Abstract
The 5R thymine glycol (5R-Tg) DNA lesion exists as a mixture of cis-(5R,6S) and trans-(5R,6R) epimers; these modulate base excision repair. We examine the 7:3 cis-(5R,6S):trans-(5R,6R) mixture of epimers paired opposite adenine in the 5′-GTgG-3′ sequence with regard to nucleotide excision repair. Human XPA recognizes the lesion comparably to the C8-dG acetylaminoflourene (AAF) adduct, whereas XPC/HR23B recognition of Tg is superior. 5R-Tg is processed by the Escherichia coli UvrA and UvrABC proteins less efficiently than the C8-dG AAF adduct. For the cis-(5R, 6S) epimer Tg and A are inserted into the helix, remaining in the Watson–Crick alignment. The Tg N3H imine and A N6 amine protons undergo increased solvent exchange. Stacking between Tg and the 3′-neighbor G•C base pair is disrupted. The solvent accessible surface and T2 relaxation of Tg increases. Molecular dynamics calculations predict that the axial conformation of the Tg CH3 group is favored; propeller twisting of the Tg•A pair and hydrogen bonding between Tg OH6 and the N7 atom of the 3′-neighbor guanine alleviate steric clash with the 5′-neighbor base pair. Tg also destabilizes the 5′-neighbor G•C base pair. This may facilitate flipping both base pairs from the helix, enabling XPC/HR23B recognition prior to recruitment of XPA.
INTRODUCTION
5,6-dihydroxy-5,6-dihydro-2′-thymine, thymine glycol (Tg), is formed by exposure to radiation and chemical oxidants (1,2). It is also formed by oxidation of 5-methylcytosine to 5-methylcytosine glycol, followed by deamination (3,4). The C5 and C6 atoms of Tg are chiral and it exists in DNA as diastereomeric pairs of epimers, the 5R cis, trans pair (5R,6S;5R,6R) and the 5S cis, trans pair (5S,6R; 5S,6S) (Scheme 1) (5–7). The 5R pair is more abundant and more stable; in either case, the cis isomers predominate at the nucleoside level (6). Human cells repair hundreds of Tg lesions per day (8,9).
Scheme 1.
(A) Interconversion of the cis-(5R,6S) and trans-(5R,6R) Tg lesions. When the 5R-Tg isomer is paired opposite dA in this 5′-GTgG-3′ sequence a 7:3 cis-(5R,6S): trans-(5R,6R) mixture is present at equilibrium, in slow exchange on the NMR timescale (12). (B) Oligodeoxynucleotide duplex used for NMR studies, indicating the numbering of the nucleotides. X6 is the cis-(5R,6S) Tg lesion.
The 5R-Tg lesion has been examined in the 5′-ATgA-3′ sequence, paired opposite dA (10). It was concluded that Tg was partially extrahelical (10). It was also reported that the structure of 5R-Tg placed opposite dA in the 5′-GTgC-3′ sequence was disordered (11). These studies (10,11) did not report cis–trans epimerization (5–7) of Tg. In DNA the complementary base modulates the cis–trans equilibrium of 5R-Tg. When paired opposite dA in the 5′-GTgG-3′ sequence a 7:3 cis (5R,6S): trans (5R,6R) mixture exists at equilibrium at 25°C, whereas when paired with dG in the same sequence, only the cis (5R,6S) epimer is observed (12). When cis-(5R,6S) Tg is mismatched with dG in the 5′-GTgG-3′ sequence it assumes the wobble orientation, and shifts toward the major groove. This increases its solvent accessible surface but it remains stacked into the helix. Intra-strand hydrogen bonding between the hydroxyl on C6 of Tg and the N7 of the 3′ purine (13) is weak (14).
The 5R-Tg lesion hinders DNA replication (15,16). It is a substrate for base excision repair. This is mediated by at least two DNA N-glycosylase/AP lyases that are influenced by the diastereoisomer of Tg, the cis–trans epimerization of each diastereoisomer, and the identity of the complementary purine (17). The 5R-Tg lesion is also repaired by nucleotide excision repair (NER), although the effects of the cis–trans epimerization of each diastereoisomer of Tg with regard to NER have not been characterized. Both randomly introduced 5R-Tg and abasic sites are substrates for the Escherichia coli UvrABC proteins (18,19). Tg is also excised in vitro by human NER (20).
Here we report on the 7:3 cis-(5R,6S):trans-(5R,6R) mixture of Tg epimers paired opposite adenine in the 5′-GTgG-3′ sequence (12) with regard to NER by the NER proteins of E. coli and the binding of the lesion by the human NER proteins XPA and XPC/HR23B. The 5R-Tg lesion is a good substrate for binding by UvrA and excision by UvrABC, although the bulky C8-dG DNA adduct of AAF is recognized and incised more efficiently. However, recognition of 5R-Tg by the human XPC/HR23B complex is superior to the bulky AAF adduct, whereas recognition by human XPA is comparable to the AAF adduct. To elucidate structure–activity relationships underlying these observations, the structure of the cis-(5R,6S) Tg epimer embedded in the same 5′-GTgG-3′ sequence and placed opposite to deoxyadenosine has also been refined. It remains in the Watson–Crick orientation with respect to the complementary dA, but the solvent accessible surface area of Tg is increased. The complementary A19 remains stacked in the helix. Significantly, the 5R-Tg lesion also destabilizes the 5′-neighbor G•C base pair. We propose that this lowers the activation barrier with respect to flipping both base pairs from the helix, at least in this particular sequence, enabling XPC/HR23B to recognize 5R-Tg prior to the recruitment of XPA.
METHODS
Sample preparation
The undamaged ND-50-bp and AAF-50-bp oligodeoxynucleotides were constructed as described (21). The Tg-modified 5′-d(GTGCGTgGTTTGT)-3′ (17) was characterized using a Voyager-DE MALDI-TOF mass spectrometer (PerSeptive Biosystems, Inc., Foster City, CA, USA). It was ligated with a 5′-32P-labeled 20-mer, and a 19-mer on the 5′- and 3′-ends, respectively, to form the 5′-32P-labeled Tg-51mer. The oligodeoxynucleotides 5′-d(GTGCGTGTTTGT)-3′ and 5′-d(ACAAACACGCAC)-3′ were purchased from the Midland Certified Reagent Co. (Midland, TX, USA) and purified by reverse phase HPLC.
Binding as measured by electrophoretic mobility shift assays
Assays for the UvrA protein were performed as described (21). For the human NER proteins XPA and XPC/HR23B, the binding assay was performed as described for XPA binding (22), with minor modifications. The radioactivity of DNA bands on gels was quantified using a Fuji FLA-5000 phosphoimager. Dissociation constants (Kd) were determined from the binding curves as a concentration of the substrate at which half of DNA was bound to protein. Three experiments were performed for each binding curve.
UVrABC incision assays
The 5′-32P-labeled substrates (2 nM) were incised by UvrABC nucleases (UvrA, 15 nM; UvrB, 250 nM; UvrC, 100 nM) in the UvrABC buffer with 1 mM ATP at 37°C and the products were resolved on a 12% polyacrylamide gel and analyzed as described (21). The initial rates were calculated by linear least-squares fits of data. The substrate incised in femtomoles was calculated based on the total molar amount of substrate employed in each reaction and the incision percentage of the substrate. Three experiments were performed to determine initial rates.
Nuclear magnetic resonance spectroscopy
Oligodeoxynucleotides were annealed in 20 mM sodium phosphate, containing 100 mM NaCl, 10 μM NaN3 and 50 μM Na2EDTA (pH 7.0). Experiments were performed at 800 MHz. NOESY spectra for the nonexchangeable protons were recorded at 30°C with mixing times of 80, 150, 200 and 250 ms using States-TPPI phase cycling, 512 real data points in the d1 dimension with 32 scans per FID, 2K real data points in the d2 dimension, sweep width of 10 p.p.m., and a relaxation delay of 2.0 s. The water resonance was suppressed using presaturation. NOESY spectra of exchangeable protons were obtained at 5°C using watergate H2O suppression (23). T1 spin–lattice relaxation experiments were collected using the inversion recovery method (24,25). T2 transverse relaxation experiments were collected using the CPMG method (24,25). 1H spectra were referenced to 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid, sodium salt (3-TMSP). The program XWINNMR (Bruker Inc., Billerica, MA, USA) was used for data processing. A skewed sinebell-squared apodization function with a 90° phase shift was used for NOESY experiments; the same function with a 10° phase shift was used for COSY experiments.
Distance restraints
NOE intensities were determined from volume integration using SPARKY (26). Intensities for the cis-(5R, 6S) epimer were corrected for the 7:3 cis-(5R,6S):trans-(5R,6R) molar ratio. These were combined with intensities generated from CORMA analysis of a B-form starting structure producing a hybrid intensity matrix (27,28), which was refined using MARDIGRAS (29–31) with the RANDMARDI function. Calculations at mixing times of 80, 150, 200 and 250 ms were run at correlation times of 2, 3, 4 and 5 ns. Distance restraints were divided into categories indicative of confidence levels. Empirical restraints were used to define Watson–Crick hydrogen bonding, but not for the Tg 6•A19 or G5•C20 pairs.
Torsion angle restraints
3J 1H coupling constants were obtained by amplitude constrained multiplet evaluation of COSY data (32). Electronegativity of substituent Karplus curves were generated and converted to phase angle space assuming a maximum pucker amplitude (Φ) of 44° (33,34). Scalar couplings were fit to the curves to determine pseudorotation ranges (P). The sugar pseudorotation and amplitude ranges were converted to restraints for the dihedral angles ν0 to ν4. Measurements of the mol fraction of sugar puckers in the N or S conformations were determined from the sum of J1'2;' and J1'2;'' scalar couplings (34). Nucleotides with <50% XS were allowed to explore both N and S conformations during rMD calculations (ρ = 0°–210°). Nucleotides with XS > 50% were restrained such that ρ = 125°–210°. Backbone torsion angles were restrained with data where available; otherwise they were restrained empirically based on canonical A-form and B-form values.
Structural refinement
Partial charges for Tg were generated using GAUSSIAN (35). Geometry optimization and ESP calculations were performed using the Hartree–Fock method with the 6-31G* basis set (12). The output was formatted using ANTECHAMBER (36). Starting structures were generated using NAB (37) and energy minimized using the SANDER module of AMBER (36). Coordinate and topology files were generated with xLEaP (36) using ff99 parameters (38). The restraint function utilized square-well potentials (39). The generalized Born model was used for simulated annealing calculations, with a salt concentration of 0.2 mM (40,41). Temperature was maintained using the Berendsen algorithm (42). Complete relaxation matrix analysis (27,28) was performed to determine agreement with 1H NOESY data. A refined structure from simulated annealing was neutralized with the addition of sodium ions and placed in a truncated octahedral TIP3P water box with periodic boundaries at a distance of 8 Å from the solute. After equilibration, a 10-ns isothermal rMD calculation was performed. The temperature was controlled using the Langevin thermostat (43,44) with a collision frequency of 1 ps−1. Electrostatic interactions were treated with the PME method (45). A 15-Å cutoff for nonbonded interactions was used. Bond lengths involving hydrogen were fixed using SHAKE (46). A structural ensemble was extracted from the isothermal trajectory using PTRAJ (36). The heavy atoms were subjected to pairwise RMSD comparisons using SUPPOSE. Helicoidal analysis was performed using CURVES (47,48). Structures were rendered using Chimera (49). Solvent-excluded and solvent-accessible surface areas of individual bases as a function of probe radius were calculated using MSMS (50).
RESULTS
Dodecamer containing the 5R-Tg lesion
The 5′-d(GTGCGXGTTTGT)-3′, X = 5R-Tg, was subjected to mass spectrometric analysis, which yielded the anticipated molecular ion peak with mass 3732 (m/z). Capillary gel electrophoretic and HPLC analysis showed that it eluted as a single peak. The adducted oligodeoxynucleotide was pure and existed as a single chromatographically separable species.
Recognition and incision of the 5R-Tg substrate by E. coli UvrABC proteins
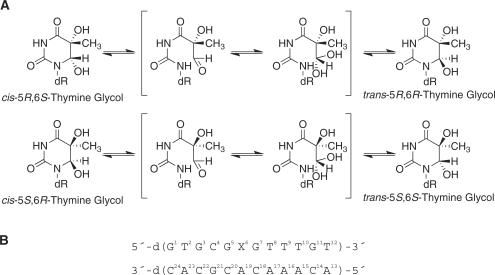
A 51-bp substrate containing 5R-Tg (Supplementary Scheme S1) was utilized. A 50-bp substrate containing an AAF adduct (AAF-50 bp) that is recognized and incised by E. coli UvrABC (21), was used for comparison. A 50-bp substrate (ND-50 bp) was used as a negative control. Figure 1A and B show gel mobility shift assays for UvrA binding to Tg and AAF substrates, respectively. The slowly migrating bands represent the formation of DNA–UvrA2 complexes. Binding isotherms (Figure 1C) showed that the affinity of UvrA to the 5R-Tg substrate was high, though the binding of UvrA to the AAF substrate was stronger (Table 1). The UvrA had a 2.4-fold lower affinity to the Tg-50-bp substrate (Kd = 24 ± 2 nM) than the AAF-51-bp substrate (Kd = 10 ± 1 nM). The Kd for UvrA binding to the AAF substrate agreed with the reported value (21). UvrA had lower affinity to the undamaged than to the damaged substrate, in agreement with previous observations (21). While the differing sequence contexts of the Tg- and AAF–DNA substrates (Scheme 2A) might affect their interactions with the UvrABC system (51–53), it was assumed that sequence specific effects were smaller than those caused by the difference in the types of DNA lesions.
Figure 1.
Binding of UvrA to the duplex containing the site-specific 5R-Tg lesion paired with dA. In (A) and (B), UvrA at the indicated concentrations was incubated with 2-nM substrates at 37°C for 20 min in UvrABC binding buffer and then analyzed on a 3.5% polyacrylamide native gel by gel mobility shift assays. (A) TG-51 bp; (B) AAF-50 bp. (C) The binding curves generated based on the titration data in (A) and (B).
Table 1.
Equilibrium dissociation constants for binding of UvrA and human NER proteins to TG-51-bp and AAF-50-bp DNA substrates at 30°Ca
| Protein | Kd for TG-51 bp (nM) | Kd for AAF-50 bp (nM) |
|---|---|---|
| UvrA | 24 ± 2 | 10 ± 1 |
| XPAb | 48 ± 4 | 44 ± 6 |
| XPC/HR23B | 18 ± 2 | 27 ± 3 |
aData represent the means ± SD of three experiments.
bFor XPA the dissociation constants were determined for XPA dimers.
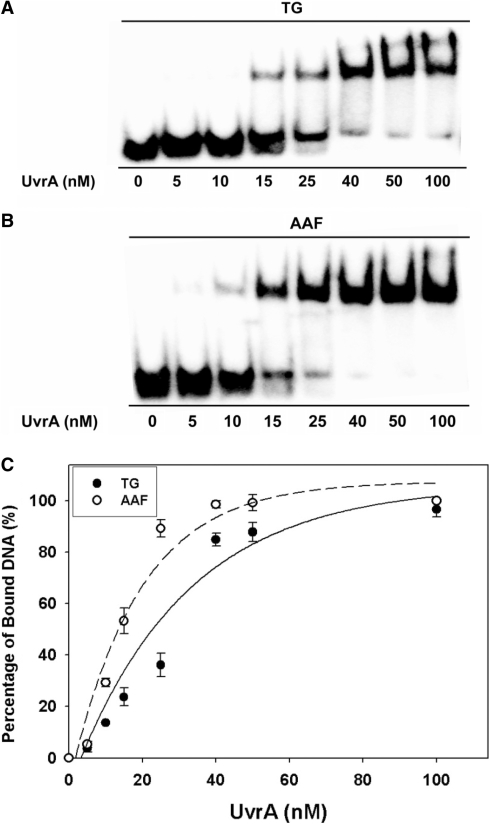
UvrC cut the damaged strand 4 or 5 nt 3′ to the lesion and then 8 nt 5′ to the lesion (Supplementary Scheme S1). The substrates were 32P-labeled at the 5′-end and the incision products of were 18-mers. Figure 2A and B show temporal data for UvrABC incision of Tg and AAF substrates, respectively. No products were observed for the ND-50-bp undamaged substrate. The initial rates were determined from the relative yields of the products (Figure 2C). For the 5R-Tg substrate the initial rate was 0.48 ± 0.04 fmol/min. For the AAF substrate the comparable rate was 0.80 ± 0.02 fmol/min. Thus, the 5R-Tg substrate was incised 1.7X less efficiently than was the AAF substrate. However, the incision rate of the 5R-Tg substrate was greater than that of the DNA helix-distorting cross-linked tandem G(8,5-Me)T lesion (21). Therefore, in this sequence 5R-Tg was a good substrate for the E. coli UvrABC proteins. The ratio of binding affinities of UvrA to the AAF- versus 5R-Tg substrate was 2.4, greater than the ratio of initial rates of UvrABC incision of the AAF- and 5R-Tg substrates, which was 1.7.
Figure 2.
Incisions of the site-specific 5R-Tg paired with dA substrate by UvrABC nuclease. In (A) and (B), 2-nM DNA substrates 5′-terminally labeled with 32P were incubated with UvrABC proteins in the UvrABC buffer in the presence of 1-mM ATP at 37°C for the indicated time periods. The incision products were analyzed on a 12% urea PAGE under denaturing conditions. (A) AAF-50 bp; (B) Tg-51 bp. (C) Kinetics of UvrABC incisions based upon the data in (A) and (B).
Binding of human NER proteins to the 5R-Tg substrate
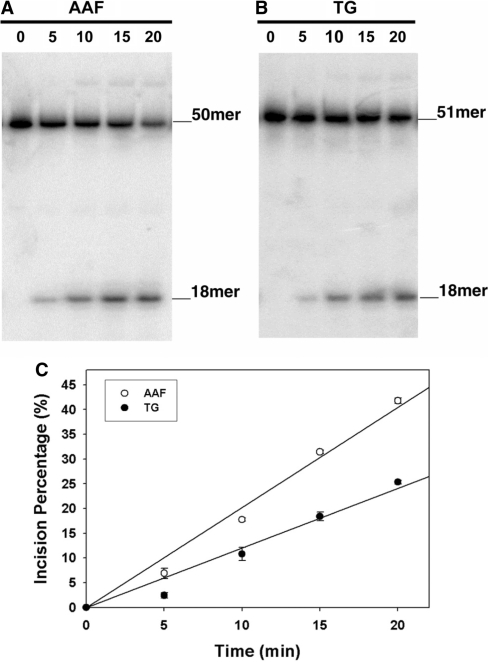
The binding of XPA and XPC/HR23B to the 5R-Tg damaged duplex was compared with the AAF-damaged duplex using EMSA (Figure 3). Neither XPA nor XPC/HR23B bound to the undamaged substrate. The shifted bands represent formation of DNA–XPA2 complexes. At low concentrations XPA bound to AAF-50 bp with a greater affinity than to Tg-51 bp, which was evident from a comparison of the 25 nM XPA lanes for 5R-Tg and AAF. At concentrations >50 nM, XPA bound to both substrates with comparable affinities (Figure 3A). The binding of XPC/HR23B to 5R-Tg and AAF-damaged DNA is shown in Figure 3B. Figure 3C shows binding isotherms for XPA interacting with the 5R-Tg- and AAF–DNA substrates. The dissociation constants estimated for the 5R-Tg and AAF substrates were similar, 48 ± 4 nM and 44 ± 6 nM, respectively (Table 1). XPC/HR23B bound to Tg-51 bp with a greater affinity than to AAF-50 bp (Figure 3D). Unlike XPC/HR23B the E. coli NER protein UvrA bound better to AAF–DNA adduct than to Tg-DNA adduct (Figure 1).
Figure 3.
Binding of human XPC/HR23B and XPA proteins to the site-specific 5R-Tg paired with dA substrate. In (A) and (B), XPA (A) or XPC/HR23B (B) proteins at the indicated concentrations were incubated with 4-nM substrates at 30°C for 30 min in XPA binding buffer and then analyzed on 3.5% polyacrylamide native gels by gel mobility shift assays (XPA, 4°C; XPC/HR23B, room temperature). (C) and (D) show the binding curves generated from the data in (A) and (B).
Nuclear magnetic resonance spectroscopy
Data were collected upon preparation of the duplex containing the 5R-Tg lesion (Scheme 1), and again after 4 weeks. Spectral changes were not observed, suggesting that the cis-(5R,6S) and trans-(5R,6R) epimers had achieved equilibrium. The cis-(5R,6S) epimer was favored 7:3 over the trans-(5R,6R) epimer, as determined by integration of Tg CH3 peaks (12). It was possible to obtain spectroscopic data for the cis-(5R,6S) epimer; the trans-(5R,6R) epimer was not present at sufficient levels to allow evaluation of its spectrum.
Nonexchangeable DNA protons
Figure S1 in the Supplementary Data shows NOESY cross-peaks between the base aromatic H6/H8 protons and the deoxyribose H1′ protons. The resonances were assigned using standard strategies (54,55). Complete sequential NOE connectivity was obtained for both the modified and the complementary strands. With the exception of several of the H4′ protons, and the stereotopic assignments of the H5′ and H5′′ sugar protons, assignments of the deoxyribose protons were made unequivocally; the resonance assignments have been reported (12).
Exchangeable DNA protons
Supplementary Figure S2 shows NOESY cross-peaks between the purine N1H and pyrimidine N3H imino protons and the amino protons of the complementary bases, involved in Watson–Crick hydrogen bonding. At the mismatched X6•A19 pair, the Tg N3H imino resonance was not identified. This was attributed to rapid exchange with solvent. The assignments of the remaining hydrogen-bonded imino and amino protons were made using standard methods (56). The G5 N1H imino resonance was broad at 5°C and disappeared when the temperature was increased to 15°C. In contrast, for an unmodified duplex, the G5 N1H imino resonance was sharp and was observed at temperatures as high as 40°C (12). There was no cross peak between the G5 N1H resonance and G21 N1H, located at base pair C4•G21. This was attributed to its exchange with solvent. The imino resonances for base pairs T2•A23, G3•C22, C4•G21, G7•C18, T8•A17, T9•A16, T10•A15 and G11•C14 were observed. The imino resonances for the terminal base pairs G1•C24 and T12•A13 were not observed, attributed to exchange broadening with water. The assignments have been reported (12).
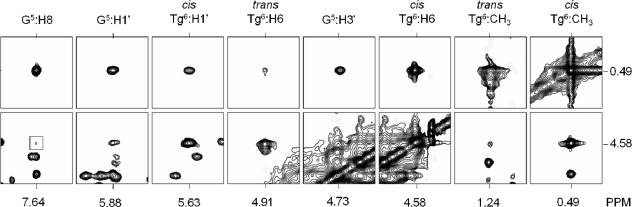
Tg protons
Figure 4 shows NOESY data obtained for the Tg CH3 and Tg H6 protons. The proximate Tg H6 and Tg CH3 protons yielded a strong Tg H6→Tg CH3 NOE at all mixing times. The G5 H1′→X6 H6 and G5 H8→X6 H6 NOEs were diagnostic of the cis-(5R,6S) configuration. The cis-(5R,6S) Tg CH3 protons exhibited a chemical shift of 0.49 p.p.m., while the Tg H6 proton resonated at 4.58 p.p.m. A total of 23 NOE cross peaks were assigned between Tg CH3 and H6 in the cis-(5R,6S) epimer and DNA (seven for Tg H6 and sixteen for Tg CH3). The trans-(5R,6R) Tg CH3 protons exhibited a chemical shift of 1.24 p.p.m., while the Tg H6 proton resonated at 4.91 p.p.m. For the trans-(5R,6R) epimer, only one NOE cross peak was observed, between Tg H6 and Tg H2′. Integration of the respective Tg H6→Tg H2′ cross peaks confirmed the 7:3 ratio of epimers. A single set of resonances was observed for G5 and G7. Thus, the equilibrium mixture of epimers did not influence the chemical shift environment of the neighboring nucleotides. The spectroscopic assignments have been reported (12).
Figure 4.
(A) NOESY data collected at a NOE mixing time of 250 ms, showing NOEs between the cis-(5R,6S) Tg lesion and the DNA that were used for structural refinement. The spectra were collected at 800 MHz at a temperature of 30°C.
Spin–lattice relaxation
The T1 relaxation times of the T2, T8, T9, T10, & T12 CH3 and X6 CH3 groups were compared to those of the unmodified duplex (Supplementary Figure S3). The X6 CH3 T1 relaxation time was 1.9 s faster than was the corresponding T6 CH3 relaxation time of the unmodified duplex containing T6•A19. The X6 CH3 relaxed an average of 1.4 s faster than other thymine CH3 groups in the X6•A19 duplex.
Spin–spin relaxation
The T2 relaxation times of the T2, T8, T9, T10, & T12 CH3 and X6 CH3 groups were compared to those of the unmodified duplex (Supplementary Figure S4). The Tg CH3 of the X6•A19 duplex relaxed ∼70 ms faster than the corresponding CH3 of the unmodified sample.
Structural refinement
Thirty starting structures were generated, of which half had the Tg CH3 group in the axial conformation and half had the Tg CH3 group in the equatorial conformation. These exhibited a maximum pairwise RMSD of 4.3 Å. The distance and torsion angle restraints used in the calculations are summarized in Supplementary Table S1. As Watson–Crick hydrogen bonding at the X6•A19 base pair was not observed, hydrogen bond restraints between X6 and A19 were not used. All but three of the 30 structures emerged from simulated annealing calculations with Tg CH3 in the axial conformation. A refined structure with Tg CH3 in the axial conformation was placed into a truncated octahedron TIP3P water box, and subjected to 10 ns of isothermal rMD calculations at 300 K (Supplementary Figure S5). At 4.38 ns the Tg CH3–C5–C6–H6 torsion angle shifted from −40° to 50°. This corresponded to a change from the Tg CH3 axial to the equatorial conformation. The glycosyl Tg O4′–H1′–N1–C2 torsion angle fluctuated between −90° and −140° during the first 4.38 ns; when the Tg CH3 group shifted from axial to equatorial conformation, this angle fluctuated between −130° and −150°.
The distances between Tg HO5→G7 N7 (Supplementary Figure S5, panel C) and Tg HO6→G7 N7 (Supplementary Figure S5, panel D) were monitored during the isothermal calculations. Analysis of trajectories indicated intra-strand hydrogen bond stabilization of the X6•A19 base pair as predicted (13). Hydrogen bond occupancy was defined as hydrogen bond donors and acceptors being within 3.5 Å with an angle cutoff of 120°. During the first 4.38 ns, in which the Tg CH3 group was in the axial conformation, the Tg HO6→G7 N7 hydrogen bond criteria were satisfied (Supplementary Figure S5, panel D). This induced altered propeller twist at the lesion site. There was 42% occupancy of the Tg OH6→G7 N7 hydrogen bond during the trajectory and the entirety of this occupancy occurred during the first 4.38 ns. In contrast, during the first 4.38 ns, in which the Tg CH3 group was in the axial conformation, the distance between Tg HO5 and G7 N7 was 4.5 Å, which was not consistent with hydrogen bonding. As a consequence of Tg CH3 reorienting to the equatorial conformation at 4.38 ns, Tg OH5 shifted to the axial conformation. This allowed for hydrogen bond formation with G7 N7. The Tg OH5→G7 N7 occupancy was 11% over 10 ns and was only observed after the conformational shift. During these isothermal calculations, the RMSD of the DNA backbone heavy atoms (Supplementary Figure S5, panel F) was 2 Å.
Structural ensembles representing both axial and equatorial conformations of Tg CH3 were extracted for analyses (Supplementary Figure S6). The axial ensemble (PDB ID 2KH5) was extracted from the 120 ps before the conformation change at 4.38 ns; this ensemble had an RMSD of 0.56 Å for the core 6 bp. The equatorial ensemble (PDB ID 2KH6) was extracted 1 ns after the conformational change at 4.38 ns. This had an RMSD of 0.69 Å for the core 6 bp.
To evaluate the accuracies of the rMD ensembles with respect to the 1H NOESY data, complete relaxation matrix analyses (27,28) were performed (Supplementary Figure S7). The ensemble with Tg CH3 in the axial conformation had an average sixth root residual R1X value of 8.12 × 10−2. The Tg lesion exhibited an R1X value of 7.19 × 10−2 for inter-nucleotide cross-peaks with G5 and 7.61 × 10−2 for intra-nucleotide cross-peaks. The ensemble with Tg CH3 in the equatorial position had an average R1X value of 8.15 × 10−2. The Tg lesion exhibited an R1X value of 1.77 × 10−1 for inter-nucleotide cross peaks with G5 and 1.01 × 10−1 for intra-nucleotide cross-peaks. This indicated that both ensembles of structures satisfied the 1H NOESY data, although at the lesion site, the agreement between the structures with the Tg CH3 axial conformation and the NOESY data was improved.
Refined structures
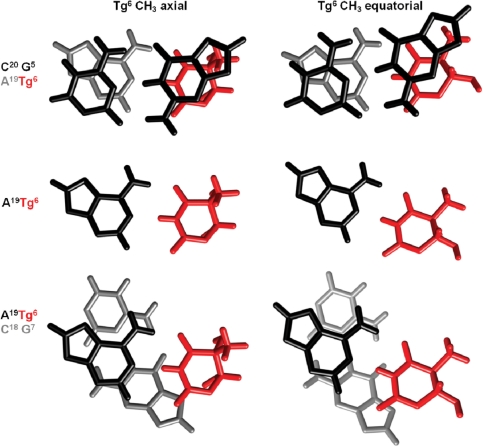
In both the Tg-axial and Tg-equatorial ensembles the cis-(5R,6S) lesion was positioned in the Watson–Crick orientation such that Tg NH3 was proximate to A19 N1 and Tg O4 was proximate to A19 N6H (Figure 5). However, for the Tg-axial ensemble, the base pair opening at X6•A19 increased such that the Tg shifted toward the major groove. For the Tg-equatorial ensemble, Tg maintained Watson–Crick positioning. Irrespective of the conformation of Tg, the complementary dA remained in the anti conformation about the glycosyl bond and stacked into the duplex (Figure 6). The Tg base stacked below the 5′-neighbor base G5, while its complement A19 stacked below the 5′-neighbor base C20. In the 3′-direction, base stacking between Tg and the G7•C18 base pair was disrupted (Figure 5). Helicoidal analyses of both the axial and equatorial conformations of Tg indicated disturbances in shear, opening, stagger and stretch at the lesion (Figure 6). For the Tg CH3 axial conformation the disturbance of the glycosyl torsion angle (χ) alleviated steric interactions between Tg CH3 and the 5′-neighbor purine G5. The solvent accessible surface of Tg in the duplex relative to that of a free Tg base was compared with bases A23, T9, and T12 (Supplementary Figure S8). Irrespective of whether the Tg CH3 group was in the axial or equatorial conformation, Tg exhibited an increased solvent accessible surface relative to T9 and A23 but was not as exposed to solvent as the terminal T12 base. The average accessible surface area of the Tg base was 19% in the CH3 axial structure and 18% in the CH3 equatorial structure.
Figure 5.
Base pair stacking interactions of the cis-(5R,6S) Tg lesion. Comparison stacking interactions in which Tg CH3 is in the axial (PDB ID 2KH5) or equatorial (PDB ID 2KH6) conformations. The top panel shows the G5• C20 base pair (black) stacked above the X6• A19 base pair (Tg is colored red and A19 is colored gray). The center panel shows the orientation of the X6 lesion (red) with respect the complementary nucleotide A19 (black). The bottom panel shows the X6• A19 base pair (Tg is colored red and A19 is colored black) stacked above the G7• C18 base pair (grey).
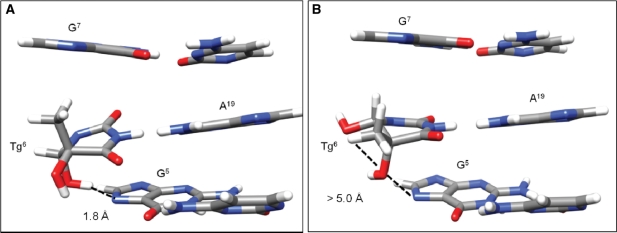
Figure 6.
The cis-(5R,6S)Tg lesion at the X6• A19 base pair as viewed from the major groove showing potential hydrogen bonding interactions as predicted from analyses of rMD trajectories. (A) The Tg OH6 formed a hydrogen bond with G5 N7 when Tg CH3 was in the axial conformation (PDB ID 2KH5). (B) When Tg CH3 was in the equatorial conformation Tg OH6 did not hydrogen bond with G5 N7, however, improved hydrogen bonding was observed with Tg OH5 (PDB ID 2KH6).
DISCUSSION
Defects in human NER are associated with the disease xeroderma pigmentosum (57,58). In human global genome NER, the XPC/HR23B complex (59–61) is the damage recognition factor. Damage-induced thermal destabilization of the helix is believed to modulate recognition of a diverse group of damages by XPC (62–66). The XPA protein is essential for NER and Yang et al. (67) reported that it exists as a homodimer either in the free state or as a complex with human RPA. It binds to mismatched DNA bubble substrates with or without DNA adducts, including the C8-dG adducts of AF, AAF and 1-nitropyrene, and the T[6,4]T photoproducts (68). XPA is likely involved in the verification of DNA damage (64,69). It also probably plays a role in recruitment of repair factors and stabilization of repair intermediate structures since it binds more efficiently to undamaged ds–ssDNA junctions with ssDNA branches (68), intermediate structures found in NER. In light of observations that the repair of 5R-Tg by DNA N-glycosylases/AP lyases is modulated by cis–trans epimerization (17) it was of interest to examine the 7:3 cis-(5R,6S):trans-(5R,6R) mixture of Tg epimers paired opposite adenine in the 5′-GTgG-3′ sequence (12) with regard to NER by the NER proteins of E. coli (18,19), and the binding of the lesion by the human NER proteins XPA and XPC/HR23B (20), and to compare these observations with the structure of the cis-(5R,6S) Tg lesion in this same sequence.
When placed opposite dA in the 5′-GTgG-3′ sequence the 5R-Tg lesion exists at 25°C as a 7:3 cis-(5R,6S):trans-(5R,6R) mixture at equilibrium; the structure of the cis-(5R,6S) epimer has been refined herein. That there is no disruption of sequential NOE connectivity for either the complementary strand or the modified strand (Supplementary Figure S2) supports the conclusion that the cis-(5R,6S) Tg lesion minimally distorts the helical backbone (12). At the X6•A19 pair, both Tg and A19 are inserted into the helix (Figure 6). The X6•A19 pair remains in a Watson–Crick type alignment in which Tg O4 is proximate to the exocyclic amine of A19, and Tg N3H is proximate to A19 N1 (Figure 5). Both the exchangeable Tg amine and A19 N6 amine resonances protons undergo increased exchange with solvent. In the 3′-direction, stacking between the cis-(5R,6S) Tg6 and the G7•C18 base pair is also disrupted (Figure 6). For the X6•A19 pair the cis (5R,6S) Tg lesion is more exposed to solvent, as compared to a Watson–Crick T•A base pair (Supplementary Figure S8). This agrees with findings in the 5′-AXA-3′ sequence when the 5R Tg lesion was placed opposite dA (10). Tg6 lesion remains stacked into the duplex and it is not flipped into the major groove (Figure 6).
Nuclear magnetic resonance (NMR) does not differentiate between axial versus equatorial conformations of the Tg CH3 group. Both are observed in the rMD calculations. The axial conformation provides a modestly improved agreement with the NOE restraints. This is consistent with the crystal structure at the nucleoside level (70). Moreover, quantum mechanical calculations of the modified base predict the axial conformation of the cis-(5R,6S) Tg epimer is favored (13). It was also predicted that the Tg•A pair should be stabilized by an intra-strand hydrogen bond between the Tg OH6 and the N7 position of a 3′ purine (13). Its presence is supported by analyses of the present rMD trajectories. This hydrogen bond is present when Tg CH3 is in the axial conformation; its occupancy in the 10-ns trajectory is 46% (Supplementary Figure S5, panel D). It induces propeller twist at the lesion site, which alleviates steric interactions between the CH3 group and the 5′-neighbor guanine. This is reflected in the perturbation of the glycosyl torsion angle χ at Tg (Supplementary Figure S5). The rMD trajectories suggest the potential for Tg OH5→G7 N7 hydrogen bond formation when the Tg CH3 group shifts from the axial to the equatorial conformation. This alleviates steric clash between the Tg CH3 group and the 5′-neighbor guanine, and cannot be ruled out. However, the rMD trajectories predict only 11% occupancy of this hydrogen bond. Overall, it seems that the axial conformation of the cis-(5R,6S) Tg epimer is favored. The increase in longitudinal relaxation for the Tg CH3 protons is consistent with this conclusion; in the axial conformation the Tg CH3 group orients toward G5. It also exhibits NOEs to G5 protons; all are sources of longitudinal relaxation. In contrast, thymine has the CH3 group facing into the major groove with fewer sources of longitudinal relaxation. Epimerization to the trans-(5R,6R) Tg lesion, for which the equatorial conformation of the Tg CH3 group is favored by 4 kcal/mol (13), probably represents the more favorable mechanism for alleviating steric strain (12). Consistent with this conclusion, in the structure of the RB69 polymerase involving a template containing the cis-(5R,6S) Tg lesion and an incoming dATP, the Tg CH3 group is in the axial conformation, despite hindering stacking of the adjacent 5′-template guanine (71). Bolton and co-workers (11) reported that a disordered structure resulted when 5R-Tg was placed into a duplex containing the 5′-GTgC-3′ sequence. This suggests that formation of an intra-strand hydrogen between the Tg C6 OH and the N7 position of a 3′ purine (13) is important in stabilizing the cis-(5R,6S) Tg lesion in duplex DNA.
The observation that the Kd values of XPC/HR23B from either the 5R-Tg- (∼18 nM) or AAF-DNA substrates (∼27 nM) are lower than those from XPA is consistent with the proposed role of XPC/HR23B as the damage recognition factor in human genomic NER (64,69). In comparison, XPC/HR23B binds to cisplatin 1,3-intrastrand adducts or 6-nt lengths of mispaired DNA with a Kd of 1–3 nM (64). The recognition of the AAF-damaged DNA by XPC/HR23B has been attributed to the adduct's; inability to base pair efficiently with cytosine (72), and an emerging consensus posits that disruption of normal base pairing and the resulting destabilization of the helix, rather than the recognition of helical distortion by bulky lesions, governs the affinity of XPC/HR23B binding (73). It should be noted, however, that while 5R-Tg can be recognized by XPC/HR23B and XPA, the efficiency of incision of this site-specific lesion by the human NER system remains to be determined.
From studies of the yeast XPC orthologue Rad4 bound to DNA containing a cyclobutane pyrimidine dimer, Min and Pavletch (74) concluded that damage recognized by Rad4 destabilizes the helix and facilitates the flipping out of two base pairs by the protein. The present data are consistent with this model. The presence of 5R-Tg in the duplex perturbs the 5′-neighbor base pair G5•C20, in addition to the damaged base pair X6•A19. As indicated in Figure 5, the imino resonance of base pair G5•C20 broadens due to solvent exchange. In fact, it disappears from the 1H NMR spectrum ∼35°C lower in the 5R-Tg modified DNA as compared to the unmodified duplex (12).
This destabilization of the two base pairs by the cis-(5R,6S) epimer may lower the activation barrier with respect to flipping both base pairs out of the helix, enabling XPC/HR23B to recognize the 5R Tg lesion prior to the recruitment of XPA. Based upon MD simulations of the Dickerson-Drew 5R-Tg-modified dodecamer d(CGCGAATgTCGCG)2, both Miller et al. (75) and Miaskiewicz et al. (76) also concluded that Tg weakened Watson–Crick hydrogen bonds of the 5′-neighbor base pair. The increase in transverse relaxation for Tg CH3 as compared to unmodified thymine CH3 protons is attributed to the puckering of the Tg six-member ring. However, increased backbone and sugar motions in tandem with the puckering of the Tg ring cannot be excluded.
The 5R-Tg lesion is also a substrate for base excision repair in E. coli and in mammalian cells (77). In E. coli, repair of Tg is initiated by endonuclease III (Nth) (78) and endonuclease VIII (Nei) (79). Yeast (80), mammalian (81,82) and human orthologs (83–85) of Nth have been characterized. Likewise, human orthologs of Nei have been characterized (86,87). The human hNTH1 exhibits a 13:1 preference for excising the 5R versus the 5S epimers, whereas hNEIL1 (86,88) shows a 1.5:1 preference for excising the 5R versus the 5S epimers (89). Similar observations have been made for prokaryotic, yeast and murine glycosylases (90). Ocampa-Hafalla et al. (17) showed that the repair of Tg by DNA N-glycosylases/AP lyases is modulated by the cis–trans epimerization of these two sets of diastereomers and that repair of 5R Tg by hNEIL1 depends upon the opposing base, with Tg•G pairs being excised much more rapidly than Tg•A pairs. Significantly, when 5-methyl cytosine is oxidatively damaged, forming 5-methycytosine glycol, hydrolytic deamination yields Tg mismatched with dG in (3). Computational studies suggest that substrates for hNEIL1 possess in common a pyrimidine-like ring and hydrogen bond donor-acceptor properties, allowing them to be accommodated within the enzyme's; binding pocket (91). Based upon structural data for the 5R-Tg lesion in a T•G mismatch, we suggested that the wobble orientation of the cis-(5R,6S) Tg base in the Tg•G pair shifts it toward the major groove, reflected in an increased solvent accessible surface and reduced barrier for intrinsic breathing of the lesion (14). While the Tg•A structure also shows an increased solvent accessible surface area, it differs from the Tg•G pair in that an efficient hydrogen bond between Tg OH6→G7 N7 is possible; this may be sufficient to reduce the barrier toward base-flipping of the Tg lesion into the active site pocket of the glycosylase.
In summary, in the 5′-GTG-3′ sequence, 5R-Tg, paired with dA, is a good binding substrate for the NER proteins from E. Coli and humans, corroborating earlier reports (18–20). It is a better substrate of the human NER proteins XPA and XPC/HR23B in comparison to the C8 dG adduct of AAF. The structure of the cis-(5R,6S) Tg lesion in this same sequence is consistent with the repair data in that the lesion causes destabilization of the 5′-neighboring G·C base pair, in addition to an increase in the solvent accessible area of Tg. It is conceivable that these thermodynamic perturbations of two base pairs contribute toward efficient recognition and NER of the 5R-Tg lesion.
ACCESSION NUMBERS
PDB ID 2KH5, PDB ID 2KH6.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health Grants R01 CA-055678 and P30 ES-000267 (to M.P.S.) and R01 ES-013324 (to A.K.B). Funding for open access charge: National Institutes of Health Grant R01 CA-55678.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Dr Markus Voehler and Dr Don Stec assisted with NMR spectroscopy. Dr Nicholas Uylanov and Dr Jarrod A. Smith assisted with structural refinement. Professor D. A. Johnson, ETSU, provided the human serum albumin. Drs Thomas M. and Constance M. Harris provided constructive comments.
REFERENCES
- 1.Teoule R, Bonicel A, Bert C, Cadet J, Polverelli M. Identification of radioproducts resulting from the breakage of thymine moiety by gamma irradiation of E. coli DNA in an aerated aqueous solution. Radiat. Res. 1974;57:46–58. [PubMed] [Google Scholar]
- 2.Frenkel K, Goldstein MS, Teebor GW. Identification of the cis-thymine glycol moiety in chemically oxidized and gamma-irradiated deoxyribonucleic acid by high-pressure liquid chromatography analysis. Biochemistry. 1981;20:7566–7571. doi: 10.1021/bi00529a035. [DOI] [PubMed] [Google Scholar]
- 3.Zuo S, Boorstein RJ, Teebor GW. Oxidative damage to 5-methylcytosine in DNA. Nucleic Acids Res. 1995;23:3239–3243. doi: 10.1093/nar/23.16.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfeifer GP. p53 mutational spectra and the role of methylated CpG sequences. Mutat. Res. 2000;450:155–166. doi: 10.1016/s0027-5107(00)00022-1. [DOI] [PubMed] [Google Scholar]
- 5.Vaishnav Y, Holwitt E, Swenberg C, Lee HC, Kan LS. Synthesis and characterization of stereoisomers of 5,6-dihydro-5,6-dihydroxy-thymidine. J. Biomol. Struct. Dyn. 1991;8:935–951. doi: 10.1080/07391102.1991.10507858. [DOI] [PubMed] [Google Scholar]
- 6.Lustig MJ, Cadet J, Boorstein RJ, Teebor GW. Synthesis of the diastereomers of thymidine glycol, determination of concentrations and rates of interconversion of their cis-trans epimers at equilibrium and demonstration of differential alkali lability within DNA. Nucleic Acids Res. 1992;20:4839–4845. doi: 10.1093/nar/20.18.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y. HPLC isolation and mass spectrometric characterization of two isomers of thymine glycols in oligodeoxynucleotides. Chem. Res. Toxicol. 2002;15:671–676. doi: 10.1021/tx0155855. [DOI] [PubMed] [Google Scholar]
- 8.Cathcart R, Schwiers E, Saul RL, Ames BN. Thymine glycol and thymidine glycol in human and rat urine: a possible assay for oxidative DNA damage. Proc. Natl Acad. Sci. USA. 1984;81:5633–5637. doi: 10.1073/pnas.81.18.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adelman R, Saul RL, Ames BN. Oxidative damage to DNA: relation to species metabolic rate and life span. Proc. Natl Acad. Sci. USA. 1988;85:2706–2708. doi: 10.1073/pnas.85.8.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kung HC, Bolton PH. Structure of a duplex DNA containing a thymine glycol residue in solution. J. Biol. Chem. 1997;272:9227–9236. doi: 10.1074/jbc.272.14.9227. [DOI] [PubMed] [Google Scholar]
- 11.Kao JY, Goljer I, Phan TA, Bolton PH. Characterization of the effects of a thymine glycol residue on the structure, dynamics, and stability of duplex DNA by NMR. J. Biol. Chem. 1993;268:17787–17793. [PubMed] [Google Scholar]
- 12.Brown KL, Adams T, Jasti VP, Basu AK, Stone MP. Interconversion of the cis-5R,6S- and trans-5R,6R-thymine glycol lesions in duplex DNA. J. Am. Chem. Soc. 2008;129:11701–11710. doi: 10.1021/ja8016544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark JM, Pattabiraman N, Jarvis W, Beardsley GP. Modeling and molecular mechanical studies of the cis-thymine glycol radiation damage lesion in DNA. Biochemistry. 1987;26:5404–5409. doi: 10.1021/bi00391a028. [DOI] [PubMed] [Google Scholar]
- 14.Brown KL, Basu AK, Stone MP. The cis-(5R,6S)-thymine glycol lesion occupies the wobble position when mismatched with dG in DNA. Biochemistry. 48:9722–9733. doi: 10.1021/bi900695e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ide H, Kow YW, Wallace SS. Thymine glycols and urea residues in M13 DNA constitute replicative blocks in vitro. Nucleic Acids Res. 1985;13:8035–8052. doi: 10.1093/nar/13.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark JM, Beardsley GP. Thymine glycol lesions terminate chain elongation by DNA polymerase I in vitro. Nucleic Acids Res. 1986;14:737–749. doi: 10.1093/nar/14.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ocampo-Hafalla MT, Altamirano A, Basu AK, Chan MK, Ocampo JE, Cummings A., Jr, Boorstein RJ, Cunningham RP, Teebor GW. Repair of thymine glycol by hNth1 and hNeil1 is modulated by base pairing and cis-trans epimerization. DNA Repair (Amst) 2006;5:444–454. doi: 10.1016/j.dnarep.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Lin JJ, Sancar A. A new mechanism for repairing oxidative damage to DNA: (A)BC excinuclease removes AP sites and thymine glycols from DNA. Biochemistry. 1989;28:7979–7984. doi: 10.1021/bi00446a002. [DOI] [PubMed] [Google Scholar]
- 19.Kow YW, Wallace SS, Van Houten B. UvrABC nuclease complex repairs thymine glycol, an oxidative DNA base damage. Mutat. Res. 1990;235:147–156. doi: 10.1016/0921-8777(90)90068-g. [DOI] [PubMed] [Google Scholar]
- 20.Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc. Natl Acad. Sci. USA. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z, Colis LC, Basu AK, Zou Y. Recognition and incision of gamma-radiation-induced cross-linked guanine-thymine tandem lesion G[8,5-Me]T by UvrABC nuclease. Chem. Res. Toxicol. 2005;18:1339–1346. doi: 10.1021/tx050147+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Liu Y, Yang Z, Utzat C, Wang G, Basu AK, Zou Y. Cooperative interaction of human XPA stabilizes and enhances specific binding of XPA to DNA damage. Biochemistry. 2005;44:7361–7368. doi: 10.1021/bi047598y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 24.Carr HY, Purcell EM. Effects of diffusion on free precession in nuclear magnetic resonance experiments. Phys. Rev. 1954;94:630. [Google Scholar]
- 25.Vold RL, Waugh JS, Klein MP, Phelps DE. Measurement of spin relaxation in complex systems. J. Chem. Phys. 1968;48:3831–3832. [Google Scholar]
- 26.Goddard TD, Kneller DG. San Francisco: University of California; 2006. SPARKY v. 3.113. [Google Scholar]
- 27.Keepers JW, James TL. A theoretical study of distance determinations from NMR – two-dimensional nuclear Overhauser effect spectra. J. Magn. Reson. 1984;57:404–426. [Google Scholar]
- 28.James TL. Relaxation matrix analysis of two-dimensional nuclear Overhauser effect spectra. Curr. Opin. Struct. Biol. 1991;1:1042–1053. [Google Scholar]
- 29.Borgias BA, James TL. Two-dimensional nuclear Overhauser effect: complete relaxation matrix analysis. Methods Enzymol. 1989;176:169–183. doi: 10.1016/0076-6879(89)76011-0. [DOI] [PubMed] [Google Scholar]
- 30.Borgias BA, James TL. MARDIGRAS – a procedure for matrix analysis of relaxation for discerning geometry of an aqueous structure. J. Magn. Reson. 1990;87:475–487. [Google Scholar]
- 31.Liu H, Spielmann HP, Ulyanov NB, Wemmer DE, James TL. Interproton distance bounds from 2D NOE intensities: effect of experimental noise and peak integration errors. J. Biomol. NMR. 1995;6:390–402. doi: 10.1007/BF00197638. [DOI] [PubMed] [Google Scholar]
- 32.Delaglio F, Wu Z, Bax A. Measurement of homonuclear proton couplings from regular 2D COSY spectra. J. Magn. Reson. 2001;149:276–281. doi: 10.1006/jmre.2001.2297. [DOI] [PubMed] [Google Scholar]
- 33.Van De Ven FJM, Hilbers CW. Nucleic acids and nuclear magnetic resonance. Eur. J. Biochem. 1988;178:1–38. doi: 10.1111/j.1432-1033.1988.tb14425.x. [DOI] [PubMed] [Google Scholar]
- 34.van Wijk J, Huckriede BD, Ippel JH, Altona C. Furanose sugar conformations in DNA from NMR coupling constants. Methods Enzymol. 1992;211:286–306. doi: 10.1016/0076-6879(92)11017-d. [DOI] [PubMed] [Google Scholar]
- 35.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Vreven T, Kudin KN, Burant JC, et al. Wallingford, CT: Gaussian, Inc.; 2004. GAUSSIAN 03. [Google Scholar]
- 36.Case DA, Cheatham TE, III, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The AMBER biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macke T, Case DA. In: Molecular Modeling of Nucleic Acids. Leontes NB, SantaLucia J Jr, editors. Vol. 682. Washington, D.C: American Chemical Society; 1998. [Google Scholar]
- 38.Wang JM, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 39.Clore GM, Brunger AT, Karplus M, Gronenborn AM. Application of molecular dynamics with interproton distance restraints to three-dimensional protein structure determination. J. Mol. Biol. 1986;191:523–551. doi: 10.1016/0022-2836(86)90146-4. [DOI] [PubMed] [Google Scholar]
- 40.Tsui V, Case DA. Theory and applications of the generalized Born solvation model in macromolecular simulations. Biopolymers. 2000;56:275–291. doi: 10.1002/1097-0282(2000)56:4<275::AID-BIP10024>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 41.Bashford D, Case DA. Generalized Born models of macromolecular solvation effects. Annu. Rev. Phys. Chem. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 42.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Phys. Chem. 1984;81:3684–3690. [Google Scholar]
- 43.Loncharich RJ, Brooks BR, Pastor RW. Langevin dynamics of peptides: the frictional dependence of isomerization rates of N-acetylalanyl-N′-methylamide. Biopolymers. 1992;32:523–535. doi: 10.1002/bip.360320508. [DOI] [PubMed] [Google Scholar]
- 44.Izaguirre JA, Catarello DP, Wozniak JM, Skeel RD. Langevin stabilization of molecular dynamics. J. Chem. Phys. 2001;114:2090–2098. [Google Scholar]
- 45.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 46.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of cartesian equations of motion of a system with constraints: molecular dynamics of N-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 47.Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic-acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 48.Ravishanker G, Swaminathan S, Beveridge DL, Lavery R, Sklenar H. Conformational and helicoidal analysis of 30 ps of molecular-dynamics on the d(CGCGAATTCGCG) double helix – curves, dials and windows. J. Biomol. Struct. Dyn. 1989;6:669–699. doi: 10.1080/07391102.1989.10507729. [DOI] [PubMed] [Google Scholar]
- 49.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 50.Sanner MF, Olson AJ, Spehner JC. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 51.Mekhovich O, Tang M, Romano LJ. Rate of incision of N-acetyl-2-aminofluorene and N-2-aminofluorene adducts by UvrABC nuclease is adduct- and sequence-specific: comparison of the rates of UvrABC nuclease incision and protein–DNA complex formation. Biochemistry. 1998;37:571–579. doi: 10.1021/bi971544p. [DOI] [PubMed] [Google Scholar]
- 52.Zou Y, Shell SM, Utzat CD, Luo C, Yang Z, Geacintov NE, Basu AK. Effects of DNA adduct structure and sequence context on strand opening of repair intermediates and incision by UvrABC nuclease. Biochemistry. 2003;42:12654–12661. doi: 10.1021/bi034446e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meneni SR, D'M;ello R, Norigian G, Baker G, Gao L, Chiarelli MP, Cho BP. Sequence effects of aminofluorene-modified DNA duplexes: thermodynamic and circular dichroism properties. Nucleic Acids Res. 2006;34:755–763. doi: 10.1093/nar/gkj480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reid BR. Sequence-specific assignments and their use in NMR studies of DNA structure. Q. Rev. Biophys. 1987;20:2–28. doi: 10.1017/s0033583500004212. [DOI] [PubMed] [Google Scholar]
- 55.Patel DJ, Shapiro L, Hare D. DNA and RNA: NMR studies of conformations and dynamics in solution. Q. Rev. Biophys. 1987;20:35–112. doi: 10.1017/s0033583500004224. [DOI] [PubMed] [Google Scholar]
- 56.Boelens R, Scheek RM, Dijkstra K, Kaptein R. Sequential assignment of imino- and amino-proton resonances in 1H NMR spectra of oligonucleotides by two-dimensional NMR spectroscopy. Application to a lac operator fragment. J. Magn. Reson. 1985;62:378–386. [Google Scholar]
- 57.Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat. Rev. Cancer. 2005;5:564–573. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]
- 58.Cleaver JE. Historical aspects of xeroderma pigmentosum and nucleotide excision repair. Adv. Exp. Med. Biol. 2008;637:1–9. doi: 10.1007/978-0-387-09599-8_1. [DOI] [PubMed] [Google Scholar]
- 59.Legerski R, Peterson C. Expression cloning of a human DNA repair gene involved in xeroderma pigmentosum group C. Nature. 1992;359:70–73. doi: 10.1038/359070a0. [DOI] [PubMed] [Google Scholar]
- 60.Masutani C, Sugasawa K, Yanagisawa J, Sonoyama T, Ui M, Enomoto T, Takio K, Tanaka K, van der Spek PJ, Bootsma D, et al. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 1994;13:1831–1843. doi: 10.1002/j.1460-2075.1994.tb06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 62.Gunz D, Hess MT, Naegeli H. Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J. Biol. Chem. 1996;271:25089–25098. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- 63.Buterin T, Hess MT, Luneva N, Geacintov NE, Amin S, Kroth H, Seidel A, Naegeli H. Unrepaired fjord region polycyclic aromatic hydrocarbon-DNA adducts in ras codon 61 mutational hot spots. Cancer Res. 2000;60:1849–1856. [PubMed] [Google Scholar]
- 64.Hey T, Lipps G, Sugasawa K, Iwai S, Hanaoka F, Krauss G. The XPC-HR23B complex displays high affinity and specificity for damaged DNA in a true-equilibrium fluorescence assay. Biochemistry. 2002;41:6583–6587. doi: 10.1021/bi012202t. [DOI] [PubMed] [Google Scholar]
- 65.Dip R, Camenisch U, Naegeli H. Mechanisms of DNA damage recognition and strand discrimination in human nucleotide excision repair. DNA Repair (Amst) 2004;3:1409–423. doi: 10.1016/j.dnarep.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 66.Buterin T, Meyer C, Giese B, Naegeli H. DNA quality control by conformational readout on the undamaged strand of the double helix. Chem. Biol. 2005;12:913–922. doi: 10.1016/j.chembiol.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 67.Yang ZG, Liu Y, Mao LY, Zhang JT, Zou Y. Dimerization of human XPA and formation of XPA2-RPA protein complex. Biochemistry. 2002;41:13012–13020. doi: 10.1021/bi026064z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang Z, Roginskaya M, Colis LC, Basu AK, Shell SM, Liu Y, Musich PR, Harris CM, Harris TM, Zou Y. Specific and efficient binding of xeroderma pigmentosum complementation group A to double-strand/single-strand DNA junctions with 3′- and/or 5′-ssDNA branches. Biochemistry. 2006;45:15921–15930. doi: 10.1021/bi061626q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Riedl T, Hanaoka F, Egly JM. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 2003;22:5293–5303. doi: 10.1093/emboj/cdg489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hruska FE, Sebastian R, Grand A, Voituriez L, Cadet J. Characterization of a gamma-radiation-induced decomposition product of thymidine. Crystal and molecular structure of the (-)cis(5R,6S) thymidine glycol. Can. J. Chem. 1987;65:2618–2623. [Google Scholar]
- 71.Aller P, Rould MA, Hogg M, Wallace SS, Doublie S. A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc. Natl Acad. Sci. USA. 2007;104:814–818. doi: 10.1073/pnas.0606648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.O'H;andley SF, Sanford DG, Xu R, Lester CC, Hingerty BE, Broyde S, Krugh TR. Structural characterization of an N-acetyl-2-aminofluorene (AAF) modified DNA oligomer by NMR, energy minimization, and molecular dynamics. Biochemistry. 1993;32:2481–2497. doi: 10.1021/bi00061a005. [DOI] [PubMed] [Google Scholar]
- 73.Sugasawa K, Shimizu Y, Iwai S, Hanaoka F. A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair (Amst) 2002;1:95–107. doi: 10.1016/s1568-7864(01)00008-8. [DOI] [PubMed] [Google Scholar]
- 74.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 75.Miller J, Miaskiewicz K, Osman R. Structure–function studies of DNA damage using ab initio quantum mechanics and molecular dynamics simulation. Ann. NY Acad. Sci. 1994;726:71–91. doi: 10.1111/j.1749-6632.1994.tb52799.x. [DOI] [PubMed] [Google Scholar]
- 76.Miaskiewicz K, Miller J, Ornstein R, Osman R. Molecular dynamics simulations of the effects of ring-saturated thymine lesions on DNA structure. Biopolymers. 1995;35:113–124. doi: 10.1002/bip.360350112. [DOI] [PubMed] [Google Scholar]
- 77.Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair (Amst) 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weiss B, Cunningham RP. Genetic mapping of Nth, a gene affecting endonuclease III (thymine glycol-DNA glycosylase) in Escherichia coli K-12. J. Bacteriol. 1985;162:607–610. doi: 10.1128/jb.162.2.607-610.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang D, Hatahet Z, Melamede RJ, Kow YW, Wallace SS. Characterization of Escherichia coli endonuclease VIII. J. Biol. Chem. 1997;272:32230–32239. doi: 10.1074/jbc.272.51.32230. [DOI] [PubMed] [Google Scholar]
- 80.Roldan-Arjona T, Anselmino C, Lindahl T. Molecular cloning and functional analysis of a Schizosaccharomyces pombe homologue of Escherichia coli endonuclease III. Nucleic Acids Res. 1996;24:3307–3312. doi: 10.1093/nar/24.17.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hilbert TP, Boorstein RJ, Kung HC, Bolton PH, Xing D, Cunningham RP, Teebor GW. Purification of a mammalian homologue of Escherichia coli endonuclease III: identification of a bovine pyrimidine hydrate-thymine glycol DNAse/AP lyase by irreversible cross linking to a thymine glycol-containing oligoxynucleotide. Biochemistry. 1996;35:2505–2511. doi: 10.1021/bi952516e. [DOI] [PubMed] [Google Scholar]
- 82.Sarker AH, Ikeda S, Nakano H, Terato H, Ide H, Imai K, Akiyama K, Tsutsui K, Bo Z, Kubo K, et al. Cloning and characterization of a mouse homologue (mNthl1) of Escherichia coli endonuclease III. J. Mol. Biol. 1998;282:761–774. doi: 10.1006/jmbi.1998.2042. [DOI] [PubMed] [Google Scholar]
- 83.Hilbert TP, Chaung W, Boorstein RJ, Cunningham RP, Teebor GW. Cloning and expression of the cDNA encoding the human homologue of the DNA repair enzyme, Escherichia coli endonuclease III. J. Biol. Chem. 1997;272:6733–6740. doi: 10.1074/jbc.272.10.6733. [DOI] [PubMed] [Google Scholar]
- 84.Aspinwall R, Rothwell DG, Roldan-Arjona T, Anselmino C, Ward CJ, Cheadle JP, Sampson JR, Lindahl T, Harris PC, Hickson ID. Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc. Natl Acad. Sci. USA. 1997;94:109–114. doi: 10.1073/pnas.94.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ikeda S, Biswas T, Roy R, Izumi T, Boldogh I, Kurosky A, Sarker AH, Seki S, Mitra S. Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys-212 as the active nucleophilic residue. J. Biol. Chem. 1998;273:21585–21593. doi: 10.1074/jbc.273.34.21585. [DOI] [PubMed] [Google Scholar]
- 86.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl Acad. Sci. USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra S, Izumi T. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J. Biol. Chem. 2002;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 88.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 89.Katafuchi A, Nakano T, Masaoka A, Terato H, Iwai S, Hanaoka F, Ide H. Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. J. Biol. Chem. 2004;279:14464–14471. doi: 10.1074/jbc.M400393200. [DOI] [PubMed] [Google Scholar]
- 90.Miller H, Fernandes AS, Zaika E, McTigue MM, Torres MC, Wente M, Iden CR, Grollman AP. Stereoselective excision of thymine glycol from oxidatively damaged DNA. Nucleic Acids Res. 2004;32:338–345. doi: 10.1093/nar/gkh190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jia L, Shafirovich V, Geacintov NE, Broyde S. Lesion specificity in the base excision repair enzyme hNeil1: modeling and dynamics studies. Biochemistry. 2007;46:5305–5314. doi: 10.1021/bi062269m. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.