Abstract
The analysis of targeted genetic loci from ancient, forensic and clinical samples is usually built upon polymerase chain reaction (PCR)-generated sequence data. However, many studies have shown that PCR amplification from poor-quality DNA templates can create sequence artefacts at significant levels. With hominin (human and other hominid) samples, the pervasive presence of highly PCR-amplifiable human DNA contaminants in the vast majority of samples can lead to the creation of recombinant hybrids and other non-authentic artefacts. The resulting PCR-generated sequences can then be difficult, if not impossible, to authenticate. In contrast, single primer extension (SPEX)-based approaches can genotype single nucleotide polymorphisms from ancient fragments of DNA as accurately as modern DNA. A single SPEX-type assay can amplify just one of the duplex DNA strands at target loci and generate a multi-fold depth-of-coverage, with non-authentic recombinant hybrids reduced to undetectable levels. Crucially, SPEX-type approaches can preferentially access genetic information from damaged and degraded endogenous ancient DNA templates over modern human DNA contaminants. The development of SPEX-type assays offers the potential for highly accurate, quantitative genotyping from ancient hominin samples.
INTRODUCTION
This study compares and contrasts polymerase chain reaction (PCR)-based approaches to ancient hominin samples (i.e. humans and extinct relatives such as Neanderthals) with a single primer extension (SPEX)-based approach to genotyping ancient DNA (aDNA) at targeted loci. In particular, we examine the ability of SPEX-type approaches to preferentially access genetic information from highly damaged and degraded aDNA templates and to genotype ancient samples with enhanced sequence reliability and depth-of-coverage.
These are important issues, as reports of the ‘retrieval’ of aDNA sequences from hominin samples via PCR amplification (1–5) can create an impression of a relatively straightforward exponential copying of surviving aDNA template molecules into PCR products at specific loci. In reality, however, non-authentic sequences are often created at significant levels during PCR amplification from degraded DNA extracts, making both sequence reliability and depth-of-coverage two key and inter-related considerations in the accurate genotyping of ancient, forensic and clinical samples.
In terms of sequence reliability, DNA damage and fragmentation makes extracts from ancient hominin samples particularly vulnerable to exponential PCR amplification from both contaminant human DNA and artefactual PCR-generated endogenous-contaminant hybrids (2,6–16, this study). In addition, due to both authentic sequence-modifying DNA damage and wholly PCR-generated sequence artefacts (17), traditional PCR amplification from aDNA templates can introduce above-background levels of all four transition base changes (8,9,18–21); even from well-preserved permafrost specimens with estimated copy numbers in the tens-of-thousands (22).
The depth-of-coverage generated by traditional PCR amplification from aDNA extracts is particularly difficult to estimate, as the numbers of discrete, intact, endogenous aDNA templates directly copied into amplification products without PCR-generated recombination (if any) cannot be accurately assessed. However, it has become clear that from aDNA extracts, PCR amplicons commonly originate from just one or a tiny number of ‘starting templates’ (1,5,23,24). Incorrect sequence inferences are inevitable where a majority of ‘starting templates’ are contaminant DNA templates, PCR-generated recombinant hybrids or result in the ‘fixation’ of a non-authentic transition base change at a key diagnostic nucleotide position.
Unfortunately, therefore, many PCR-based analyses of human sub-fossil material have produced results that are difficult, if not impossible, to authenticate (6,25–27). The most widely accepted results from PCR-based hominin studies have been those that targeted genetic loci at which Neanderthals and some ancient human individuals appear to have highly distinctive DNA sequence motifs that are rare or absent in modern human populations (6,8,10,28–31). Venturing into wider genomic regions has generally been restricted to exceptional samples where levels of exogenous human DNA contamination are reckoned to be low, using high-throughput shotgun sequencing approaches that have the potential to generate a sufficient depth-of-coverage to overcome aDNA-derived transition base changes and maximize the accuracy of inferred consensus sequences at loci-of-interest (32).
In contrast to PCR, the SPEX-type approach used in this study does not impose a pre-defined target size based on a pair of primers. An initial copying step uses a single biotinylated primer to create primer extension products of different length, followed by the removal of all other potential DNA templates via streptavidin beads, homopolymer tailing and only then nested PCR amplification. As a result, intact DNA templates can be quantifiably amplified from just one of the strands at a target locus, with the introduction of non-authentic C > T, A > G and T > C transition base changes reduced to the background levels generated during the amplification of ‘modern’ DNA (17). These unique attributes enabled a SPEX-type approach to resolve the long-standing question of the biochemical nature of ‘miscoding lesions’ in aDNA by providing direct proof that C > U-type base modifications are overwhelmingly the cause of authentic sequence-modifying DNA damage (17, contrast 33,34).
This study evaluates the ability of SPEX-type approaches to accurately genotype pre-handled and pre-contaminated hominin samples through the following key novel attributes: the ability to (i) generate highly accurate sequence data from damaged and degraded DNA templates; (ii) reduce PCR-generated recombinant template switching (and therefore the creation of non-authentic endogenous-contaminant hybrids and other sequence artefacts) to below detectable levels; and (iii) preferentially access surviving fragments of damaged and degraded endogenous aDNA over the pervasive exogenous human contaminant templates that can be efficiently co-amplified by traditional PCR-based approaches.
MATERIALS AND METHODS
Samples and experimental rationale
This study uses DNA extracted from museum and archaeological hominin samples covering a range of ages, source materials, storage conditions and geographical source locations. Following both traditional PCR and SPEX amplification, cloned sequence data are investigated in terms of: the generation of non-authentic sequence data; the levels of PCR-generated recombinant template switching; the accuracy of typing single nucleotide polymorphisms (SNPs) at targeted loci; the depth-of-coverage generated; and any relative preferences for accessing genetic information from endogenous aDNA versus contaminant human DNA. The dedicated Ancient Biomolecules Centre (ABC) at Oxford University was used to extract DNA from ancient samples and to set up PCR and SPEX reactions. The ABC is physically isolated, subject to stringent anti-contamination procedures, equipped with positive air pressure and UV lighting, and has a DNA laboratory and equipment (glove box, instruments, full body suits, protective masks, etc.) dedicated solely to ancient hominin samples. The thermal cycling reactions and subsequent downstream work take place in a separate laboratory located in the Department of Zoology.
Specifically, the samples that underwent DNA extraction are the La Quina 4 Neanderthal specimen from the collections of the Musée de l'H;omme in Paris (LQ4; single tooth >54 000 years old); a colonial era Asian individual from the Natural History Museum in London (PE168; ∼150-year-old tooth); two pre-Columbian Amerindian archaeological samples (PE40, PE187; both ∼1000-year-old teeth); the Tyrolean Iceman—a.k.a. Ötzi (HB50; ∼5000-year-old bone sample from the ileum). DNA was also extracted from a modern sample (PEM33; hair tips, which therefore had prior exposure to similar environmental conditions as the sweat, flakes of skin, dandruff, etc., expected to be the predominant sources of modern human contaminant DNA). Detailed explanations of the background behind (i) the samples chosen; (ii) the particular SNPs targeted by SPEX- and PCR-based approaches; and (iii) the quantitative PCR (qPCR) experiments performed are given in Supplementary Figure S1, Section S1 and, where required, in the ‘Results and Discussion’ sections.
DNA extraction and PCR/SPEX amplification
DNA extractions used Proteinase K digestion and purification by the phenol–chloroform method (35) following Endicott et al. (30). For teeth, material was removed from dental canals using a dental drill. Hair samples were processed by the keratin digestion method of Maniatis et al. (35). PCR amplification of targets within the first hypervariable section (HVS1) of the human mitochondrial DNA (mtDNA) control region used a two-stage process with oligonucleotide primers listed in Supplementary Table S1 and the following conditions: 94°C for 10 min; followed by 35 cycles of 94°C for 30 s, 58°C for 30 s and 72°C for 30 s; then 10 min at 72°C followed by a 4°C hold. Each 25-μl reaction mix consisted of 1–3 μl of aDNA extract; 0.25 U of AmpliTaq Gold; 1× Geneamp PCR Buffer II (without MgCl2); 2 mM MgCl2; 250 μM dNTPs; and 250 nM of each primer. Products from the first-stage PCR were visualized on 4% agarose gels under UV light, excised, purified on a Qiagen MinElute column using the recommended protocol and eluted in EB buffer. Each purified PCR product was then diluted 1:99 in water and used to seed a second-stage 30-cycle PCR using the same primers and conditions. For the Neanderthal LQ4 extract, amplifications were made in one-stage reactions with the same conditions as above, but for 40 cycles, using 5 μl of extract plus 2 μl of 10 mg/ml bovine serum albumin (BSA; Sigma) to help overcome polymerase inhibitors as previously described (17). After a further MinElute column purification step, PCR products were cloned using the Topo TA Cloning kit with F′ competent cells and amplified as per the manufacturer’s instructions (Invitrogen). The processing of clones and sequencing on an ABI 377 or 3700 sequencer was performed as described by Endicott et al. (30).
Single primer approaches originally developed to amplify full-length cDNA molecules (36,37) can be adapted to the amplification of targeted aDNA sequences (17). SPEX amplification experiments were carried out as described previously (17) using oligonucleotides listed in Supplementary Table S1, according to the rationale set out in Supplementary Figure S2. Following single biotinylated primer extension, the biotinylated reaction products are isolated on strepavidin beads, all DNA templates (aDNA and any contaminants) are then removed via stringency washes, bead-isolated single-stranded DNA molecules are permanently ‘trapped’ by polyC-tailing (using terminal transferase/dCTP) and nested PCR can then amplify what are now (effectively) freshly synthesised, polyC-tailed, first-generation copies of just one of the template DNA strands at a targeted locus (thereby minimizing any potential creation and exponential amplification of the kinds of PCR-generated recombinants and other sequence artefacts commonly produced from aDNA).
RESULTS
Accessing genetic information from endogenous hominin aDNA over modern human DNA contaminants: a quantitative side-by-side comparison between PCR and SPEX-type approaches
When differently sized target templates share the same pair of PCR primer-binding sites, shorter templates are amplified more efficiently than longer ones (38). As the products of SPEX primer extensions on aDNA templates are generally significantly shorter than the lengths targeted by traditional two-primer PCR approaches to aDNA (17), we quantitatively investigated the possibility that a SPEX-type approach might be able to preferentially access genetic information from highly damaged and degraded aDNA templates. A portion of the Iceman HB50 aDNA extract was deliberately ‘contaminated’ with the ‘modern’ PEM33 hair-tip DNA extract and the resulting mixture was amplified by both SPEX and traditional PCR. Relative qPCR was performed on a dilution series of both extracts using an effective PCR amplifiable size of 61 bp (Supplementary Table S1; Section S1). Dilutions of the HB50 Iceman extract (European mtDNA haplogroup K) and the PEM33 extract (East Asian mtDNA haplogroup M7b) giving an observed equal ‘starting copy number’ of templates were then combined, homogenized and used in side-by-side PCR and SPEX amplifications. Amplification products were cloned and sequenced to accurately determine the numbers and relative proportions of amplicons derived from ‘ancient’ (HB50) or ‘modern’ (PEM33) mtDNA templates via two directly adjacent SNP differences: 16223T (M7b) and 16224C (K) (Supplementary Figure S1c; Section S1).
Traditional PCR amplification targeted 86 bp (Supplementary Figure S1c) of the control region, encompassing both the haplogroup (hg) K diagnostic SNP T16224C and the directly adjacent hg M7b diagnostic SNP at C16223T. Cloning and sequencing showed that 42/93 (45%) of PCR amplicons were derived from modern ‘contaminant’ (PEM33) DNA and 51/93 (55%) from endogenous Iceman (HB50) aDNA templates, thereby confirming that qPCR had estimated the effective ‘starting template’ copy numbers of the extracts for PCR amplification in the relevant ∼60- to 90-bp range within an acceptable level of accuracy (Supplementary Figure S3). SPEX amplification (Supplementary Figure S1c) from the same homogenized mix produced markedly different proportions. Of 183 cloned amplicons whose original SPEX event had traversed the two directly adjacent diagnostic SNP sites at nucleotide positions (nps) 16223 and 16224, only 3/183 (1.6%) were derived from modern ‘contaminant’ templates; whereas 180/183 (98.4%) were derived from endogenous Iceman aDNA templates within the HB50 extract (Supplementary Figure S3). This heavily skewed ratio clearly demonstrates the ability of a SPEX-type approach to preferentially access genetic information from endogenous aDNA templates; even in the presence of significant levels of human contaminant DNA.
Accessing genetic information from endogenous aDNA over modern human DNA contaminants within extensively handled ancient hominin samples: side-by-side comparisons between PCR and SPEX-type approaches
In order to investigate a more realistic aDNA research scenario (i.e. hominin museum samples that have been washed and extensively handled), two such aDNA extracts were used in side-by-side PCR and SPEX amplifications. One extract (PE168) was made from the ∼150-year-old tooth of an Asian human individual from the Natural History Museum in London. At the other extreme of the aDNA scale, an extract (LQ4) was made from a >54 000-year-old tooth of the La Quina 4 Neanderthal specimen from the collections of the Musée de l'H;omme in Paris. PCR amplifications from both extracts generated products with sample-appropriate SNPs. The PE168 aDNA extract produced all three expected hg N9a SNPs (at nps 16223, 16257 and 16261) in 10/12 cloned amplicons (Figure 1a). The LQ4 aDNA extract demonstrably contains amplifiable Neanderthal mtDNA through the presence of the Neanderthal marker motif 16263.1A-C16262T (6,8–11) amongst cloned PCR amplicons (Figure 2a). However, PCR amplification from both aDNA extracts also reveals the presence of at least one other population of PCR-amplifiable human mtDNA templates.
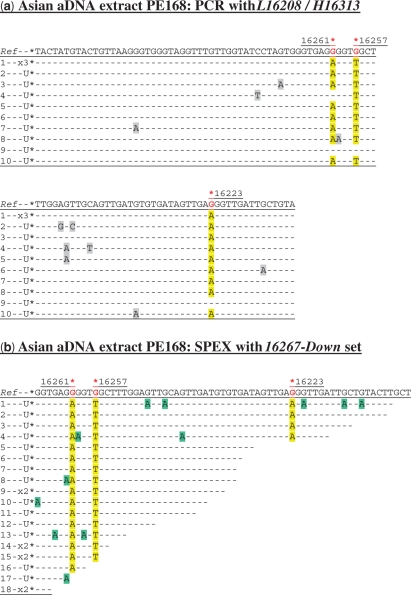
Figure 1.
Cloned sequence data following (a) PCR and (b) SPEX amplification from the Asian aDNA extract PE168. U = a unique sequence found in only one clone from that particular amplification reaction; X1, X2, etc. = identical sequences independently cloned (once, twice, etc). The L16208/H16313 PCR primer pair was used (Supplementary Figure S1a and Table S1). The 16267-Down SPEX primer set (Supplementary Table S1) was used in the amplification strategy described in Supplementary Figure S1a. In relation to the rCRS, hg N9a SNPs are C16223T, C16257A and C16261T. As the 16267-Down SPEX primer set targets the light mtDNA strand, hg N9a-specific SNPs are highlighted by a red asterisk above the reverse complement of the reference sequence (Ref). N9a-specific SNPs in PCR- and SPEX-generated sequence data are highlighted in yellow. In this and subsequent PCR-derived sequence data, base changes at non-diagnostic nucleotide positions—arising from sequence-modifying DNA damage and PCR-generated sequence artefacts—are highlighted in grey. In this and subsequent SPEX-derived sequence data, G > A transition base changes (arising from C > U-type miscoding lesion sequence-modifying DNA damage on template strands) are highlighted in green, with any other non-authentic base changes highlighted in grey. The final 3′-most positions of SPEX amplicons (i.e. the position immediately 5′ to the polyC-tail) do not necessarily reflect underlying template strand sequences, due to potential polymerase-mediated ‘non-templated’ nucleotide addition (NTNA) following SPEX (17). The 3′-most position is therefore not presented, or analysed, in SPEX-generated sequence data.
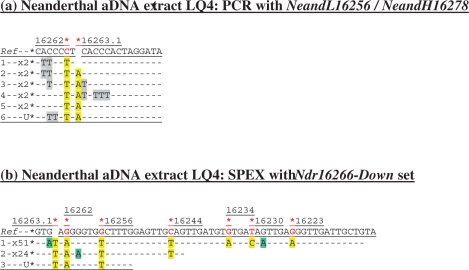
Figure 2.
Cloned sequence data following (a) PCR, and (b) SPEX amplification from the Neanderthal LQ4 aDNA extract. The NeandL16256/NeandH16278 PCR primer pair was used (Supplementary Figure S1b and Table S1). The Ndr16266-Down SPEX primer set (Supplementary Table S1) was used in the amplification strategy described in Supplementary Figure S1b. Nucleotide positions of expected Neanderthal-specific SNPs are each highlighted by a red asterisk above: (a) the reference sequence (Ref); or (b) the reverse complement of the reference sequence (Ref). Presumed Neanderthal-specific SNPs in PCR- and SPEX-generated sequence data are highlighted in yellow. Given above-background levels of C > U-derived G > A transitions in SPEX data, an absence of np 16228 SNPs in published Neanderthal data, and a depth-of-coverage of just one generated for this site by SPEX, the G > A transition base change here is assumed to be due to miscoding lesion DNA damage.
From PE168, trace nine of Figure 1a has the ancestral allele at all three diagnostic sites. The presence of both C > T and G > A transition base changes within trace four indicates PCR-generated recombinant template switching between complementary template DNA strands with independent C > U-type damage events (13,14,17,23). The creation of a non-authentic endogenous-contaminant recombinant hybrid might also account for an A > G reversion to the ancestral allele at np 16261 in the same amplicon. Alternatively, this could represent another example of the kinds of transition base change artefacts that can be generated at significant levels during the PCR amplification of aDNA (8,9,18–22; Figures 1a and 2a; Supplementary Figure S4). Whatever the underlying cause, only 10/12 PCR amplicons (83%) from PE168 display the endogenous hg N9a SNP at np 16261.
From LQ4, despite a target size of just 66 bp and the use of two Neanderthal-specific PCR primers, recombinant template switching has still generated non-authentic hybrid human-Neanderthal sequences (Figure 2a). (For example, note the absence of the diagnostic Neanderthal insertion at np 16263.1A alongside the C16262T SNP in trace one, but the additional presence of identical C > T transitions and A > T transversions in both traces one and two, which together made up more than one-third of the cloned PCR amplicon sequences.) Repeated C > T base change motifs (in 8/9 cloned sequences) also indicate that a very small pool of endogenous Neanderthal aDNA templates with extensive sequence-modifying miscoding lesion DNA damage contributed to the PCR-amplified products.
For both PE168 and LQ4 aDNA extracts, a complete lack of mtDNA amplification from extraction blanks or PCR controls indicates that (i) these much-handled museum samples are now irretrievably contaminated with exogenous human DNA; and (ii) PCR has co-amplified these exogenous human DNA templates alongside endogenous aDNA templates whilst also creating a third, wholly non-authentic category of endogenous-contaminant hybrid sequences. In contrast, every SPEX amplicon from both the PE168 and LQ4 aDNA extracts displays the appropriate endogenous SNP at every diagnostic site traversed by SPEX primer extension (Figures 1b and 2b). Unlike the PCR-generated sequence data, not only is there no evidence for the amplification of intact exogenous contaminant sequences by the SPEX-type approach, but there is also no evidence for the generation of ‘jumping-PCR’ type template switching artefacts in the SPEX-generated data; either in the form of non-authentic endogenous-contaminant recombinant hybrid sequences or in the form of duplicated (C > U derived) G > A miscoding lesion damage motifs (compare Figures 1a and 2a with Figures 1b and 2b).
SPEX can SNP-type ancient samples with a quantifiable depth-of-coverage
In the above SPEX versus PCR studies, the use of ancient samples with distinctive clustered SNP combinations allowed sequence data from endogenous aDNA to be distinguished from exogenous contaminants and non-authentic recombinant hybrids. However, a serious problem for PCR-based hominin aDNA authentication arises in the much more common scenario where endogenous aDNA templates possess SNPs that are common within contemporary human populations. For the 5000-year-old Tyrolean Iceman, there have now been six independent studies (15,39–43). Despite this, an accurate characterization of the Tyrolean Iceman’s mtDNA sequence by PCR has proven problematic (compare 15,39,41–43) and conflicting assessments of the exact mtDNA haplotype of the Iceman have been produced.
A SNP at np 16362 (T16362C) characterized by two recent studies of the Iceman (42,43) was not identified by the original Iceman aDNA investigations (15,39). Due to the importance of np 16362 in recent arguments concerning the accurate phylogenetic placement of the Iceman’s endogenous mtDNA within hg K (41–43), this key SNP was targeted by SPEX amplification from heavy mtDNA template strands in the HB50 Iceman aDNA extract. Extracts from two pre-Columbian Amerindian tooth samples (PE40 and PE187; both ∼1000 years old) were also genotyped at np 16362 for control purposes, with endogenous aDNA from both expected to carry the allele equivalent to the revised Cambridge reference sequence (rCRS) at this site (44).
Whatever the number of clones sequenced, any individual PCR amplification from an aDNA extract cannot be assumed to represent more than a single ‘starting template’ and therefore to have a depth-of-coverage greater than one (1,5,23,24). In contrast, from a single SPEX-type amplification reaction, single primer extension events that produce cloned SPEX amplicons with discrete lengths and/or discrete G > A miscoding lesion damage patterns are highly likely to have originated on discrete endogenous aDNA templates (17). Nineteen such discrete SPEX clones from the Iceman HB50 aDNA extract generated 100% sequence traces with 16362C (Figure 3).
Figure 3.
Cloned sequence data following SPEX amplification from the Amerindian (PE40, PE187) and Iceman (HB50) aDNA extracts using the 16360-Up SPEX primer set (Supplementary Table S1) as for the amplification strategy described in Supplementary Figure S1c. A red asterisk above the reference sequence (Ref) highlights np 16362. The diagnostic site in SPEX-generated sequence data is highlighted in yellow; C > U-derived G > A transitions are highlighted in green; any other base changes are highlighted in grey.
Since T > C transition artefacts are introduced at background rates during SPEX amplification (17; Figures 1b, 2b, 3 and 4), the recovery of 19/19 discrete amplicons with 16362C must result from SPEX amplification of 19 discrete mtDNA templates from a single individual with an authentic in vivo substitution event at np 16362 (i.e. a 19-fold depth-of-coverage with no evidence for any SPEX-amplified contaminant human DNA sequences). Therefore, up to 19 individual PCR amplifications would have been required to approach the depth-of-coverage generated by a single SPEX assay, and even then T > C transitions can also be introduced at levels significantly higher than background during PCR amplification from aDNA extracts (8,9,18–22).
Figure 4.
Cloned sequence data following SPEX amplification from the Amerindian (PE40, PE187) and Iceman (HB50) aDNA extracts using the 16225-Down SPEX primer set (Supplementary Table S1), as for the amplification strategy described in Supplementary Figure S1c. Nucleotide positions of the hg K (np 16224) and hg B4b (np 16217) SNPs are each highlighted by a red asterisk above the reverse complement of the reference sequence (Ref). Diagnostic hg K (np 16224) and hg B4b (np 16217) sites in SPEX-generated sequence data are highlighted in yellow; C > U-derived G > A transitions are highlighted in green; any other base changes are highlighted in grey.
SPEX amplification from the Amerindian aDNA extracts provided 15- and 17-fold coverage, respectively, at np 16362 (Figure 3), with 100% of clones returning the expected 16362T ancestral allele. From PE168, SPEX provided a 16-fold depth-of-coverage for the diagnostic hg N9a SNP at np 16261 (Figure 1b). From the LQ4 extract, three discrete SPEX primer extension lengths (with distinct G > A transition motifs from sporadic C > U-type DNA damage events on discrete Neanderthal templates either side of the 16263.1A insertion) provides a depth-of-coverage of just three (Figure 2b); confirming the inference made from the PCR-generated data of extremely low copy number endogenous Neanderthal mtDNA in the LQ4 extract.
SPEX can SNP-type aDNA as accurately as modern DNA
As SPEX amplifies only one of the duplex DNA strands, the choice of targeted template strand is a key part of the design strategy. G > A transitions (from C > U-type damage events on template strands) are the only base changes observed at above-background levels in SPEX-amplified aDNA (17; Figures 1b, 2b, 3 and 4). This means that if the DNA template strand that does not carry a C at a key SNP-of-interest is interrogated by SPEX, potential DNA damage-derived SNP-mistyping is reduced to background rates of polymerase misincorporation. For this reason, the above experiments were able to generate 100% of SPEX amplicons with the expected endogenous SNPs at the relevant key diagnostic sites: on the targeted PE168 light mtDNA strand, the endogenous SNPs were 16223T-16257A-16261T; on the LQ4 Neanderthal light mtDNA strand 16223T-16230G-16234T-16244A-16256A-C16262T-16263.1A; and on the Iceman and Amerindian heavy mtDNA strands 16362G and 16362A, respectively.
Targeting a DNA template strand that does carry a C at a key diagnostic SNP site highlights this issue. Both Amerindian individuals belong to hg B4b, with mtDNA SNPs A13590G and T16217C (45). Endogenous aDNA from the Iceman HB50 aDNA extract should carry a diagnostic T16224C hg K SNP (Figure S1c), since this SNP has been identified by every study of the Iceman’s mtDNA so far (15,39,41–43). Targeting the light mtDNA strand from both Amerindian aDNA extracts (PE40 and PE187), SPEX amplification produced the expected A > G transition at np 16217 (equating to the T16217C SNP typical of hg B4b on the rCRS) from 16/17 and 20/22 templates, respectively (Figure 4), whilst invariably returning the ancestral allele at np 16224. As expected, SPEX amplification from the HB50 Iceman aDNA extract returned the ancestral allele at np 16217 (from 18 discrete templates), with the diagnostic hg K SNP at np 16224 generating an A > G transition from 20/21 templates (reflecting T16224C relative to the rCRS). PCR-generated sequence data from the Iceman HB50 aDNA extract is presented elsewhere (43), whilst comparative PCR-generated data from the two Amerindian aDNA extracts are presented in Supplementary Figure S4.
As A > G transition artefacts are introduced at background rates by SPEX (17; Figures 1b, 2b, 3 and 4), the great majority of amplicons in the multi-fold depths-of-coverage generated here overwhelmingly reflect the predicted underlying endogenous aDNA sequences. Given the 100% SNP-typing accuracy (Figures 1b, 2b and 3) observed in this study when C is avoided at diagnostic sites on the target strand (17), amplicons with the ancestral (rCRS) allele (A) at np 16217 (from the Amerindians) or (A) at np 16224 (from the Iceman) are therefore almost certainly due to sporadic C > U miscoding lesion DNA damage (i.e. C16217U or C16224U) on the targeted light mtDNA strands of endogenous aDNA templates (observed as apparent G > A transitions back to the ancestral allele in first-generation SPEX copies). In particular, because of the 100% correct SNP-typing results described at np 16362 from the Amerindian and Iceman extracts, any argument that these ancestral alleles could have arisen due to low-level SPEX amplification from contaminant human DNA templates can be discounted.
DISCUSSION
Preferential access to genetic information from endogenous aDNA via SPEX
The SPEX-type approach used in this study preferentially accessed genetic information from endogenous aDNA over exogenous human DNA contaminants; both in controlled scenarios and from extensively handled, apparently irretrievably contaminated ancient hominin samples. In contrast to PCR, which can co-amplify modern human DNA contaminants and non-authentic PCR-generated artefacts more efficiently than degraded aDNA templates (15,46–48, this study), SPEX-type approaches impose no pre-defined target size based on a primer pair. Therefore, genetic information can be accessed from any DNA templates that cover the nested SPEX primers (generally approximately 40 bases); a substantially shorter minimum read-frame than that achievable via traditional PCR-based approaches.
Moreover, SPEX amplification also appears to be strongly skewed towards shorter molecules. As one moves towards shorter initial single primer extension lengths, the frequency of cloned SPEX amplicons in that size range exponentially increases (17). (For example, only ∼1% of 1669 cloned SPEX amplicons represents an initial SPEX event on a DNA template with an effective amplifiable size of 100 bases or more). Figure 3 shows that 75/101 SPEX amplicons genotyping np 16362 represent targeted heavy mtDNA templates with an effective amplifiable size between 43 and 60 bases in length (the nested SPEX primers cover 40 bases and primer extension of a further three bases is required to genotype np 16362).
Given the same primer binding sites, shorter PCR amplicons are amplified more efficiently than longer ones in the same reaction (38). Several hominin studies have also shown that larger PCR targets generate higher ratios of contaminant/endogenous sequences than shorter amplicons derived from the same samples (15,46–48). This suggests that (i) contaminant DNA templates are generally less damaged and fragmented than endogenous aDNA templates; (ii) their average effective amplifiable lengths are therefore longer. As PCR target size is increased, the numbers of directly amplifiable aDNA templates would be expected to decline (17), with amplification therefore increasingly tending to be skewed towards less damaged, more intact, more efficiently amplified molecules (such as contaminant human DNA templates and exponentially amplifiable PCR-generated recombinants that possess both PCR primer binding sites).
To further illustrate this point, consider PCR amplification with a typical hominin aDNA target size of approximately 80 bp. The PCR primer pair can exponentially amplify from ‘starting templates’ with an effective size 80 bases or more, including contaminants and recombinant artefacts which might have average effective amplifiable sizes far greater than 80 bases in length. Highly damaged and degraded templates with an average amplifiable size less than 80 bases could not be amplified directly by PCR, but only following some kind of ‘jumping-PCR’ recombinant event. In contrast, SPEX can target all templates approximately 40 bases and above, greatly increasing the range of accessible damaged and degraded endogenous aDNA templates. Crucially, however, because SPEX does not impose a pre-defined target length, SPEX primer extensions on more damaged and fragmented templates (such as endogenous aDNA) will tend to produce shorter SPEX amplicons that are likely to be amplified more efficiently than SPEX primer extensions on longer and less damaged templates (such as contaminant human DNA templates).
This is clearly seen in SPEX-generated sequence data, which show that shorter amplicons are preferentially amplified, and this may be a plausible explanation for the preferential access to genetic information from endogenous aDNA repeatedly observed via SPEX in this study. Presumably, there is some kind of hypothetical limit where human DNA contaminants could become just as damaged and fragmented as endogenous hominin aDNA templates. However (contra 46), this study did not detect such a situation even with side-by-side PCR versus SPEX experiments on museum samples that have been pre-contaminated via washing and extensive handling over many years. The data so far suggest that in many research scenarios, at least some degree of preferential access to endogenous aDNA over contaminant DNA might reasonably be expected via a SPEX-based approach; particularly when it is widely anticipated that in most ancient extracts, there will be a higher proportion of damaged and fragmented (i.e. effectively shorter) endogenous aDNA templates compared with modern contaminant human DNA templates over the kinds of size ranges demonstrably preferentially amplified by SPEX.
Increased depth-of-coverage and sequence reliability via SPEX
Depth-of-coverage and sequence reliability are of paramount importance for the accurate interpretation of hominin aDNA. In both respects, SPEX assays have proved successful. Regarding depth-of-coverage, amplification without a pre-defined target size, from just one of the DNA strands at target loci, means that a single SPEX assay can identify individual intact DNA ‘starting template’ molecules via distinct primer extension lengths and/or patterns of authentic miscoding lesion DNA damage transitions. Any single PCR amplification from aDNA cannot be assumed to have generated a greater depth-of-coverage than one (1,17,23), meaning individual SPEX assays can provide a substantially greater depth-of-coverage at key SNP sites. This is generally more than 10-fold for all but the most marginal aDNA extracts (Figures 1b, 2b, 3 and 4), thereby providing a critical advantage over the use of multiple, repeated PCR amplification reactions. SPEX-type approaches can therefore simultaneously increase genotyping accuracy whilst decreasing the consumption of valuable, often irreplaceable, samples and aDNA extracts.
SPEX can also maximize sequence reliability. In PCR-derived aDNA data, all four transition base changes can be generated at above-background levels (e.g. 8,9,18–22). C > U-type base modifications can create both C > T and G > A (‘Type 2’) transitions because both template strands are targeted by the PCR primer pair. PCR amplification from damaged and degraded DNA templates can also introduce sequence artefacts into PCR amplicons at significant levels. For example, T > C and A > G transition base changes—so-called ‘Type 1’ aDNA damage (19,20)—which have been shown to be non-authentic, wholly PCR-generated, artefacts (17). The percentage of all transition base changes corresponding to these kinds of ‘Type 1’ PCR-generated artefacts can vary widely between aDNA studies utilizing a variety of species and specific amplification conditions: e.g. 20% (21); 29% (9); 35% (8); 39% (20); and 42% (18). In contrast, SPEX creates direct first-generation copies from just one of the DNA strands at target loci and G > A transitions (from C > U-type miscoding lesions on DNA templates) are the only base changes observed at above-background levels (17, this study). These properties of SPEX significantly reduce sequence interpretation difficulties following amplification from aDNA extracts.
PCR-generated recombinant template switching artefacts can also create non-authentic sequences at significant levels (6–16,25), particularly from hominin aDNA extracts due to the ever-present risk of human DNA contamination (49). Our PCR-generated data from samples with clustered multiple diagnostic SNP differences clearly produced non-authentic recombinant artefacts (Figures 1a and 2a). In common with many previous studies that demonstrate significant levels of ‘jumping PCR’ between endogenous and contaminant templates (e.g. 2,7–11,13, 15), these artefacts appear to be an unavoidable mechanistic consequence of ‘two-primer’ PCR amplification from poor-quality DNA templates. However, true levels of PCR-generated recombinant template switching must generally be significantly higher than those estimated, since endogenous–endogenous and contaminant–contaminant recombinant hybrids would not be so easily identified, and nor would endogenous–contaminant hybrids when there are not multiple diagnostic SNP differences between the two.
The existence of such ‘hidden’ events might nevertheless contribute to the generation of non-authentic sequences by PCR. Many DNA polymerases can add a single nucleotide to the 3′-end of both fully extended and damage-halted primer extensions by non-templated nucleotide addition (NTNA). Often, an A is added following full extension on a template molecule, but also G, T and C at abasic sites and other damage-derived lesions (17,50–53). Although further investigation is clearly required, some combination of partial primer extension, NTNA and template switching events during early PCR cycles offers the potential for the generation and ‘fixation’ of non-authentic base changes into amplicons (1,17,23).
The SPEX approach, by removing all other potential DNA templates (via bead washes following single primer extension) and permanently blocking the 3′-end of primer extensions (via polyC-tailing), appears to strongly suppress the subsequent creation of non-authentic hybrids and other sequence artefacts. Whatever mechanisms create PCR-generated artefacts, there is a striking absence of above-background levels of ‘Type 1’ T > C and A > G artefacts in studies where recombinant template switching is largely precluded, as in SPEX or 454-based studies (17,32,33,54, this study). So perhaps, the statistically significant Types 1 and 2 transition ‘hotspots’ described by some PCR-based studies (19,20) might now be reconsidered as hotspots of PCR-generated artefacts, rather than being ascribed to aDNA ‘damage hotspots’.
Genotyping ancient hominin samples reliably
By providing a multi-fold depth-of-coverage, combined with a reduction of potential SNP-mistyping to background levels at key target sites, SPEX-generated sequence data from the Iceman aDNA extract strongly suggests that T16362C is the endogenous mtDNA SNP. This conclusion supports the findings of two other recent studies (42,43). In marked contrast, previous PCR-based studies (15,39,41) had defined the Iceman’s control region motif as 16223C-16311C only (i.e. lacking the T16362C polymorphism).
However, interpretations of why this might have occurred differ markedly; Ermini et al. (42) suggested that the original study (15) might have failed to identify an authentic polymorphism at np 16362 amongst their hg K mtDNA sequences due to PCR-generated recombinant template switching events. This explanation appears highly unlikely because it would have required the total elimination of T16362C SNPs—by template switching—from every PCR amplicon cloned from multiple-independent reactions. Nor does this proposal explain the parallel absence of the T16362C polymorphism in the consensus sequence inferred following independent replication (15,39). A more plausible explanation for the repeated failure to identify the Iceman’s T16362C polymorphism is that both studies had been compromised by non-endogenous hg K contaminant DNA (43).
Only the combination of a sustained and continuing interest in the celebrated Iceman, the highly fortuitous circumstances of him having an uncommon genotype (currently unique within hg K) and the availability of a number of relatively ‘unhandled’ alternate sources for DNA extraction led to the discovery that the original studies had not actually genotyped the Iceman. This cautionary tale highlights the precarious nature of traditional PCR-based approaches to hominin aDNA—even where the individual concerned is considered to be generally well preserved (41) and every criterion-of-authenticity (such as dedicated aDNA facilities, multiple PCR amplifications, cloning, independent replication, etc.) appears to be fully satisfied.
Unfortunately, however, the luxury of distinctive/uncommon genotypes and unhandled/uncontaminated sample material cannot be expected to be the norm with the majority of archaeological and museum hominin samples. The original Iceman investigations clearly demonstrate that whenever low-level (but highly amplifiable) contaminants are likely to have identical, or very similar, sequences to underlying endogenous hominin aDNA, it may ultimately prove impossible for PCR-based aDNA approaches to tease out the relative contributions from endogenous aDNA, contaminant human DNA, template switching recombinant hybrids and other non-authentic sequence artefacts (particularly over the necessarily short regions targeted).
In scenarios like these, there are clearly considerable advantages to the use of SPEX-type assays. By targeting template DNA strands that do not have C as one of the alleles at diagnostic polymorphisms, SPEX-type approaches can quantitatively genotype aDNA as accurately as modern DNA (17, this study). As the SPEX-based genotyping of the Amerindian samples at np 16362 shows, a SPEX-type approach can even begin to meaningfully tackle aDNA extracts with sequences that are commonplace in the modern human populations likely to contribute contaminant DNA via field workers, museum curators, laboratory personnel, etc.
CONCLUSIONS
This study builds on initial SPEX data from animal aDNA and establishes proof-of-principle that SPEX-type approaches to ancient hominin samples could be a highly useful addition to the aDNA methodological repertoire. The potential shortcomings of traditional PCR-based approaches to hominin aDNA highlighted by this study, and many previous studies, appear to be a mechanistic consequence of the methodology itself. The clear failure to correctly genotype the Iceman’s endogenous mtDNA via high profile, independently replicated, PCR-based aDNA studies serves as a demonstration of how unsuited ‘two-primer’ exponential amplification can be when targeting damaged and degraded endogenous aDNA in the presence of highly amplifiable human contaminant templates.
In sharp contrast, this study has shown that SPEX-type approaches can (i) preferentially access highly damaged and degraded endogenous aDNA templates over contaminant human DNA; (ii) reduce non-authentic recombinant template switching events to below detectable levels; and (iii) reduce potential sources of non-authentic polymorphisms from DNA damage and amplification-generated sequence artefacts to background levels. These attributes mean that ancient hominin samples could begin to be quantitatively genotyped as accurately as modern ones. The cases of the Iceman and La Quina 4 individuals, in particular, suggest that future SPEX-type approaches offer the potential to accurately recover authentic aDNA sequence data from a far wider range of archaeological samples. This would not only include samples carefully excavated ‘uncontaminated’, as used in many current PCR- and metagenomic-based approaches, but also the far more numerous extensively pre-handled hominin samples in archaeological storage facilities and museums.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The SPEX work was supported by the UK NERC, Wellcome and Leverhulme Trusts (grants to A.C.); National Environment Council of Great Britain and Magdalen College Oxford (to P.E.). Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Peter Pramstaller, the Natural History Museum London, the Peruvian National Institute of Culture, and the Mexican National Museum of Anthropology, for providing the human samples for this study. PB developed the concept of amplifying damaged/degraded aDNA templates via single primer extension (SPEX) based approaches and designed and performed the SPEX-based experiments. PE provided ancient hominin DNA extracts, and appropriate target SNPs for both PCR and SPEX assays. PE and JJS designed the PCR-based experiments, which were performed by PE. Experimental data was analysed by PB, JJS, AC & PE. PB, JJS, AC & PE wrote the manuscript.
REFERENCES
- 1.Pääbo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, Hofreiter M. Genetic analyses from ancient DNA. Annu. Rev. Genet. 2004;38:645–679. doi: 10.1146/annurev.genet.37.110801.143214. [DOI] [PubMed] [Google Scholar]
- 2.Handt O, Krings M, Ward RH, Pääbo S. The retrieval of ancient human DNA sequences. Am. J. Hum. Genet. 1996;59:368–376. [PMC free article] [PubMed] [Google Scholar]
- 3.Höss M, Jaruga P, Zastawny TH, Dizdaroglu M, Pääbo S. DNA damage and DNA sequence retrieval from ancient tissues. Nucleic Acids Res. 1996;24:1304–1307. doi: 10.1093/nar/24.7.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krause J, Lalueza-Fox C, Orlando L, Enard W, Green RE, Burbano HA, Hublin J-J, Bertranpetit J, Hänni C, de la Rasilla M, et al. The derived FOXP2 variant of modern humans was shared with Neanderthals. Curr. Biol. 2007;17:1908–1912. doi: 10.1016/j.cub.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Lalueza-Fox C, Römpler H, Caramelli D, Stäubert C, Catalano G, Hughes D, Rohland N, Pilli E, Longo L, Condemi S, et al. A melanocortin 1 receptor allele suggests varying pigmentation among Neanderthals. Science. 2007;318:1453–1455. doi: 10.1126/science.1147417. [DOI] [PubMed] [Google Scholar]
- 6.Serre D, Langaney A, Chech M, Teschler-Nicola M, Paunovic M, Mennecier P, Hofreiter M, Possnert G, Pääbo S. No evidence of Neandertal mtDNA contribution to early modern humans. PLoS Biol. 2004;2:e57. doi: 10.1371/journal.pbio.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouwman AS, Chilvers ER, Brown KA, Brown TA. Brief communication: identification of the authentic ancient DNA sequence in a human bone contaminated with modern DNA. Am. J. Phys. Anthropol. 2006;131:428–431. doi: 10.1002/ajpa.20411. [DOI] [PubMed] [Google Scholar]
- 8.Krings M, Stone A, Schmitz RW, Krainitzki H, Stoneking M, Pääbo S. Neandertal DNA sequences and the origin of modern humans. Cell. 1997;90:19–30. doi: 10.1016/s0092-8674(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 9.Krings M, Geisert H, Schmitz RW, Krainitzki H, Pääbo S. DNA sequence of the mitochondrial hypervariable region II from the Neandertal type specimen. Proc. Natl Acad. Sci. USA. 1999;96:5581–5585. doi: 10.1073/pnas.96.10.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ovchinnikov IV, Götherström A, Romanova GP, Kharitonov VM, Lidén K, Goodwin W. Molecular analysis of Neandertal DNA from the northern Caucasus. Nature. 2000;404:490–493. doi: 10.1038/35006625. [DOI] [PubMed] [Google Scholar]
- 11.Lalueza-Fox C, Sampietro ML, Caramelli D, Puder Y, Lari M, Calafell F, Martínez-Maza C, Bastir M, Fortea J, de la Rasilla M, et al. Neandertal evolutionary genetics: mitochondrial DNA data from the Iberian peninsula. Mol. Biol. Evol. 2005;22:1077–1081. doi: 10.1093/molbev/msi094. [DOI] [PubMed] [Google Scholar]
- 12.Meyerhans A, Vartanian JP, Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;18:1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pääbo S, Irwin DM, Wilson AC. DNA damage promotes jumping between templates during enzymatic amplification. J. Biol. Chem. 1990;265:4718–4721. [PubMed] [Google Scholar]
- 14.Ruano G, Brash DE, Kidd KK. PCR: the first few cycles. Amplifications. 1991;7:1–4. [Google Scholar]
- 15.Handt O, Richards M, Trommsdorff M, Kilger C, Simanainen J, Georgiev O, Bauer K, Stone A, Hedges R, Schaffner W, et al. Molecular genetic analyses of the Tyrolean Ice Man. Science. 1994;264:1775–1778. doi: 10.1126/science.8209259. [DOI] [PubMed] [Google Scholar]
- 16.Wang GC, Wang Y. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl. Environ. Microbiol. 1997;63:4645–4650. doi: 10.1128/aem.63.12.4645-4650.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brotherton P, Endicott P, Sanchez JJ, Beaumont M, Barnett R, Austin J, Cooper A. Novel high-resolution characterization of ancient DNA reveals C > U-type base modification events as the sole cause of post mortem miscoding lesions. Nucleic Acid Res. 2007;35:5717–5728. doi: 10.1093/nar/gkm588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen A, Willerslev E, Wiuf C, Mourier T, Arctander P. Statistical evidence for miscoding lesions in ancient DNA templates. Mol. Biol. Evol. 2001;18:262–265. doi: 10.1093/oxfordjournals.molbev.a003800. [DOI] [PubMed] [Google Scholar]
- 19.Gilbert MTP, Hansen AJ, Willerslev E, Rudbeck L, Barnes I, Lynnerup N, Cooper A. Characterization of genetic miscoding lesions caused by post-mortem damage. Am. J. Hum. Genet. 2003;72:48–61. doi: 10.1086/345379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert MTP, Shapiro BA, Drummond A, Cooper A. Post mortem DNA damage hotspots in Bison (Bison bison and B. bonasus) provide supporting evidence for mutational hotspots in human mitochondria. J. Archaeol. Sci. 2005;32:1053–1060. [Google Scholar]
- 21.Lalueza-Fox C, Krause J, Caramelli D, Catalano G, Milani L, Sampietro ML, Calafell F, Martínez-Maza C, Bastir M, García-Tabernero A, et al. Mitochondrial DNA of an Iberian Neandertal suggests a population affinity with other European Neandertals. Curr. Biol. 2006;16:R629–R630. doi: 10.1016/j.cub.2006.07.044. [DOI] [PubMed] [Google Scholar]
- 22.Binladen J, Wiuf C, Gilbert MTP, Bunce M, Larson G, Barnett R, Hansen AJ, Willerslev E. Comparing miscoding lesion damage in mitochondrial and nuclear ancient DNA. Genetics. 2006;172:733–741. doi: 10.1534/genetics.105.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofreiter M, Jaenicke V, Serre D, von Haeseler A, Pääbo S. DNA sequences from multiple amplifications reveal artefacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 2001;29:4793–4799. doi: 10.1093/nar/29.23.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lalueza-Fox C, Gigli E, de la Rasilla M, Fortea J, Rosas A, Bertranpetit J, Krause J. Genetic characterization of the ABO blood group in Neandertals. BMC Evol. Biol. 2008;8:342. doi: 10.1186/1471-2148-8-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bandelt HJ. Mosaics of ancient mitochondrial DNA: positive indicators of nonauthenticity. Eur. J. Hum. Genet. 2005;13:1106–1112. doi: 10.1038/sj.ejhg.5201476. [DOI] [PubMed] [Google Scholar]
- 26.Caramelli D, Lalueza-Fox C, Vernesi C, Lari M, Casoli A, Mallegni M, Chiarelli B, Dupanloup I, Bertranpetit J, Barbujani G, et al. Evidence for a genetic discontinuity between Neandertals and 24,000-year-old anatomically modern Europeans. Proc. Natl Acad. Sci. USA. 2003;100:6593–6597. doi: 10.1073/pnas.1130343100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.García-Bour J, Pérez-Pérez A, Alvarez S, Fernández E, López-Parra AM, Arroyo-Pardo E, Turbón D. Early population differentiation in extinct aborigines from Tierra del Fuego-Patagonia: ancient mtDNA sequences and Y-chromosome STR characterization. Am. J. Phys. Anthropol. 2004;123:361–370. doi: 10.1002/ajpa.10337. [DOI] [PubMed] [Google Scholar]
- 28.Endicott P, Gilbert MT, Stringer C, Lalueza-Fox C, Willerslev E, Hansen AJ, Cooper A. The genetic origins of the Andaman Islanders. Am. J. Hum. Genet. 2003;72:178–184. doi: 10.1086/345487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haak W, Forster P, Bramanti B, Matsumura S, Brandt G, Tanzer M, Villems R, Renfrew C, Gronenborn D, Alt KW, et al. Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science. 2005;310:1016–1018. doi: 10.1126/science.1118725. [DOI] [PubMed] [Google Scholar]
- 30.Endicott P, Metspalu M, Stringer C, Macaulay V, Cooper A, Sanchez JJ. Multiplexed SNP typing of ancient DNA clarifies the origin of Andaman mtDNA haplogroups amongst South Asian tribal populations. PLoS ONE. 2006;1:e81. doi: 10.1371/journal.pone.0000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barik SS, Sahani R, Prasad BVR, Endicott P, Metspalu M, Sarkar BN, Bhatacharya S, Annapooma PCH, Sreenath J, Sun D, et al. Detailed mtDNA genotypes permit a reassessment of the settlement and population structure of the Andaman Islands. Am. J. Phys. Anthropol. 2008;136:19–27. doi: 10.1002/ajpa.20773. [DOI] [PubMed] [Google Scholar]
- 32.Green RE, Krause J, Ptak SE, Briggs AW, Ronan MT, Simons JF, Du L, Egholm M, Rothberg JM, Paunovic M, et al. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- 33.Stiller M, Green RE, Ronan M, Simons JF, Du L, He W, Egholm M, Rothberg JM, Keates SG, Ovodov ND, et al. Patterns of nucleotide misincorporations during enzymatic amplification and direct large-scale sequencing of ancient DNA. Proc. Natl Acad. Sci. USA. 2006;103:13578–13584. doi: 10.1073/pnas.0605327103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilbert MT, Binladen J, Miller W, Wiuf C, Willerslev E, Poinar H, Carlson JE, Leebens-Mack JH, Schuster SC. Recharacterization of ancient DNA miscoding lesions: insights in the era of sequencing-by-synthesis. Nucleic Acids Res. 2007;35:1–10. doi: 10.1093/nar/gkl483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts by amplification using a single gene-specific oligonucleotide primer. Proc. Natl Acad. Sci. USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Hara O, Dorit RI, Gilbert W. One-sided PCR: the amplification of cDNA. Proc. Natl Acad. Sci. USA. 1989;86:5673–5677. doi: 10.1073/pnas.86.15.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh PS, Erlich HA, Higuchi R. Preferential PCR amplification of alleles: mechanisms and solutions. PCR Methods Appl. 1992;1:241–250. doi: 10.1101/gr.1.4.241. [DOI] [PubMed] [Google Scholar]
- 39.Richards M, Sykes B, Hedges R. Authenticating DNA extracted from ancient skeletal remains. J. Arch. Sci. 1995;22:291–299. [Google Scholar]
- 40.Thomas MG, Jones AL, Osman F, Weale ME, Egarter-Vigl E, Pramstaller PP. Bolzano: The Iceman, Sud-Tyrol Archaeological Museum; 2001. Y-chromosome Variation in the Iceman and Modern Europeans. [Google Scholar]
- 41.Rollo F, Ermini L, Luciani S, Marota I, Olivieri C, Luiselli D. Fine characterization of the Iceman’s mtDNA haplogroup. Am. J. Phys. Anthropol. 2006;130:557–564. doi: 10.1002/ajpa.20384. [DOI] [PubMed] [Google Scholar]
- 42.Ermini L, Olvieri C, Rizzi E, Corti G, Bonnal R, Soares P, Luciani S, Marota I, De Bellis G, Richards MB, et al. Complete mitochondrial genome sequence of the Tyrolean Iceman. Curr. Biol. 2008;18:1–7. doi: 10.1016/j.cub.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 43.Endicott P, Sanchez JJ, Pichler I, Brotherton P, Brooks J, Egarter-Vigl E, Cooper A, Pramstaller P. Genotyping human ancient mtDNA control and coding region polymorphisms with a multiplexed single-base-extension assay: the singular maternal history of the Tyrolean Iceman. BMC Genet. 2009;10:29. doi: 10.1186/1471-2156-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamm E, Kivisild T, Reidla M, Metspalu M, Smith DG, Mulligan CJ, Bravi CM, Rickards O, Martinez-Labarga C, Khusnutdinova EK, et al. Beringian standstill and spread of Native American founders. PLoS ONE. 2007;2:e829. doi: 10.1371/journal.pone.0000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kong QP, Yao YG, Sun C, Bandelt HJ, Zhu CL, Zhang YP. Phylogeny of East Asian mitochondrial DNA lineages inferred from complete sequences. Am. J. Hum. Genet. 2003;73:671–676. doi: 10.1086/377718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sampietro ML, Gilbert MTP, Lao O, Caramelli D, Lari M, Bertranpetit J, Lalueza-Fox C. Tracking down human contamination in ancient human teeth. Mol. Biol. Evol. 2006;23:1801–1807. doi: 10.1093/molbev/msl047. [DOI] [PubMed] [Google Scholar]
- 47.Malmstrom H, Svensson EM, Gilbert MTP, Willerslev E, Gotherstrom A, Holmlund G. More on contamination: the use of asymmetric molecular behaviour to identify authentic ancient human DNA. Mol. Biol. Evol. 2007;24:998–1004. doi: 10.1093/molbev/msm015. [DOI] [PubMed] [Google Scholar]
- 48.Wall JD, Kim SK. Inconsistencies in Neanderthal genomic DNA sequences. PLoS Genet. 2007;3:1862–1866. doi: 10.1371/journal.pgen.0030175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malmström H, Storå J, Dalén L, Holmlund G, Götherström A. Extensive human DNA contamination in extracts from ancient dog bones and teeth. Mol. Biol. Evol. 2005;22:2040–2047. doi: 10.1093/molbev/msi195. [DOI] [PubMed] [Google Scholar]
- 50.Clark JM. Novel non-templated nucleotide addition reactions catalyzed by prokaryotic and eukaryotic DNA polymerases. Nucleic Acids Res. 1988;16:9677–9686. doi: 10.1093/nar/16.20.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strauss BS. The ‘A rule’ of mutagen specificity: a consequence of DNA polymerase bypass of non-instructional lesions? Bioessays. 1991;13:79–84. doi: 10.1002/bies.950130206. [DOI] [PubMed] [Google Scholar]
- 52.Shibutani S, Takeshita M, Grollman AP. Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the “A rule”. J. Biol. Chem. 1997;272:13916–13922. doi: 10.1074/jbc.272.21.13916. [DOI] [PubMed] [Google Scholar]
- 53.Berthet N, Roupioz Y, Constant JF, Kotera M, Lhomme J. Translesional synthesis on DNA templates containing the 2′-deoxyribonolactone lesion. Nucleic Acids Res. 2001;29:2725–2732. doi: 10.1093/nar/29.13.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Green RE, Malaspinas AS, Krause J, Briggs AW, Johnson PL, Uhler C, Meyer M, Good JM, Maricic T, Stenzel U, et al. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell. 2008;134:416–426. doi: 10.1016/j.cell.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.