF is no longer translated, and polyamine levels fall (Figure 1C). Upstream ORFs are found in the 5′ leader sequences of AdoMetDC mRNAs from diverse organisms in the metazoa, Pezizomycotina fungi, land plants, green and red algae and diatoms, apparently emerging on at least three separate occasions. The tiny uORF seems to have arisen within the chlorophytan algae and is present in Chlamydomonas and Volvox but absent in Chlorella and prasinophyte algae. Within the green algae, the tiny uORF is longer and overlaps to a greater extent with the small uORF than in the land plants (Supplementary Figures S1 and S2). It is notable that the amino acid sequence of the uORF always contains a penultimate proline residue at the C-terminus except for the vertebrate and sea squirt AdoMetDC mRNAs.
Antizyme mRNA: Ribosomal frameshifting from the uORF to the main ORF
The main posttranslational regulation of ODC in eukaryotes is mediated by the protein antizyme (20,21). Antizyme binds to ODC, resulting in disruption of the ODC homodimer and sequestration of ODC monomers. Antizyme presents the ODC monomer to the 26S proteasome for degradation without the involvement of ubiquitin, and the antizyme monomer is recycled (20). Separately, antizyme can also inhibit the cellular polyamine transporter (22,23). The translational regulation of antizyme itself can be considered a special case of regulation through a uORF. In almost all cases antizyme is encoded by two partially overlapping ORFs. Although the biochemical properties of antizyme described above are encoded by the downstream ORF, that coding region can only be translated after initiation at the upstream ORF, orf1. Protein sequencing and mutagenesis studies revealed that a proportion of ribosomes that initiate at the start of orf1 switch to the +1 reading frame at its last codon and proceed to decode orf2 and synthesize functional antizyme (24). This ribosomal frameshift is modulated by the level of free polyamines in the cell. Elevated levels of polyamines increase the proportion of ribosomes that switch to the orf2 frame and thereby synthesize antizyme, whereas reduced levels decrease the proportion resulting in less synthesis of the negative regulator of polyamine synthesis and uptake (21).
Antizyme inhibitor and other ODC homolog mRNAs: uORFs rich in non-aug initiators
Antizyme is negatively regulated by a protein called antizyme inhibitor (AZI). Antizyme inhibitor is a homolog of ODC that has lost the decarboxylation activity. Binding between AZI and antizyme is even stronger than that between ODC and antizyme. As a result AZI tends to sequester the intracellular pool of antizyme which leads to more free ODC protein, higher ODC activity and higher levels of polyamines (Figure 2). Mammals have two paralogs of AZI which have diverged from ODC in two independent lineages. The more abundant paralog, also known as AZI1, is present in all vertebrates and appears to have diverged from ODC in early vertebrate evolution. The second mammalian paralog, also known as antizyme inhibitor 2, AZI2 or ODCp, emerged just prior to mammalian radiation (I.P.I., unpublished results).
Recent findings suggest that ODC homologs in many species are, or might be, regulated by uORFs with unusual properties (25). Bioinformatic analysis identified upstream conserved coding regions (uCCs) in the 5′ leaders of ODC homologs in animals, the Pezizomycota subphylum of Ascomycota (e.g. N. crassa, A. nidulans, etc.), Basidiomycota (e.g. in mushrooms) and in Zygomycota. There is strong circumstantial evidence that in each of those four groups the uCCs emerged independently of the others. In the different groups the uCC varies in length from 100 codons in Pezizomycota to as few as 22 codons in Zygomycota. Although there is little amino acid conservation between uCCs from different phyla almost all examples have two adjacent proline residues near the C-terminus. In animals belonging to six phyla the uCC ends with another proline followed by a serine—a dipeptide sequence identical to the C-terminus of the ‘short’ uORF regulating the expression of AdoMetDC in plants. The presence of the two adjacent nearly universally conserved prolines is significant because of the known effects of this amino acid on translation termination when encoded by the penultimate or last sense codon of an ORF (25). Within each of the four orthologous uCC groups there are additional amino acid positions, largely concentrated near the C-terminus, that are also well conserved and therefore could be functionally significant.
The most unusual and intriguing feature of the uCC in the 5′ mRNA leaders of ODC homologs, in all but a small minority of cases, is that they appear to be translated after initiation at non-AUG codons. Only in AZI1 mRNA has the initiation codon been verified experimentally (26). In that case translation is initiated at an absolutely conserved AUU codon that is present in a ‘Kozak consensus’. In many of the other uCCs a highly conserved in-frame non-AUG codon in a good ‘Kozak consensus’ can be identified as a putative initiation codon. In most cases this appears to be an AUU codon just like in AZI1 mRNA, but putative UUG and ACG initiation codons could also be identified. It is salient that polyamines also influence initiation from the inefficient GUG initiation codon of the cra mRNA in E. coli (27).
The uCC of AZI1 mRNA represses expression of the main ORF. This repression is enhanced in the presence of spermidine and is dependent on the wild-type amino acid sequence near the C-terminus. Putting the last 10 codons of the uCC out-of-frame leads to complete loss of the polyamine dependant repression. A different set of experiments show that translation initiated at the AUU codon of the uCC is preferentially enhanced in the presence of spermidine relative to a mutation that changes the AUU to a standard AUG initiation codon. These results suggest that both the non-AUG initiation of the uCC and the amino acid sequence near the C-terminus, the sequence that includes the adjacent prolines and ends with proline-serine residues, are key components of the polyamine dependant regulation of the system.
Since non-AUG codons serve as inefficient initiation sites, most scanning ribosomes would skip translation of the uCC and instead initiate at the AUG of the main ORF. Spermidine appears to preferentially stimulate initiation of the AUU codon of mammalian AZI1 uCC compared to an AUG codon. Under increasing spermidine conditions translation of the uCC would disproportionally increase at the expense of main ORF translation (Figure 1C). If the C-terminally conserved portion of the uCC can induce ribosome stalling the polyamine repression of the main ORF would be further multiplied.
Although so far only the role of the uCC of AZI1 mRNA in regulation has been investigated directly there is reason to believe that the uCC in Pezizomycota ODC plays a similar role. Experiments with the 5′ leader of ODC mRNA in N. crassa showed that a region that contains the uCC causes close to 30-fold repression of expression of ODC protein (28). Approximately 3-fold of this could be directly attributed to translational repression of the main coding sequence, and this translational repression is partially relieved by polyamine depletion. The authors were unaware of the presence of a uCC and its role was not interrogated directly.
In addition to a uCC, many 5′ leaders of ODC homologs have one or more conventional uORFs. For example vertebrate orthologs of AZI1 mRNA have two to three such uORFs the position of which relative to the uCC, are highly conserved suggesting they probably modulate its activity in some way. Experimental results so far do not show what the effect of these uORFs is on the expression of the main ORF.
Intriguingly, among the various homologs of ODC in vertebrates only the orthologs of AZI1 have a non-AUG initiated uCC. The orthologs of ODC and AZI2 have an AUG initiated version of the uCC and a uCC is completely absent from the orthologs of ODC in mammals and birds. Induction and repression of AZI1 protein would have the same effect on polyamine levels as induction and repression of ODC itself. Induction of AZI1 would increase the levels of polyamines by sequestering antizyme and repression would lead to reduction of polyamines by leaving a larger pool of antizyme. Like the ODC homologs from most other vertebrates and animals, mammalian ODC mRNA has a long 5′ UTR. Multiple reports have demonstrated that it represses translation of the main ORF and some suggest that at least part of this 5′ UTR-dependant repression is enhanced in the presence of polyamines. Although mammalian ODC mRNA 5′ leaders lack a uCC sequence, all contain a standard relatively short uORF. Most reports suggest that this uORF represses translation of the main ORF in vitro and in vivo (29,30) though one report contradicts this conclusion (31). Ironically in the latter case the authors ‘eliminated’ the uORF by changing its AUG initiation codon to an UUG codon, a codon we now believe initiates endogenous uCC repressive translation in several ODC homologs.
Predicted overlapping uORFs in SpmSyn and SSAT mRNAs
Human spermine synthase (SpmSyn) is a fusion of an N-terminal domain whose structure is highly similar to a bacterial AdoMetDC monomer and a downstream spermidine synthase-like domain. The human SpmSyn N-terminal AdoMetDC-like domain does not retain any sequence similarity to the bacterial AdoMetDC and has no corresponding AdoMetDC activity. However, this domain is essential for dimer formation and enzymatic activity of human SpmSyn (32). We have detected in all vertebrate SpmSyn mRNAs an uORF that overlaps with the 5′-end of the main SpmSyn ORF (Supplementary Figure S3). In the human and other mammalian SpdSyn mRNA 5′ leaders, the overlap between the uORF and main ORF is 70 nt. If the uORF were to be recognized and translated by the scanning 43S preinitiation complex and if the post-termination ribosome were able to reinitiate translation at an AUG only 33 nt downstream of the uORF termination codon, then the N-terminally truncated SpmSyn would be enzymatically inactive. It is also unlikely that there would be backward ribosomal scanning after reinitiation at the main ORF downstream internal AUG over such a large distance to the authentic SpmSyn initiation codon. The SpmSyn mRNA uORF therefore constitutes yet another uORF configuration for possible translational control of a polyamine biosynthetic enzyme. If the SpmSyn uORF is a polyamine sensor, the effector is most probably spermine.
The presence of uORFs in polyamine pathway mRNAs is not confined to those encoding biosynthetic enzymes and their regulators but also extends to a catabolic enzyme. Spermidine/spermine N1-acetyltransferase is a key enzyme for regulation of intracellular polyamine levels (33). It acetylates the aminopropyl ends of spermine and spermidine thereby altering their charges and biochemical function. The acetylpolyamines are then either oxidized by polyamine oxidase or excreted from the cell. We have noticed that the SSAT mRNAs from vertebrate species contain an uORF that overlaps the SSAT main ORF (Supplementary Figure S4). In addition, most vertebrate SSAT mRNAs contain a discrete uORF (uORF1) upstream of the overlapping uORF (uORF2). It would seem that the vertebrate SSAT mRNA is also a potential example of an uORF-mediated translational regulation mechanism. With the human SSAT mRNA, uORF1 encodes 4 amino acids and is separated from the downstream uORF2 by 28 nt. The uORF2 encodes 44 amino acids, initiates 74 nt upstream of the SSAT main ORF and overlaps it by 16 nucleotides. All the vertebrate overlapping uORFs are in the +1 reading frame relative to the SSAT main ORF.
Perspective
Notwithstanding occurrences of internal ribosome entry and shunting, the hallmark feature of eukaryotic initiation is dependence on 5′cap-mediated scanning. Consequently, the great majority of eukaryotic mRNAs are generally considered to be monocistronic even though leaky scanning sometimes results in utilization of a secondary start site for the main coding sequence. However, to an increasing extent, knowledge of utilized uORFs is modifying the generality of this perception and the cases presented here add to this. Parallels with bacterial polycistronic mRNAs are becoming interesting, e.g. polarity in bacteria affecting the levels of distal gene expression versus stalled ribosomes blocking scanning dependent main ORF expression, even though the mechanisms are different. The polyamine pathway seems to have an inordinate fondness for uORF-mediated mechanisms of polyamine responsive regulation. Extensive use of ribosomal recognition of uORFs for relaying information about polyamine levels may indicate that non-optimal polyamine levels adversely affect translation initiation in general. The uORF mechanisms employed in polyamine homeostasis are a highly sensitive way of detecting changes to general ribosome behavior before they adversely affect translation of the global cellular pool of mRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health (grant R01 GM079523); Science Foundation Ireland (to J.F.A.); Institute Development Fellowship (BB/E024467/1); from the Biotechnology and Biological Sciences Research Council, UK (to A.J.M.). Funding for open access charge: Science Foundation Ireland.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Gary Loughran for his input to this work.
REFERENCES
- 1.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powell ML, Napthine S, Jackson RJ, Brierley I, Brown TD. Characterization of the termination-reinitiation strategy employed in the expression of influenza B virus BM2 protein. RNA. 2008;14:2394–2406. doi: 10.1261/rna.1231008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol. Cell. Biol. 1989;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promoted translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Levic S, Gratton MA, Doyle KJ, Yamoah EN, Pegg AE. Spermine synthase deficiency leads to deafness and a profound sensitivity to alpha-difluoromethylornithine. J. Biol. Chem. 2009;284:930–937. doi: 10.1074/jbc.M807758200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe S, Kusama-Eguchi K, Kobayashi H, Igarashi K. Estimation of polyamine binding to macromolecules and ATP in bovine lymphocytes and rat liver. J. Biol. Chem. 1991;266:20803–20809. [PubMed] [Google Scholar]
- 7.Sinoneau AR, Gerner EW, Nagle R, Ziogas A, Fujikawa-Brooks S, Yerushalmi H, Ahlering TE, Lieberman R, McLaren CE, Anton-Culver H, et al. The effect of difluoromethylornithine on decreasing prostate size and polyamines in men: results of a year-long phase IIb randomized placebo-controlled chemoprevention trial. Cancer Epidemiol. Biomarkers Prev. 2008;17:292–299. doi: 10.1158/1055-9965.EPI-07-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pegg AE. Regulation of ornithine decarboxylase. J. Biol. Chem. 2006;281:14529–14532. doi: 10.1074/jbc.R500031200. [DOI] [PubMed] [Google Scholar]
- 9.Atkins JF, Lewis JB, Anderson CW, Gesteland RF. Enhanced differential synthesis of proteins in a mammalian cell-free system by addition of polyamines. J. Biol. Chem. 1975;250:5688–5695. [PubMed] [Google Scholar]
- 10.Tabor CW, Tabor H. Polyamines. Annu. Rev. Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 11.Law GL, Raney A, Heusner C, Morris DR. Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J. Biol. Chem. 2001;276:38036–38043. doi: 10.1074/jbc.M105944200. [DOI] [PubMed] [Google Scholar]
- 12.Raney A, Law GL, Mize GJ, Morris DR. Regulated translation termination at the upstream open reading frame in S-adenosylmethionine decarboxylase mRNA. J. Biol. Chem. 2002;277:5988–5994. doi: 10.1074/jbc.M108375200. [DOI] [PubMed] [Google Scholar]
- 13.Cruz-Vera LR, New A, Squires C, Yanofsky C. Ribosomal features essential for tna operon induction: tryptophan binding at the peptidyl transferase center. J. Bacteriol. 2007;189:3140–3146. doi: 10.1128/JB.01869-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang P, Spevak CC, Wu C, Sachs MS. A nascent polypeptide domain that can regulate translation elongation. Proc. Natl Acad. Sci. USA. 2004;101:4059–4064. doi: 10.1073/pnas.0400554101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaba A, Wang Z, Krishnamoorthy T, Hinnebusch AG, Sachs MS. Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J. 2001;20:6453–6463. doi: 10.1093/emboj/20.22.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franceschetti M, Hanfrey C, Scaramagli S, Torrigiani P, Bagni N, Burtin D, Michael AJ. Characterization of monocot and dicot plant S-adenosyl-l-methionine decarboxylase gene families including identification in the mRNA of a highly conserved pair of upstream overlapping open reading frames. Biochem. J. 2001;353(Pt 2):403–409. doi: 10.1042/0264-6021:3530403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanfrey C, Franceschetti M, Mayer MJ, Illingworth C, Michael AJ. Abrogation of upstream open reading frame-mediated translational control of a plant S-adenosylmethionine decarboxylase results in polyamine disruption and growth perturbations. J. Biol. Chem. 2002;277:44131–44139. doi: 10.1074/jbc.M206161200. [DOI] [PubMed] [Google Scholar]
- 18.Franceschetti M, Perry B, Thompson B, Hanfrey C, Michael AJ. Expression proteomics identifies biochemical adaptations and defense responses in transgenic plants with perturbed polyamine metabolism. FEBS Lett. 2004;576:477–480. doi: 10.1016/j.febslet.2004.08.085. [DOI] [PubMed] [Google Scholar]
- 19.Hanfrey C, Elliott KA, Franceschetti M, Mayer MJ, Illingworth C, Michael AJ. A dual upstream open reading frame-based autoregulatory circuit controlling polyamine–responsive translation. J. Biol. Chem. 2005;280:39229–39237. doi: 10.1074/jbc.M509340200. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi S, Murakami Y, Matsufuji S. Ornithine decarboxylase antizyme: a novel type of regulatory protein. Trends Biochem. Sci. 1996;21:27–30. [PubMed] [Google Scholar]
- 21.Ivanov IP, Atkins JF. Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Res. 2007;35:1842–1858. doi: 10.1093/nar/gkm035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell JL, Judd GG, Bareyal-Leyser A, Ling SY. Feedback repression of polyamine transport is mediated by antizyme in mammalian tissue-culture cells. Biochem. J. 1994;299:19–22. doi: 10.1042/bj2990019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoshino K, Momiyama E, Yoshida K, Nishimura K, Sakai S, Toida T, Kashiwagi K, Igarashi K. Polyamine transport by mammalian cells and mitochondria: role of antizyme and glycosaminoglycans. J. Biol. Chem. 2005;280:42801–42808. doi: 10.1074/jbc.M505445200. [DOI] [PubMed] [Google Scholar]
- 24.Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins JF, Gesteland RF, Hayashi S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssen BD, Hayes CS. Kinetics of paused ribosome recycling in Escherichia coli. J. Mol. Biol. 2009 doi: 10.1016/j.jmb.2009.09.020. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivanov IP, Loughran G, Atkins JF. uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc. Natl Acad. Sci. USA. 2008;105:10079–10084. doi: 10.1073/pnas.0801590105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terui Y, Higashi K, Taniguchi S, Shigemasa A, Nishimura K, Yamamoto K, Kashiwagi K, Ishihama A, Igarashi K. Enhancement of the synthesis of RpoN, Cra, and H-NS by polyamines at the level of translation in Escherichia coli cultured with glucose and glutamate. J. Bacteriol. 2007;189:2359–2368. doi: 10.1128/JB.01562-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoyt MA, Broun M, Davis RH. Polyamine regulation of ornithine decarboxylase synthesis in Neurospora crassa. Mol. Cell. Biol. 2000;20:2760–2773. doi: 10.1128/mcb.20.8.2760-2773.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manzella JM, Blackshear PJ. Regulation of rat ornithine decarboxylase mRNA translation by its 5′-untranslated region. J. Biol. Chem. 1990;265:11817–11822. [PubMed] [Google Scholar]
- 30.Shantz LM, Hu RH, Pegg AE. Regulation of ornithine decarboxylase in a transformed cell line that overexpresses translation initiation factor eIF-4E. Cancer Res. 1996;56:3265–3269. [PubMed] [Google Scholar]
- 31.Van Steeg H, Van Oostrom CT, Hodemaekers HM, Peters L, Thomas AA. The translation in vitro of rat ornithine decarboxylase mRNA is blocked by its 5′ untranslated region in a polyamine-independent way. Biochem. J. 1991;274:521–526. doi: 10.1042/bj2740521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H, Min J, Zeng H, McCloskey DE, Ikeguchi Y, Loppnau P, Michael AJ, Pegg AE, Plotnikov AN. Crystal structure of human spermine synthase: implications of substrate binding and catalytic mechanism. J. Biol. Chem. 2008;283:16135–16146. doi: 10.1074/jbc.M710323200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pegg AE. Spermidine/spermine-N(1)-acetyltransferase: a key metabolic regulator. Am. J. Physiol. Endocrinol. Metab. 2008;294:E995–E1010. doi: 10.1152/ajpendo.90217.2008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.