Abstract
In this study, we demonstrate the identification of an internal ribosome entry site (IRES) within the 5′-untranslated region (5′-UTR) of the mouse mammary tumor virus (MMTV). The 5′-UTR of the full-length mRNA derived from the infectious, complete MMTV genome was cloned into a dual luciferase reporter construct containing an upstream Renilla luciferase gene (RLuc) and a downstream firefly luciferase gene (FLuc). In rabbit reticulocyte lysate, the MMTV 5′-UTR was capable of driving translation of the second cistron. In vitro translational activity from the MMTV 5′-UTR was resistant to the addition of m7GpppG cap-analog and cleavage of eIF4G by foot-and-mouth disease virus (FMDV) L-protease. IRES activity was also demonstrated in the Xenopus laevis oocyte by micro-injection of capped and polyadenylated bicistronic RNAs harboring the MMTV-5′-UTR. Finally, transfection assays showed that the MMTV-IRES exhibits cell type-dependent translational activity, suggesting a requirement for as yet unidentified cellular factors for its optimal function.
INTRODUCTION
Mouse mammary tumor virus (MMTV), the prototype of betaretrovirus, causes breast cancer in mice (1). MMTV is transmitted as an exogenous virus through the milk of infected female mice to newborn pups or through endogenous proviruses (Mtvs) integrated within the germline (2). Interestingly, MMTV can infect, replicate and spread in human cells (3,4), while various MMTV-like sequences have been identified in human breast cancers (5–7) and in liver in a wide range of hepatic disorders (8). These findings highlight a possible role for MMTV, or MMTV-like viruses in human disease. Strikingly, and although MMTV was discovered more than half a century ago (9), the biology of this virus remains poorly understood. In this study, we contribute to the understanding of MMTV by presenting evidence to demonstrate that, akin to other retroviruses, the MMTV full-length mRNA harbors an internal ribosome entry site (IRES).
Translation initiation of the vast majority of eukaryotic mRNAs occurs by a cap-dependent mechanism. This mechanism involves the recognition of the 5′-cap structure (m7GpppN) by eukaryotic translation initiation factors (eIFs), followed by binding of the 40S ribosomal subunit and scanning downstream to the initiation codon [reviewed in (10)]. A number of eIFs participate in both mRNA binding and scanning by the small ribosomal subunit of the 5′-untranslated region (5′-UTR). The cap-binding protein eIF4E together with the ATP-dependent RNA helicase eIF4A and translation IF eIF4G form the protein complex eIF4F, which binds and mediates recruitment of the 40S ribosomal subunit to the 5′-cap structure. 1988 heralded the discovery that translation of the uncapped picornaviral mRNA is mediated by an RNA structure, the IRES, which allows assembly of the translational machinery at a position close to or directly at the initiation codon (11,12). Since the initial characterization of IRES elements in Picornaviridae, viruses from other families including several members of the Retroviridae have been shown to initiate translation via an IRES (13). An interesting feature, common to many retroviruses found to harbor an IRES, is that their viral-encoded proteases cleave eIF4G, hydrolysis that leads to the inhibition of cap-dependent but not IRES-mediated translation (14–17). Interestingly, the MMTV protease eIF4GI both in intact cells and in cell-free systems, inhibiting cap-dependent translation initiation (14). Intrigued by these findings, we set out in this study to evaluate if the 5′-UTR of the MMTV mRNA was capable of initiating translation using a cap-independent mechanism. Results presented herein describe an IRES element present within the 5′-UTR of MMTV.
MATERIALS AND METHODS
Cell culture
NIH3T3 (CRL-1658TM), HeLa (CCL-2TM), 293T (CRL-11268TM) and NMuMG (CRL-1636TM) cell lines were grown in Dulbecco modified Eagle medium (DMEM, Gibco BRL, Life Technologies Corporation, Carlsbad, CA, USA) containing 10% bovine fetal serum (HyClone, Logan, UT, USA) and 1% of penicillin–streptomycin (Gibco BRL). T47D (HTB-133TM) cells were grown in RPMI medium without phenol red (HyClone) containing 10% bovine fetal serum, 1% of penicillin–streptomycin (Gibco BRL) and 2 mg/ml insulin (Novo Nordisk A/S, Bagsvaerd, Denmark). All cell lines were grown at 37°C in a 5% CO2 atmosphere.
Plasmid constructions
For the generation of the bicistronic vector dl 5′-UTR-MMTV, the 5′-UTR of MMTV (320 bp) was recovered by PCR from plasmid pGR102 (kindly provided by W. H. Günzburg and B. Salmons, Institute of Virology, University of Veterinary Sciences, Vienna, Austria) using primers MMTV5′ (5′-CCCGGAATTCGCAACAGTCCTAACATTCACC-3′) and MMTV3′ (5′-ACCCCCATGGTCCAATGGCTCACCGTAAC-3′). Plasmid pGR102 harbors a biologically active MMTV genomic clone and has been previously described (18). The MMTV3′ primer was designed to eliminate an additional AUG sequence found upstream from the original Gag AUG present only in the pGR102 plasmid and not in wild-type virus (genbank accession: M15122). Two cysteine residues were added to the original sequence to allow in-frame cloning into the dual luciferase vector by creating a NcoI restriction site as previously described (19). Nucleotide modifications with respect to the pGR102 sequence introduced during PCR are indicated above in bold italics within the sequence of the primer MMTV3′. The amplicon was digested with EcoRI and NcoI (both restriction sites added by PCR) and inserted into the intercistronic region of dl HIV-1 IRES plasmid (19) previously digested with the same enzymes (Fermentas, Vilnius, Lithuania). The final vector, dl 5′-UTR-MMTV, was verified by sequence analysis (Macrogen Corp, Rockville, MD, USA). The dl ΔEMCV, dl HIV-1 IRES and dl HCV IRES plasmids were previously described (19–21). To generate plasmids without the SV40 mammalian promoter, the dl 5′-UTR-MMTV and dl ΔEMCV vectors were digested with HindIII and MluI followed by a ligation using T4 DNA Ligase (Fermentas).
In vitro transcription
Uncapped mRNAs were synthesized with T7 RNA polymerase. In brief, plasmids linearized with the requisite enzyme were used for in vitro transcription in a 25-µl final volume (Ribomax T7, Promega Corporation, Madison, WI, USA). Template DNA was digested with DNase I, and RNA was precipitated with 2.5 M LiCl. RNA was resuspended in diethyl pyrocarbonate (DEPC)-treated water. Capped mRNAs were synthesized using the mMESSAGE mMACHINE High Yield Capped RNA Transcription Kit (Applied Biosystems/Ambion, Austin, TX, USA) following manufacturer’s specifications. The poly(A) tailing kit (Applied Biosystems/Ambion) was used to add poly(A) tail to the capped mRNAs according to the manufacturer’s specifications. RNA concentrations were determined spectrophotometrically (NanoDrop Technology, Wilmington, DE, USA) and RNA integrity was monitored by electrophoresis on denaturing agarose gels.
In vitro translation
In vitro translations were carried out in nuclease-treated Rabbit Reticulocyte Lysate or the Flexi® rabbit reticulocyte system (RRL, Promega), at 35% (v/v), supplemented with 20 µM amino acids (Promega), 8 U/μl Ribolock® Rnase inhibitor (Fermentas), potassium acetate (KOAc) (in the range of 40–200 mM as indicated in the figure legend or text), magnesium acetate (MgOAc2) (in the range of 0.25–1.5 mM as indicated in the figure legend) and RNA at final concentration of 1–6.25 ng/μl. For cap-analog experiments, Flexi® Rabbit Reticulocyte System was pre-incubated for 15 min at 30°C with 250 or 500 µM of the Ribo-m7G-Cap Analog (Promega), then bicistronic mRNAs (6.25 ng/μl) were added for in vitro translation. Reactions were conducted in 80 mM KOAc and Mg2+ were adjusted for each experimental condition by addition of MgOAc2. Reactions that lacked cap analog were conducted in a final concentration of 0.75 mM Mg2+, reactions with 0.25 mM of m7GpppG were conducted in a final concentration of 1.125 mM Mg2+ and those with 0.5 mM of m7GpppG were conducted in a final concentration of 1.5 mM Mg2+, as suggested by others (22). For the foot-and-mouth disease virus (FMDV) L-protease assays, uncapped RNA of the FMDV L-protease was in vitro translated in nuclease-treated RRL as described above from the pLb plasmid (a kind gift of Dr G. Belsham, Institute for Animal Health, Pirbright, UK) (23). In brief, 220 ng/μl of the FMDV L-protease uncapped RNA was translated in 35% (v/v) of RRL for 90 min. The L-protease-RRL was diluted in fresh RRL 35% (v/v) in a ratio of 1:2 and 1:4. The diluted L-protease-RRL was added to the fresh RRL used for the translation of the bicistronic mRNAs to a final concentration of 2 and 4% (v/v) of Protease L. Translation reactions were incubated at 30°C for 90 min. Firefly luciferase (FLuc) and Renilla luciferase (RLuc) activities were measured using the DLR™ Assay System (Promega) according to manufacturer’s instructions on a Sirius Single Tube Luminometer (Berthold Detection Systems GmbH, Pforzheim, Germany).
Western blot
eIF4GI cleavage by FMDV L-protease was assessed by 5–20% gradient SDS–PAGE of RRL (10 μl), followed by transfer to nitrocellulose (Pierce Biotechnology, Inc. Rockford, IL, USA), and eIF4GI detection using a mixture of well-characterized polyclonal antibodies against the N- and C-terminal fragments of eIF4GI (kindly provided by Dr L. Carrasco, Centro de Biología Molecular Severo Ochoa, Madrid, Spain), as previously described (24,25).
DNA transfection
Cells were seeded at 1 × 105 cell/well in 12-well culture plates. DNA transfection was performed at 60% confluency by the JetPei system (Polyplus-transfection SA, Illkirch, France) according to manufacturer’s protocols. After 24 h, the culture medium was removed and the cells were directly mixed with the Passive Lysis buffer supplied with the DLR™ Assay System (Promega). FLuc and RLuc activities were measured as described above. The activity of β-galactosidase was measured in the same cells using the Beta-Glo™ Assay System according to manufacturer’s protocols (Promega). Protein concentration was determined by a Bradford assay using the Bio-Rad Protein Assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
RNA, DNA extraction, PCR and RT-PCR
Cells in culture were trypsinized (Gibco BRL) and resuspended in DMEM (Gibco BRL) containing 10% bovine fetal serum (HyClone). Cells were collected by centrifugation at 710 g for 5 min and washed three times in phosphate-buffered s aline (PBS, 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 · 7H2O, 1.4 mM KH2PO4, pH 7.4) at 4°C. For RNA extraction, the cell pellets were directly lysed in solution D (4 M guanidine thiocyanate, 25 mM sodium citrate pH 7.0, 0.55% w/v sarkosyl, 0.1 M β-mercaptoethanol) at 500 µl/105 cells, and 1/10 volume of sodium acetate 3 M pH 4.2, 1 volume of water-saturated phenol and 1/10 volume of chloroform/isoamyl alcohol (24:1) were added, vortexed for 15 s and incubated 15 min on ice. The mixture was centrifuged for 15 min at 16 000g at 4°C, the aqueous phase was transferred to a new tube, precipitated with 500 µl of isopropanol at −20°C for 3 h and centrifuged for 30 min at 16 000g at 4°C. The pellet was washed with 100 µl of 75% ethanol, centrifuged for 5 min at 16 000g at 4°C and resuspended in 20 µl of DEPC–water and was DNase-treated and re-extracted as described above. RNA integrity was monitored by electrophoresis on denaturing agarose gels and quantified by spectrophotometry (NanoDrop Technology). DNA was extracted from cells using the EZNA™ kit (Omega Bio-Tek, Inc., GA, USA) according to the manufacture manual. DNA concentration was determined by spectrophotometry (NanoDrop Technology).
The RT-PCR assay was carried out using the SuperScript™ III One-Step RT-PCR System with Platinum® Taq DNA polymerase kit (Invitrogen, Life Technologies Corporation, Carlsbad, CA, USA) according to manufacturer’s protocol, using 3 µg of total RNA and primers p2anti (5′-TCTCTTCATAGCCTTATGCAGTTG-3′) and Pforluc (5′-CATGACTTCGAAAGTTTATGATC-3′). The PCR assay was conducted using primers p2anti and Pforluc, 100 ng of total DNA and the Go Taq® Green Master mix (Promega), according to the manufacturer’s protocol.
Oocyte harvesting and RNA microinjection
Oocytes were isolated from Xenopus laevis ovarian fragments by manual dissection as previously described (26). Oocytes were incubated at 15°C for 24 h in standard Barth’s solution supplemented with 10 IU/l penicillin–streptomycin and 2 mM pyruvate (26). To evaluate viral IRES activity in oocytes, 6.25 ng of in vitro transcribed capped and polyadenylated RNA was microinjected into each oocyte with glass micropipettes calibrated to deliver a final volume of 50 nl (27,28). After 3, 6, 12 or 24 h (refer to the figure legend) oocytes were lysated in Passive Lysis buffer (Promega), and centrifuged at 16 000g for 5 min, and 1–5 μl of the supernatant was used in the detection assay using the kit DLR™ Assay System (Promega).
HIV-1 Rev and MMTV Rem plasmid constructs for protein expression
The HIV-1 rev gene was derived from the plasmid pCMVsrevBF and inserted into the commercially available prokaryotic expression vector pET-3a (Stratagene, La Jolla, CA, USA) to create the intermediate plasmid pETsRev and thereafter pETsRevCHis, wherein six histidine residues (His) were fused in-frame immediately downstream of rev to generate a C-terminal His tag. These plasmids have been previously described (29).
The MMTV rem gene used in these studies has been previously described in detail (30) and was obtained as a kind gift from Dr S. Indik (Institute for Virology, University of Veterinary Medicine Vienna, Austria). To insert the rem gene into the pET-3a vector for prokarytotic protein expression, the rev gene in the vector pETsRev was removed by restriction digestion with NdeI and BamHI and replaced by an NdeI/BamHI rem PCR fragment to create the intermediate cloning plasmid pETRem. Using a PCR mutagenesis method that has been previously described (31), a C-terminal His tag was then inserted immediately downstream of rem by means of overlapping, matching, sense and anti-sense ends. Briefly, by annealing the two products together in a reaction without primers, the His tag became inserted in-frame immediately upstream of the rem stop codon. In a third PCR step, using only outside primers, a complete RemCHis fragment was then amplified and could be re-inserted into pETRem vector using again the NdeI and BamHI sites. The resulting plasmid was termed pETRemCHis. A final plasmid was then created for protein expression of the active, signal peptide (SP) region component of the full-length rem protein (RemSP). RemSP is the nucleocytoplasmic shuttling RNA transport factor component of Rem (30,32). Long-template PCR (Expand Kit from Roche Molecular Systems, Inc., Branchburg, NJ, USA) was performed to remove the C-terminal region of Rem, leaving only the SP region and the product was ligated to reform the fusion with the C-terminal His tag. The resulting plasmid pETRemSPCHis was sequenced to confirm the correct, tagged RemSP sequence.
Production of recombinant Rev and Rem protein
pETsRevCHis and pETsRemCHis were transformed by heat shock into BL21(DE3) bacteria (Stratagene) according to the manufacturer’s instructions. To ensure optimal activity, purity and quantity, Rev was obtained under denaturing and refolding conditions as previously described in detail (29). In contrast to Rev, the majority of RemSP protein was expressed as a soluble protein after 3 h at 37°C with 1 mM isopropyl-β-d-thio-galactopyranoside stimulation. Stimulated bacterial pellet was resuspended in PR buffer (20 mM phosphate buffer pH 7.4, 0.5 M NaCl, 20 mM Na-EDTA) and spiked with protein inhibitor complex for His-tagged proteins (Sigma Aldrich, St Louis, MO, USA) as per manufacturer’s instructions. Lysozyme (Sigma), dissolved in 10 mM phosphate buffer with 0.5 M NaCl, was added to a final concentration of 2 mg/ml and the sample was subjected to two sonication and vortexing cycles on ice to disrupt bacteria. To destroy nucleic acids, 500 U Benzonase (Sigma) were added per gram of pellet in the presence of a final concentration of 20 mM MgCl2. The sample was centrifuged at high speed to remove remaining debris and the supernatant used for protein purification using HisTrap FF Crude (fast flow) chelating high-performance (HP) sepharose columns (GE Healthcare, Piscataway, NJ, USA) and the native protein, step elution programme of the ÄKTAprime protein purification unit (GE Healthcare) according to the manufacturer’s guidelines. To localize RemSP protein in eluted fractions, aliquots of the fractions were analyzed by SDS-PAGE followed by silver staining and the positive fractions containing only pure protein were concentrated via ultrafiltration using a regenerated cellulose membrane with a molecular weight cut-off of 5000 D Centricon units (Millipore, Billerica, MA, USA). Protein concentration, typically ∼5 µg/µl, was determined by the Bradford assay (Bio-Rad Laboratories). An aliquot of the final refolded, purified and concentrated RemSP protein was tested by western blot to confirm correct identity of the protein. Western blot was performed using either an anti-p14 (RemSP) antisera (a kind gift from Dr J. Hochman, the Hebrew University of Jerusalem, Jerusalem, Israel) or a commercially available anti-His-tag antisera. Coomassie staining also showed the protein to be of high integrity and >99% purity. RemSP aliquots were frozen at −80°C in 15% glycerol. Activity was assayed by electro-mobility gel-shift assay (EMSA).
EMSA
For the production of radio-labeled test RNAs (RRE), 10% of the total CTP in an in vitro transcription reaction (RiboMax T7, Promega) was substituted for [α-32P]CTP (GE Healthcare). RNA-protein binding reactions were carried out in DEPC-treated PBS containing 50 μg/ml BSA, 10 μg of eukaryotic tRNA and 10 U of RNaseOut (Invitrogen). First, 1–2 μg of Rev or RemSP was added to the reaction mix and left to equilibrate for 15 min at room temperature before 32P-labeled RNA probe was added and incubated for a further 15 min. Samples were then subjected to 5% 60:1 native TBE PAGE. For competition experiments, unlabeled RNA was DNase treated, purified via phenol/chloroform extraction and tested for integrity by agarose gel electrophoresis.
RESULTS
The MMTV 5′-UTR drives translation when in the context of a bicistronic mRNA
The full-length mRNA of some retroviruses can initiate protein synthesis through both a cap-dependent and cap-independent mechanism (13,33). With the exception of HIV-2 (34), IRES activity in retroviruses is mainly found within the 5′-UTR. In HIV-1 and SIV, a second IRES is found within the Gag-coding region (19,35–37). At present, an IRES element is defined solely by functional criteria and cannot yet be predicted by the presence of characteristic RNA sequences or structural motifs (38). Thus, to evaluate the possibility of an IRES element within MMTV, the 5′-UTR of its full-length mRNA was cloned into a dual luciferase (dl) reporter construct containing an upstream Renilla luciferase gene (RLuc) and a downstream firefly luciferase gene (FLuc). The 5′-UTR of MMTV was recovered from the pGR102 plasmid (18), which harbors the infectious, complete MMTV genome, as indicated in the ‘Material and Methods’ section. To ensure that the two cistrons are independently translated, a defective encephalomyocarditis virus IRES (ΔEMCV), known to inhibit ribosome reinitiation and readthrough (19,21), was inserted upstream of the MMTV 5′-UTR (Figure 1). In this context, the translational activity of the MMTV 5′-UTR was monitored using FLuc activity as the readout, while the RLuc reporter gene serves as an upstream translational control. In vitro, IRES-mediated translation initiation is highly dependent on the concentration of potassium and magnesium present within the translation mixture (19,35). Thus, in the first series of experiments, the putative IRES activity of the MMTV 5′-UTR was assessed by programming the nuclease-treated rabbit RRL with 1 ng/μl of RNA for translation in the presence of a range of KOAc or MgOAc2 concentrations (Figure 2A). RLuc and FLuc activities obtained in RRL (40 mM KOAc and 0.25 mM MgOAc2) without additional salt supplementation were arbitrarily set to 1 (±SEM). The data demonstrate that, when in the context of a bicistronic mRNA translation driven by the MMTV 5′-UTR bicistronic mRNA was substantial in RRL programmed with 1 ng/μl of RNA at 60 mM KOAc (Figure 2A, compare dl 5′UTR-MMTV and dlΔEMCV). Interestingly, and in keeping with what has been described for other retroviral IRESes (19,35), both cistrons showed a distinct response to varying K+ and Mg2+ concentrations, which itself is symptomatic of independent translation for RLuc and FLuc cistrons. Therefore, the MMTV 5′-UTR emerges as a good candidate for harboring an IRES element.
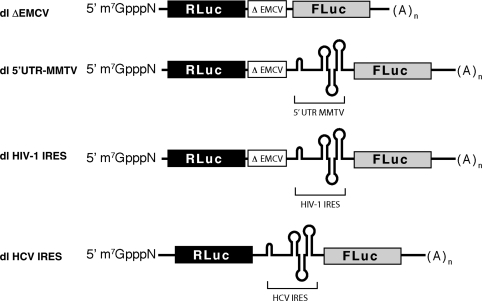
Figure 1.
Schematic representation of the bicistronic mRNAs used in this study. Bicistronic mRNAs dl ΔEMCV (19,21), dl HIV-1 IRES (19) and dl HCV IRES (20) have been previously described. RNAs correspond to the dual luciferase (dl) reporter construct containing an upstream Renilla luciferase gene (RLuc) and a downstream firefly luciferase gene (FLuc). In these RNAs, ΔEMCV corresponds to a defective encephalomyocarditis virus internal ribosome entry site known to inhibit ribosome re-initiation and readthrough (19,21), while HIV-1 IRES and HCV IRES correspond to the IRESes described in the HIV-1 5′-UTR (19) and the HCV 5′-UTR (20).
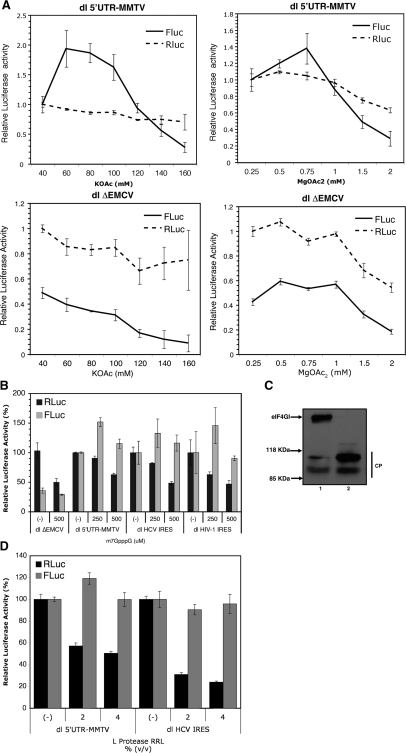
Figure 2.
Assessment of IRES activity within the MMTV-5′-UTR in nuclease-treated rabbit RRL. (A) dl 5′-UTR MMTV and dl ΔEMCV in vitro transcripts with 5′-caps were synthesized using T7 RNA polymerase and translated in RRL in the presence of different concentrations of KOAc and MgOAc2. (B) In vitro transcribed capped bicistronic RNA corresponding to dl ΔEMCV (19,21), dl 5′UTR-MMTV, dl HCV IRES (20) or dl HIV-1 IRES (19) were translated in salt-optimized RRL in the absence (−) or presence of 250 or 500 µM of m7GpppG cap-analog. In these assays, Mg2+ ions were adjusted to optimal concentrations as indicated in ‘Materials and Methods’ section (22). RLuc and FLuc luciferase activities [relative light units (RLUs)] were measured as indicated in ‘Materials and Methods’ section. The relative RLuc and FLuc activities for each RNA in the absence of cap-analog were arbitrarily set to 100% (±SEM). RLuc and FLuc values for the dl ΔEMCV RNA were compared against the dl 5′UTR-MMTV RNA. Values are the mean ± SEM from three independent experiments each conducted in triplicates. (C) Analysis of eIF4GI cleavage. RRL translation reactions (10 µl) with (lane 2) or without (lane 1) FMDV L protease (2% v/v) were resolved by SDS gradient 5–20% PAGE, transferred to nitrocellulose paper and incubated with a mixture of polyclonal antibodies against the N- and C-terminal fragments of eIF4GI as described in ‘Materials and Methods’ section. Positions of molecular mass standards (in kilo daltons) are shown. Polyclonal antibodies against N- and C-terminal peptides of eIF4GI were described previously (24). The cleavage products, CP in the figure, obtained with the FMDV-L protease treatment have been previously characterized (41). (D) Capped bicistronic RNA dl 5′UTR-MMTV or dl HCV IRES (20) were translated in RRL in the absence (−) or presence of 2 or 4% v/v of FMDV-L protease. The relative RLuc and FLuc activities for each RNA in the absence of FMDV-L protease were arbitrarily set to 100% (±SEM). Values are the mean ± SEM from three independent experiments each conducted in triplicates.
The MMTV 5′-UTR directs translation in RRL when cap-dependent translation initiation is inhibited
We then evaluated the translational activity of the MMTV 5′-UTR in salt-optimized RRL when cap-dependent translation initiation was inhibited, beginning with the addition of m7GpppG cap-analog to the in vitro translation reaction. The rational behind this approach lies in the competitive binding by cap-analog of the translation IF eIF4E, the cap-binding protein of the translation initiation complex (39). We used previously characterized bicisctronic mRNAs harboring IRES elements from either HIV-1 or HCV as experimental controls (Figure 1) (19,20). In these experiments, the dl ΔEMCV vector, containing only the defective encephalomyocarditis virus IRES inserted upstream of the FLuc reporter (Figure 1), was used as a negative control (19,20). The relative RLuc and FLuc activities for each RNA in the absence of cap-analog were arbitrarily set to 100% (±SEM). RLuc and FLuc values for the dl ΔEMCV dl RNA were compared against the dl 5′UTR-MMTV RNA (Figure 2B). m7GpppG cap-analog negatively impacted on the translation of RLuc, the first cistron, in all bicistronic RNAs, while it did not affect synthesis of FLuc driven by the MMTV 5′-UTR, the HIV-1 IRES or the HCV IRES (Figure 2B). These experiments were conducted in optimal salt conditions (Figure 1). Considering chelation of Mg2+ ion by m7GpppG cap-analog (22), ion concentration was adjusted accordingly as indicated in ‘Materials and Methods’ section. Interestingly, inhibition of cap-dependent translation by the addition of cap-analog initially increases translation driven from the MMTV 5′-UTR (Figure 2B), observations which endorse previous reports describing a stimulation of IRES-dependent translation when cap-dependent translation is inhibited in vitro (19,35,40). Intriguingly, at high concentrations of cap-analog (500 µM) activity from the HIV-1 IRES was not increased (Figure 2B). Even though these observations cannot be readily explained, they are consistent with what has been previously described for the HIV-1 IRES under conditions known to inhibit cap-dependent translation initiation [see Figure 5 of (19)].
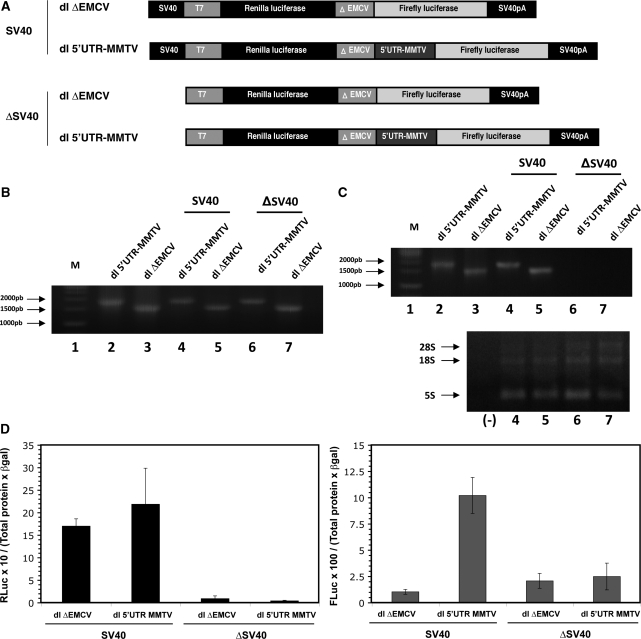
Figure 5.
Analysis of a promoter-less bicistronic construct containing the MMTV 5′-UTR sequence. (A) Schematic representation of the bicistronic constructs. The SV40 promoter from dl ΔEMCV (lane 3) or dl 5′UTR-MMTV was removed to generate the equivalent promoter-less (ΔSV40) vectors. (B) NMuMG cells were transfected with DNA (200 ng) corresponding to the vectors depicted in (A) together with the pcDNA3.1 lacZ (50 ng) plasmid. Total DNA was extracted from transfected NMuMG cells and the presence of the transfected plasmids was confirmed by PCR. Plasmids (100 ng) dl ΔEMCV (lane 3) or dl 5′UTR-MMTV (lane 2) were used as amplification controls. (C) Total RNA was extracted from transfected NMuMG cells and the presence of transcripts for the dl ΔEMCV (lane 3), the dl 5′UTR-MMTV, the Δ SV40-dl ΔEMCV and Δ SV40-dl 5′UTR-MMTV was evaluated by a one-step RT-PCR designed to detect the bicistronic RNA (depicted in Figure 3B). In vitro transcribed RNA (100 ng) generated from plasmids dl ΔEMCV (lane 3) or dl 5′UTR-MMTV (lane 2) were used as amplification controls. The presence of template RNA was confirmed in parallel by loading the total RNA (10 µg) used in the RT-PCR onto a 0.7% denaturing agarose gel. (D) NMuMG cell were co-transfected with either the SV40 or Δ SV40 version of dl ΔEMCV (200 ng) or dl 5′UTR-MMTV (200 ng) plasmids together with the pcDNA3.1 lacZ (50 ng) plasmid. Cells were processed, and β-galactosidase and total proteins were quantified. RLuc and FLuc activities were measured, and data are presented as [RLuc/(total protein) × (β-galactosidase)] (left panel) and [FLuc/(total protein) × (β-galactosidase)] (right panel).
Next we examined the impact of eIF4GI cleavage on the translation of dl-mRNAs harboring the 5′-UTR MMTV or the HCV IRES. The L-protease from FMDV cleaves eIF4GI into an N-terminal (one-third of the molecule) and a C-terminal (two-thirds of the molecule) domain (41). This proteolytic cleavage of eIF4GI resulted in inhibition of cap-dependent translation, while IRES-driven translation was unaffected or sometimes even stimulated (42,43). FMDV L-protease was prepared in RRL as previously described (44), and 2 or 4% v/v of RRL-L protease was added to fresh RRL as indicated in the ‘Materials and Methods’ section. Dual luciferase mRNAs were translated in the presence or absence of L-protease. Cleavage of eIF4GI was monitored (2% v/v of RRL-L protease; Figure 2C) by immunoblotting as previously described (24,25). The relative RLuc and FLuc activities for each RNA in the absence of L-protease were arbitrarily set to 100% (±SEM). Our results show that the cleavage of eIF4GI (Figure 2C) negatively impacts on translation of RLuc, the first cistron, from the dl 5′UTR-MMTV RNA, without affecting the synthesis of FLuc (Figure 2D). Similar behavior was observed with the dl HCV IRES mRNA (Figure 2D). Together, these data strongly suggest that the 5′-UTR of the MMTV full-length mRNA harbors an IRES.
The MMTV 5′-UTR mediates cap-independent translation initiation in X. laevis oocytes
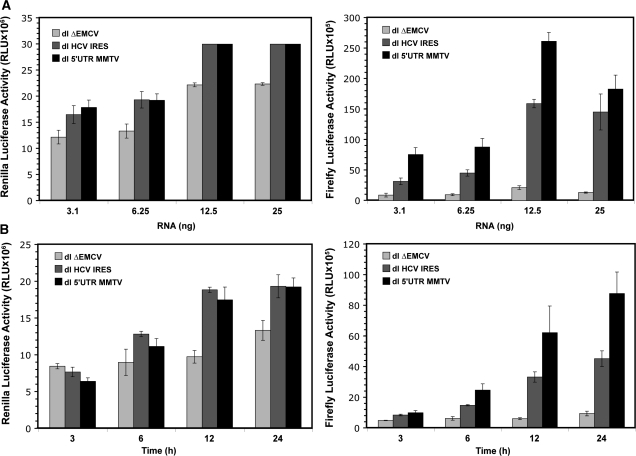
To further challenge our findings, we next evaluated if the 5′-UTR of MMTV could drive cap-independent translation in X. laevis oocytes, an experimental system which has been proven useful for the study of viral IRESes (45,46). Different concentrations of in vitro synthesized capped and polyadenylated RNA corresponding to dlΔEMCV, dl HCV IRES and dl 5′UTR-MMTV vectors were microinjected into the cytoplasm of X. laevis oocytes. RLuc and FLuc activities were measured 24 h post-injection (Figure 3A). RNA concentrations of ≥12.5 ng gave rise to saturating RLuc activity 24 h after injection. Thus, experiments were repeated using 6.25 ng of each RNA and RLuc and FLuc activities were now measured at different times post-injection (Figure 3B). The RLuc levels from the dl HCV IRES and the dl 5′UTR-MMTV mRNAs were comparable at all time points (Figure 3B), suggesting not only that cap-dependent translation initiation is functional in oocytes, but also confirming that similar amounts of RNA were microinjected in all experiments. A low level of expression from the second cistron of the dlΔEMCV reporter RNA, considered to be background, was also observed (Figure 3). Notably, FLuc was expressed above the dlΔEMCV background levels, from the dl 5′UTR-MMTV RNA (Figure 3), demonstrating that the MMTV 5′-UTR is capable of driving cap-independent translation in X. laevis oocytes. As an additional control dl ΔEMCV, dl HCV IRES and dl 5′UTR-MMTV DNA plasmids were microinjected into the cytoplasm of X. laevis oocytes, in all cases the RLuc and FLuc activity could not be detected (data not shown) discarding the possibility of de novo RNA synthesis, cryptic promoter activity and RNA splicing starting from a DNA template. Together, these data provide substantial evidence supporting the presence of an IRES within the MMTV 5′-UTR. Furthermore, these data also suggest that additional viral proteins are not required for MMTV-IRES activity in X. laevis oocytes.
Figure 3.
Assessment of IRES activity present within the MMTV-5′-UTR in X. laevis oocytes. (A) Different concentrations (3.1, 6.25, 12.5 or 25 ng) of capped and polyadenylated RNA corresponding to dl ΔEMCV, dl 5′UTR-MMTV or dl HCV IRES vectors were microinjected into X. laevis oocyte as described in ‘Material and Methods’ section. Oocytes were harvested 24 h after the microinjection and processed. Renilla luciferase (RLuc) and Firefly luciferase (FLuc) activities were determined (in RLUs). The RLuc (left panel) and FLuc (right panel) activities for each RNA in each concentration are shown. Oocytes injected with 12.5 and 25 ng of RNA showed saturating RLuc values. RLU values for oocytes injected with 25 ng RNA were obtained by diluting samples (1/4 volumes) prior to measurement. Each value is the mean ± SEM from at least nine oocytes obtained from two different animals. (B) Capped and polyadenylated RNA corresponding to the dl ΔEMCV, dl 5′UTR-MMTV or dl HCV IRES vectors (6.25 ng) were microinjected into X. laevis oocyte as described in ‘Material and Methods’ section. Oocytes were harvested 3, 6, 12 or 24 h after the microinjection and processed. RLuc and FLuc activities were determined (in RLU). The RLuc (left panel) and FLuc (right panel) activities for each RNA in each concentration are shown. Each value is the mean ± SEM from at least three oocytes obtained from different animals.
MMTV 5′-UTR-mediated internal initiation exhibits cell type-dependent translational activity
The precise molecular mechanism by which the host translational apparatus recognizes retroviral IRESes is unknown (13). However, the study of picornaviral IRESes has revealed that cap-independent translation initiation uses both canonical eIFs, as well as specific cellular proteins known as IRES trans-acting factors (ITAFs) (47). The presence or absence of particular ITAFs alters IRES activity in different cellular backgrounds (48,49). Thus, having established the presence of an IRES within the 5′-UTR of the MMTV RNA and knowing that IRES efficiencies depend on the cellular background, we next evaluated MMTV IRES activity in a range of cell types. Cells used in these experiments included NMuMG (normal murine mammary gland), HeLa (human cervical cancer cells), 293-T (human embryonic kidney cells), T47D (human breast carcinoma) and NIH 3T3 (mouse embryonic fibroblast). Cells were co-transfected with either dl ΔEMCV or dl 5′UTR-MMTV and pcDNA3.1 lacZ plasmids. The expression from both cistrons was assessed and normalized against the transfection control β-galactosidase. The normalized relative translation efficiency (RTE) [(FLuc/RLuc)/(total protein) × (β-galactosidase)] ratio, depicted in the figures as (FLuc/RLuc) was used as an index of MMTV IRES activity. In accordance to what has been described for other IRESes (48,49), the RTE of MMTV IRES-driven translation varied widely between cell lines (Figure 4A). The MMTV IRES showed greatest activity in NMuMG cells and HeLa cells (Figure 4A). These observations suggest that for optimal activity, the MMTV IRES requires as yet unidentified cell factors.
Figure 4.
A comparison of the efficiency of MMTV-IRES-initiated translation in cell lines of different origin. (A) NMuMG, 293-T, HeLa, T47D and NIH 3T3 cells were co-transfected with either dl ΔEMCV (200 ng) or dl 5′UTR-MMTV (200 ng) and pcDNA3.1 lacZ (50 ng) plasmids. β-Galactosidase and total proteins were quantified as indicated in ‘Materials and Methods’ section. RLuc and FLuc activities were measured and normalized by the total protein content and the β-galactosidase activity. Thus, the [(FLuc/RLuc)/(total protein) × (β-galactosidase)] ratio was used as an index of IRES activity. The ratio obtained with the dl ΔEMCV and the dl 5′UTR-MMTV plasmids are independently depicted. Values are the mean ± SEM from three independent experiments each conducted in triplicates. (B) NMuMG, 293-T, HeLa, T47D and NIH 3T3 cells were co-transfected with either dl ΔEMCV (200 ng) or dl 5′UTR-MMTV (200 ng) and pcDNA3.1 lacZ (50 ng) plasmids. Total RNA was extracted from each transfected cell type and quantified. Extracted RNA (3 µg) was used as template in a one-step RT-PCR designed to specifically detect the dl ΔEMCV and dl 5′UTR-MMTV bicistronic RNA (bottom panel). A schematic representation of the experimental procedure showing the primers and the size of the expected amplicons is presented (top panel). In vitro transcribed FLuc monocictronic RNA (200 ng; lane 2), RLuc monocistronic RNA (200 ng; lane 3) or an equimolar mixture of both the FLuc and RLuc monocistronic RNAs (lane 4) were included as negative controls. An additional water control (−) for the RT-PCR was included (lane 5).
FLuc expression from the dl 5′-UTR MMTV plasmid in NMuMG cells is not due to cryptic promoter activity within the MMTV 5′-UTR
Studies to identify internal initiation in isolated viral or cellular UTR segments utilizing the transfection of bicistronic reporter plasmids have been strongly criticized. It is recognized that a caveat to the above described approach is false-positive IRES activity attributable to cryptic promoter activity or splicing of the tested sequence within cells. Alternative splicing would potentially bypass the first cistron, while allowing the expression of the second cistron from the SV40 promoter. Even though experiments shown in Figure 3 addressed the possibility of cryptic promoter activity and of alternative splicing, data are limited to X. laevis oocytes. Thus, we verified the presence of the full-length bicistronic RNA in transfected cells. To achieve this, total RNA was extracted from cells and analyzed by RT-PCR as described by others (34,48,50,51). Results confirmed the presence of the full-length bicistronic mRNA for both constructs, dl ΔEMCV and dl 5′UTR-MMTV, in all studied cell lines (Figure 4B).
To exclude the possibility that the FLuc synthesized from the dl 5′UTR-MMTV RNA is associated with cryptic promoter activity, we removed the SV40 promoter from the bicistronic constructs (Figure 5A). This approach has been suggested and used by others for a similar purpose (52). In this setting, expression of the second transgene implies that the DNA coding for the IRES possesses cryptic promoter activity. Promoter-less bicistronic constructs were then transfected into NMuMG cells, and 24 h later cells were processed as indicated in the ‘Materials and Methods’ section and DNA, RNA and protein analyses were conducted. PCR confirmed that all cells were positively transfected with the dl plasmids (Figure 5B). RNA analysis confirmed the expression of the bicistronic (RLuc-FLuc) RNA only in cells transfected with the constructs that contained an active SV40 promoter (Figure 5C). Analysis of RLuc and FLuc activities confirmed that the proteins were not expressed for the dl 5′UTR-MMTV lacking the SV40 promoter (Figure 5D). We thereby exclude the possibility that expression of FLuc from the dl 5′UTR-MMTV constructs observed in NMuMG cells is due to the production of an additional transcript encoding FLuc via a cryptic promoter. Consequently, we conclude that the 5′-UTR of the full-length mRNA of MMTV harbors an IRES.
In vitro, activity of the MMTV IRES is not modulated by the MMTV Rem or HIV-1 Rev proteins
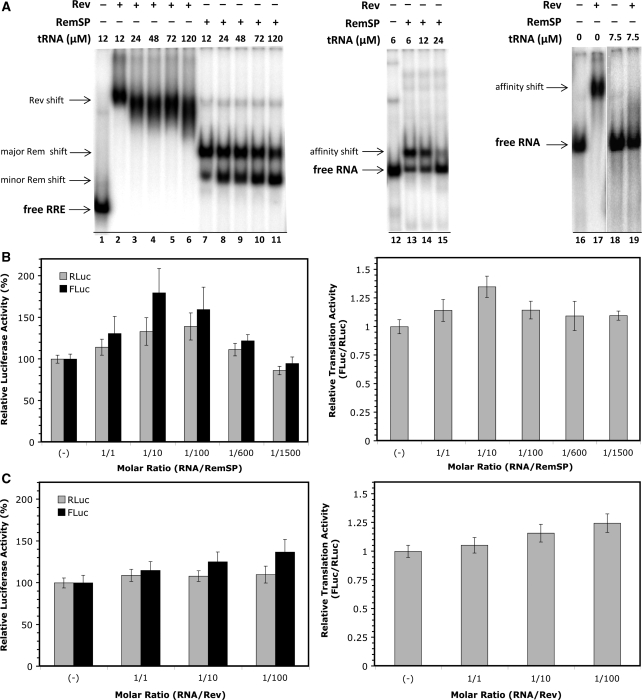
MMTV encodes a functional homolog of the HIV-1 nuclear export protein Rev (Rem) (30,53). Rem is a 33-kDa protein that is encoded by a doubly spliced mRNA. The N-terminal portion of Rem contains nuclear and nucleolar localization signals as well as an arginine-rich motif similar to the HIV-1 Rev RNA export protein (30,53). Rev is a 116 amino acid nuclear–cytoplasmic shuttling RNA-binding protein absolutely required for the cytoplasmic accumulation of intron-containing HIV-1 mRNAs (54). Rev achieves this by binding in a highly specific manner to an RNA secondary structure found in the envelope gene (env) intron called the Rev responsive element (RRE) (55). Several reports indicate, however, that the role of Rev is not restricted to the nuclear export of RRE-containing mRNA (56). The protein is also responsible, for example, for mRNA localization to polysomes, enhancing their use by the cellular translational machinery (57–59). Additionally, Rev has been shown to bind the 5′-UTR of HIV-1 mRNA (60), binding that is implicated in a direct modulation of translation initiation of the HIV-1 mRNA (61). This translational modulation is achieved in a RRE-independent fashion. Interestingly, Rev is capable of interacting with the MMTV Rem responsive element (RmRE), while Rem can interact with the HIV-1 RRE, in both cases increasing expression of transcripts carrying viral sequences (29,32,62). Based on these findings, we next sought to establish if Rem and Rev could modulate translation from the MMTV IRES in the context of a bicistronic RNA lacking the MMTV-RmRE or HIV-RRE sequences. These experiments were conducted in RRL using in vitro transcribed bicistronic RNA and recombinant MMTV Rem (RemSP) or HIV-1 Rev proteins. In order to test the functionality of the recombinant Rev and RemSP proteins in terms of their ability to bind RNA, an RNA EMSA was performed (Figure 6A). Recombinant Rev clearly binds to and shifts HIV-1 RRE RNA (compare lane 1 with 2). Also, recombinant RemSP clearly binds to and shifts HIV-1 RRE (compare lane 1 with 7). These observations confirm that the recombinant proteins conserve their ability to bind the HIV-1 RRE. With increasing concentrations of competitor RNA (tRNA), a shift can still be detected in the case of Rev (lanes 2–6) and RemSP (lanes 7–11) showing that both interactions are of high specificity. Both Rev and RemSP are capable of binding random RNA sequences, no doubt due to their general RNA-binding affinity (compare lanes 12 and 13 with 16 and 17, respectively); however, this interaction is disrupted at relatively low concentrations of competitor RNA (lanes 15 and 19, respectively). Based on the above evidence, we conclude that the recombinant Rev and RemSP proteins utilized herein were fully competent in their ability to interact with RNA.
Figure 6.
Neither the MMTV Rem nor the HIV-1 Rev proteins are directly implicated in translation driven from the MMTV-IRES. (A) RNA-binding activity as a measure of functionality for recombinant Rev and RemSP proteins was determined by radio-labeled RNA EMSA. Recombinant Rev binds to and shifts HIV-1 RRE RNA (compare lane 1 with 2). Recombinant RemSP binds to and shifts HIV-1 RRE (compare lane 1 with 7). The specificity of the interaction of Rev (lanes 2–6) and RemSP (lanes 7–11) with the target RNA was evaluated by adding increasing concentrations of competitor RNA (tRNA). The use of random RNA sequences as control confirmed that both RemSP and Rev can bind RNA at high protein concentrations (compare lanes 12 and 13 with 16 and 17, respectively). Binding is lost when relatively low concentrations of competitor RNA are present (lanes 15 and 19, respectively), i.e. there is no specificity for binding to the control RNA. The concentration of radio-labeled test RNA in all test samples was ∼0.5 µM. (B and C) T7 polymerase-generated capped and polyadenylated dl 5′UTR-MMTV RNAs were mixed with different molar concentrations of recombinant RemSP (B) or Rev (C) protein. The mean RLuc and FLuc luciferase activities in absence of the recombinant protein were arbitrarily set at 100% (±SEM). Values are the mean ± SEM from three independent experiments each conducted in triplicates. RLuc and FLuc values are relative to the 100% activity in the absence of recombinant protein (left panel). The (FLuc/RLuc) ratio was used as an index of IRES activity (right panel).
In the next set of experiments, capped and polyadenylated RNA generated from the dl 5′UTR-MMTV plasmid was mixed with different concentrations of recombinant RemSP protein. Proteins and RNA were added to the RRL at the indicated molar ratios (Figure 6B). The mean luciferase activity in the absence of the recombinant protein was arbitrarily set at 100% (±SEM). The mean FLuc/RLuc ratio was then used as an index of IRES activity where the ratio in the absence of recombinant protein was arbitrarily defined as 1 (Figure 6B). In our hands, this form of recombinant RemSP did not modulate translation from the MMTV IRES in RRL, as both cistrons in the bicistronic mRNA were affected similarly (Figure 6B; right panel). Similar results were obtained when Rev recombinant protein was added to the translation mix (Figure 6C; right panel). Collectively, our data suggest, at least under these experimental conditions in RRL, that HIV-1 Rev protein does not exert translational control over the MMTV-IRES.
DISCUSSION
Much like eukaryotic mRNAs, retroviral mRNAs are capped at their 5′-ends and contain a 3′-poly(A) tail. Interestingly, the presence of IRES elements within the viral mRNA has been described in a large number of retroviruses and retrotransposons [reviewed in (13)]. Retroviral IRESes have generally been identified by using heterologous bicistronic mRNAs. Nevertheless, the real requirement for IRES activity in the context of the full-length viral mRNA during the viral replication cycle still remains highly controversial (13). In eukaryotic mRNAs, the translation initiation complex is recruited to the vicinity of the 5′-cap structure [reviewed in (10)]. However, IRES-mediated translation initiation of viral mRNAs has emerged as a unique mechanism for bypassing the requirement of the 5′-cap structure (10,63,64). This strategy would enable viral mRNAs to overcome cellular translational control normally exerted at the stage of translation initiation, conferring virus mRNA a translational advantage over cellular mRNAs during viral infection, under stress conditions, or during very specific phases of the cell cycle (10,63,64). An alternative mechanism of translation initiation would thus ensure that viral protein synthesis is maintained in an efficient and, in some instances, a cell type-specific manner when global cap-dependent translation initiation is repressed (10,64). For instance, the requirement for an IRES element can potentially be rationalized in the case of HIV-1 given the constraint to translate its full-length mRNA during the G2/M phase of the cell cycle (19,65), where cap-dependent translation initiation is suppressed (66). A model biological requirement for IRES function in other retroviruses has yet to be established.
In this study, we show that the 5′-UTR of the full-length MMTV mRNA harbors an IRES. Translational activity of the MMTV-IRES was demonstrated in three different experimental systems; RRL, X. laevis oocytes and in cells (Figures 2–4). We show that translation from the MMTV 5′-UTR continues even when eIF4F function is disrupted (Figure 2B and C). Furthermore, we rule out that the expression of the second cistron in our bicistronic construct is exclusively due to alternative splicing or cryptic promoter activity (Figures 3–5). Interestingly, our data suggest that translation driven from the MMTV IRES does not require additional viral proteins (Figure 3) and is not modulated by, at least, the MMTV RemSP protein (Figure 6). Strikingly, the data do indicate that optimal MMTV IRES activity requires as yet unidentified cell-specific ITAFs as IRES activity strongly differs in different cellular backgrounds (Figure 4). Alternatively, data may indicate dissimilar degrees of MMTV IRES-specific suppression in different cell types, i.e. in cell exhibiting relatively high-IRES activity an inhibitory factor(s) might be lacking.
As for most retroviral IRES elements, the biological significance of the MMTV IRES within the viral context remains obscure. Yet, it is tempting to speculate that the presence of an IRES within the 5′-UTR of the MMTV full-length mRNA confers this transcript with a clear recruitment advantage over cellular mRNAs during tumorigenesis. Both molecular and epidemiological data have indicated a possible role for the MMTV, associated with breast cancer and T-cell lymphomas in mice, in a certain percentage of human breast tumors (67). This notion has been recently strengthened by data demonstrating that MMTV infects and rapidly propagates in cultured human breast cells (3,4). Yet, there is no definitive evidence to date as to whether MMTV is causal or merely an innocuous infection in humans.
Translation regulation plays a key role in cancer development and progression, impacting on tumor cell proliferation and growth, response to stresses such as hypoxia and stimulation by mitogenic signals and cell hormone receptors (68). The presence of IRES elements in so many genes that are involved in cell growth, proliferation, apoptosis and angiogenesis raises the possibility that deregulated internal (cap independent) initiation may contribute toward tumorigenesis (http://iresite.org/ or http:www.rangueil.inserm.fr/iresdatabase/). In keeping with this notion, aberrant translational regulation of the c-myc IRES has been associated with human neoplasia multiple myeloma (69–71). In this context, a recent report established that a switch from cap-dependent to cap-independent mRNA translation, mediated by hypoxia, in large advanced breast cancers is required for promoting angiogenesis, tumor survival and progression (72). The translational switch is the result of over-expression of 4E-BP1, the eIF4E-binding protein, resulting in the inhibition of cap-dependent translation (72). Concomitant with the reduction in cap-mediated translation initiation, over-expression of 4E-BP1 enhances IRES-dependent translation from mRNAs encoding HIF1a, VEGF and Bcl2, proteins required for growth under hypoxic conditions (72). It thus seems plausible to propose that under hypoxic conditions present in mammary tumors, MMTV IRES-mediated protein synthesis allows efficient virus gene expression, thus increased virus production during hypoxia. If so, our data would provide the mechanism by which MMTV proteins are produced during breast tumor development.
FUNDING
Comisión Nacional de Investigación Científica y Tecnológica, Gobierno de Chile, CONICYT (Grants FONDECYT N° 1060655 and 1090318 to M.L.L., FONDAP 13980001 to the Centre for Cell Regulation and Pathology and PFB12/2007 to J.P.H.-T); CONICYT Doctoral Fellowship (to M.V. and F.V.); MIFAB Institute (to F.R.); MECE(2)SUP-UNAB Doctoral Fellowship (to P.R.). Funding for open access charge: Comisión Nacional de Investigación Científica y Tecnológica, Gobierno de Chile, CONICYT (Grants FONDECYT N° 1090318 to M.L.L.). Author Loyalty Discount (Code G513XR7HP990).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr M. Rau for critical reading and editing of the manuscript; Drs G. Belsham (Institute for Animal Health, Pirbright, UK), W. H. Günzburg, B. Salmons and S. Indik (University of Veterinary Medicine Vienna, Austria) for kindly providing the plasmids used in this study; Drs L. Carrasco (Centro de Biología Molecular Severo Ochoa, Madrid, Spain) for kindly providing the polyclonal antibodies against N-terminal and C-terminal peptides of eIF4GI; and J. Hochman (The Hebrew University of Jerusalem, Israel) for kindly providing anti-p14 antisera. M.V. conducted this work as part of her PhD thesis, programa de Doctorado en Ciencias Médicas, Facultad de Medicina, Pontificia Universidad Católica de Chile. P.R. is a student of the programa de Doctorado en Biotecnología, Universidad Andrés Bello. F.V.-E. and F.E.R. are the students of the programa de Doctorado en Microbiología and Doctorado en Biotecnología, Universidad de Santiago de Chile, respectively.
REFERENCES
- 1.Callahan R. MMTV-induced mutations in mouse mammary tumors: their potential relevance to human breast cancer. Breast Cancer Res. Treat. 1996;39:33–44. doi: 10.1007/BF01806076. [DOI] [PubMed] [Google Scholar]
- 2.Cohen JC, Varmus HE. Endogenous mammary tumour virus DNA varies among wild mice and segregates during inbreeding. Nature. 1979;278:418–423. doi: 10.1038/278418a0. [DOI] [PubMed] [Google Scholar]
- 3.Indik S, Gunzburg WH, Kulich P, Salmons B, Rouault F. Rapid spread of mouse mammary tumor virus in cultured human breast cells. Retrovirology. 2007;4:73. doi: 10.1186/1742-4690-4-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Indik S, Gunzburg WH, Salmons B, Rouault F. Mouse mammary tumor virus infects human cells. Cancer Res. 2005;65:6651–6659. doi: 10.1158/0008-5472.CAN-04-2609. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Holland JF, Bleiweiss IJ, Melana S, Liu X, Pelisson I, Cantarella A, Stellrecht K, Mani S, Pogo BG. Detection of mammary tumor virus env gene-like sequences in human breast cancer. Cancer Res. 1995;55:5173–5179. [PubMed] [Google Scholar]
- 6.Pogo BG, Melana SM, Holland JF, Mandeli JF, Pilotti S, Casalini P, Menard S. Sequences homologous to the mouse mammary tumor virus env gene in human breast carcinoma correlate with overexpression of laminin receptor. Clin. Cancer Res. 1999;5:2108–2111. [PubMed] [Google Scholar]
- 7.Yin H, Medstrand P, Kristofferson A, Dietrich U, Aman P, Blomberg J. Characterization of human MMTV-like (HML) elements similar to a sequence that was highly expressed in a human breast cancer: further definition of the HML-6 group. Virology. 1999;256:22–35. doi: 10.1006/viro.1998.9587. [DOI] [PubMed] [Google Scholar]
- 8.Johal H, Scott GM, Jones R, Camaris C, Riordan S, Rawlinson WD. Mouse mammary tumour virus-like virus (MMTV-LV) is present within the liver in a wide range of hepatic disorders and unrelated to nuclear p53 expression or hepatocarcinogenesis. J. Hepatol. 2009;50:548–554. doi: 10.1016/j.jhep.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 9.Bittner JJ. Some possible effects of nursing on the mammary gland tumor incidence in mice. Science. 1936;84:162. doi: 10.1126/science.84.2172.162. [DOI] [PubMed] [Google Scholar]
- 10.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 13.Balvay L, Lopez Lastra M, Sargueil B, Darlix JL, Ohlmann T. Translational control of retroviruses. Nat. Rev. Microbiol. 2007;5:128–140. doi: 10.1038/nrmicro1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alvarez E, Menendez-Arias L, Carrasco L. The eukaryotic translation initiation factor 4GI is cleaved by different retroviral proteases. J. Virol. 2003;77:12392–12400. doi: 10.1128/JVI.77.23.12392-12400.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohlmann T, Prevot D, Decimo D, Roux F, Garin J, Morley SJ, Darlix JL. In vitro cleavage of eIF4GI but not eIF4GII by HIV-1 protease and its effects on translation in the rabbit reticulocyte lysate system. J. Mol. Biol. 2002;318:9–20. doi: 10.1016/S0022-2836(02)00070-0. [DOI] [PubMed] [Google Scholar]
- 16.Perales C, Carrasco L, Ventoso I. Cleavage of eIF4G by HIV-1 protease: effects on translation. FEBS Lett. 2003;533:89–94. doi: 10.1016/s0014-5793(02)03764-x. [DOI] [PubMed] [Google Scholar]
- 17.Ventoso I, Blanco R, Perales C, Carrasco L. HIV-1 protease cleaves eukaryotic initiation factor 4G and inhibits cap-dependent translation. Proc. Natl Acad. Sci. USA. 2001;98:12966–12971. doi: 10.1073/pnas.231343498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salmons B, Groner B, Calberg-Bacq CM, Ponta H. Production of mouse mammary tumor virus upon transfection of a recombinant proviral DNA into cultured cells. Virology. 1985;144:101–114. doi: 10.1016/0042-6822(85)90309-5. [DOI] [PubMed] [Google Scholar]
- 19.Brasey A, Lopez-Lastra M, Ohlmann T, Beerens N, Berkhout B, Darlix JL, Sonenberg N. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 2003;77:3939–3949. doi: 10.1128/JVI.77.7.3939-3949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barria MI, Gonzalez A, Vera-Otarola J, Leon U, Vollrath V, Marsac D, Monasterio O, Perez-Acle T, Soza A, Lopez-Lastra M. Analysis of natural variants of the hepatitis C virus internal ribosome entry site reveals that primary sequence plays a key role in cap-independent translation. Nucleic Acids Res. 2009;37:957–971. doi: 10.1093/nar/gkn1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson JE, Powell MJ, Hoover SE, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Draper DE. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medina M, Domingo E, Brangwyn JK, Belsham GJ. The two species of the foot-and-mouth disease virus leader protein, expressed individually, exhibit the same activities. Virology. 1993;194:355–359. doi: 10.1006/viro.1993.1267. [DOI] [PubMed] [Google Scholar]
- 24.Aldabe R, Feduchi E, Novoa I, Carrasco L. Efficient cleavage of p220 by poliovirus 2Apro expression in mammalian cells: effects on vaccinia virus. Biochem. Biophys. Res. Commun. 1995;215:928–936. doi: 10.1006/bbrc.1995.2553. [DOI] [PubMed] [Google Scholar]
- 25.Gradi A, Svitkin YV, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl Acad. Sci. USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Acuna-Castillo C, Villalobos C, Moya PR, Saez P, Cassels BK, Huidobro-Toro JP. Differences in potency and efficacy of a series of phenylisopropylamine/phenylethylamine pairs at 5-HT(2A) and 5-HT(2C) receptors. Br. J. Pharmacol. 2002;136:510–519. doi: 10.1038/sj.bjp.0704747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altafaj X, Joux N, Ronjat M, De Waard M. Oocyte expression with injection of purified T7 RNA polymerase. Methods Mol. Biol. 2006;322:55–67. doi: 10.1007/978-1-59745-000-3_5. [DOI] [PubMed] [Google Scholar]
- 28.Gamarnik AV, Andino R. Exploring RNA virus replication in Xenopus oocytes. Methods Mol. Biol. 2006;322:367–378. doi: 10.1007/978-1-59745-000-3_26. [DOI] [PubMed] [Google Scholar]
- 29.Dangerfield JA, Hohenadl C, Egerbacher M, Kodajova P, Salmons B, Gunzburg WH. HIV-1 Rev can specifically interact with MMTV RNA and upregulate gene expression. Gene. 2005;358:17–30. doi: 10.1016/j.gene.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 30.Indik S, Gunzburg WH, Salmons B, Rouault F. A novel, mouse mammary tumor virus encoded protein with Rev-like properties. Virology. 2005;337:1–6. doi: 10.1016/j.virol.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 31.Vallette F, Mege E, Reiss A, Adesnik M. Construction of mutant and chimeric genes using the polymerase chain reaction. Nucleic Acids Res. 1989;17:723–733. doi: 10.1093/nar/17.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullner M, Salmons B, Gunzburg WH, Indik S. Identification of the Rem-responsive element of mouse mammary tumor virus. Nucleic Acids Res. 2008;36:6284–6294. doi: 10.1093/nar/gkn608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ricci EP, Soto Rifo R, Herbreteau CH, Decimo D, Ohlmann T. Lentiviral RNAs can use different mechanisms for translation initiation. Biochem. Soc. Trans. 2008;36:690–693. doi: 10.1042/BST0360690. [DOI] [PubMed] [Google Scholar]
- 34.Herbreteau CH, Weill L, Decimo D, Prevot D, Darlix JL, Sargueil B, Ohlmann T. HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nat. Struct. Mol. Biol. 2005;12:1001–1007. doi: 10.1038/nsmb1011. [DOI] [PubMed] [Google Scholar]
- 35.Ohlmann T, Lopez-Lastra M, Darlix JL. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J. Biol. Chem. 2000;275:11899–11906. doi: 10.1074/jbc.275.16.11899. [DOI] [PubMed] [Google Scholar]
- 36.Buck CB, Shen X, Egan MA, Pierson TC, Walker CM, Siliciano RF. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 2001;75:181–191. doi: 10.1128/JVI.75.1.181-191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicholson MG, Rue SM, Clements JE, Barber SA. An internal ribosome entry site promotes translation of a novel SIV Pr55(Gag) isoform. Virology. 2006;349:325–334. doi: 10.1016/j.virol.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 38.Jackson RJ, Hunt SL, Reynolds JE, Kaminski A. Cap-dependent and cap-independent translation: operational distinctions and mechanistic interpretations. Curr. Top. Microbiol. Immunol. 1995;203:1–29. doi: 10.1007/978-3-642-79663-0_1. [DOI] [PubMed] [Google Scholar]
- 39.Sonenberg N. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochem. Cell Biol. 2008;86:178–183. doi: 10.1139/O08-034. [DOI] [PubMed] [Google Scholar]
- 40.Svitkin YV, Herdy B, Costa-Mattioli M, Gingras AC, Raught B, Sonenberg N. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol. Cell Biol. 2005;25:10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 42.Ohlmann T, Rau M, Pain VM, Morley SJ. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- 43.Borman AM, Kirchweger R, Ziegler E, Rhoads RE, Skern T, Kean KM. elF4G and its proteolytic cleavage products: effect on initiation of protein synthesis from capped, uncapped, and IRES-containing mRNAs. RNA. 1997;3:186–196. [PMC free article] [PubMed] [Google Scholar]
- 44.Ohlmann T, Rau M, Morley SJ, Pain VM. Proteolytic cleavage of initiation factor eIF-4 gamma in the reticulocyte lysate inhibits translation of capped mRNAs but enhances that of uncapped mRNAs. Nucleic Acids Res. 1995;23:334–340. doi: 10.1093/nar/23.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gamarnik AV, Andino R. Replication of poliovirus in Xenopus oocytes requires two human factors. EMBO J. 1996;15:5988–5998. [PMC free article] [PubMed] [Google Scholar]
- 46.Keiper BD, Rhoads RE. Cap-independent translation initiation in Xenopus oocytes. Nucleic Acids Res. 1997;25:395–402. doi: 10.1093/nar/25.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belsham GJ, Sonenberg N. Picornavirus RNA translation: roles for cellular proteins. Trends Microbiol. 2000;8:330–335. doi: 10.1016/s0966-842x(00)01788-1. [DOI] [PubMed] [Google Scholar]
- 48.Kazadi K, Loeuillet C, Deutsch S, Ciuffi A, Munoz M, Beckmann JS, Moradpour D, Antonarakis SE, Telenti A. Genomic determinants of the efficiency of internal ribosomal entry sites of viral and cellular origin. Nucleic Acids Res. 2008;36:6918–6925. doi: 10.1093/nar/gkn812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoneley M, Subkhankulova T, Le Quesne JP, Coldwell MJ, Jopling CL, Belsham GJ, Willis AE. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 2000;28:687–694. doi: 10.1093/nar/28.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silvera D, Arju R, Darvishian F, Levine PH, Zolfaghari L, Goldberg J, Hochman T, Formenti SC, Schneider RJ. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat. Cell Biol. 2009;11:903–908. doi: 10.1038/ncb1900. [DOI] [PubMed] [Google Scholar]
- 51.Rivas-Aravena A, Ramdohr P, Vallejos M, Valiente-Echeverria F, Dormoy-Raclet V, Rodriguez F, Pino K, Holzmann C, Huidobro-Toro JP, Gallouzi IE, et al. The Elav-like protein HuR exerts translational control of viral internal ribosome entry sites. Virology. 2009;392:178–185. doi: 10.1016/j.virol.2009.06.050. [DOI] [PubMed] [Google Scholar]
- 52.Han B, Zhang JT. Regulation of gene expression by internal ribosome entry sites or cryptic promoters: the eIF4G story. Mol. Cell Biol. 2002;22:7372–7384. doi: 10.1128/MCB.22.21.7372-7384.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mertz JA, Simper MS, Lozano MM, Payne SM, Dudley JP. Mouse mammary tumor virus encodes a self-regulatory RNA export protein and is a complex retrovirus. J. Virol. 2005;79:14737–14747. doi: 10.1128/JVI.79.23.14737-14747.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hope TJ. The ins and outs of HIV Rev. Arch. Biochem. Biophys. 1999;365:186–191. doi: 10.1006/abbi.1999.1207. [DOI] [PubMed] [Google Scholar]
- 55.Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 56.Groom HC, Anderson EC, Lever AM. Rev: beyond nuclear export. J. Gen. Virol. 2009;90:1303–1318. doi: 10.1099/vir.0.011460-0. [DOI] [PubMed] [Google Scholar]
- 57.Arrigo SJ, Chen IS. Rev is necessary for translation but not cytoplasmic accumulation of HIV-1 vif, vpr, and env/vpu 2 RNAs. Genes Dev. 1991;5:808–819. doi: 10.1101/gad.5.5.808. [DOI] [PubMed] [Google Scholar]
- 58.D’Agostino DM, Felber BK, Harrison JE, Pavlakis GN. The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol. Cell Biol. 1992;12:1375–1386. doi: 10.1128/mcb.12.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kimura T, Hashimoto I, Nishikawa M, Fujisawa JI. A role for Rev in the association of HIV-1 gag mRNA with cytoskeletal beta-actin and viral protein expression. Biochimie. 1996;78:1075–1080. doi: 10.1016/s0300-9084(97)86732-6. [DOI] [PubMed] [Google Scholar]
- 60.Gallego J, Greatorex J, Zhang H, Yang B, Arunachalam S, Fang J, Seamons J, Lea S, Pomerantz RJ, Lever AM. Rev binds specifically to a purine loop in the SL1 region of the HIV-1 leader RNA. J. Biol. Chem. 2003;278:40385–40391. doi: 10.1074/jbc.M301041200. [DOI] [PubMed] [Google Scholar]
- 61.Groom HC, Anderson EC, Dangerfield J, Lever AM. Rev regulates translation of Human Immunodeficiency Virus Type-1 RNAs. J. Gen. Virol. 2009 doi: 10.1099/vir.0.007963-0. [DOI] [PubMed] [Google Scholar]
- 62.Mertz JA, Lozano MM, Dudley JP. Rev and Rex proteins of human complex retroviruses function with the MMTV Rem-responsive element. Retrovirology. 2009;6:10. doi: 10.1186/1742-4690-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bushell M, Sarnow P. Hijacking the translation apparatus by RNA viruses. J. Cell Biol. 2002;158:395–399. doi: 10.1083/jcb.200205044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lopez-Lastra M, Rivas A, Barria MI. Protein synthesis in eukaryotes: the growing biological relevance of cap-independent translation initiation. Biol. Res. 2005;38:121–146. doi: 10.4067/s0716-97602005000200003. [DOI] [PubMed] [Google Scholar]
- 65.Thierry S, Marechal V, Rosenzwajg M, Sabbah M, Redeuilh G, Nicolas JC, Gozlan J. Cell cycle arrest in G2 induces human immunodeficiency virus type 1 transcriptional activation through histone acetylation and recruitment of CBP, NF-kappaB, and c-Jun to the long terminal repeat promoter. J. Virol. 2004;78:12198–12206. doi: 10.1128/JVI.78.22.12198-12206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pyronnet S, Dostie J, Sonenberg N. Suppression of cap-dependent translation in mitosis. Genes Dev. 2001;15:2083–2093. doi: 10.1101/gad.889201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amarante MK, Watanabe MA. The possible involvement of virus in breast cancer. J. Cancer Res. Clin. Oncol. 2009;135:329–337. doi: 10.1007/s00432-008-0511-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schneider RJ, Sonenberg N. Translational Control in Biology and Medicine. Cold Spring Harbor, USA: Cold Spring Harbor Press; 2007. Translational control in cancer development and progression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chappell SA, LeQuesne JP, Paulin FE, deSchoolmeester ML, Stoneley M, Soutar RL, Ralston SH, Helfrich MH, Willis AE. A mutation in the c-myc-IRES leads to enhanced internal ribosome entry in multiple myeloma: a novel mechanism of oncogene de-regulation. Oncogene. 2000;19:4437–4440. doi: 10.1038/sj.onc.1203791. [DOI] [PubMed] [Google Scholar]
- 70.Evans JR, Mitchell SA, Spriggs KA, Ostrowski J, Bomsztyk K, Ostarek D, Willis AE. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene. 2003;22:8012–8020. doi: 10.1038/sj.onc.1206645. [DOI] [PubMed] [Google Scholar]
- 71.Paulin FE, West MJ, Sullivan NF, Whitney RL, Lyne L, Willis AE. Aberrant translational control of the c-myc gene in multiple myeloma. Oncogene. 1996;13:505–513. [PubMed] [Google Scholar]
- 72.Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, Cangiarella J, Arju R, Formenti SC, Schneider RJ. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol. Cell. 2007;28:501–512. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]