Abstract
DNA replication, similar to other cellular processes, occurs within dynamic macromolecular structures. Any comprehensive understanding ultimately requires quantitative data to establish and test models of genome duplication. We used two different super-resolution light microscopy techniques to directly measure and compare the size and numbers of replication foci in mammalian cells. This analysis showed that replication foci vary in size from 210 nm down to 40 nm. Remarkably, spatially modulated illumination (SMI) and 3D-structured illumination microscopy (3D-SIM) both showed an average size of 125 nm that was conserved throughout S-phase and independent of the labeling method, suggesting a basic unit of genome duplication. Interestingly, the improved optical 3D resolution identified 3- to 5-fold more distinct replication foci than previously reported. These results show that optical nanoscopy techniques enable accurate measurements of cellular structures at a level previously achieved only by electron microscopy and highlight the possibility of high-throughput, multispectral 3D analyses.
INTRODUCTION
The spatiotemporal coordination of genome duplication is an essential biological process that is still far from being understood. Precise measurement of replication foci (RF) size and their total number in the cell nucleus are a crucial prerequisite for establishing and testing of quantitative models of the functional organization of the genome and its duplication at each cell division cycle. There are several methods for RF visualization, including labeling of the replication machinery or labeling of the newly synthesized DNA in live cells (1–3) or by immunofluorescence in fixed cells (e.g. refs. 4–7) and nano-gold labeling for electron microscopy (EM) (8–10). The subnuclear distribution of RF during S-phase of the cell cycle has been shown to change over time giving rise to varying numbers of RF with different sizes. Such variability in numbers and sizes is mostly related to the imaging methodology used and in part also dependent on the sample labeling and preparation procedure. Importantly, light microscopy (LM) combined with deconvolution analysis (1) and, more recently, EM (10,11) have uncovered complex substructures within the larger RF corresponding to heterochromatin replicating in late S-phase. From the nucleotide-labeled RF measured using EM, an average size of 110–120 nm was calculated (11), well below the size measurable by conventional LM techniques. However, estimating RF foci size with EM combined with in vivo immunogold labeling of GFP-PCNA (10) yielded much larger RF sizes of ∼200–350 nm. Importantly, either living cells with green fluorescent protein (GFP)-tagged proliferating cell nuclear antigen (PCNA)-labeled RF or fixed cells that had been pulse labeled with modified nucleotides were analyzed, but both labeling strategies were not directly compared using the same imaging method.
In this study, we investigated and compared the characteristics of sites of DNA replication in mammalian cells using novel nanoscale optical microscopy techniques—spatially modulated illumination (SMI) microscopy and 3D-structured illumination microscopy (3D-SIM). We employed hypotonic treatment of the cells to increase the separation of the RF followed by flattening of the cells and analysis of RF size by SMI. In order to precisely estimate the numbers of replication structures in the entire cell nucleus, we also used 3D-SIM in 3D preserved samples.
Super-resolution SMI microscopy is a light microscopic method that uses axial structured illumination in the form of a standing wave to allow high-precision measurements of the size and position of subresolution objects. The technique has previously been described in detail, including experimental construction, proof-of-concept measurements on fluorescent beads and first biological applications to specific chromatin regions and molecular complexes (12–18). We have thus applied SMI microscopy to the comprehensive optical analysis of the size of RF in mammalian cells and have found that their average size is about 120 nm and that this remains conserved throughout S-phase, in agreement with results from EM (11). In addition, we have shown that both methods of visualizing replication—labeling of the replication machinery with GFP-tagged forms of PCNA, and direct labeling of newly replicated DNA—give a similar RF size (∼125 nm). The ability to measure a vast number of foci by LM has allowed us to show that there is considerable variation in the size distribution of RF in both cases. This result would be difficult to obtain with the comparatively low-throughput technique of EM.
The SMI microscope lacks optical sectioning ability and is consequently incapable of separating objects which overlap along the z-axis. This limits the application of SMI microscopy to objects <200 nm thick (19). For sparse objects, where the probability that objects are located above one another approaches zero, this does not present a significant problem. For more densely packed structures, such as RF, especially in late S-phase nuclei, the limitations of optical sectioning can be overcome through sample preparation, as shown in the present study and/or using 3D-SIM. Although 3D-SIM does not offer the same size-measurement accuracy, it is not limited in 3D (20,21). We thus used 3D-SIM to measure the numbers of RF throughout S-phase in 3D preserved samples and found an average of ∼4000 RF; three to five times higher numbers than the numbers estimated using conventional LM.
In this study, we have compared two novel optical nanoscopy methods, applied to the measurement of replication structures and correlate the data with previously reported ultrastructural EM analysis. We additionally evaluated two sample preparation approaches (ultrathin cryosections and hypotonically treated cells) and compared detection of the replication machinery (GFP-tagged PCNA or antibody stained) with replicated DNA (using antibody detection of incorporated nucleotide analogs). These studies assess and demonstrate the capability of new nanoscopy methods to provide quantitative data on macromolecular structures involved in the catalysis and coordination of biological processes.
MATERIALS AND METHODS
Cell culture
C2C12 mouse myoblast cells (22) and C2C12 cells stably expressing GFP-tagged PCNA (1) were cultured using DMEM supplemented with 20% fetal calf serum and 50 µg/ml gentamycin at 37°C in a humidified atmosphere of 5% CO2.
In situ replication labeling
DNA synthesis foci were labeled by adding the thymidine analog BrdU to the cell culture medium at a final concentration of 10–4 M for 10 min.
Hypotonic treatments
Nucleotide-labeled C2C12 cells were trypsinized, the cell suspension was washed twice with cold phosphate-buffered saline (PBS) and resuspended in a small volume (∼50 µl) of PBS. Excess of prewarmed 50 mM KCl solution was added to the suspension and the cells were allowed to swell at 37°C for another 10 min. The cells were pelleted again and resuspended in 30 µl of the hypotonic solution. Freshly made ice-cold methanol:acetic acid mixture (3:1) was added drop wise and the cells were kept for at least 30 min at −20°C. The resulting nuclei were pelleted and excess of the fixative was removed. Nuclei were gently pipetted in the remaining volume, transferred to microscope slides and air-dried. The slides were kept at −20°C until immunostaining.
For time lapse analysis, GFP–PCNA expressing C2C12 cells were cultured in eight-well LabTech cover slip chambers. After acquiring a z-stack of images, the cell culture medium in a well was carefully substituted by prewarmed 35 mM KCl solution and further z-stacks of images were acquired at various times. The parameters of acquisition were adjusted taking into account that upon treatment a cell (nucleus) started expanding, mainly in the z direction and, hence, the fluorochrome signal (GFP–PCNA) decreased.
Cryosections
Cells were fixed in 4% freshly depolymerized paraformaldehyde in 250 mM HEPES (pH 7.6, 10 min, 4°C), refixed in 8% paraformaldehyde in 250 mM HEPES (pH 7.6, 2 h, 4°C), scraped and pelleted during the latter fixation [30]. Cell pellets were embedded (2 h) in 2.1 M sucrose in PBS, transferred onto a copper block, frozen and stored in liquid nitrogen. Cryosections (100–150 nm thick) were cut with a glass knife using an ultracryomicrotome (UltraCut UCT52, Leica, UK), captured on drops of 2.1 M sucrose in PBS, transferred to 0.17-mm-thick glass coverslips or Cellocate coverslips (Eppendorf) and stored at –20°C.
Anti-BrdU immunostaining
All incubations were at room temperature, unless otherwise stated. Briefly, formaldehyde fixed cells were permeabilized by 10-min incubation in PBS containing 0.5% Triton X-100, followed by blocking for 30 min in PBS containing 0.2% fish skin gelatin or 2% bovine serum albumin. Hypotonically treated methanol:acetic acid fixed cells were not further permeabilized and directly blocked in PBS with 0.2% gelatin. Incorporated BrdU was detected by incubating for 30 min to 1 h with anti-BrdU mouse monoclonal antibody (6 µg/ml; clone B44, Becton Dickinson or clone IU-4, CALTAG Labs) at 37°C in a solution (33 mM Tris–Cl pH 8.1, 0.33 mM MgCl2, 0.5 mM beta-mercaptoethanol, 1% BSA) containing DNase I (20 U/ml) to expose the epitope. Detection was performed with an antimouse immunoglobulin G (IgG) antibody conjugated with FITC (Jackson Immuno Research), Alexa Fluor 488 or Alexa Fluor 555 (Invitrogen). For 3D-SIM cells were typically postfixed with 2% formaldehyde in PBS in order to minimize background fluorescence and floating particles or antibody precipitates and counterstained with 100 ng/ml 4′,6-diamidino-2-phenylindole (DAPI; Sigma). Samples were mounted in Mowiol or Vectashield for confocal and conventional microscopy and in Vectashield (Vector Laboratories) for high-resolution microscopy.
Anti-GFP immunostaining
Cryosections were rinsed (3×) in PBS, incubated (15 min) in 20 mM glycine in PBS, rinsed (3×) in PBS, treated (10 min) with 0.1% Triton X-100 in PBS, blocked (30 min) with PBS+ (PBS supplemented with 1% BSA, 0.2% fish skin gelatin, 0.1% casein; pH 7.6), incubated (2 h) with rabbit antibodies anti-GFP (diluted 1:50 in PBS+; affinity-purified rabbit polyclonal antibodies raised against full-length GFPS65T), washed (5×, over 1 h) in PBS+, incubated (1 h) with Alexa Fluor 488-conjugated antibodies against rabbit IgGs (in PBS+; raised in donkey; Molecular Probes), rinsed (3×, 30 min) in PBS+, rinsed (3×) in PBS, counterstained (45 min) with TOTO-3 (2 µM, Molecular Probes) in 0.05% Tween-20 in PBS, rinsed successively in 0.05% Tween-20 in PBS and then PBS, before coverslips were mounted in Vectashield (Vector Laboratories) on glass slides coated with 0.1 µm FluoresbriteTM Carboxylate YG microspheres (Polysciences). The long incubation times used allow for antibody accessibility throughout the thickness of cryosections, providing the highest sensitivity (23).
Confocal laser scanning and wide field fluorescence microscopy
Confocal optical sections of cells were obtained with a Zeiss LSM510Meta microscope using a 63× NA1.4 Plan-Apochromat oil immersion phase contrast objective heated to 370 C. Excitation of GFP/FITC, TexasRed and Cy5 was performed with 488 nm (Argon laser), 543 nm and 633 nm (He-Ne lasers) laser lines, respectively. The parameters of the system were adjusted to avoid saturation. Alternatively, confocal image stacks were recorded with a TCS SP5 confocal laser scanning microscope (Leica) using a 63×/1.4 NA HXA Plan-Apochromat oil immersion objective. Fluorochromes were excited with the 488 nm line of an argon laser and a 561-nm diode-pumped solid-state (DPSS) laser. Settings used were: 512 × 512 pixels frame size; 40 or 50-nm pixel size, 125-nm z-distances between sections; 1 Airy unit pinhole diameter; 800-Hz scan speed; and 6-times averaging.
Wide field images of the hypotonically treated cell preparations were obtained with a Zeiss Axiovert 200M microscope using a 63× NA1.4 Plan-Apochromat oil immersion phase contrast objective and a bandpass fluorescein filter set (excitation 480/20 nm, dichroic 495 nm, emission 510/20 nm; Chroma). Images were acquired with a cooled charge coupled device (CCD) camera (SensiCam: 1280 × 1024 pixels; pixel size 6.8 µm) using the Axiovision 3 software (Zeiss) and time of exposure per image varied between 0.4 and 4 s.
Images were assembled and annotated using ImageJ 1.33 and Adobe Photoshop software.
SMI microscopy
Image stacks of cells were taken using the SMI microscope [experimental setup described in (12,13,24)]. The experiments presented here were performed using a 100× 1.4NA oil immersion objective from Leica, a 1.25× tube lens and a SensicamQE cooled CCD camera from PCO. The camera pixels were binned (2 × 2) resulting in an effective pixel size in the object of 108 nm in both x and y directions, and a maximum image size of 688 × 512 pixels, although this was typically reduced through use of a region of interest (ROI). Each stack consisted of a series of between 80 and 150 images taken while moving the object along the optical axis with a step size of 40 nm. The exposure time for each image was between 0.06 and 0.2 s for the BrdU labeling, and between 0.4 and 2 s for the GFP–PCNA labeling.
SMI image analysis
The SMI data were analyzed using a custom software package developed in MATLAB. This package uses a combination of filtering and the use of multiple thresholds to find ‘point-like’ objects within an SMI data stack. Once the position of each object is known, an axial profile through the raw data is extracted at that point, averaging over a 5 × 5 lateral ROI and subtracting the average over a 15 × 15 ROI as a background estimate. A model function is fitted to this profile using a nonlinear least squares fit, which allows the depth of the modulation to be extracted. As the depth of the modulation corresponds to the extent to which the original illumination structure has been ‘blurred’ by the object, it is possible to estimate the size of the object from the depth of the remaining modulation. In order to make a quantitative size estimate, one must assume an object form. In this case, a spherical object was assumed. The principles of SMI size estimation are described in more detail in (24,25). A schematic illustrating the results of the object identification and fitting procedure is shown in Figure 2. The curve showing the relation between modulation depth and object size, along with examples for measurements on calibration beads are shown in the Supplementary Data (Figure S4).
Figure 2.
SMI imaging of RF. After acquisition of the raw data (A), a filtering and thresholding procedure was used to identify the positions of individual foci. Once the positions of the foci have been established (blue boxed area shown in greater detail in B), a model function is fitted at the location of each focus to extract the object size and a more accurate position estimate. The results of this fitting procedure were visualized by rendering a sphere at each object position, which was colored according to the object size. This resulted in the 3D representation shown in (C). Objects that could not be fitted, due to either insufficient signal level or a size outside the effective range (∼40–200 nm) for SMI measurements were colored black. Scale bar = 5μm. As well as a size estimate, the fitting procedure delivered the 3D positions of the foci. The flatness of the resulting preparations could thus be measured using these 3D positions. The 3D representation (C) was rotated by 90° to obtain a side on view (D) showing the vertical extent of the cell. The axial spread in the centers of the foci was measured to be ∼160 ± 70 nm. Note that the tilt in the cell resulted from a small misalignment in the experimental setup. This alignment error did not otherwise affect the operation of the microscope.
The object identification uses a range of thresholds, with the upper threshold automatically selected, and the lower threshold controlled by a user-selectable parameter. For the labeling of PCNA in cryosections, the lower threshold was manually controlled for each cell to ensure that all points were detected. For the BrdU labeling, it was difficult to find a value where all foci would be detected without including some noise peaks. The lower threshold was thus set to a very low value such that both foci and noise peaks would be detected. Noise peaks generally do not result in a fit when analyzed. The data was then analyzed, and a threshold chosen a posteriori from the range of thresholds used. This threshold was chosen such that 95% of all objects were included which would have fitted had the whole threshold range been used. This compromise allows us to achieve a reasonable balance between the exclusion of real foci and the inclusion of spurious noise. The exclusion of a few percent of the foci is unlikely to have a large effect on the overall distribution, a fact confirmed by the color-coded histogram in Figure 3Aiv, which shows the contribution of each threshold level to the overall distribution.
Figure 3.
SMI measurement of RF size. (A) The application of the data analysis scheme results in a size distribution for each cell. Shown here are typical examples from each of the three different preparation methods used in our experiments. From left to right are: a hypotonic specimen labeled using BrdU incorporation and antibody staining, a cryosection labeled using GFP–PCNA and a GFP–PCNA specimen with subsequent anti-GFP antibody amplification. The size distribution obtained is shown below the corresponding cell. The distribution for the BrdU foci is, in addition, separated according to the contribution of each intensity threshold level to the total distribution. The brightest objects are red and the weakest blue. Scale bars = 5 μm. (B) Summary of the collected results for all cells in each preparation method categorized according to S-phase stage. From left to right are the BrdU-labeled hypotonic preparation, GFP–PCNA cryosections and GFP–PCNA cryosections with antibody amplification. From top to bottom are a line histogram, cumulative histogram and box-and-whisker plot. The cumulative histogram represents the integral of the distribution, and avoids the binning effects present in a normal histogram.
3D-SIM
Super-resolution imaging was performed on an OMX microscope prototype using a 100×, 1.4 NA, oil-immersion objective (Olympus). The setup has previously been described in detail (21). In short, light from one of three high-power diode lasers (405 nm, 488 nm and 532 nm) was transmitted through a piezo-driven optical grating that generated a 3D diffraction pattern of interfering light on the focal plane of the sample with laterally spaced sinusoidal stripes of ∼0.2 µm. For 3D-SIM recording, image stacks with a z-distance of 0.125 µm were acquired. For each plane, 15 raw images were recorded with different grating positions: five phases were recorded sequentially by translating the diffraction grating between exposures and three subsequent z-stacks were recorded with the angular orientation of the grating rotated by 60°. Emitted light passes through a set of specially designed dichroic mirrors directing it to three independently controlled iXon 87 back-illuminated EMCCD cameras with 512 × 512 pixels and >90% quantum efficiency (Andor Technology). Exposure times were between 100 and 200 ms, yielding typically 3000–10 000 counts in a raw image of 16-bit dynamic range.
3D-SIM image processing and quantitative image analysis
3D-SIM raw images were saved to disk and processed to reconstruct high-resolution information as described (20). Postprocessing and quantitative evaluation of 3D image stacks was performed with Volocity 5 software (Improvision). For deconvolution of confocal data sets a maximum-likelihood estimation algorithm was applied using theoretic point spread functions and a maximum number of 10 iterations. To determine numbers and volumes of RF a threshold-based segmentation was applied using the object separation option of Volocity. The procedure works by finding local maxima and separating touching objects in 3D. Volume estimates were used to control that the segmentation procedure worked properly. Despite the slightly bigger RF sizes within dense clusters possibly due to incomplete separation, the average volume of RF was found to correspond well to the 2-fold increase in resolution that 3D-SIM provides over confocal laser scanning microscopy (CLSM).
Statistical analysis
After categorization, the distributions were compared using a Kolmogorov–Smirnov test, which is independent of distribution form and can be used on arbitrary distributions. As our distributions are approximately Gaussian, a t-test gives similar results.
As a tool for visualizing distributions, we use a box-and-whisker plot on several occasions throughout the paper. The box-and-whisker plot depicts the distribution’s median, spread (upper and lower quartiles) and total range.
RESULTS
Labeling replication structures in mammalian cells with replication factors and by nucleotide incorporation
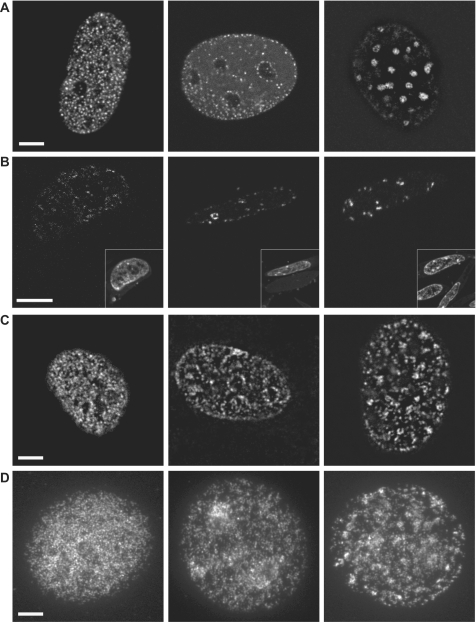
To compare the localization of protein and DNA components of RF (through GFP-PCNA and nucleotide labeling of nascent DNA, respectively) during S-phase, we collected optical sections of live cells labeled with GFP-PCNA or immunoassayed BrdU labeled nascent DNA after formaldehyde fixation (Figure 1). All three major characteristic replication patterns—early, mid- and late S-phase (4,26)—could be discriminated in GFP–PCNA expressing mouse cells as directly confirmed by live cell microscopy (Figure 1A). The early replication pattern appeared as many intranuclear foci of small uniform size. During mid- S-phase this pattern changed to a localization at the nuclear and nucleolar peripheries, and in late S-phase larger replication domains became visible with many small foci still existing at the nuclear periphery. Similar patterns were observed in formaldehyde fixed cells after immunostaining of the incorporated thymidine analog, BrdU (Figure 1C). However, after fixation the cells became flatter (a smaller number of optical sections were required to cover the whole nucleus; data not shown), and thus a higher number of smaller foci were detected away from nuclear and nucleolar periphery in mid- and late S-phase patterns (Figure 1C).
Figure 1.
Characteristic RF patterns in mammalian cells and effect of various treatments. (A) The three main patterns of RF that appear in course of S-phase are presented. From left to right: early, middle and late S-phase. Representative confocal optical sections of RF in untreated live GFP–PCNA expressing mouse cells. (B) Appearance of the same S-phase patterns in cryosections is presented. The reduced amount of visible GFP–PCNA RF is due to the fact that thickness of the cryosections (100–200 nm) is several times smaller than z-axis optical resolution at 520 nm (GFP). Inserts show images of the same cell stained with DAPI before making the sections. (C) BrdU-labeled (10 min), formaldehyde-fixed cells were immunostained with anti-BrdU antibody and mounted in Mowiol. The fixation/mounting of the cells provided for the decrease in nuclei thickness and hence for poorer resolution of RF. (D) Cells labeled 10 min with BrdU were trypsinized and the cells in suspension were treated with the hypotonic solution and fixed with methanol:acetic acid mixture followed by immunostaining with anti-BrdU antibody and mounting in Vectashield. Scale bars = 5 µm.
Hypotonic treatment of cells has lead to observation of an increased number of labeled sites suggesting the existence of multiple constituent units within each RF (7,11). However, after hypotonic treatment all three major patterns could still be identified (Figure 1D). Importantly, hypotonic treatment of GFP–PCNA expressing cells followed by in vivo imaging, showed that the complexity of RF was maintained after the treatment (Figure S1). In vivo hypotonic treatment lead to swelling of the cells and ‘opening’ of late S-phase RF as compared to mid- and early S-phase ones (see Figure S1 for details). Importantly, further replacement of the hypotonic solution with culture medium lead to reverse compaction of RF and no cell cycle effects were detected.
Analysis of RF labeled with GFP–PCNA in ultrathin cryosections (∼100–150 nm thick) revealed similar patterns of replication (Figure 1B). Sizes of RF appeared to be uniform during early S-phase although clustering was observed in mid- to late stages. The low resolution of widefield microscopy methods could explain why late S-phase RF clusters might have been initially taken as larger RF (4). GFP–PCNA patterns in cryosections confirm these results.
Hypotonically treated nuclear preparations and cryosections of cells were deemed the most suitable for the comprehensive analysis of RF sizes by SMI microscopy.
Quantitative measurement of RF size
We first investigated the size of RF in BrdU-labeled, hypotonically treated cells. The amplification of BrdU labeling by indirect immunofluorescence provided a strong signal, allowing SMI measurements to be made with a good signal-to-noise ratio and, hence, accuracy. Most foci identified in each cell could be automatically recognized (up to ∼2000/cell; Figure 2A and B), and more than 80% of all foci identified gave SMI profiles with modulation that contained quantifiable size information (Figure 2C). The remaining sites were either very weak (∼100 times weaker than the brightest foci) or could not be resolved from an adjacent focus. Within each cell, a wide distribution of estimated foci sizes was obtained. The axial positions of RF in hypotonically treated cells were found to be within 160 ± 70 nm, consistent with flattening of the cell and increased separation of the foci in xy (Figure 2D).
We next compared the size distribution of RF labeled in hypotonically treated cells from different stages of the cell cycle after BrdU incorporation (Figure 3Ai, iv and Bi, iv, vii). Sizes ranging from 40 to 210 nm were found at all stages of the cell cycle with average ∼126 nm (n > 40 000). The parameters of the size distributions shown in Figure 3Bi, iv, vii are summarized in Table 1. While the small (∼5 nm) difference between the early and late S-phase distributions is statistically highly significant (Kolmogorov–Smirnov test, P = 3e-38), it is unlikely that, given the ∼30-nm standard deviation of the individual distributions, this difference has biological significance. Indeed, it is possible that this difference could be the result of small systematic errors such as would be introduced when our assumption of optical isolation is not entirely fulfilled, which is more likely to occur in late S-phase when other RF that are close by can affect measurement precision.
Table 1.
Summary of SMI measurements of BrdU-labeled foci
| RF pattern | Mean size (nm)] | SD (nm)] | # foci |
|---|---|---|---|
| Early | 123.5 | 32.2 | 17 572 |
| Mid | 127.1 | 31.5 | 13 041 |
| Late | 128.6 | 31.1 | 10 897 |
To obtain measurements of RF in ultrastructurally preserved nuclei (27), we next analyzed RF labeled with GFP–PCNA in ultrathin cryosections (Figure 3Aii, v). As RF intensities were lower than the antibody-based BrdU labeling, signal-to-noise ratios were lower, and hence it was not possible to analyze all foci (50% of the foci could be measured). In spite of these difficulties, the size distribution obtained from these measurements (Figure 3Av and Bii, v, viii) was in agreement with that obtained from BrdU-labeled RF and confirmed the uniformity of sizes during the different S-phase stages.
To increase the signal-to-noise ratio of GFP–PCNA foci analyzed in ultrathin cryosections, we indirectly immunolabeled GFP with antibodies (Figure 3Aiii, vi). Only a small improvement in signal was obtained, which resulted in an increase in the portion of foci that could be detected and measured to about 60%. Perhaps surprisingly, the amplification of signal with antibodies resulted in smaller average RF sizes (Figure 3Avi and Biii, vi, ix; 105 nm relative to 125 nm obtained with either BrdU or directly by GFP–PCNA).
The results of the different labeling methods are compared in detail in Figure S2 and Table S1. Unexpectedly, this suggests that the immunolabeling procedure may underestimate the size of RF. A possible explanation for this observation could be quenching of GFP signal by the primary antibody against GFP, without a measurable trade-off in FITC signal in the secondary antibody. Cross-linking effects due to the bivalency of whole IgGs may also cause the immunolabeling complexes that mark each RF to shrink relative to the whole size of the structure (28).
Quantitative measurement of RF numbers
To determine the number of RF at different stages of S-phase, we first used data obtained from nucleotide labeled, hypotonically treated, cells analyzed by SMI microscopy (see also Figure 2A and B). The number of foci identified within each cell by this approach was highly variable (Figure S3), and very likely underestimated in view of the poor performance of SMI in resolving objects overlapping along the z-axis (19). Therefore, we used 3D-SIM, a different high-resolution optical microscopy technique, which allows a better resolution of objects in 3D (20).
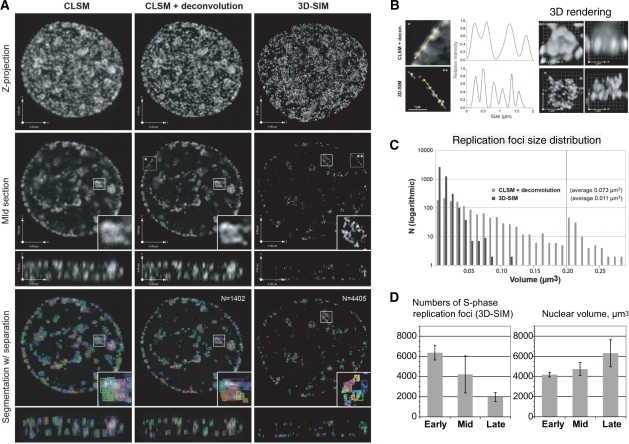
Compared to confocal laser scanning microscopy 3D-SIM has a 2-fold better resolution in xyz (Figure 4A and B). Application of 3D-SIM led to measurement of RF sizes of about 100–120 nm (Figure 4B), which concurred with the SMI data (Figure 3). Calculation of RF numbers using a threshold-based segmentation algorithm and separation of closely positioned objects yielded a 3- to 5-fold increase in RF numbers relative to previous reports (6,7,29,30). The same image processing and analysis steps were also applied on deconvolved images of the cells obtained using CLSM and gave roughly 3-fold less RF than with 3D-SIM approach (Figure 4A). The threshold level of 10% intensity of normalized image stacks was selected on the basis of visual inspection of the images. To ensure the segmentation algorithm produced sensible results, we also controlled the object size distribution and found that after the segmentation RF had a 7-fold smaller average volume corresponding to about 0.011 µm3 (∼10 voxel; Figure 4C). Such a difference in the size of RF is consistent with the 2-fold better xyz resolution provided by 3D-SIM. Hence, the object calculation algorithm used was unlikely to overestimate the RF numbers due to incorrect object separation.
Figure 4.
3D-SIM super-resolution imaging and counting of RF numbers. (A) Mouse C2C12 cells were BrdU pulse labeled (30 min) and sites of BrdU incorporation were visualized using BrdU-specific antibodies after DNase I denaturation. Nuclei with replication pattern typical for mid–late S-phase are shown. To improve signal-to-noise ratio a constrained iterative deconvolution was applied to the confocal (CLSM) image stack using a maximum-likelihood estimation algorithm (column 2). Large RF at chromocenters consisted of ∼100–120-nm-sized subdomains that could only be resolved with 3D-SIM (insets in second row), which leads to dramatic increase in apparent RF numbers. To determine numbers and volumes of RF a threshold-based segmentation was applied combined with an object separation routine (third row). (B) Intensity plot profile yielded about 2-fold smaller peak width (FWHM) of RF recorded with 3D-SIM compared to deconvolved confocal images. 3D volume rendering of a replicating chromocenter demonstrated the increased resolution in x, y and z-direction with 3D-SIM. (C) The segmented foci (CLSM + deconvolution n ∼ 1400; 3D-SIM n ∼ 4400) were characterized by lower size heterogeneity and about 7-fold smaller average volume in accordance with the 2-fold better xyz resolution of 3D-SIM. (D) Quantitative analysis of the RF segmented in the 3D-SIM data sets gave 3- to 5-fold higher RF numbers that have been previously detected (see also RF numbers detected in the deconvolved CLSM image stack in A and Table 2). The RF numbers decreased from early to late S-phase cells.
In summary, the application of 3D-SIM microscopy on 3D preserved cells yielded an average of ∼4000 RF, which decreased during the course of S-phase (∼6800 in early S-phase decreasing to ∼2000 objects in late S-phase; Figure 4D and Table 2).
Table 2.
Summary of 3D-SIM measurements of BrdU-labeled foci
| RF Pattern | Mean number | SD | Volume ± SD (µm) |
|---|---|---|---|
| Early | 6371 | 726 | 0.009 ± 0.002 |
| Mid | 4206 | 1828 | 0.011 ± 0.001 |
| Late | 1973 | 442 | 0.015 ± 0.002 |
DISCUSSION
We have used SMI and 3D-SIM microscopy to provide a quantitative estimate of the size and numbers of RF in mammalian cells at a level of accuracy previously inaccessible to light microscopic methods. The use of LM employing wide field detection makes it feasible to acquire the number of images required for good statistics in a comparatively short time span relative to EM. Using the SMI approach in total over 40 000 foci were included in the analysis of nucleotide-labeled RF and compared with RF labeled with GFP-tagged replication proteins. EM analysis, however, requires more complicated sample preparation and is more time consuming. There are also indications that a combination of the staining and fixation procedures required for EM immunogold labeling may result in a lower labeling density than is achievable using fluorescence techniques which could translate into a reduced accuracy of size determination.
SMI measurements, as with any measurement, will be affected by some bias, including the assumption that objects are spherical, the labeling procedure (we see a significant difference between the pure GFP and antibody amplified GFP cryosection samples), and the sample preparation. SMI microscopy is limited to sizes in the range from ∼40 nm to 200 nm. The measurements of objects <40 nm are imprecise, and sizes approximately >220 nm cannot be measured since no further correlation between the axial modulation and the size of the object can be detected (15), resulting in a truncation of the distribution at this value. However, we estimate that in the present study only a small number (<<5%) of foci were outside of the above range; inclusion of these structures would be unlikely to significantly affect the results. However, relative changes as a result of, for example, S-phase stage, should be preserved and any possible bias of the method is incorporated within the 30-nm standard deviation of RF size distributions (Figure 3B).
To bypass the poor z resolution of SMI microscopy, we adapted two sample preparation procedures to maximize the number of structures analyzed. In one, ultrathin cryosections of cells were employed as used previously for the SMI measurements of transcription factories in mammalian cells (14); this allowed the use of cells expressing GFP-tagged replication proteins. However, the signal strength was limiting and hence the numbers of RF measured were lower than with the nucleotide incorporation approach. The second method involved incubation of cells in hypotonic solution followed by cytospinning onto glass coverslips. This method de facto created flattened cells and increased the separation between the RF structures, i.e. it created an enlarged 2D projection of all RF in the cell nucleus. The hypotonic treatment was also possible for living cells and with GFP-tagged replication factors but the second step to flatten the cells using methanol:acetic acid and centrifugation was not compatible with the GFP in vivo labeling. Thus, for this sample preparation approach, only samples with nucleotide labeling were measured. This approach generated large numbers of measured RF. The RF sizes determined by both sample preparation approaches and replication visualization methods were similar and, importantly, comparable to the ones obtained using EM (11).
Since SMI microscopy yielded very variable numbers of RF probably due to the above-mentioned limitations, we evaluated another super-resolution microscopy method—3D-SIM. This method allows increased resolution in all three directions and direct use of 3D preserved samples stained using the same conditions as for conventional epifluorescence microscopy. These properties make it particularly amenable for such measurements and allow direct comparison with conventional LM data. Combination of 3D-SIM imaging with object separation algorithms allowed discrimination of several thousands of RF in most of the S-phase cells. Interestingly, the observed RF numbers appeared to be dependent on the S-phase stage. Between ∼6800 (in early S-phase) and ∼2000 (late S-phase) objects could be identified with a volume of about 0.011 µm3. Application of the same image processing and analysis steps on CLSM images of the cells followed by deconvolution gave roughly three times less RF than with 3D-SIM approach.
Altogether, our studies show that the difference between the size distributions of RF at different points throughout S-phase is very small. Both the absence of a dependence on S-phase stage and the estimated size are in agreement with previous EM studies (11). We have, however, achieved an improved level of statistics, obtained direct whole-cell 3D data and extended the analysis to the PCNA core component of the replication machinery, showing that the RF size distributions are similar for both, the replicated DNA and the replication machinery. Finally, the outcome of our measurement of total RF numbers of ∼4000 per cell definitely indicates that the numbers calculated so far (6,7,29,30) have been significantly underestimated. We have previously determined that the C2C12 mouse cells used in this study have near triploid DNA content (∼8.5 Gbp, unpublished data). Assuming a 1-hour, or less, average RF lifetime (30–32) and an average S-phase duration of 10 h, this implies ∼40 000 RF form during S-phase. Comparison of this number with the total genome size of C2C12 cells gives ∼210 kb per RF, which can be completely synthesized by a pair of replication forks traveling at ∼1.8 kb/min (7) within 1 h. These nanoscopic measurements in combination with established parameters of DNA replication suggest the existence of microscopically visible single replicon units as the basis for the functional organization and duplication of the genome in mammalian cells. This study shows that nanoscopic analysis of macromolecular structures can provide new insights and quantitative data for the modeling of biological processes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The BioImaging Network and the Nanosystems Initiative Munich (to H.L.); the Medical Research Council (to A.P. and S.M.); and the Deutsche Forschungsgemeinschaft (to H.L., C.C. and M.C.C.). Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Danny Nowak for excellent technical assistance with antibody generation and characterization and John W. Sedat for giving us access to his OMX microscope and for helpful advice.
REFERENCES
- 1.Leonhardt H, Rahn HP, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso MC. Dynamics of DNA replication factories in living cells. J. Cell Biol. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Somanathan S, Suchyna T, Siegel A, Berezney R. Targeting of PCNA to sites of DNA replication in the mammalian cell nucleus. J. Cell Biochem. 2001;81:56–67. doi: 10.1002/1097-4644(20010401)81:1<56::aid-jcb1023>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 3.Sadoni N, Cardoso MC, Stelzer EH, Leonhardt H, Zink D. Stable chromosomal units determine the spatial and temporal organization of DNA replication. J. Cell Sci. 2004;117:5353–5365. doi: 10.1242/jcs.01412. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura H, Morita T, Sato C. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp. Cell Res. 1986;165:291–297. doi: 10.1016/0014-4827(86)90583-5. [DOI] [PubMed] [Google Scholar]
- 5.Aten JA, Bakker PJ, Stap J, Boschman GA, Veenhof CH. DNA double labelling with IdUrd and CldUrd for spatial and temporal analysis of cell proliferation and DNA replication. Histochem. J. 1992;24:251–259. doi: 10.1007/BF01046839. [DOI] [PubMed] [Google Scholar]
- 6.Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng P, Meng C, Berezney R. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J. Cell Biol. 1998;143:1415–1425. doi: 10.1083/jcb.143.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazzotti G, Rizzoli R, Galanzi A, Papa S, Vitale M, Falconi M, Neri LM, Zini N, Maraldi NM. High-resolution detection of newly synthesized DNA by anti-bromodeoxyuridine antibodies identifies specific chromatin domains. J. Histochem. Cytochem. 1990;38:13–22. doi: 10.1177/38.1.2403578. [DOI] [PubMed] [Google Scholar]
- 9.Jaunin F, Visser AE, Cmarko D, Aten JA, Fakan S. A new immunocytochemical technique for ultrastructural analysis of DNA replication in proliferating cells after application of two halogenated deoxyuridines. J. Histochem. Cytochem. 1998;46:1203–1209. doi: 10.1177/002215549804601014. [DOI] [PubMed] [Google Scholar]
- 10.Kireev I, Lakonishok M, Liu W, Joshi VN, Powell R, Belmont AS. In vivo immunogold labeling confirms large-scale chromatin folding motifs. Nat. Methods. 2008;5:311–313. doi: 10.1038/nmeth.1196. [DOI] [PubMed] [Google Scholar]
- 11.Koberna K, Ligasova A, Malinsky J, Pliss A, Siegel AJ, Cvackova Z, Fidlerova H, Masata M, Fialova M, Raska I, et al. Electron microscopy of DNA replication in 3-D: evidence for similar-sized replication foci throughout S-phase. J. Cell Biochem. 2005;94:126–138. doi: 10.1002/jcb.20300. [DOI] [PubMed] [Google Scholar]
- 12.Failla AV, Spoeri U, Albrecht B, Kroll A, Cremer C. Nanosizing of fluorescent objects by spatially modulated illumination microscopy. Appl. Opt. 2002;41:7275–7283. doi: 10.1364/ao.41.007275. [DOI] [PubMed] [Google Scholar]
- 13.Failla AV, Albrecht B, Spöri U, Schweitzer A, Kroll A, Hildenbrand G, Bach M, Cremer C. Nanostructure analysis using spatially modulated illumination microscopy. Complexus. 2003;1:77–88. [Google Scholar]
- 14.Martin S, Failla AV, Spori U, Cremer C, Pombo A. Measuring the size of biological nanostructures with spatially modulated illumination microscopy. Mol. Biol. Cell. 2004;15:2449–2455. doi: 10.1091/mbc.E04-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathee H, Baddeley D, Wotzlaw C, Fandrey J, Cremer C, Birk U. Nanostructure of specific chromatin regions and nuclear complexes. Histochem. Cell Biol. 2006;125:75–82. doi: 10.1007/s00418-005-0096-7. [DOI] [PubMed] [Google Scholar]
- 16.Baddeley D, Batram C, Weiland Y, Cremer C, Birk UJ. Nanostructure analysis using spatially modulated illumination microscopy. Nat. Protoc. 2007;2:2640–2646. doi: 10.1038/nprot.2007.399. [DOI] [PubMed] [Google Scholar]
- 17.Hildenbrand G, Rapp A, Spori U, Wagner C, Cremer C, Hausmann M. Nano-sizing of specific gene domains in intact human cell nuclei by spatially modulated illumination light microscopy. Biophys. J. 2005;88:4312–4318. doi: 10.1529/biophysj.104.056796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batram C, Baddeley D, Kreth G, Cremer C. High precision size measurement of centromere 8 and the 8q24/c-myc gene region in metaphase and interphase human fibroblasts indicate differential condensation. J. Struct. Biol. 2008;164:293–303. doi: 10.1016/j.jsb.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Reymann J, Baddeley D, Gunkel M, Lemmer P, Stadter W, Jegou T, Rippe K, Cremer C, Birk U. High-precision structural analysis of subnuclear complexes in fixed and live cells via spatially modulated illumination (SMI) microscopy. Chromosome Res. 2008;16:367–382. doi: 10.1007/s10577-008-1238-2. [DOI] [PubMed] [Google Scholar]
- 20.Gustafsson MG, Shao L, Carlton PM, Wang CJ, Golubovskaya IN, Cande WZ, Agard DA, Sedat JW. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 23.Branco MR, Xie SQ, Martin S, Pombo A. In: Cell Imaging (Methods Express Series) Stephens D, editor. Bloxham: Scion Publishing Ltd.; 2006. pp. 201–217. [Google Scholar]
- 24.Failla AV, Cavallo A, Cremer C. Subwavelength size determination by spatially modulated illumination virtual microscopy. Appl. Opt. 2002;41:6651–6659. doi: 10.1364/ao.41.006651. [DOI] [PubMed] [Google Scholar]
- 25.Wagner C, Spöri U, Cremer C. High-precision SMI microscopy size measurements by simultaneous frequency domain reconstruction of the axial point spread function. Optik. 2005;116:15–21. [Google Scholar]
- 26.O'K;eefe RT, Henderson SC, Spector DL. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J. Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guillot PV, Xie SQ, Hollinshead M, Pombo A. Fixation-induced redistribution of hyperphosphorylated RNA polymerase II in the nucleus of human cells. Exp. Cell Res. 2004;295:460–468. doi: 10.1016/j.yexcr.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 28.Iborra FJ, Cook PR. The size of sites containing SR proteins in human nuclei. Problems associated with characterizing small structures by immunogold labeling. J. Histochem. Cytochem. 1998;46:985–992. doi: 10.1177/002215549804600901. [DOI] [PubMed] [Google Scholar]
- 29.Gotoh E. Visualizing the dynamics of chromosome structure formation coupled with DNA replication. Chromosoma. 2007;116:453–462. doi: 10.1007/s00412-007-0109-5. [DOI] [PubMed] [Google Scholar]
- 30.Muck J, Zink D. Nuclear organization and dynamics of DNA replication in eukaryotes. Front. Biosci. 2009;14:5361–5371. doi: 10.2741/3600. [DOI] [PubMed] [Google Scholar]
- 31.Manders EM, Stap J, Brakenhoff GJ, van Driel R, Aten JA. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J. Cell Sci. 1992;103:857–862. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- 32.Sporbert A, Gahl A, Ankerhold R, Leonhardt H, Cardoso MC. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol. Cell. 2002;10:1355–1365. doi: 10.1016/s1097-2765(02)00729-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.