Abstract
Methylation of lysine residues on the tails of histone proteins is a major determinant of the transcription state of associated DNA coding regions. The interplay among methylation states and other histone modifications to direct transcriptional outcome is referred to as the histone code. In addition to histone methyltransferases and demethylases which function to modify the methylation state of lysine sidechains, other proteins recognize specific histone methylation marks essentially serving as code readers. While these interactions are highly specific with respect to site and methylation state of particular lysine residues, they are generally weak and therefore difficult to monitor by traditional assay techniques. Herein, we present the design and implementation of a homogeneous, miniaturizable, and sensitive assay for histone methylation-dependent interactions. We use AlphaScreen, a chemiluminescence-based technique, to monitor the interactions of chromodomains (MPP8, HP1β and CHD1), tudor domains (JMJD2A) and plant homeodomains (RAG2) with their cognate trimethyllysine histone partners. The utility of the method was demonstrated by profiling the binding specificities of chromo- and tudor domains toward several histone marks. The simplicity of design and the sensitive and robust nature of this assay should make it applicable to a range of epigenetic studies, including the search for novel inhibitors of methylation-dependent interactions.
INTRODUCTION
Regulation of gene expression is closely tied to post-translational modifications of histones. Several types of covalent histone modifications have been identified, including acetylation, phosphorylation, ubiquitination, sumoylation and methylation (1). The spectrum and coordination of these post-translational modifications has been proposed to dictate transcriptional activity and downstream cellular processes, giving rise to the ‘histone code’ hypothesis (2). Methylation of lysine has been of particular interest due to its slow turnover in vivo and association with several disease phenotypes, including cancer (3,4).
At least six lysine residues are methylated in the core histones H3 and H4. The lysine residues can be mono-, di- or tri-methylated, thus adding another layer of complexity. Whereas acetylation generally leads to transcriptional activation, the physiological outcome of histone methylation is more ambiguous. For example, H3K4me2 is a signature of actively transcribed genes, while H3K9 methylation is a hallmark of constitutive heterochromatin (5,6). Methylation itself is not sufficient to effect changes in transcriptional activity (7), indicating the need for coordination with effector or recognition proteins. Specific domains have been identified in chromatin-associated proteins that bind to methylated Lys of histones H3 or H4 in a site- and degree-specific manner. These domains include chromodomains, tudor domains, plant homeodomain (PHD) modules and ankyrin repeats (3,8–14). Proteins harboring these modules may function to recruit chromatin remodeling and histone-modifying proteins, stabilize chromatin complexes, or directly affect chromatin structure (1). These ‘readers’ of the post-translational modifications are gaining attention as potential therapeutic targets for diseases associated with epigenetic regulation. Disruption of a specific protein–protein interaction at a covalently modified histone residue is a potentially more attractive target than inhibition of the enzymes that may promiscuously add or remove these modifications at more than one histone residue.
Several methods have been used to detect binding of epigenetic partners, including fluorescence spectroscopy (15), fluorescence polarization (FP; 16) and fluorescence resonance energy transfer (FRET) (17). While these are established methods, they generally require large sample sizes and/or are subject to interference and high background levels that make them difficult to use with the low-affinity interactions of methyl-binding domains with Kd values in the micromolar range (18–20). Surface plasmon resonance (SPR) (21) has the advantage in its ability to detect low-affinity interactions but it is severely restricted in its high-throughput capabilities. In vitro pull-down assays have been used to identify binding of protein domains to post-translationally modified histone peptides. This technique was modified to increase throughput as a CADOR (chromatin-associated domain array) chip (13). The CADOR chip is useful for initial screens of binding partners, but for high-throughput screening (HTS) it is limited by its qualitative nature, requirement for multiple wash steps and the expense of production.
Here, we introduce a simple, homogeneous method for measuring binding of epigenetic partners using AlphaScreen technology. AlphaScreen is a colloidal bead-based assay founded on luminescent oxygen channeling chemistry (22). This format is flexible and may be utilized by multiple assay paradigms, including detection of protein–protein interactions (23). The homogeneous, sensitive and scalable AlphaScreen assay presented here is an appropriate method to measure binding of covalently modified peptides to a wide range of histone-associated proteins. We present the design and optimization of binding assays for chromodomains CHD1, MPP8 and HP1β, as well as for the JMJD2A tudor domains, and the PHD domain of RAG2. Furthermore, the assay was used to interrogate the binding specificity of the above domains to a series of peptides representing various methylation states and methyllysine loci.
MATERIALS AND METHODS
General reagents
Assay buffer consisted of PBS, pH 7.4, containing 0.01% Tween-20. Microplates used were 384- or 1536-well white solid-bottom type from Greiner Bio-One (Monroe, NC). Peptides were synthesized by the Tufts Medical School Department of Physiology Core Facility (Boston, MA, USA). AlphaScreen detection was performed with PerkinElmer Glutathione S-Transferase (GST) and Histidine (Nickel Chelate) Detection Kits (Waltham, MA, USA).
Histone-binding proteins
Mouse heterochromatin protein 1-beta (NM_007622) chromodomain-containing region of amino acids 1–185 was expressed as a recombinant protein with N-terminal GST tag (GST-HP1β). Human M-phase phosphoprotein (NM_017520) chromodomain-containing region of amino acids 50–120 was expressed as a recombinant protein with either an N-terminal GST tag (GST-MPP8) or N-terminal His6 tag (His-MPP8). Human chromodomain-containing protein, Y-linked, 2B (NM_001001722) chromodomain-containing region of amino acids 5–101 was expressed as a recombinant protein with N-terminal GST tag (GST-CDY2B). Human chromodomain helicase DNA-binding protein 1 (NM_001270) chromodomain-containing region of amino acids 251–467 was expressed as a recombinant protein with N-terminal GST tag (GST-CHD1). Human jumonji domain containing 2A protein (NM_014663) was expressed with an N-terminal His6 tag (His-JMJD2A) from amino acids 890–1031 to include the double tudor domain. Human recombination activating gene 2 (NM_000536) PHD-containing region of amino acids 397–493 was expressed as a recombinant protein with N-terminal GST tag (GST-RAG2).
AlphaScreen assay in 384-well format
A 2-fold dilution series of biotinylated glutathione S-transferase (b-GST), final 0.1–50 nM, was plated in a 384-well plate. Dilutions of b-GST were incubated with 20 µg/ml streptavidin-conjugated donor and anti-GST acceptor beads for 30 min at room temperature. Samples were read in an EnVision Multilabel Plate Reader (PerkinElmer, Waltham, MA, USA) using the 384 Plate HTS AlphaScreen aperture (35-ms excitation time, 100-ms measurement time), and values were corrected with an internal crosstalk optimization.
A panel of chromodomain proteins was assayed to measure binding of trimethylated histone H3 peptides. Chromodomain proteins, tagged with N-terminal GST, were incubated at a final concentration of 100 nM with 100 nM biotinylated-H3 peptide trimethylated at either Lys4 or Lys9 (b-H3K4me3 or b-H3K9me3, respectively). Reactions were incubated for 30 min at room temperature in 384-well plates in a volume of 30 µl. AlphaScreen streptavidin-conjugated donor and anti-GST acceptor beads (10 µl) were added to wells at a final concentration 20 µg/ml and incubated for 10 min at room temperature. Plates were read in the EnVision reader as described above.
Assay miniaturization
GST-HP1β was incubated at a final concentration of 100 nM with either b-H3K4me3 or b-H3K9me3 peptides (final 20–200 nM). Incubations in 384-well plates consisted of 30-µl samples. Following a 30 min incubation of protein and peptide at room temperature, 10 µl of AlphaScreen streptavidin-conjugated donor and anti-GST acceptor beads (final 20 µg/ml each) were added to sample wells and incubated for 10 min. Plates were read in an EnVision microplate reader as previously described.
Complementary reactions in 1536-well plates were achieved by employing a BioRAPTR (Beckman Coulter, Fullerton, CA, USA) flying reagent dispenser (FRD) for automated addition of reagents. To each well, 0.2–2.0 µl of biotinylated peptide was dispensed in combination with 1 µL GST-HP1β and varying volume of buffer for a final volume of 3 µl. Plates were centrifuged for 1 min at 1000 r.p.m. and incubated at room temperature for 30 min. AlphaScreen streptavidin-conjugated donor and anti-GST acceptor beads were added in a 1 µL FRD dispense for final 20 µg/mL concentration of each bead. Plates were centrifuged again for 1 min at 1000 r.p.m. and incubated for 10 min at room temperature. Samples were measured with an EnVision multilabel plate reader equipped with a 1536 Plate HTS AlphaScreen aperture (80-ms excitation time, 240-ms measurement time), and values were corrected with an internal crosstalk optimization. Data are expressed as the mean of four replicate measurements.
Peptide–protein binding matrices
Interactions of methyl-binding domain proteins with trimethyllysine histone peptides were measured in matrix titrations. The concentration of GST- or His-tagged protein was varied from 5–1000 nM as the concentration of biotinylated peptide was varied from 10–1000 nM. Incubations were performed in duplicate in 384-well plates for 30 min at room temperature. AlphaScreen streptavidin-conjugated donor and acceptor beads (anti-GST and nickel-chelate acceptor beads for GST- and His-tagged proteins, respectively) were added for final 20 µg/ml each bead and final volume of 40 µl. Samples were incubated for 10 min at room temperature and read with an EnVision plate reader.
Competitive binding of unlabeled peptides in 1536-well plate format
Competition of biotinylated histone H3 peptides with unlabeled peptides of varying sequence and post-translational modification was performed to measure the specificity and affinity of binding to chromatin-associated domains. Proteins were preincubated with the corresponding biotinylated peptide for 30 min at room temperature. Competitor peptide dilutions were plated in a 384-well plate. A CyBi-Well (CyBio Inc., Woburn, MA) 384-channel simultaneous pipettor was then used to transfer 3 µl of peptide from the 384-well plate into an empty 1536-well plate in quadruplicate (final 60 nM–1 mM unlabeled peptide). Preincubated protein-peptide solution was dispensed (1 µl) into these wells with a BioRAPTR FRD. Reactions were incubated at room temperature for 30 min. AlphaScreen beads (1 µl, final 20 µg/ml) were dispensed and reactions were incubated for 10 min prior to an EnVision read. AlphaScreen signal (counts per second, c.p.s.) was plotted versus log[Peptide, M] as the mean of four replicates for each peptide concentration. Nonlinear curve fitting was carried out using Prism 4 software (GraphPad, La Jolla, CA, USA).
RESULTS
Assay principle and validation
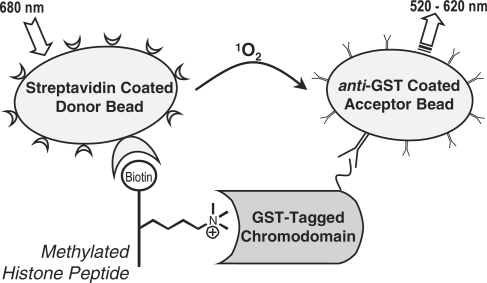
The AlphaScreen assay we employ is a proximity-based reporting system used to measure binding between two cognate partners. As shown in Scheme 1, laser excitation (680 nm) of a donor bead releases a flow of singlet oxygen. Acceptor beads in close proximity (<200 nm) utilize this singlet oxygen to generate a narrow chemiluminescent signal of 520–620 nm. Donor and acceptor beads are conjugated with functional groups that allow bioconjugation with specific analytes. Here, we have configured an AlphaScreen assay by using streptavidin-coated donor beads and anti-GST antibody coated acceptor beads to measure binding of a GST-tagged chromodomain protein to a biotinylated histone peptide.
Scheme 1.
AlphaScreen assay of chromodomain protein binding to methylated histone peptides.
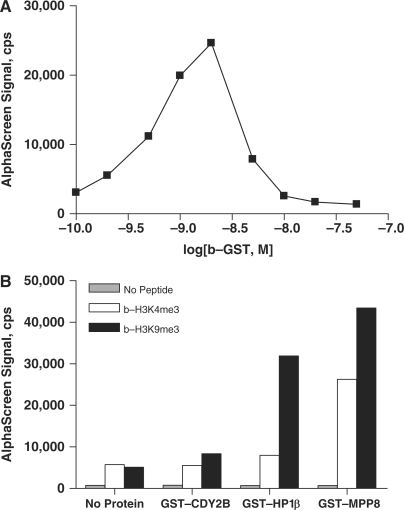
Biotinylated-GST (b-GST) was titrated with 20 µg/ml streptavidin-conjugated donor beads and anti-GST acceptor beads to validate AlphaScreen detection and assess the signal window (Figure 1A). AlphaScreen signal reached a maximum at 2 nM b-GST of ∼25 000 c.p.s. Higher concentrations of b-GST decreased the signal, the so-called ‘hook effect’ (www.perkinelmer.com) whereby b-GST concentration surpasses the binding capacity of the AlphaScreen beads. Determining where high concentrations of analyte lead to nonproductive binding and loss of signal is essential in the optimization of AlphaScreen assays.
Figure 1.
(A) Biotinylated-GST titration curve was used to identify the ‘hook’ point of AlphaScreen assay. (B) Chromodomain proteins CDY2B, HP1β, and MPP8 demonstrated binding selectivity for biotinylated histone H3 peptides trimethylated at Lys9 over peptides trimethylated at Lys4. GST-tagged proteins (100 nM) were incubated with either 100 nM b-H3K4me3 (white) or b-H3K9me3 (black). Assay background was low, as no signal was observed in the absence of peptide (gray).
To assess the applicability of the AlphaScreen format to measure chromodomain interactions, initial binding studies were performed with 100 nM GST-tagged chromodomain proteins and 100-nM biotinylated-histone H3 peptides that were trimethylated at either K4 or K9 (Figure 1B). All chromodomain proteins tested showed selectivity for b-H3K9me3 over b-H3K4me3, except for CDY2B where the assay signal was too low to allow a definitive conclusion. These data are in agreement with previous reports of HP1β selectivity (13,18,24), while recognition of specific histone lysine methylation by the MPP8 chromodomain has not been reported to date. Under these conditions, GST-HP1β and GST-MPP8 also showed interaction with b-H3K4me3. The affinity of GST-MPP8 for b-H3K4me3 was greater than that of GST-HP1β (26 000 and 7800 c.p.s., respectively). However, selectivity for trimethylation at H3K9 versus at H3K4 was greater for GST-HP1β than for GST-MPP8, 3.3- versus 1.7-fold.
Miniaturization of chromodomain-binding assay
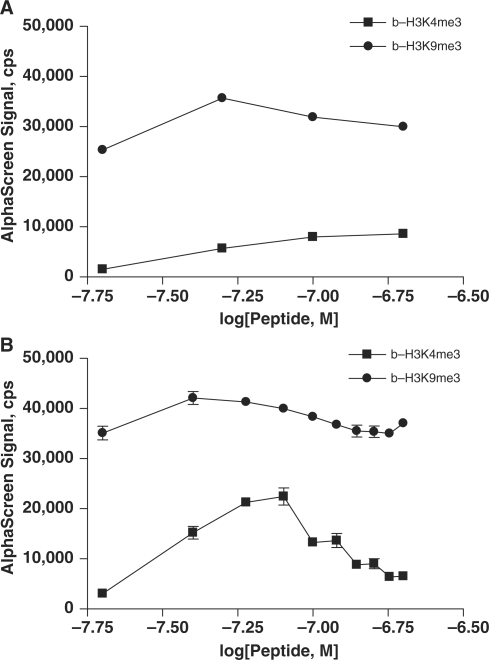
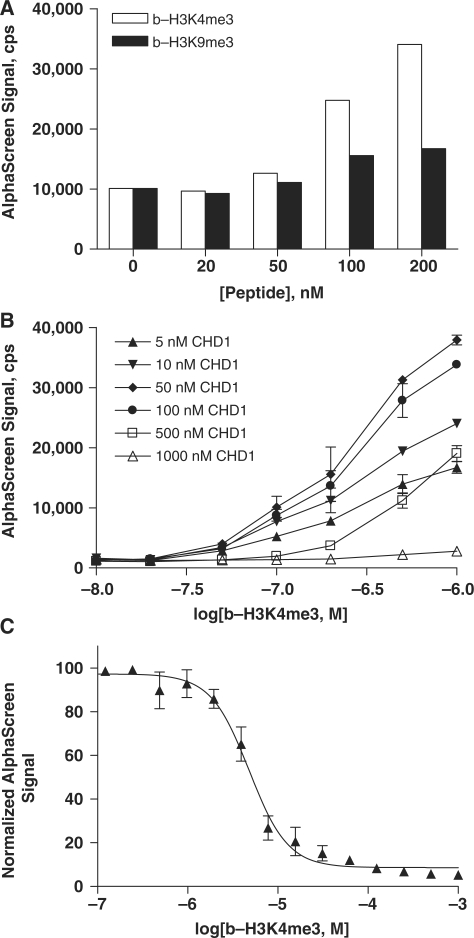
To determine the scalability of the assay, HP1β–b-H3K4me3 and HP1β–b-H3K9me3 binding reactions were performed in 384- and 1536-well microplates. A range of both protein and peptide concentrations was used. In both plate types, binding patterns were nearly identical (Figure 2A and B). HP1β was again shown to be selective for binding b-H3K9me3 over b-H3K4me3. The hook effect was observed in both 384- and 1536-well plates at concentrations of b-H3K9me3 peptide >50 nM. These data indicate this AlphaScreen peptide–protein-binding assay is amenable to HTS at the low final assay volume of 3 µl per well.
Figure 2.
AlphaScreen assay was miniaturized to a volume of 3 µl/well for high-throughput screening. Comparable binding of 20–200 nM b-H3K4me3 (squares) and b-H3K9me3 (circles) peptides to 100 nM GST-HP1β was observed in 384-well (A) and 1536-well (B) plate formats. Data in 1536-well plate format (B) are shown as the mean and standard deviation of four replicate measurements.
MPP8 chromodomain assay optimization
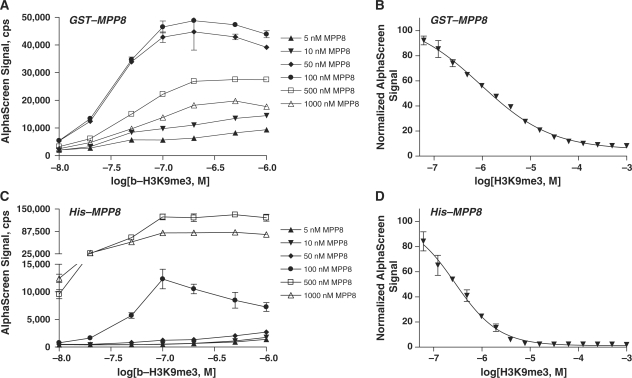
Binding of H3K9me3 to the MPP8 chromodomain was observed using a wide range of concentrations of both GST-MPP8 (5–1000 nM protein) and b-H3K9me3 (10–1000 nM peptide) (Figure 3A). The ‘hook effect’ was observed at peptide concentrations >100 nM, and at protein concentrations >100 nM GST-MPP8. We thus selected 50 nM peptide and 50 nM GST-MPP8 to use in our studies. To demonstrate the reporting capability of the new assay, H3K9me3 peptide was titrated into preincubated complex of 50 nM GST-MPP8 and b-H3K9me3 (Figure 3B). The unlabeled peptide displaced binding of its cognate biotinylated peptide to the MPP8 chromodomain with an EC50 of 0.92 µM.
Figure 3.
A range of GST-tagged MPP8 protein concentrations was titrated with 10–1000 nM b-H3K9me3 peptide (A) (B) Unlabeled H3K9me3 peptide competed with 50 nM b-H3K9me3 for binding to 50 nM GST-MPP8. (C) Binding of b-H3K9me3 to MPP8 was further validated in an alternative AlphaScreen assay format where GST-tagged protein was replaced with His6-MPP8 in a matrix titration with 10–1000 nM b-H3K9me3. (D) Unlabeled H3K9me3 peptide competed with 50 nM b-H3K9me3 for binding to 10 nM His-MPP8 protein. Data are shown as the mean and standard deviation of two or four replicate measurements for matrix titrations (A, C) and peptide competition assays (B, D), respectively.
To assess the generality of the chromodomain binding assay initially developed with GST-tagged chromodomain proteins, we replaced GST-MPP8 with its His6-tagged version, His-MPP8, and measured its binding to b-H3K9me3 by using nickel chelate-functionalized acceptor beads. Titration of biotinylated-His6 revealed a higher binding capacity for nickel-chelate acceptor beads compared to anti-GST acceptor beads (data not shown). As shown in Figure 3C, protein and b-H3K9me3 peptide were titrated in 384-well plates. While AlphaScreen signal was higher for His-MPP8, patterns were similar for both protein tags. Titration of unlabeled H3K9me3 peptide into preincubated complex of 10 nM His-MPP8 and 50 nM b-H3K9me3 resulted in displacement of biotinylated peptide with an EC50 of 0.28 µM (Figure 3D). The observed ∼3-fold difference in EC50 values between H3K9me3 competitions against the GST- and His6-based MPP8 assay systems was to be expected based on the difference in protein level in the assays and was within the typical error of EC50 determination.
Application of assay to other trimethylated lysines
Thus far, we have demonstrated the assay for chromatin-associated domains that target histones trimethylated at K9. To investigate alternative methyllysine marks, specificity of CHD1 chromodomain binding was investigated using b-H3K4me3 and b-H3K9me3 peptides (Figure 4A). AlphaScreen signal was 2-fold greater for 50 nM GST-CHD1 interaction with 200 nM peptides trimethylated at H3K4 versus H3K9. Titration of GST-CHD1 with b-H3K4me3 peptide in 1536-well plate format confirmed binding of the protein–peptide pair (Figure 4B). No hook point was observed for the b-H3K4me3 peptide in the concentration range examined, but concentrations of CHD1 >50 nM protein resulted in decreased AlphaScreen signal. Unlabeled H3K4me3 peptide displaced binding of its cognate biotinylated peptide (200 nM) to GST-CHD1 (20 nM) with an EC50 of 4.8 µM (Figure 4C).
Figure 4.
The assay for chromodomain binding of histones is applicable to other trimethylated lysines. (A) GST-CHD1 (50 nM) is selective for binding b-H3K4me3 (white) compared to b-H3K9me3 (black). GST-tagged CHD1 protein was titrated with 10–1000 nM b-H3K4me3 peptide in duplicate measurements (B). Data are shown as the mean and standard deviation for each peptide concentration. Unlabeled H3K4me3 peptide competed with its cognate biotinylated peptide (200 nM) for interaction with 20 nM CHD1 (C). Data are shown as the mean and standard deviation of four replicate measurements.
Extending the assay beyond chromodomains
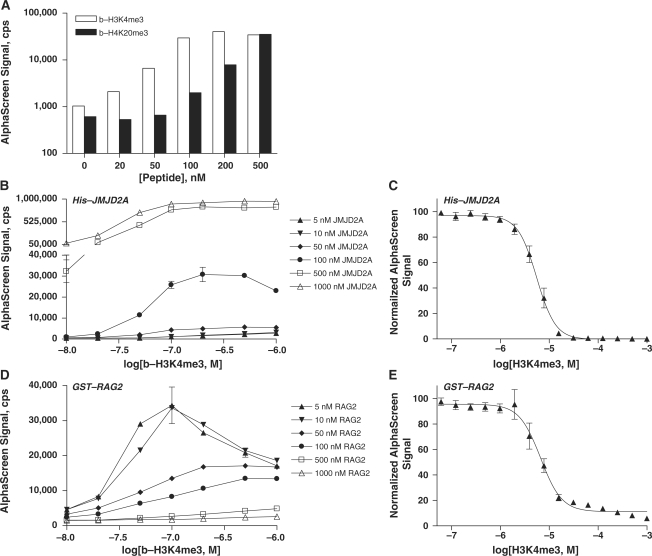
The AlphaScreen assay described is applicable not only to chromodomain proteins but also to all domains that bind modified histones. Here, we demonstrate that the double tudor domain-containing protein JMJD2A binds a histone H3K4me3 peptide, in addition to a histone H4K20me3 peptide (Figure 5A). The signal for b-H3K4me3 interaction leveled off around 100 nM peptide, whereas the signal for b-H4K20me3 binding continued to increase in the concentration range tested.
Figure 5.
The AlphaScreen assay is adaptable to measuring binding of proteins harboring other chromatin-associated domains. The double tudor domain-containing JMJD2A protein (100 nM) interacts with b-H3K4me3 (white), and b-H4K20me3 (black) peptides (A). His6-tagged JMJD2A protein was titrated with 10–1000 nM b-H3K4me3 peptide (B). Unlabeled H3K4me3 peptide competed with 100 nM b-H3K4me3 for binding to 100 nM His-JMJD2A (C).The PHD-containing RAG2 protein was similarly titrated with 10–1000 nM b-H3K4me3 peptide (D). Unlabeled H3K4me3 peptide competed with b-H3K4me3 (50 nM) for binding to 5 nM GST-RAG2 (E). Data are shown as the mean and standard deviation of two or four replicate measurements for matrix titrations (A, C) and peptide competition assays (B and D), respectively.
Binding of the JMJD2A/H3K4me3 epigenetic pair was measured over a range of both peptide and protein concentrations (Figure 5B). Conditions of 100 nM His-JMJD2A and 100 nM b-H3K4me3 were selected for peptide competition experiments following the observation of AlphaScreen hook points at ∼500 nM protein and 200 nM peptide. Unlabeled H3K4me3 displaced b-H3K4me3 peptide from the His-JMJD2A double tudor domain with an EC50 of 5.5 µM (Figure 5C).
Binding of histone methyllysine marks was also examined with the PHD-domain containing protein RAG2. Interaction of b-H3K4me3 with GST-tagged RAG2 was measured in a matrix titration, using a wide range of both protein and peptide concentrations (Figure 5D). Concentrations of GST-RAG2 and b-H3K4me3 > 5 nM and 200 nM, respectively, resulted in decreased AlphaScreen signal. Unlabeled H3K4me3 competed with b-H3K4me3 (50 nM) for binding to GST-RAG2 (5 nM) with an EC50 of displacement equal to 6.3 µM (Figure 5E).
Peptide competition assays
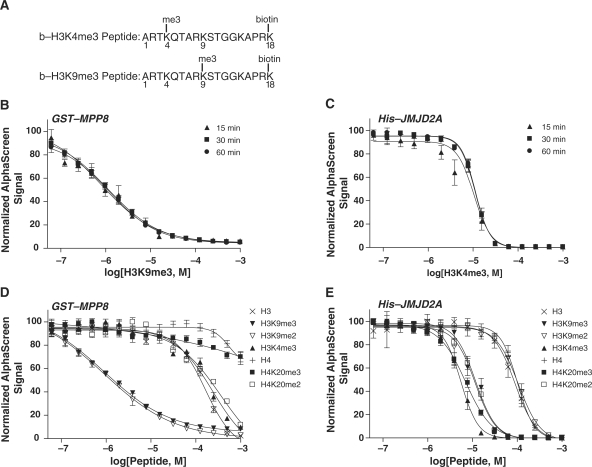
With the assay at hand, we proceeded to investigate the binding interactions of chromo- and tudor domains with peptides representing key histone marks. Biotinylated and trimethylated histone peptides, shown in Figure 6A, were co-incubated with their GST- or His-tagged epigenetic binding partner. EC50 values for displacement of biotinylated H3K9me3 from a preformed complex with GST-MPP8 were determined to be 0.94, 1.1 and 1.2 µM following 15-, 30- and 60-min incubations with unlabeled peptide, respectively (Figure 6B). Biotinylated H3K4me3 interaction with His-JMJD2A was disrupted with EC50 values of 10.3, 11.1 and 11.3 µM following 15-, 30- and 60-min incubations with unlabeled peptide, respectively (Figure 6C). We selected an optimal incubation time of 30 min with unlabeled competitor peptides for use in our studies.
Figure 6.
Specificity and affinity of binding to specific histone methyllysines was determined in a competitive binding assay. Unlabeled histone peptides displaced biotinylated histone H3 peptides, b-H3K4me3 and b-H3K9me3 (A), from methyl-binding domains. Unlabeled peptides were titrated into incubations containing 50 nM GST-MPP8 and 50 nM b-H3K9me3 (B and D), or 100 nM His-JMJD2A and 100 nM b-H3K4me3 (C and E). Varying incubation times of 15 min (triangles), 30 min (squares) and 60 min (circles) were examined for disruption of preformed biotinylated peptide/protein complex with unlabeled peptide (B and C). Displacement of b-H3K9me3 from GST-MPP8 and b-H3K4me3 from His-JMJD2A was measured following 30-min incubation with a range of unlabeled histone peptides (D and E, respectively). Data are expressed as the mean and standard deviation of four replicate measurements.
Unlabeled peptides varied in sequence, methylation state and methylation site were employed to displace these interactions to measure specificity and relative affinities of binding. Unlabeled H3K9me3 peptide displaced binding of its cognate biotinylated peptide, b-H3K9me3, to MPP8 protein with an EC50 of 0.92 µM (Figure 6D, Table 1). While not a direct measure of binding affinity, this value is in the range of Kd values reported for other chromodomain proteins (18–20). The selectivity of MPP8 binding to histone H3K9me3 over H3K4me3, observed in Figure 1B, was confirmed here with a higher EC50 of displacement measured with H3K4me3 than with H3K9me3 (240 µM and 0.92 µM, respectively). Furthermore, H4K20me3 was also ruled out as a binding partner for MPP8. Finally, H3 and H4 peptides lacking methylation at Lys residues did not significantly disrupt MPP8 interaction with b-H3K9me3, indicating MPP8 chromodomain binding is specific for covalently modified histones. Interestingly, the H3K9me2 peptide had approximately the same EC50 value as its trimethylated form (0.90 µM and 0.92 µM, respectively). To our knowledge, this represents the first characterization of MPP8 binding of histone methyllysine marks.
Table 1.
Peptides used in competitive binding assay and their EC50 values
| Peptide | Length | Sequencea | GST-MPP8/b-H3K9me3 | His-JMJD2A/b-H3K4me3 |
|---|---|---|---|---|
| EC50 (μM) | EC50 (μM) | |||
| H3 | 18 | ARTKQTARKSTGGKAPRK | 160 | 89 |
| H3K9me3 | 18 | ARTKQTARK***STGGKAPRK | 0.92 | 12 |
| H3K3me2 | 18 | ARTKQTARK**STGGKAPRK | 0.90 | 110 |
| H3K4me3 | 18 | ARTK***QTARKSTGGKAPRK | 240 | 5.5 |
| H4 | 18 | GKGGAKRHRKVLRDNIQG | 550 | 100 |
| H4K20me3 | 18 | GKGGAKRHRK***VLRDNQG | >104 | 7.0 |
| H4K20me2 | 18 | GKGGAKRHRK**VLRDNIQG | 360 | 12 |
aSequence where double asterisk indicates dimethylation of Lys and triple asterisk indicates trimethylation of Lys.
Peptide competition assays were also used to measure binding of several histone peptides to the JMJD2A double tudor domain protein (Figure 6E and Table 1). Unlabeled H3K4me3 displaced its biotinylated form, b-H3K4me3, with an EC50 of 5.5 µM, the lowest value of the peptides tested. In agreement with our previous observations (Figure 5A), H4K20me3 displaced b-H3K4me3 by competing for binding to His-JMJD2A (EC50 7.0 µM). H3K9me3 and H4K20me2 were also found to compete effectively for JMJD2A tudor binding with low EC50 values of 12 µM. In contrast to the near-equal effect of H3K9me3 and H3K9me2 on MPP8 (Figure 6B and Table 1), the same pair of peptides displayed very different competition capacity in the JMJD2A tudor assay, with the trimethylated peptide showing ∼10-fold greater affinity than its dimethylated counterpart. Further, the JMJD2A tudor domain displayed a greater discriminating power against the lysine methylation state at the H3K9 locus than it did against the H4K20 site. The rank order of JMJD2A binding affinity was H3K4me3 > H4K20me3 > (H3K9me3, H4K20me2) >> (H3, H4, H3K9me2). These data are largely in accordance with previous reports of recognition of methylated histones by the JMJD2A tudor domain (13,25,26).
DISCUSSION
Here, we describe the development and application of a novel assay for detection of methyl-binding domain interactions. We utilize biotinylated peptide fragments representing histone epigenetic marks and make use of the common affinity purification tags existing on the methyl-dependent protein binders to configure a homogeneous, sensitive and highly modular assay operating on the AlphaScreen detection principle. Laser excitation and chemiluminescent detection allow measurement of low-affinity interactions on the scale from Kd 1–50 µM observed for histone binding to chromodomain and tudor domain proteins. This robust assay requires only nanomolar concentrations of protein, a distinct advantage over other techniques used to study similar peptide–protein interactions. For example, FP measurements require 0.1 µM–1 mM protein (20) and the direct dependence of FP signal on the molar fraction of all fluorescent species present in the assay makes that technique incapable of detecting low-abundance bound ligand in the presence of high-abundance unbound counterpart.
The data presented here indicate that this assay can be used to study multiple post-translational modifications as well as various methyl-binding domains. The assay was initially demonstrated with GST-tagged chromodomains and biotinylated histone H3 peptide trimethylated at Lys9, b-H3K9me3. However, substitution of the functional GST tag with an N-terminal His6-tag retained assay outcome, thus validating its design and extending its scope beyond a single affinity tag. We demonstrated versatility of the assay through chromodomain binding of other peptides, b-H3K4me3 and b-H4K20me3. Finally, we performed the assay with a JMJ2DA double tudor domain protein and a RAG2 PHD protein to demonstrate application to other methyl-binding domains. Thus, it appears that the assay is flexible enough to allow its application to be extended to additional methyl-dependent interactors, beyond the domains studied here. The relative ease of configuring an AlphaScreen assay is in part due to the low sensitivity of its signal strength to the location of the bead binding sites, which is in contrast with FRET-based detection schemes where a careful optimization of the label placement positions is often required.
Our mechanistic study of chromodomain-binding specificity involved multiple peptide competition assays performed in a highly miniaturized format. As anticipated, unlabeled H3K9me3 displaced its cognate biotinylated peptide by competing for binding to the MPP8 chromodomain. At the same time, an off-target trimethylated peptide H4K20me3 failed to compete off the cognate H3K9me3 peptide, further proving that the present assay faithfully reports on the relevant domain–histone interactions. Dimethylated peptide, H3K9me2, was found to be a novel MPP8 binding partner, displacing b-H3K9me3 with an EC50 approximately equal to that of trimethylated peptide. Binding partners of the JMJD2A double tudor domain were also investigated using the present assay. JMJD2A tudor was found to bind H3K4me3, H4K20me3 and, to a lesser extent, H3K9me3 and H4K20me2 and only marginally with H3K9me2 and unmethylated histone peptides. We conclude that this assay is capable of accurately mapping the binding of chromatin-associated domains to methylated histone peptides, as the profiling data obtained are in agreement with previous reports (13,26).
The simple design and homogeneous nature of the assay presented here should make it amenable to HTS for small molecules that inhibit binding of chromatin-associated proteins to methylated histones, as evidenced by its implementation here into a 1536-well format. Excitation at a long wavelength and emission at a shorter wavelength circumvents the high background encountered with fluorescence-based assays. Importantly, recent fluorescence spectroscopic profiling of the National Institutes of Health Chemical Genomics Center library, containing >70 000 compounds, found dramatic decreases in autofluorescence with relatively small shifts (100 nm) to longer excitation wavelengths (27). In addition, the availability of dual-recognition AlphaScreen reagents (e.g. biotinylated GST or biotinylated hexahistidine, which bind to both the donor and acceptor beads) provides the opportunity to perform an immediate counterscreen in order to exclude possible false positives acting on the detection system.
The past two decades have seen an explosion of epigenetic-related clinical and molecular biological discoveries, highlighted by the global hypomethylation of DNA found in human tumors, hypermethylation of tumor suppressor genes and inactivation of microRNA genes by DNA methylation (28–30). Almost all cancers show abnormal epigenetic features, and epigenetic factors are commonly disrupted in debilitating mental retardations. Early development, and subsequent susceptibility to disease, is also in part regulated epigenetically (31,32). Furthermore, the regulatory pathways that underpin the maintenance of stem cell pluripotency also rely on chromatin-based epigenetic memory systems.
Histone deacetylase and DNA methyltransferase inhibitors are currently used in the clinic to treat various cancers through reversal of tumor suppressor silencing incurred in disease development (33). Histone methylation became a target of epigenetic therapy following the association of lysine methyltransferase and demethylase enzymes with cancer (25,34–36). Recently, mutations and dysregulation of methyl-binding domains have been identified as contributing to human disease (37). Disruption of methyl-binding domain interactions at specific methylated histone resides may represent the future of epigenetic therapy. However, to fully exploit the potential that epigenetics as a target area holds, our understanding of underlying complexities, e.g. the combinatorial nature of histone modifications, has to increase. It is anticipated that the method described here will be an important tool for the identification of epigenetic interactions and the discovery of small molecules to be used as chemical probes or starting points for drug discovery.
FUNDING
The Molecular Libraries Initiative of the NIH Roadmap for Medical Research; the Intramural Research Program of NHGRI, NIH; a Welch Foundation Grant (G-1495 to M.T.B.), an NIDA grant (DA025800 to M.T.B.) and a pilot project on grant P30ES007784 (to M.T.B.); and The German Research Foundation (SP1262/1-1 to A.S.). The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research and the Wellcome Trust. Funding for open access charge: NIH Roadmap for Medical Research, National Institutes of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Alice Grabbe and Michelle Daniel for provision of purified JmjD2A Tudor proteins.
REFERENCES
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 4.Schneider R, Bannister AJ, Kouzarides T. Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem. Sci. 2002;27:396–402. doi: 10.1016/s0968-0004(02)02141-2. [DOI] [PubMed] [Google Scholar]
- 5.Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 6.Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, Weisenberger DJ, Egger G, Takai D, Gonzales FA, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl Acad. Sci. USA. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 8.Collins RE, Northrop JP, Horton JR, Lee DY, Zhang X, Stallcup MR, Cheng X. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat. Struct. Mol. Biol. 2008;15:245–250. doi: 10.1038/nsmb.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 10.Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, III, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 11.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 13.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng SS, Yue WW, Oppermann U, Klose RJ. Dynamic protein methylation in chromatin biology. Cell Mol. Life Sci. 2009;66:407–422. doi: 10.1007/s00018-008-8303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eftink MR. Fluorescence methods for studying equilibrium macromolecule-ligand interactions. Methods Enzymol. 1997;278:221–257. doi: 10.1016/s0076-6879(97)78013-3. [DOI] [PubMed] [Google Scholar]
- 16.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 17.Lin CW, Jao CY, Ting AY. Genetically encoded fluorescent reporters of histone methylation in living cells. J. Am. Chem. Soc. 2004;126:5982–5983. doi: 10.1021/ja038854h. [DOI] [PubMed] [Google Scholar]
- 18.Fischle W, Franz H, Jacobs SA, Allis CD, Khorasanizadeh S. Specificity of the chromodomain Y chromosome family of chromodomains for lysine-methylated ARK(S/T) motifs. J. Biol. Chem. 2008;283:19626–19635. doi: 10.1074/jbc.M802655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs SA, Fischle W, Khorasanizadeh S. Assays for the determination of structure and dynamics of the interaction of the chromodomain with histone peptides. Methods Enzymol. 2004;376:131–148. doi: 10.1016/S0076-6879(03)76009-1. [DOI] [PubMed] [Google Scholar]
- 21.Sun B, Hong J, Zhang P, Dong X, Shen X, Lin D, Ding J. Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36. J. Biol. Chem. 2008;283:36504–36512. doi: 10.1074/jbc.M806564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ullman EF, Kirakossian H, Switchenko AC, Ishkanian J, Ericson M, Wartchow CA, Pirio M, Pease J, Irvin BR, Singh S, et al. Luminescent oxygen channeling assay (LOCI): sensitive, broadly applicable homogeneous immunoassay method. Clin. Chem. 1996;42:1518–1526. [PubMed] [Google Scholar]
- 23.Cauchon E, Liu S, Percival MD, Rowland SE, Xu D, Binkert C, Strickner P, Falgueyret JP. Development of a homogeneous immunoassay for the detection of angiotensin I in plasma using AlphaLISA acceptor beads technology. Anal. Biochem. 2009;388:134–139. doi: 10.1016/j.ab.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 26.Lee J, Thompson JR, Botuyan MV, Mer G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat. Struct. Mol. Biol. 2008;15:109–111. doi: 10.1038/nsmb1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simeonov A, Jadhav A, Thomas CJ, Wang Y, Huang R, Southall NT, Shinn P, Smith J, Austin CP, Auld DS, et al. Fluorescence spectroscopic profiling of compound libraries. J. Med. Chem. 2008;51:2363–2371. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 28.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 29.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 31.Seckl JR, Holmes MC. Mechanisms of disease: glucocorticoids, their placental metabolism and fetal ‘programming’ of adult pathophysiology. Nat. Clin. Pract. Endocrinol. Metab. 2007;3:479–488. doi: 10.1038/ncpendmet0515. [DOI] [PubMed] [Google Scholar]
- 32.Weaver IC. Epigenetic programming by maternal behavior and pharmacological intervention. Nature versus nurture: let's; call the whole thing off. Epigenetics. 2007;2:22–28. doi: 10.4161/epi.2.1.3881. [DOI] [PubMed] [Google Scholar]
- 33.Cortez CC, Jones PA. Chromatin, cancer and drug therapies. Mutat. Res. 2008;647:44–51. doi: 10.1016/j.mrfmmm.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 35.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 36.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 37.Baker LA, Allis CD, Wang GG. PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks. Mutat. Res. 2008;647:3–12. doi: 10.1016/j.mrfmmm.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]