Abstract
The repair of DNA double-strand breaks (DSBs) is essential to maintain genomic integrity. In higher eukaryotes, DNA DSBs are predominantly repaired by non-homologous end joining (NHEJ), but DNA ends can also be joined by an alternative error-prone mechanism termed microhomology-mediated end joining (MMEJ). In MMEJ, the repair of DNA breaks is mediated by annealing at regions of microhomology and is always associated with deletions at the break site. In budding yeast, the Mre11/Rad5/Xrs2 complex has been demonstrated to play a role in both classical NHEJ and MMEJ, but the involvement of the analogous MRE11/RAD50/NBS1 (MRN) complex in end joining in higher eukaryotes is less certain. Here we demonstrate that in Xenopus laevis egg extracts, the MRN complex is not required for classical DNA-PK-dependent NHEJ. However, the XMRN complex is necessary for resection-based end joining of mismatched DNA ends. This XMRN-dependent end joining process is independent of the core NHEJ components Ku70 and DNA-PK, occurs with delayed kinetics relative to classical NHEJ and brings about repair at sites of microhomology. These data indicate a role for the X. laevis MRN complex in MMEJ.
INTRODUCTION
In every living organism, the integrity of the genome is threatened by exogenous or endogenous factors that generate a diverse range of DNA lesions. DNA double-strand breaks (DSBs) are perhaps the most hazardous form of DNA damage, occurring as a result of ionizing radiation, oxidative free radicals, DNA replication across a nick and, in lymphocytes, from V(D)J recombination. Unrepaired DSBs give rise to broken chromosomes, while misrepair of DSBs can produce genomic rearrangements with the potential to induce transformation and carcinogenesis (1,2). The two main pathways used to repair DNA DSBs in eukaryotes are homologous recombination (HR) and non-homologous end joining (NHEJ). The HR pathway, dependent on the members of the RAD52 epistasis group (Rad51, Rad54, Rad59, XRCC2/3 and BRCA1/2) and the MRE11/RAD50/NBS1 (MRN) complex (3,4), repairs DNA with high fidelity using an undamaged homologous DNA template to restore the original sequence at the break (5). This requirement for a homologous donor sequence limits the HR pathway to the S and G2 phases of the cell cycle. The NHEJ pathway which, unlike HR, is not constrained by the need for extensive sequence homology can occur throughout the cell cycle and is the predominant mechanism for DSB repair in G1 and G0 cells (6).
Classical NHEJ effects the repair of DSBs by processing DNA ends to reveal short stretches (1–4 nt) of complementary sequence on either side of the break. Following alignment of these complementary sequences, nucleolytic trimming or gap filling occur in order to generate a ligatable structure. During this process, nucleotides can often be inserted or lost at the repair junction, thus NHEJ is inherently more error-prone than HR (7). Seven core NHEJ factors have been identified: Ku70, Ku80, DNA-PKcs, Artemis, XRCC4 and Ligase IV and XLF/Cernunnos. The Ku heterodimer binds to DNA ends and recruits the serine/threonine kinase DNA-PKcs and probably Artemis to the break site (8,9). The Artemis:DNA-PKcs complex possesses an endonuclease activity that cleaves 5′- or 3′-overhangs. Subsequent ligation of the processed ends is catalysed by a complex of XRCC4 and Ligase IV (10). XLF/Cernunnos associates with the XRCC4/Ligase IV to promote NHEJ (11,12), while two DNA polymerases, pol µ and pol λ, are involved in gap filling of NHEJ intermediates (13–15). The majority of DNA DSBs in G0/G1 cells are repaired within minutes via the canonical DNA-PK-dependent NHEJ process but in cells where this pathway is inactivated, either chemically or genetically, an alternative DNA-PK-independent NHEJ mechanism can be seen to operate (16–19). This end joining pathway, termed as microhomology-mediated end joining (MMEJ), operates with 20- to 30-fold slower kinetics than DNA-PK-dependent NHEJ, requires four or more bases of microhomology and is error-prone, generating deletions at the break site (20,21). MMEJ-like activities have been identified in a number of systems including budding yeast, fission yeast, Drosophila and Xenopus laevis egg extracts as well as in mammalian cells. In Saccharomyces cerevisiae, MMEJ operates independent of the RAD52 epistasis group of genes but requires a number of proteins normally involved in other repair pathways including the MRX complex, the Rad1-Rad10 3′-flap endonuclease, Nej1 and Sae2 (22,23). In mammalian cells DNA ligase I, DNA ligase III, PARP-1, the ERCC1-XPF endonuclease and CtBP-interacting protein (CtIP) have all been implicated in MMEJ, while a recent study of alternative end joining of V(D)J recombination intermediates revealed a role for NBS1 in this process (24–28).
MRN comprises a conserved multi-subunit nuclease with multiple roles in the cellular response to DNA damage (29,30). The MRN complex is required for DNA DSB detection, checkpoint signalling and for the resection of DNA ends to allow HR repair of DSBs (4,31). MRN is also important for chromatin remodelling at DSBs and has been demonstrated to have a role in the induction of apoptosis (32–34). The critical importance of MRN in orchestrating this response to DSBs is highlighted by the fact that MRE11, RAD50 and NBS1 are all essential genes in higher eukaryotes (35–37). Hypomorphic mutations in MRE11 or NBS1 give rise to Ataxia telangiectasia-like disorder (ATLD) or Nijmegen breakage syndrome, respectively, both of which are associated with clinical features such as radiosensitivity, chromosomal instability and increased cancer predisposition (38,39).
Although the requirement for the MRN complex in HR repair is well documented, the involvement of MRN in NHEJ is more controversial. In S. cerevisiae, the analogous MRX complex has been shown to be important for NHEJ-mediated repair of DNA DSBs (40). In contrast, mutation of MRN components in Schizosaccharomyces pombe did not reveal any significant defects in efficiency or fidelity of plasmid end joining (41). No significant DSB repair deficiency was found in human ATLD cells, as judged by pulse-field gel electrophoresis of irradiated DNA, but since these hypomorphic Mre11 mutants still retain some activity a role in NHEJ could not be ruled out (38). Aberrant DSB rejoining has, however, been described for NBS1-deficient human cells (42). In addition, an NHEJ defect has been reported for NBS1 and ATLD cells using γ-H2AX focus formation as an assay for unrepaired DSBs (43). In vitro studies using mammalian cell extracts have indicated a requirement for MRN in NHEJ (44,45). Moreover, human cells lacking Nbs1 function are reported to have defects in class-switch recombination, which depends on NHEJ activities (46–48) although cells harbouring hypomorphic NBS1 mutations are still capable of normal V(D)J recombination (49,50). In contrast, targeted disruption of NBS1 and MRE11 in chicken DT40 cells resulted in a reduction in HR but no apparent NHEJ defect in this system (51,52), while the conditional deletion of Nbs1 in mouse cells led to an increase in NHEJ implying a role for Nbs1 in repression of the NHEJ pathway (53).
Xenopus laevis egg extracts have also been used to analyse eukaryotic NHEJ. Xenopus laevis cell-free extracts exhibit highly efficient, accurate end joining in which the DNA DSB break is precisely repaired in a Ku-dependent manner (54,55) as well as an error-prone microhomology-mediated NHEJ pathway (56). Using a plasmid DSB repair assay to analyse the role of Mre11 in NHEJ, a study by Di Virgilio and Gautier (57) concluded that neither Ku-dependent or Ku-independent end joining is affected by the absence of Mre11 in X. laevis. However, the enzymatically derived repair substrates used in this study, although highly informative, are not representative of the DSB termini usually produced in vivo as a consequence of ionizing radiation. To investigate a potential role for the MRN complex in the repair of DSBs that are more representative of IR-induced lesions, we have used internally radio-labelled plasmid substrates with either 3′-hydroxyl (3′-OH) (normal) or 3′-phosphoglycolate (3′-PG) (damaged) termini (58) to study end joining in X. laevis egg extracts. We demonstrate that, in fact, there is no specific requirement for the MRN complex in the repair of 3′-PG termini; however, this assay system revealed a role for XMRN in resection-based end joining at a region of microhomology. Depletion of XMre11, XNbs1 or XRad50 from X. laevis egg extracts abolishes this resection-based end joining and stimulates Ku70-dependent accurate end joining. These data indicate that the MRN complex does function in DNA end joining in X. laevis and support the critical role that has been proposed for the MRN complex in determining the choice of DSB repair pathway.
MATERIALS AND METHODS
Identification and cloning of the Xenopus homologues of the MRN complex
tBLASTn searches using the human amino acid sequences of Rad50 and Nbs1 identified multiple X. laevis expressed sequence tags showing a significant level of identity (>60%) to the human proteins (XRAD50 IMAGE 3380700; XNBS1 IMAGE 4056851, 5157068, 6865382, 6318196 and 405685). Following sequence analysis of these clones (obtained from UK HGMP/Geneservice Ltd), oligonucleotide primers were designed for polymerase chain reaction (PCR) amplification of the complete open reading frame (ORF) of XNBS1 (2.1 kb) and a 2.3-kb 5′-fragment of XRAD50 from X. laevis cDNA. The DNA sequence for XNBS1 was submitted to Genbank (accession number AY312176). You and co-workers (59) subsequently submitted a similar sequence to our independently isolated clone (accession number AY999019). The complete XMRE11 ORF was isolated by PCR from X. laevis cDNA using oligonucleotide primers designed to the sequence deposited in Genbank (accession number AF134569) (60).
Generation of antibodies to XMre11, XNbs1 and XRad50
DNA fragments corresponding to amino acid residues 541–710 of XMre11, 1–447 of XNbs1 and 1–490 of XRad50 were cloned into vector pET16b (Novagen) and expressed in Escherichia coli BL21 cells (Novagen). Protein was purified using Nickel-NTA agarose (Qiagen) under denaturing conditions and used to generate polyclonal antisera in rabbits (Eurogentec). Where necessary, antisera were affinity purified against the respective antigen immobilized on Amino link+ resin (Perbio) using the manufacturer’s instructions.
Preparation of X. laevis egg extract
Xenopus laevis egg extract was made according to the method of Felix (61). Briefly, eggs were dejellied [20 mM Tris, pH 8.5, 5 mM dithiothreitol (DTT) and 110 mM NaCl), washed in ¼× MMR (5 mM HEPES, pH 7.5, 100 mM NaCl, 0.5 mM KCl, 0.25 mM MgS04, 0.5 mM CaCl2 and 0.025 mM ethylenediaminetetraacetic acid (EDTA)] and activated in ¼× MMR containing 0.25 µg/ml ionophore A23187 (Roche) for 2 min. Eggs were washed five times with ice-cold extraction buffer (100 mM KOAc, 2.5 mM Mg(OAc)2, 60 mM EGTA, 250 mM sucrose and 1 mM DTT, pH 7.4) resuspended in 5 ml Felix buffer containing 10 µg/ml aprotinin and 50 µg/ml cytochalasin B and transferred to 2 ml microcentrifuge tubes. Eggs were packed by a brief spin (6000 r.p.m. for 10 s) and excess buffer removed before centrifugation at 15 000 r.p.m. for 10 min. The cytoplasmic layer was transferred to 5 ml ultracentrifuge tubes (Beckman) and centrifuged at 48 000 r.p.m. for 2 h at 4°C (Ti55 rotor). The clear cytosol was removed by side puncture of the tube with a 21 gauge needle. This high-speed supernatant egg extract was snap frozen in liquid nitrogen.
Immunodepletion of X. laevis egg extract
Immunodepletion of X. laevis egg extract was performed using affinity-purified antibodies against XMre11, XNbs1 and XRad50 covalently cross-linked to protein A sepharose 4B beads (GE Healthcare). For mock depletions, nonspecific rabbit IgGs (Sigma, Poole, UK) were coupled to protein A sepharose 4B beads at the same concentration. For XKu70-depletion, human Ku70 antibody (Covance) was coupled to Protein G Sepharose beads (Sigma) in 2:1 ratio. Extracts were depleted by mixing with the appropriate antibody beads (50% v/v) for 45 min at 4°C. Two rounds of depletion were routinely performed.
NHEJ assay
The linear DNA templates, labelled 3′-OH and 3′-PG overhang ends, were produced as previously described (58,62). Twenty microlitres of egg extract was combined with 12.5 ng of linear DNA template and 1 µl of NHEJ mix (1 mM ATP, 1 mM MgCl2 and 50 μM dNTPs) (63) and incubated at 21°C. Samples were then processed for analysis by agarose or acrylamide gel electrophoresis.
Analysis of end joining products on agarose gel
Reactions were treated with proteinase K mix [1 mg/ml proteinase K, 200 mM NaCl, 30 mM EDTA, 50 mM Tris and 0.5% (v/v) sodium dodecyl sulfate (SDS)] and incubated at 37°C for at least an hour. Following phenol/chloroform extraction, the DNA was precipitated with 3 mM sodium acetate, 100% ethanol and 2.5 µg linear acrylamide (Ambion) at −20°C. DNA samples were resolved on a 0.7% (w/v) agarose gel in Tris–borate–EDTA buffer at 30 V. The gel was then dried and exposed to a phosphorimager screen. Quantification was performed using ImageQuant software (Molecular Dynamics).
Southern blotting
DNA was transferred to a nylon transfer membrane (Hybond-N+, Amersham Biosciences) by Southern blotting. A nonradioactive probe, prepared using the ECL Random Prime labeling kit version II (Amersham Biosciences), was incubated with the membrane at 60°C overnight. After washing, membranes were incubated with anti-fluorescein antibody conjugated to horseradish peroxidase. Repair products were visualized using the Amersham ECL detection system according to the manufacturer’s instructions.
Electrophoretic analysis of end joining products at the nucleotide level
NHEJ reaction mixtures were incubated in lysis buffer [0.3 M NaCl, 2 mM Tris, pH 6.7 (HCl), 10 mM EDTA, 1% (w/v) SDS and 1 mg/ml proteinase K] at 65°C for 3 h. Following phenol/chloroform extraction, the DNA was ethanol precipitated, washed once with ethanol and dissolved in TE buffer. DNA was digested with 10 U BstXI enzyme (New England Biolabs) at 55°C for 3 h and 20 U Taq∝I (New England Biolabs) at 65°C for 4 h and ethanol precipitated. The recovered DNA was dried at 37°C then dissolved in loading buffer (20 mM EDTA, bromophenol blue, in formamide). Samples were resolved on a 20% sequencing gel (SequaGel National Diagnostics) for 3 h at 40 Watts and exposed to a phosphorimager screen at −20°C. Screens were scanned on a Typhoon 9410 (Amersham Biosciences).
Sequence analysis of repair junctions
NHEJ reaction mixtures were subjected to PCR using the following oligonucleotides (5′-AATGCGCTCATCGTCATCC-3′ and 5′-GCTTCTTCCTTAAATCCTGGT-3′) in order to amplify an approximately 430-bp fragment spanning the repair junction. PCR products were cloned into pGEM-T-easy (Promega) and analysed by restriction digest with BsaHI and MluI or by sequencing (MWG Biotech).
SDS–PAGE and Western blotting
Protein samples were run on 8% acrylamide gels and transferred onto nitrocellulose membrane (Nitrobind, Osmonics) by semi-dry transfer. The membrane was blocked in Blotto [phosphate buffered saline (PBS), 5% non-fat milk powder and 0.5% Tween 20] for 1 h and was incubated with primary antibodies at a dilution of 1/2000 in Blotto overnight at 4°C. After washing with PBS, 0.5% Tween, the membrane was incubated with a peroxidase-conjugated anti-rabbit secondary antibody (Dako) at a dilution of 1/5000 before washing and detection using enhanced chemiluminescence.
RESULTS
Identification and isolation of X. laevis Mre11, Rad50 and Nbs1 orthologues
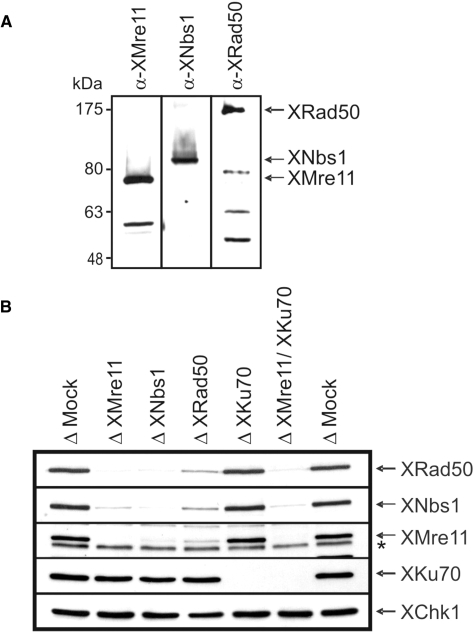
The Xenopus orthologues of Mre11, Rad50 and Nbs1 were identified and cloned by a combination of database searches (tBLASTn) and PCR (the ‘Materials and Methods’ section). The predicted amino acid sequences of all three proteins show a high degree of identity (65–70%) with the respective human sequences. Polyclonal antisera were raised against bacterially expressed fragments of each protein (the ‘Materials and Methods’ section). Western blotting confirmed that these antibodies recognized proteins of the appropriate molecular weights in X. laevis egg extract (XMre11 ∼85 kDa, XNbs1 ∼95 kDa and XRad50 ∼150 kDa) (Figure 1A). Conditions for the immunodepletion of XMre11, XRad50 and XNbs1 from X. laevis extract were established (the ‘Materials and Methods’ section). XMre11 was no longer detectable by Western blotting following depletion with the XMre11 antibody (Figure 1B). XNbs1 and XRad50 were co-depleted with XMre11 confirming that XMre11, XNbs1 and XRad50 form a stable complex in X. laevis egg extract. Depletion of XNbs1 or XRad50 similarly co-depleted all three components of the MRN complex, although the XRad50 antibody is less effective for immunodepletion than the XMre11 or XNbs1 antisera. As expected, we did not observe any interaction between the MRN complex and the core NHEJ factor Ku70 in X. laevis extract (Figure 1B).
Figure 1.
Immunodepletion of the MRN complex and Ku70 from X. laevis egg extract. (A) Xenopus laevis egg extract was analysed by Western blotting with antisera against XMre11, XNbs1 and XRad50. (B) Extract was subjected to three rounds of depletion using protein A sepharose coupled to non-specific rabbit IgGs (Δ Mock), anti-XMre11 antibodies (Δ XMre11), anti-XNbs1 antibodies (Δ XNbs1) or anti-XRad50 antibodies (Δ XRad50) or using protein G sepharose coupled to antibodies against Ku70 (Δ XKu70). Double depletion of XMre11 and XKu70 was achieved by two rounds of depletion with α-Ku70 beads followed by one round with α-XMre11 beads. Depletion efficiency was analysed by Western blotting with the appropriate antisera. XChk1 was used as a loading control. A non-specific band is denoted by asterisk.
XMre11 depletion alters the profile of end joining repair products
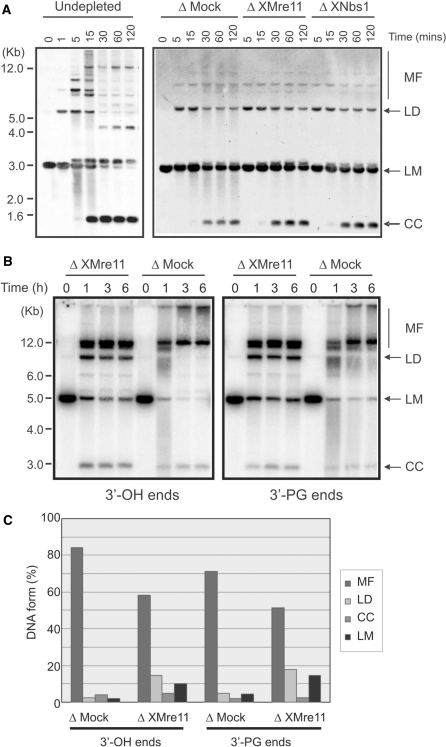
In order to directly investigate a requirement for the MRN complex in NHEJ, we used a plasmid-based NHEJ assay based on that of Aoufouchi et al. (63) to determine NHEJ efficiency in X. laevis extract immunodepleted for XMre11 or XNbs1. The NHEJ substrate was produced by digesting pUC19 plasmid with PstI and SmaI generating a linear DNA with blunt and 3′-overhang ends. Accurate re-circularization of the linearized plasmid, recovering the PstI site, will only result from NHEJ rather than from simple ligation. A typical NHEJ repair profile in undepleted X. laevis egg extract is presented in Figure 2A (left panel). Within 1 min, multimeric forms (MFs) of the plasmid can be detected above the 5-kb marker. These MFs are representative of intermolecular NHEJ (55). Linear monomers (LMs) begin to be modified into closed circular DNA supercoiled monomer (CC; detected below the 1.6 Kb marker) within 15 min. The generation of this closed circular plasmid results from intra-molecular NHEJ. Digestion of the resulting repair products showed them to be sensitive to PstI digestion and resistant to SmaI digestion, indicating that the PstI site has been restored (data not shown). A large proportion of the products have therefore been repaired accurately without deletion or insertion of nucleotides. Closed circular plasmid formation and multimerization were both observed in XMre11- and XNbs1-depleted extracts with comparable kinetics to that of mock-depleted extracts (Figure 2A, right panel). These data indicate that the MRN complex is not required for efficient NHEJ in X. laevis extracts, consistent with a study by Di Virgilio and Gautier (57), using a similar NHEJ assay, which concluded that XMre11 was not required for the efficiency, kinetics or fidelity of DSB repair by NHEJ.
Figure 2.
NHEJ in XMRN-depleted extracts. (A) Southern blot analysis of NHEJ products generated by incubation of linearized pUC19 template (1 ng/μl) in the indicated egg extracts. Repair products were detected using a fluorescein-labelled pUC19 probe. LMs were converted into MFs, linear dimers (LDs) and closed circular DNA monomers (CC). (B) Agarose gel analysis of repair products resulting from the incubation of radio-labelled pSV56 linear substrates with defined termini in mock-depleted (Δ Mock) or XMre11-depleted (Δ XMre11) extract. Left panel shows end joining of 3′-OH ends. Right panel shows repair of 3′-PG termini. (C) Quantification of DNA forms after 6 h end joining reactions using pSV56 defined linear substrates. The amount of each DNA form is expressed as a percentage of the total signal for each individual lane.
Although the enzymatically digested plasmids we have used in this experiment do mimic the presence of DNA DSBs in extracts, it should be noted that most radiation-induced DSBs represent a more complex substrate for the NHEJ machinery. Radiation-induced DNA DSBs are formed by fragmentation of closely opposed deoxyribose moieties, typically leaving in each strand a one-base gap with 5′-phosphate and either 3′-phosphate or 3′-PG termini (58). In order to assess a possible role for the MRN complex in the processing of such complex termini, we used defined substrates generated by the ligation of 5′-[32P]-labelled oligomers at the ends of a linearized pSV56 plasmid (62). One substrate, used as a control, carried 3′-OH overhang ends, while the other substrate carried termini modified to a 3′-PG overhang. Using these linear substrates, we analysed the repair products resulting from the end joining reaction in XMre11-depleted X. laevis egg extract and compared it with repair in mock-depleted extract.
Following a 1-h incubation of either 3′-OH (Figure 2B, left panel) or 3′-PG (Figure 2B, right panel) substrates in mock- or XMre11-depleted extract, closed circular plasmids (3 kb) and MFs were observed, suggesting that these substrates were processed via efficient intra- and intermolecular end joining processes, respectively. In fact, using either of the radio-labelled pSV56 substrates we see an increased proportion of intermolecular end-joining compared with the repair of linearized pUC19 (Figure 2A) or pBS KSII (57). End joining of 3′-OH substrates was more efficient than the processing of 3′-PG substrates in both mock- and XMre11-depleted extracts. This observation is consistent with previous studies in X. laevis egg extracts showing that PG ends are processed with a reduced level of efficiency when compared with hydroxyl ends, probably because the 3′-terminal blocking groups have to be removed prior to repair (64). Indeed, Gu and co-workers (64) noted an even greater effect on repair proficiency using PG ends, since the blunt-ended PG-termini used in that study are processed much less efficiently than the overhanging 3′-PG ends we have used here. XMre11-depleted extract does not show any specific defect in processing 3′-PG ends relative to mock-depleted extract (Figure 2B).
The generation of covalently closed circular DNA repair products was largely unaffected by depletion of XMre11 using either of the two repair substrates (Figure 2B and C). We did note, however, an increase in linear dimer formation (6-fold for 3′-OH ends and 4-fold for 3′-PG ends) as well as a 30% reduction in multimeric repair products in XMre11-depleted extract compared with mock-depleted extracts (Figure 2B and C). The proportion of unprocessed LMs was also greater in XMre11-depleted extract than in mock-depleted extracts (∼5-fold). These data suggest that XMre11-depleted extracts are, in fact, less proficient at intermolecular DNA end joining than mock-depleted extracts. In particular, the conversion of linear dimers to higher multimers appears to be rate-limiting in the absence of XMre11. The increased level of unrepaired substrate in XMre11-depleted extract may also result from reduced degradation of the DNA in the absence of XMre11. In fact, we observed considerably more DNA smearing in undepleted and mock-depleted extracts than with XMre11- and XNbs1-depleted extracts using either the pSV56 or pUC19 DNA templates (Figure 2A and B), suggesting that removal of the MRN complex protects the DNA ends from nucleolytic degradation in egg extracts.
Analysis of NHEJ repair products at the nucleotide level
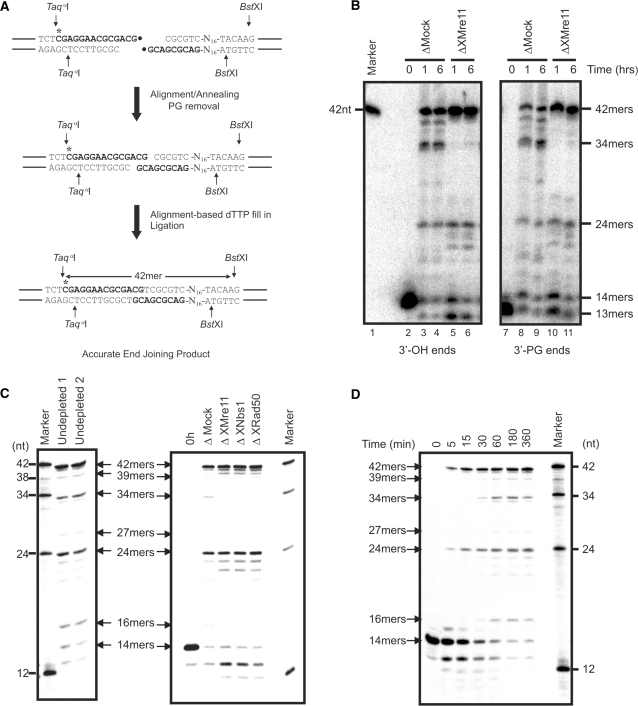
Using these [32P]-radio-labelled 3′-OH and 3′-PG end substrates, we have been able to examine the involvement of XMre11 in NHEJ using a more sensitive assay. In order to analyse the processing of these defined DNA templates at the single nucleotide level, we performed the NHEJ reaction as before, by incubation of the [32P]-radio-labelled substrates in X. laevis egg extracts, then isolated the repair products. The DNA was digested with TaqαI (on the 5′-side of the repair site) and BstXI (on the 3′-side) to produce short DNA fragments that could be analysed on denaturing acrylamide gels (Figure 3A) (62). In this way, it was possible to analyse the fidelity of repair at the nucleotide level, detecting even single nucleotide deletions or insertions.
Figure 3.
Analysis of DNA end joining at the nucleotide level. (A) Formation of accurate end joining products from 3′-OH or 3′-PG end substrates during NHEJ reactions. Following the removal of the 3′-PG moeity (•), the 3′-terminal CG residues align and the one-base gap opposite A is filled in by a DNA polymerase. Ligation of the ends generates accurate end joined products which, following treatment with the restriction enzymes TaqαI and BstXI, can be observed on a sequencing gel as 42-bp oligomers. (B) Sequencing gel analysis of NHEJ in X. laevis egg extracts using 3′-OH- and 3′-PG-defined substrates. pSV56 linear substrates with either 3′-OH or 3′-PG ends (1 ng/μl) were incubated in mock-depleted (Δ Mock) or XMre11-depleted (Δ XMre11) extract at 21°C for the indicated times. DNA was digested with TaqαI and BstXI, then resolved on a 20% denaturing polyacrylamide gel. (C) Linearized pSV56 with 3′-OH termini was incubated at 1 ng/μl in undepleted, mock-depleted and XMre11-, XNbs1- or XRad50-depleted extracts for 6 h at 21°C before digestion and analysis as described above. Substrate added to mock-depleted extract at 1 ng/μl and processed immediately serves as a control (0 h). (D) Linearized pSV56 with 3′-OH ends was incubated at 1 ng/μl in X. laevis egg extract at 21°C for the indicated times. Accurate repair occurs at earlier time points than resection-based repair.
We analysed NHEJ activity in XMre11- and mock-depleted X. laevis extract using this method. The linear substrates were initially observed as 14-mers (14-mer-OH) (Figure 3B, lane 2) or apparent 13-mers (14-mer-PG), since the 14-mer-PG comigrates with the 13-mer-OH (Figure 3B, lane 7). After incubation in mock-depleted egg extract, both the 3′-OH and 3′-PG substrates gave rise to 42-nt oligomers resulting from a head-to-tail end joining, which presumably generated either re-circularized DNA plasmid or head-to-tail dimers (Figure 3B). This 42-mer formation occurs following an error-free process identified as accurate end joining. A number of lower molecular weight oligomers were also observed. The oligomers detected below the 14-mer-OH (Figure 3B, lanes 3–6) likely represent 3′-resection products (about 13 nt), while the oligomers detected above the 14-mer-PG (Figure 3B, lanes 8–11) likely correspond to PG-removed products. Both are unligated products. Head-to-head end joining products were discerned at 24 nt. In addition, several oligomers estimated to range from 34 to 39 nt resulted from end joining of either substrate in mock-depleted extract, with the 34-nt product being most abundant. These 34- to 39-mers represent inaccurately repaired products in which end joining was accompanied by the resection of several nucleotides. Incubation of either the 3′-OH or 3′-PG substrates in XMre11-depleted extract gave strikingly different results to those seen with mock-depleted extract. In both cases, a significant increase in 42-mers was observed in XMre11-depleted extract, while 34- to 39-mers were no longer detected. These observations indicate that in XMre11-depleted extract accurate rejoining was promoted while resection-based joining was reduced. Although the 3′-PG ends were processed less efficiently than 3′-OH ends, in both mock- and XMre11-depleted extract, there was no significant difference in the pattern of repair products generated from the two DNA templates. The X. laevis MRN complex is not specifically required for the repair of these 3′-PG termini, since accurate repair of the 3′-PG ends occurred even in the absence of XMre11.
In order to confirm that the loss of resection-based end joining we observed in XMre11-depleted extracts was specific for the XMRN complex, we also tested XNbs1- and XRad50-depleted extracts for their proficiency in resection-based end joining. For effective depletion of the XMRN complex with each of the different antibodies, three rounds of depletion were required in this experiment. The repair activity is somewhat compromised by this treatment resulting in some loss of resected repair products in mock-depleted extract in comparison with extract which has only undergone two rounds of depletion. Nevertheless, in undepleted and mock-depleted extracts, we observed the accurate repair product at 42 nt as well as smaller resected end joining products, predominantly at 34 nt but also at 39, 27 and 16 nt (Figure 3C). Immunodepletion of XMre11, XNbs1 or XRad50 resulted in an increased proportion of accurately repaired 42-mer as well as an increase in 40- to 41-nt repair products that have undergone inaccurate fill-in and ligation. The resected repair products at 34, 27 and 16 nt were, however, completely abolished by immunodepletion of each of the XMRN components (Figure 3C). These data indicate a specific requirement for the XMRN complex in this resection-based DNA end joining process.
Resection-based end joining is a late event during NHEJ in X. laevis egg extracts
NHEJ occurs rapidly in X. laevis egg extracts with intermolecular end joining detectable after only 30 s and intramolecular end joining within 30 min at 13°C (55). In this study, using the linearized pUC19 repair substrate, intermolecular end joining is evident after 1 min and intramolecular end joining detected by 5 min at 21°C (Figure 2A) consistent with other recent studies (57). Using the defined 3′-OH pSV56 substrate, the 42-mer accurate end joining product is detected within 5 min of incubation in mock-depleted extract and increases over the course of 15 min (Figure 3D). The XMRN-specific resected products, in contrast, only become apparent after 30 min and increase up until 3 h. This resection-based repair pathway therefore represents a late event during DNA end joining in X. laevis cell-free extracts.
Accurate end joining in X. laevis extracts is DNA-PK dependent but resection-based end joining is Ku70 independent
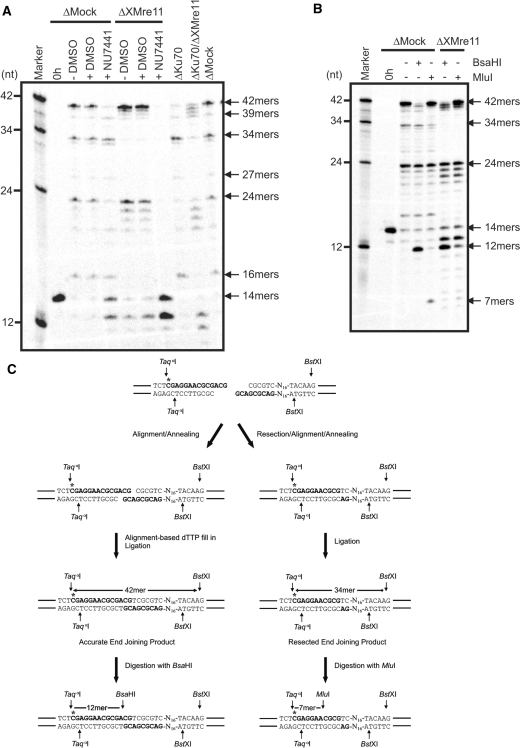
DNA-PK is essential for efficient NHEJ in higher eukaryotes. Using a plasmid-based repair assay to assess the effect of a specific inhibitor of DNA-PKcs on NHEJ, Di Virgilio and Gautier (57) demonstrated that both intra- and intermolecular end joining are dependent on DNA-PK catalytic activity in X. laevis extracts. However, the Ku heterodimer does not appear to be universally required for NHEJ in this system, since immunodepletion of Ku70 only inhibited intramolecular end joining, while multimerization was promoted. We, therefore, examined the involvement of DNA-PKcs and Ku70 in accurate and resection-based NHEJ using the defined 3′-OH radio-labelled substrate. Addition of 8 μM NU7441, a potent DNA-PKcs inhibitor (65), to mock-depleted extract significantly inhibited the formation of 42-mer repair product but had no effect on resection-based end joining products as compared with the addition of dimethyl sulfoxide (DMSO) alone (Figure 4A). This concentration of NU7441 also prevented 42-mer formation in XMre11-depleted extract. Inhibition of DNA-PKcs activity, therefore, prevents accurate but not XMRN-dependent resection-mediated end joining in this system.
Figure 4.
Resection-based end joining is Ku70 independent and occurs at sites of microhomology. (A) pSV56 3′-OH substrate was incubated in mock-, XMre11-, Ku70- and XMre11/Ku70 double-depleted extracts for 6 h. DMSO was added to a final concentration of 0.4% and NU7441 was added to a final concentration of 8 µM as indicated. (B) pSV56 3′-OH substrate was incubated in mock- or XMre11-depleted extract at 21°C for 6 h. DNA was cut with TaqαI and BstXI then further digested with BsaHI or MluI as indicated. (C) Alignment of the 3′-terminal CG residues, dTTP fill-in and ligation generates a BsaHI restriction site (5′-GRCGYC-3′) 12-bp downstream of the TaqαI site in the accurately repaired 42-mer. The 3′–5′ resection of 7 bp on either side of the DNA break creates 4-bp CGCG overhangs that anneal and are ligated without a requirement for further synthesis to yield the 34-mer resected end joining product. A diagnostic MluI restriction site (ACGCGT) is generated 7-bp downstream of the TaqαI site by repair in this manner.
To further investigate the relationship between NHEJ components in this system, the repair of the defined 3′-OH substrate was analysed in XKu70-immunodepleted egg extract. Immunodepletion of XKu70 dramatically inhibited the formation of accurately repaired 42-mers (and 24-mer head-to-head products) but had no effect on the formation of 34-, 27- and 16-mers (Figure 4A). XKu70 is, therefore, required for accurate end joining but is dispensable for resection-based end joining of the pSV56 repair substrate. In the XKu70/XMre11 double depletion, both the accurate and resection-based repair products are diminished as expected. Once again the removal of XMre11 promotes the formation of 40- to 41-mers demonstrating that these inaccurately repaired products do not depend on XKu70 or XMre11. These 40- to 41-mers are not observed in XMre11-depleted extract when Nu7441 is used to inhibit DNA-PKcs activity, perhaps because inactive DNA-PK at the DNA ends prevents access for other factors (66). Taken together, these observations suggest that classical NHEJ factors and the MRN complex are involved in two independent end joining pathways in X. laevis extracts. The existence of a further Ku- and XMre11-independent mechanism is also implied.
Restriction analysis of the repair products
Accurate end joining of the pSV56 repair substrate to yield the 42-bp TaqαI-BstXI restriction fragment requires alignment of the terminal CG residues and single-base gap-filling opposite adenine. This generates a BsaHI restriction site 12-bp downstream of the TaqαI cleavage site (Figure 4C). In order to confirm the accurate nature of Ku70-dependent repair in both mock- and XMre11-depleted extract, we subjected the TaqαI-BstXI-cut repair products to further digestion with BsaHI. In each case, the Ku70-dependent 42-mer was cleaved to yield a radio-labelled 12-mer as anticipated (Figure 4B, lanes 4 and 6). Moreover, since the generation of the 42-mer accurate end joining product depends on single-base gap-filling with dCTP, the addition of ddTTP to extracts should inhibit the fill-in mechanism and prevent ligation. We have found that addition of ddTTP (200–600 µM) prevents the appearance the 42-mer accurate repair product, but has no effect on formation of the 34-mer resected repair as expected (Supplementary Figure S1).
Since the XMRN-dependent end joining process we have identified bears many of the hallmarks of an alternative MMEJ pathway, we anticipate that the smaller, XMRN-dependent repair products are produced as a result of single-stranded resection and alignment at regions of microhomology. The predominant 34-nt end joining product may be generated by 5′–3′ resection of 4 nt on both sides of the break leaving a 4-bp region of microhomology for alignment and annealing followed by flap-trimming and ligation (Figure 4C). Alternatively, 3′–5′ resection of 7 bp on either side of the break would leave the same 4-bp microhomologous sequence for annealing and subsequent ligation. Resection and repair at the site of microhomology in either manner would create a MluI cleavage site 7-bp downstream of the TaqαI restriction site. We therefore examined the repair junction of the 34-mer by digestion of the TaqαI-BstXI-cut repair products with MluI. MluI digestion of the repair products from mock-depleted extract resulted in a significant reduction in 34-mers relative to the undigested control concomitant with the appearance of radio-labelled 7-mers (Figure 4B, lane 5). In contrast, MluI digestion of the end joining products derived from XMre11-depleted extract, in which 34-mers were abolished, did not give rise to this 7-bp fragment. These data support the conclusion that the 34-mer repair junction is generated through resection of a short stretch of nucleotides followed by annealing at a region of microhomology. Similarly, the minor 39-nt end joining product is likely formed by misalignment of the 3′-terminal CG dinucleotide with the GCGC microhomology creating a MluI site at the repair junction. MluI digestion accordingly results in loss of the 39-mer repair product contributing to the appearance of the radio-labelled 7-nt fragment (Figure 4B).
We further examined the repair junctions formed in X. laevis egg extracts by restriction digestion and sequencing of cloned repair products. Restriction analysis of repair products derived from mock-, XMre11- and Ku70-depleted extracts indicated that accurate repair (BsaHI-sensitive repair products) was enhanced in the absence of XMre11 and impaired when Ku70 was removed (Table 1). Conversely, repair products harbouring a MluI site were formed at a higher frequency in Ku70-depleted extract and reduced in XMre11-depleted extract. Sequencing of the repair junctions derived from mock-depleted extract confirmed that the BsaHI-sensitive clones were accurately repaired to produce a 42-bp TaqαI-BstXI fragment (Table 2). The majority of 34-bp TaqαI-BstXI sequences (6/8) exhibited resection and annealing at the CGCG microhomology to yield a MluI site as predicted. The two remaining 34-bp TaqαI-BstXI sequences also indicated resection and annealing at the CGCG sequence but inaccurate fill-in DNA synthesis has destroyed the MluI site in both cases, accounting for the MluI-resistant population of 34-mers noted previously (Figure 4B). In one further repair product, a 21-bp sequence has been deleted following resection and alignment at a CG dinucleotide. These sequencing data provide compelling evidence that the XMRN-dependent repair products result from a microhomology-mediated process.
Table 1.
Restriction analysis of repair junctions formed in mock-, XMre11- and Ku70-depleted extracts
| RE sensitivity | ΔMock | ΔXMre | ΔKu |
|---|---|---|---|
| Clones (%) | Clones (%) | Clones (%) | |
| BsaHI | 11/22 (50) | 14/20 (70) | 2/16 (12.5) |
| MluI | 8/22 (36.4) | 4/20 (20) | 8/16 (50) |
| – | 3/22 (13.7) | 2/20 (10) | 6/16 (37.5) |
Table 2.
Nucleotide sequence of repair junctions formed in mock-depleted extract
| TaqαI-BstX1 fragment | Joining product | No. | RE sensitivity | Derivation |
|---|---|---|---|---|
| 42-mer | TCTCGAGGAACGCGACGTCGCGTCG-N15-A | 9/18 | BsalH1 | Accurate repair |
| 34-mer | TCTCGAGGAACGCGTCG-N15-A | 6/18 | MluI | Resection to CGCG |
| 34-mer | TCTCGAGGGTCGCGTCG-N15-A | 1/18 | – | Resection to CGCG with inaccurate fill-in |
| 34-mer | TCTCGAGGATCGCGTCG-N15-A | 1/18 | – | Resection to CGCG with inaccurate fill-in |
| 21-mer | TCTCG-N15-A | 1/18 | – | Resection to CG |
DISCUSSION
In mammalian cells, most DNA DSBs are repaired by NHEJ. Classical NHEJ acts rapidly to rejoin the vast majority of DNA DSBs in a process dependent on LIG4-XRCC4 and Ku and facilitated by DNA-PKcs. A subset of more complex radiation-induced DSBs that require end processing by Artemis are repaired more slowly by a mechanism that requires the core NHEJ factors and, in addition, ATM, H2AX, 53BP1 and the MRN complex (43). There is also considerable evidence for the existence of an alternative end joining mechanism that operates in the absence of the core NHEJ factors (16–19,67,68). This alternative error-prone end joining pathway operates with significantly slower kinetics than DNA-PK-dependent NHEJ and gives rise to deletions at the repair junctions. Since these deletions occur at sites of short sequence homologies (4–25 bp), this repair mechanism has been termed as MMEJ. The use of microhomology is a feature shared with both classical NHEJ and single-strand annealing (SSA), however, NHEJ typically uses shorter homologies (1–4 bp) than MMEJ, while SSA requires longer stretches of homologous sequence (≥30 bp) (20).
The MRN complex functions in a number of processes to facilitate DSB repair and cell survival, however, the importance of MRN for NHEJ remains unclear since conflicting evidence has been reported for a variety of experimental systems. In order to investigate any requirement for the MRN complex in NHEJ in X. laevis, we initially conducted a plasmid-based NHEJ assay (63) using X. laevis egg extracts immunodepleted for XMre11 or XNbs1. Southern blot analysis of the end joining products indicated that NHEJ is unaffected in extracts lacking XMRN activity. These data are in accordance with the results of Di Virgilio and Gautier, who used a similar plasmid-based system to test end joining of a range of different 5′- and 3′-termini generated using different combinations of restriction endonucleases. A colony formation assay combined with DNA sequencing of repair junctions indicated that the loss XMre11 activity had no effect on the efficiency or the accuracy of intra- and intermolecular NHEJ in X. laevis egg extracts (57).
However, using internally radio-labelled pSV56-plasmid substrates with defined 3′-OH or 3′-PG termini, we have shown that XMre11 does actually influence DNA end joining. Immunodepletion of XMre11 from X. laevis egg extracts resulted in an altered profile of end joining repair products, suggesting that intermolecular end joining is impaired in the absence of XMre11. In fact, using this assay, we see much higher levels of inter- rather than intramolecular end joining as compared with the Southern blot analysis of pUC19 end joining, which may be one reason that we are able to detect a difference in XMre11-depleted extract in this system. This increase in intermolecular repair may be due to the loss of the internal radiolabel in closed circular repair forms that have undergone a significant level of resection. It is also possible that the short regions of microhomology near to the break site in the linearized pSV56 substrate promote repair through a different mechanism that gives rise to more intermolecular products. It is interesting to note, however, that a study using fractionated mammalian cell extracts that did not support intramolecular end joining also showed a defect in intermolecular end joining when Rad50 activity was inhibited (45).
We were able to use these defined pSV56 templates to analyse the processing of DNA ends at the single nucleotide level to determine repair fidelity. The analysis of TaqαI-BstXI-digested NHEJ products on denaturing acrylamide gels distinguished two categories of end joining following incubation of either 3′-OH or 3′-PG substrate in X. laevis extract. An accurate form of NHEJ processed DNA ends without gain or loss of nucleotides, while inaccurate NHEJ, also called resection-based end joining, resulted from the ligation of DNA ends after nucleolytic resection. In this assay, XMre11-depleted egg extract supported an increased formation of the 42-nt oligomers but no longer sustained the formation of 34-mers, indicating that depletion of XMre11 promoted accurate end joining and abolished resection-based end joining. No specific requirement for XMre11 in the processing of 3′-PG ends was observed. Depletion of XNbs1 or XRad50, like XMre11-depletion, eliminated resection-based end joining and promoted accurate end joining of the pSV56 linear template, confirming that this effect is specific to the XMRN complex. We have not, thus far, been able to rescue the XMRN-dependent resection-mediated repair by the addition of purified recombinant human MRN to depleted egg extracts (data not shown). It is possible that depletion of the XMRN complex using any of the three different antibodies may have specifically co-depleted some additional XMRN-interacting factor required for resection-based end joining. One possible candidate is the CtIP that interacts directly with Nbs1, is required for HR-mediated DSB repair and has recently been shown to play a role in MMEJ in mammalian cells and chicken DT40 cells (27,69–71).
Having established that the MRN complex in X. laevis is required for an inaccurate resection-based mechanism of DNA end joining, we went on to further characterize this pathway. When we examined the timing of NHEJ using the 3′-OH pSV56 substrate, we found that resection-based repair is a late event relative to the accurate form of NHEJ, like the alternative end joining process described in mammalian cells. Moreover, although accurate NHEJ is dependent on the core NHEJ factors DNA-PKcs and Ku70, we have shown that XMRN-dependent resection-based repair is DNA-PK independent. Since the Ku-independent end joining pathways studied to date in mammalian cells are associated with deletions at sites of microhomology, we predicted that this would also be the case in X. laevis. Through a combination of restriction analysis and sequencing of the repair junctions to test for resection and annealing at a 4-bp region of microhomology close to the break site, we were able to confirm this. Our findings are, therefore, consistent with a role for the MRN complex in an alternative microhomology-dependent end joining pathway and not in classical NHEJ in X. laevis.
In fact, depletion of XMRN actually promoted Ku-dependent accurate end joining, suggesting some degree of competition between the two end joining mechanisms. Competition between Ku-dependent NHEJ and MMEJ has also been observed in S. cerevisiae using a nuclease-dead mre11 mutant that is unable to support MMEJ but stimulates Ku-dependent NHEJ (23), while in mammalian cells the suppression of error-prone MMEJ by recruitment of Ku and DNA-PKcs has been observed (19). Our findings lend further support to the view that DSB repair in vertebrates, in the absence of a homologous template, is a balance between NHEJ and MMEJ. Although MMEJ has been widely regarded as a ‘back up’ pathway for DSB repair when NHEJ fails, recent findings suggest that MMEJ is, in fact, a surprisingly robust process that contributes to DSB repair even when NHEJ is operative. Class switch recombination and V(D)J recombination proceed, to an unexpected degree, through an alternative microhomology-directed mechanism in the absence of core NHEJ proteins (72–75). Moreover, substantial MMEJ-directed V(D)J recombination was observed in cells with functional NHEJ machinery. The balance between error-prone MMEJ and classical NHEJ has important implications for carcinogenesis. Alternative end joining has been implicated in the increased incidence of chromosomal translocations seen in classical NHEJ-deficient cells (75,76). Moreover, error-prone microhomology-associated end joining has been shown to be the predominant mechanism for repair in human bladder tumours and may contribute to the increased genomic instability seen in bladder cancer (77).
In S. cerevisiae, MMEJ has been demonstrated to be independent of KU70 and RAD52 (40,78) but shares a number of factors in common with NHEJ and SSA, including the Rad1/Rad10 structure-specific endonuclease, Pol4, Sae2, Srs2, Nej1 and, most notably, the MRX complex (22,23). In mammalian cells, alternative end joining processes have thus far been shown to involve poly(ADP-ribose) polymerase-1 (PARP-1), the XRCC1-DNA ligase III complex, ligase I, FEN-1 and CtIP (24,25,27,79). It is not entirely clear, however, whether all these activities function in a single MMEJ pathway or whether several Ku-independent mechanisms may operate on different repair substrates or in different cell types. As an MMEJ factor, the MRN complex provides an attractive candidate since it can bind to DNA ends, promote their synapsis and effect 3′–5′ resection, pausing when a region of microhomology is detected (80). The small, but statistically significant, reduction in end joining in Nbs1-deficient human cells noted by Howlett and co-workers (81) for templates containing microhomologies of more than 4 nt indicated that the MRN complex may indeed play a role in MMEJ in higher eukaryotes (81). Moreover, recent data regarding alternative end joining of V(D)J recombination intermediates revealed that NBS1 is required for alternative end joining of hairpin-coding ends (28). Our evidence of a role for the MRN complex in resection-based end joining at a region of microhomology, in X. laevis cell-free extracts, confirms that MRN involvement in MMEJ is conserved in vertebrates and should provide a useful model for the analysis of other factors involved, template sequence requirements and the mechanistic details of MMEJ.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Medical Research Council, Biotechnology and Biological Sciences Research Council (grant BB\E015662/1); North West Cancer Research Fund (grant CR782); National Cancer Institute (grant USDHHS,CA40615). Funding for open access charge: Lancaster University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge Dr Chris Ford, Dr Simon Morley, Prof. Tony Carr and Dr Sarah Allinson for their invaluable support and assistance during this work.
REFERENCES
- 1.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 2.van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- 3.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell. Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 5.West SC. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell. Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 6.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair. 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 8.Dynan WS, Hoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 10.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 11.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 12.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 13.Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, Wang Z, Povirk LF. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J. Biol. Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 14.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 15.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Cheong N, Perrault AR, Wang H, Wachsberger P, Mammen P, Jackson I, Iliakis G. DNA-PK-independent rejoining of DNA double-strand breaks in human cell extracts in vitro. Int. J. Radiat. Biol. 1999;75:67–81. doi: 10.1080/095530099140825. [DOI] [PubMed] [Google Scholar]
- 17.Kabotyanski EB, Gomelsky L, Han JO, Stamato TD, Roth DB. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nevaldine B, Longo JA, Hahn PJ. The scid defect results in much slower repair of DNA double-strand breaks but not high levels of residual breaks. Radiat. Res. 1997;147:535–540. [PubMed] [Google Scholar]
- 19.Wang H, Perrault AR, Takeda Y, Qin W, Iliakis G. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res. 2003;31:5377–5388. doi: 10.1093/nar/gkg728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhfittig-Kulle S, Feldmann E, Odersky A, Kuliczkowska A, Goedecke W, Eggert A, Pfeiffer P. The mutagenic potential of non-homologous end joining in the absence of the NHEJ core factors Ku70/80, DNA-PKcs and XRCC4-LigIV. Mutagenesis. 2007;22:217–233. doi: 10.1093/mutage/gem007. [DOI] [PubMed] [Google Scholar]
- 21.McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176:2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 25.Liang L, Deng L, Chen Y, Li GC, Shao C, Tischfield JA. Modulation of DNA end joining by nuclear proteins. J. Biol. Chem. 2005;280:31442–31449. doi: 10.1074/jbc.M503776200. [DOI] [PubMed] [Google Scholar]
- 26.Ahmad A, Robinson AR, Duensing A, van Drunen E, Beverloo HB, Weisberg DB, Hasty P, Hoeijmakers JHJ, Niedernhofer LJ. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol. Cell. Biol. 2008;28:5082–5092. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deriano L, Stracker TH, Baker A, Petrini JHJ, Roth DB. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol. Cell. 2009;34:13–25. doi: 10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D’Amours D, Jackson SP. The MRE11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell. Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 30.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell. Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 31.Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair. 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 32.Difilippantonio S, Celeste A, Kruhlak MJ, Lee Y, Difilippantonio MJ, Feigenbaum L, Jackson SP, McKinnon PJ, Nussenzweig A. Distinct domains in Nbs1 regulate irradiation-induced checkpoints and apoptosis. J. Exp. Med. 2007;204:1003–1011. doi: 10.1084/jem.20070319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stracker TH, Morales M, Couto SS, Hussein H, Petrini JH. The carboxy terminus of NBS1 is required for induction of apoptosis by the MRE11 complex. Nature. 2007;447:218–221. doi: 10.1038/nature05740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao Y, Weaver DT. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo G, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A, Petrini JH. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl Acad. Sci. USA. 1999;96:7376–7381. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 2001;11:105–109. doi: 10.1016/s0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]
- 38.Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 39.Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanova E, Cooper PR, Nowak NJ, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 40.Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manolis KG, Nimmo ER, Hartsuiker E, Carr AM, Jeggo PA, Allshire RC. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J. 2001;20:1–12. doi: 10.1093/emboj/20.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Girard P-M, Foray N, Stumm M, Waugh A, Riballo E, Maser RS, Phillips WP, Petrini J, Arlett CF, Jeggo PA. Radiosensitivity in Nijmegen Breakage Syndrome cells is due to a repair defect and not cell cycle checkpoint defects. Cancer Res. 2000;60:4881–4888. [PubMed] [Google Scholar]
- 43.Riballo E, Kuhne M, Rief N, Doherty AJ, Smith GCM, Recio M-J, Reis C, Dahm K, Fricke A, Krempler A, et al. A pathway of double strand break rejoining dependent upon ATM, Artemis and proteins locating to γ-H2AX foci. Mol. Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 44.Huang J, Dynan WS. Reconstitution of the mammalian DNA double-strand break end-joining reaction reveals a requirement for an Mre11/Rad50/NBS1-containing fraction. Nucleic Acids Res. 2002;30:667–674. doi: 10.1093/nar/30.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong Q, Boyer TG, Chen PL, Lee WH. Deficient nonhomologous end-joining activity in cell-free extracts from Brca1-null fibroblasts. Cancer Res. 2002;62:3966–3970. [PubMed] [Google Scholar]
- 46.Lahdesmaki A, Taylor AM, Chrzanowska KH, Pan-Hammarstrom Q. Delineation of the role of the Mre11 complex in class switch recombination. J. Biol. Chem. 2004;279:16479–16487. doi: 10.1074/jbc.M312796200. [DOI] [PubMed] [Google Scholar]
- 47.Kracker S, Bergmann Y, Demuth I, Frappart PO, Hildebrand G, Christine R, Wang ZQ, Sperling K, Digweed M, Radbruch A. Nibrin functions in Ig class-switch recombination. Proc. Natl Acad. Sci. USA. 2005;102:1584–1589. doi: 10.1073/pnas.0409191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reina-San-Martin B, Nussenzweig MC, Nussenzweig A, Difilippantonio S. Genomic instability, endoreduplication, and diminished Ig class-switch recombination in B cells lacking Nbs1. Proc. Natl Acad. Sci. USA. 2005;102:1590–1595. doi: 10.1073/pnas.0406289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harfst E, Cooper S, Neubauer S, Distel L, Grawunder U. Normal V(D)J recombination in cells from patients with Nijmegen breakage syndrome. Mol. Immunol. 2000;37:915–929. doi: 10.1016/s0161-5890(01)00008-6. [DOI] [PubMed] [Google Scholar]
- 50.Yeo TC, Dong X, Sabath DE, Sperling K, Gatti RA, Concannon P, Willerford DM. V(D)J rearrangement in Nijmegen breakage syndrome. Mol. Immunol. 2000;37:1131–1139. doi: 10.1016/s0161-5890(01)00026-8. [DOI] [PubMed] [Google Scholar]
- 51.Yamaguchi-Iwai Y, Sonoda E, Sasaki MS, Morrison C, Haraguchi T, Hiraoka Y, Yamashita YM, Yagi T, Takata M, Price C, et al. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 1999;18:6619–6629. doi: 10.1093/emboj/18.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tauchi H, Kobayashi J, Morishima K, van Gent DC, Shiraishi T, Verkaik NS, vanHeems D, Ito E, Nakamura A, Sonoda E, et al. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature. 2002;420:93–98. doi: 10.1038/nature01125. [DOI] [PubMed] [Google Scholar]
- 53.Yang YG, Saidi A, Frappart PO, Min W, Barrucand C, Dumon-Jones V, Michelon J, Herceg Z, Wang ZQ. Conditional deletion of Nbs1 in murine cells reveals its role in branching repair pathways of DNA double-strand breaks. EMBO J. 2006;25:5527–5538. doi: 10.1038/sj.emboj.7601411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Labhart P. Ku-dependent nonhomologous DNA end joining in Xenopus egg extracts. Mol. Cell. Biol. 1999;19:2585–2593. doi: 10.1128/mcb.19.4.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pfeiffer P, Vielmetter W. Joining of nonhomologous DNA double strand breaks in vitro. Nucleic Acids Res. 1988;16:907–924. doi: 10.1093/nar/16.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gottlich B, Reichenberger S, Feldmann E, Pfeiffer P. Rejoining of DNA double-strand breaks in vitro by single-strand annealing. Eur. J. Biochem. 1998;258:387–395. doi: 10.1046/j.1432-1327.1998.2580387.x. [DOI] [PubMed] [Google Scholar]
- 57.Di Virgilio M, Gautier J. Repair of double-strand breaks by nonhomologous end joining in the absence of Mre11. J. Cell. Biol. 2005;171:765–771. doi: 10.1083/jcb.200506029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen S, Hannis JC, Flora JW, Muddiman DC, Charles K, Yu Y, Povirk LF. Homogeneous preparations of 3′-phosphoglycolate-terminated oligodeoxynucleotides from bleomycin-treated DNA as verified by electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Anal. Biochem. 2001;289:274–280. doi: 10.1006/abio.2000.4936. [DOI] [PubMed] [Google Scholar]
- 59.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell. 2001;8:137–147. doi: 10.1016/s1097-2765(01)00294-5. [DOI] [PubMed] [Google Scholar]
- 61.Felix MA, Pines J, Hunt T, Karsenti E. A post-ribosomal supernatant from activated Xenopus eggs that displays post-translationally regulated oscillation of its cdc2+ mitotic kinase activity. EMBO J. 1989;8:3059–3069. doi: 10.1002/j.1460-2075.1989.tb08457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen S, Inamdar KV, Pfeiffer P, Feldmann E, Hannah MF, Yu Y, Lee JW, Zhou T, Lees-Miller SP, Povirk LF. Accurate in vitro end joining of a DNA double strand break with partially cohesive 3′-overhangs and 3′-phosphoglycolate termini: effect of Ku on repair fidelity. J. Biol. Chem. 2001;276:24323–24330. doi: 10.1074/jbc.M010544200. [DOI] [PubMed] [Google Scholar]
- 63.Aoufouchi S, Patrick T, Lindsay HD, Shall S, Ford CC. Post-translational activation of non-homologous DNA end-joining in Xenopus oocyte extracts. Eur. J. Biochem. 1997;247:518–525. doi: 10.1111/j.1432-1033.1997.00518.x. [DOI] [PubMed] [Google Scholar]
- 64.Gu XY, Bennett RA, Povirk LF. End-joining of free radical-mediated DNA double-strand breaks in vitro is blocked by the kinase inhibitor wortmannin at a step preceding removal of damaged 3′ termini. J. Biol. Chem. 1996;271:19660–19663. doi: 10.1074/jbc.271.33.19660. [DOI] [PubMed] [Google Scholar]
- 65.Leahy JJJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, Smith GCM. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg. Med. Chem. Lett. 2004;14:6083–6087. doi: 10.1016/j.bmcl.2004.09.060. [DOI] [PubMed] [Google Scholar]
- 66.Calsou P, Frit P, Humbert O, Muller C, Chen DJ, Salles B. The DNA-dependent protein kinase catalytic activity regulates DNA end processing by means of Ku entry into DNA. J. Biol. Chem. 1999;274:7848–7856. doi: 10.1074/jbc.274.12.7848. [DOI] [PubMed] [Google Scholar]
- 67.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 68.Guirouilh-Barbat JE, Rass E, Plo I, Bertrand P, Lopez BS. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc. Natl Acad. Sci. USA. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen L, Nievera CJ, Lee AY-L, Wu X. Cell cycle-dependent complex formation of BRCA1{middle dot}CtIP{middle dot}MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 71.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong S-Y, Arnal S, Holub AJ, Weller GR, Pancake BA, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 73.Pan-Hammarstrom Q, Jones A-M, Lahdesmaki A, Zhou W, Gatti RA, Hammarstrom L, Gennery AR, Ehrenstein MR. Impact of DNA ligase IV on nonhomologous end joining pathways during class switch recombination in human cells. J. Exp. Med. 2005;201:189–194. doi: 10.1084/jem.20040772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Soulas-Sprauel P, Le Guyader G, Rivera-Munoz P, Abramowski V, Olivier-Martin C, Goujet-Zalc C, Charneau P, de Villartay J-P. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J. Exp. Med. 2007;204:1717–1727. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 76.Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 77.Bentley J, Diggle CP, Harnden P, Knowles MA, Kiltie AE. DNA double strand break repair in human bladder cancer is error prone and involves microhomology-associated end-joining. Nucleic Acids Res. 2004;32:5249–5259. doi: 10.1093/nar/gkh842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu X, Gabriel A. Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics. 2003;163:843–856. doi: 10.1093/genetics/163.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liang L, Deng L, Nguyen SC, Zhao X, Maulion CD, Shao C, Tischfield JA. Human DNA ligases I and III, but not ligase IV, are required for microhomology-mediated end joining of DNA double-strand breaks. Nucleic Acids Res. 2008;36:3297–3310. doi: 10.1093/nar/gkn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Paull TT, Gellert M. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc. Natl Acad. Sci. USA. 2000;97:6409–6414. doi: 10.1073/pnas.110144297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Howlett NG, Scuric Z, D’Andrea AD, Schiestl RH. Impaired DNA double strand break repair in cells from Nijmegen breakage syndrome patients. DNA Repair. 2006;5:251–257. doi: 10.1016/j.dnarep.2005.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.