Abstract
Translation termination in eukaryotes is governed by the concerted action of eRF1 and eRF3 factors. eRF1 recognizes the stop codon in the A site of the ribosome and promotes nascent peptide chain release, and the GTPase eRF3 facilitates this peptide release via its interaction with eRF1. In addition to its role in termination, eRF3 is involved in normal and nonsense-mediated mRNA decay through its association with cytoplasmic poly(A)-binding protein (PABP) via PAM2-1 and PAM2-2 motifs in the N-terminal domain of eRF3. We have studied complex formation between full-length eRF3 and its ligands (GDP, GTP, eRF1 and PABP) using isothermal titration calorimetry, demonstrating formation of the eRF1:eRF3:PABP:GTP complex. Analysis of the temperature dependence of eRF3 interactions with G nucleotides reveals major structural rearrangements accompanying formation of the eRF1:eRF3:GTP complex. This is in contrast to eRF1:eRF3:GDP complex formation, where no such rearrangements were detected. Thus, our results agree with the established active role of GTP in promoting translation termination. Through point mutagenesis of PAM2-1 and PAM2-2 motifs in eRF3, we demonstrate that PAM2-2, but not PAM2-1 is indispensible for eRF3:PABP complex formation.
INTRODUCTION
Translation termination occurs when a stop codon enters the ribosomal A site and signals for polypeptide chain release from the peptidyl-tRNA located in the ribosomal P site. In eukaryotes, this process is facilitated by two proteins: eRF1 (1) and eRF3 (2). eRF1 is the class I release factor that recognizes the stop codon in the A site and stimulates nascent peptide chain release. The class II release factor eRF3 is a GTPase, and it facilitates peptide release by eRF1.
Complex formation between eRF3 and eRF1 promotes GTP binding to eRF3 (3–6), with eRF1 acting as GTP Stabilizing Factor (GSF) (7). GTP hydrolysis on eRF3 is activated by the ribosome and eRF1, and is required for fast and efficient termination of translation in eukaryotes (8,9).
In addition to its role in termination, eRF3 is involved in normal and Nonsense-Mediated mRNA Decay (NMD) via two different pathways: through its association with cytoplasmic poly(A)-binding protein (PABP) (10–13) and Upf1 (14,15). The former interaction is considered as a regulator of PABP interactions with the 3′-poly(A) tail of mRNAs, suggesting that eRF3 may also play an important role in the degradation of mRNAs and/or the regulation of translation efficiency mediated through initiation factors (11,16). Our current understanding of the eRF3 GTPase cycle regulation via interactions with these factors is far from complete. Complex formation between eRF3 and PABP, as well as eRF3 and Upf1, is GTP/GDP insensitive, as opposed to the eRF1/eRF3 interaction (15). Quantitative data regarding eRF3 interactions with PABP are virtually absent, as well as any information regarding the interplay between eRF1, G nucleotides and PABP binding to eRF3.
Mammalian eRF3 is divided into at least two regions: the N- and C-terminal regions. The C-terminal domains are homologous to those of elongation factor EF1A, and both proteins are close relatives in the translational GTPase (trGTPase) superfamily (17). The C-terminal region of eRF3 is responsible for translation termination activity and is essential for viability (2,18). In contrast, the N-terminal domain shows little conservation (17,19,20) and is dispensable for the termination process. However, it has been demonstrated to be important for binding to PABP (10–13) as well as several other factors, e.g. Sla1 (21) and Itt1 (22). Interaction with PABP, which is the focus of the present study, is mediated by a PAM2 sequence motif in the eRF3 N-terminal domain (23,24).
Importantly, the majority of the available biochemical data on the eRF3 GTPase cycle were obtained using an eRF3 variant lacking the N-domain (3–6), which could potentially lead to severe artifacts. Based on the available X-ray structure of the N-terminally deleted eRF3 (25), it has been suggested that the N-domain may block the eRF1-binding site thus potentially regulating eRF1 and GTP binding (26). Preparation of full-length eRF3 using an Escherichia coli over-expression system has never been achieved, although the truncated version of the factor was first purified almost 15 years ago (2). Over-expression in insect cells was successful, but the yields obtained precluded detailed biochemical investigations (27). Thus, rigorous biochemical investigations of the eRF1:eRF3:PABP:G nucleotides interaction network have so far been impossible.
In order to overcome this problem, we constructed an over-expression construct based on a modern protein expression vector pETM-20, which allowed over-expression and purification of the full-length eRF3. We have for the first time studied complex formation between full-length eRF3 and its ligands (GDP, GTP, eRF1 and PABP) using Isothermal Titration Calorimetry (ITC) and investigated the temperature dependence of these interactions. We have revealed the structural rearrangements in the interacting partners via thorough thermodynamic analysis. Through sequence analysis of the eRF3 N-domain, we demonstrate that the two overlapping PAM2 minidomains in human eRF3 (23) that are responsible for the eRF1–PABP complex formation, are present in a wide distribution of metazoa. Using point mutagenesis we studied the role of both of the motifs (PAM2-1 and PAM2-2) in eRF3:PABP interaction, and showed that PAM2-2, but not PAM2-1, as was suggested earlier (28), is indispensible for complex formation. This provides a tool to dissect the roles of eRF3 in translation termination and NMD.
MATERIALS AND METHODS
Cloning and mutagenesis of human eRF3a
The full-length cDNA encoding eRF3a (GSPT1, sequence corresponding to NCBI GI number 119605536) was fused with the 109aa Trx•Tag™ thioredoxin protein followed by 6His for affinity purification. The mutagenesis procedure was performed according to the PCR-based ‘megaprimer’ method (29). Two rounds of PCR were performed with the same PCR mixture. For all the mutants, the direct primer 5′-ATCCCATGGATCCGGGCAGTGGC-3′ (the NcoI site underlined) together with one of the reverse primers for these mutants were used in the first round of PCR. In the second round the ‘megaprimer’ synthesized in the first round served as the direct primer together with the reverse primer 5′-GCGCTCGAGTTAGTCTTTCTCTGGAACCAGTTTC-3′ (XhoI site underlined). The resulting 1914 bp PCR product was purified in 1% agarose gel using a DNA extraction kit (GE Healthcare), hydrolyzed with NcoI and XhoI, ligated with pETM-20 plasmid and treated with the same endonucleases. The ligated mixture was used for the transformation of E. coli, strain Top10. The cloned DNAs were sequenced and appropriate clones were used for the expression of the mutant eRF3a variants.
Bioinformatics
eRF3 sequences from a wide sample of eukaryotes were retrieved from trGTPbase, a database of translational GTPases (GCA, http://www.GTPbase.org.uk). Multiple sequence alignment was carried out with MAFFT v6.234b with strategy L-INS-I (30). The data set was reduced to a representative set of sequences that showed homology in the N-terminal domain in the region of the PAM2 minidomain. The alignment was adjusted by eye, using Bioedit, http://www.mbio.ncsu.edu/BioEdit/bioedit.html (31). Consensus sequences were computed with the Python program ConsensusFinder (GCA).
Protein expression and purification
Human 6His eRF1 was expressed and purified according to (8) with minor modifications.
For preparation of the full-length (FL) human eRF3 BL21 cells were transformed with the pETM-20 eRF3FL plasmid and grown in 2xLB, supplemented with 100 mg/ml ampicillin at 37°C until the OD reached 0.6 (A600 nm). Cells were then transferred to ice for 30 min, and induced with IPTG at a final concentration of 0.25 mM. An additional 50 mg/ml ampicillin was added and induced cells were grown at 20°C for 18 h and then harvested.
Cells were lyzed by sonication in the buffer containing 70 mM Tris pH 8.0, 10% glycerol, 30 mM KCl, 1% Triton X100, 1 mM beta-mercaptoethanol (BME), 1 mM PMSF, 1 g/l lyzosyme and 0.2 g/l DNAse A. Cell debris was removed by centrifugation at 18 krpm for 20 min, two times, and cleared lyzate was loaded onto a 5 ml NiNTA FF column (GE Healthcare). Bound column was washed with wash buffer containing 50 mM Tris pH 7.5, 10% glycerol, 1 mM BME and consecutively decreasing amounts of KCl: 2 M (20 CV), 0.8 M (5 CV) and 0.1 M (5 CV). The protein was step-eluted with the wash buffer containing 0.1 M KCl and 100 mM imidazole.
Eluted protein was further purified on a 5 ml Resource Q FF column (GE Healthcare) using a linear gradient (0.1–0.8 M KCl) with buffer containing 70 mM Tris pH 8.0, 10% glycerol, 1 mM BME. The eRF3 peak was collected and dialyzed overnight with TEV protease added to eRF3 in 1:25 ratio against the storage buffer containing 10% glycerol, 1 mM DTT, 25 mM potassium phosphate (pH 7.4), 0.1 M KCl and 2 mM MgCl2. The TEV protease and the cleaved off Trx tag were removed by incubation with NiNTA–agarose (Qiagen), and purified protein was used for the experiments directly without freezing.
Human 6His PABP was over-expressed and purified in the same way as 6His eRF3, except for the absence of the TEV protease treatment.
ITC
The thermodynamic parameters of eRF3 binding to eRF1, G nucleotides and PABP were measured using a MicroCal VP-ITC and AutoITC instruments (MicroCal, Northampton, MA) as previously described (5). Experiments were carried out in phosphate buffer (25 mM K2HPO4, 10% glycerol, 1 mM DTT and 0.1 M KCl), at pH 7.5. Ligands of 15-µl aliquots were injected from a 296-µl syringe into the 1.42-ml cell containing protein solution to achieve a complete binding isotherm. Protein concentration in the cell ranged from 4 to 15 μM and ligand concentration in the syringe ranged from 40 to 300 μM. The heat of dilution was measured by injecting the ligand into the buffer solution or by additional injections of ligand after saturation; the values obtained were subtracted from the heat of reaction to obtain the effective heat of binding. The resulting titration curves were fitted using MicroCal Origin software, assuming one set of sites. Affinity constants (Ka), binding stoichiometry and enthalpy variations (ΔH) were determined by a non-linear regression fitting procedure. Consequently, the Gibbs energy (ΔG) and the entropy variations (ΔS) were calculated from the relation ΔG = −RT ln Ka = ΔH − TΔS.
To investigate protonation effects on eRF3 interactions with eRF1, experiments were performed in two buffers, phosphate (see above) and Tris (50 mM Tris–HCl, 10% glycerol, 5 mM β-mercaptoethanol and 0.15 M KCl) with different ionization enthalpy (1 versus 11 kcal/mol) as described (5).
RESULTS
Preparation of full-length human eRF3
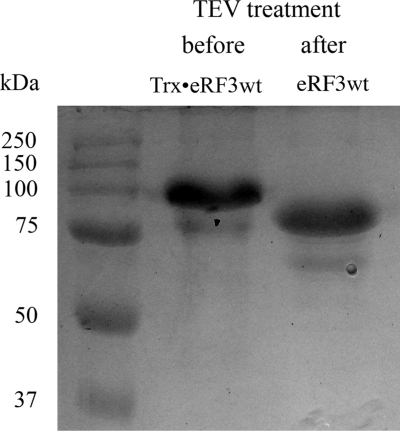
Numerous previously unsuccessful attempts of over-expression of the full-length eRF3 without stabilizing fusion proteins were undertaken by us and others, all being foiled by low levels of soluble protein. Therefore we took another approach, overexpressing eRF3 as a fusion protein in pETM-20 vector (32) (EMBL, Heidelberg, Germany) with subsequent removal of the thioredoxin + 6His tag by TEV protease cleavage (see Materials and methods section for details). The thioredoxin fusion considerably improved protein stability and solubility and was crucial for full-length eRF3 over-expression. TEV cleavage was specific and resulted in the production of a single band at apparent molecular weight of about 85 kDa (Figure 1), similarly to eRF3 over-expressed in insect cells using viral vectors (27).
Figure 1.
SDS–PAGE analysis of the full-length human eRF3 preparation. The protein was initially expressed as a fusion with a thioredoxin + 6His tag (Trx•eRF3wt), which was specifically cleaved off by TEV protease, resulting in full-length eRF3wt.
Effect of N-domain of eRF3 on binding to eRF1, GDP and GTP
In our previous work, we determined the thermodynamic parameters of interactions of an N-domain deleted variant of eRF3 (eRF3Cp) with eRF1, GDP and GTP (3–5). However, the effects of the N-domain on eRF3 GTPase function were not investigated. Here we used isothermal titration calorimetry to measure the affinities to G nucleotides and eRF1 for full-length eRF3.
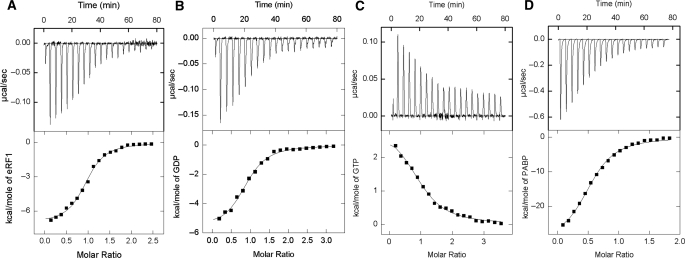
The results are summarized in Tables 1 and 2, and a typical set of ITC data for binding of eRF1 and GDP to eRF3 and GTP to eRF3:eRF1 complex at 25°C is shown in Figure 2A–C. The binding curves were fitted assuming one set of sites. Our quantitative results demonstrate that interactions among eRF1, eRF3 and G nucleotides are weakly affected by the presence of the N-domain. For free eRF3, we observed somewhat increased (four times) affinity to GDP in comparison with the eRF3Cp variant, and for the eRF3:eRF1 complex, affinities to GTP (Kd = 0.6 µM) and GDP (Kd = 1.1 µM) changed only slightly.
Table 1.
Thermodynamic parameters of eRF3 binding to eRF1, GDP, GTP and PABP at 25°C, pH 7.5 determined by isothermal titration calorimetrya
| Sample | Ligand | Kа × 10−5b (M−1) | Kdc (µM) | ΔHd (kcal/mol) | TΔS (kcal/mol) | ΔG (kcal/mol) |
|---|---|---|---|---|---|---|
| eRF3 | GDP | 20 (5.6) | 0.5 (1.9) | −5.7 (−9.2) | 2.9 (−1.4) | −8.6 (−7.8) |
| eRF3 | eRF1 | 44 (14) | 0.2 (0.7) | −6.1 (−7.2)e | 3.0 (1.2) | −9.1 (−8.4) |
| eRF3:GDP | eRF1 | 11 (49) | 0.9 (0.2) | −10.8 (−3.1)e | −2.6 (6.0) | −8.2 (−9.1) |
| eRF1:eRF3 | GDP | 9.3 (5.1) | 1.1 (2.0) | −5.8 (−11.8) | 2.3 (−4.0) | −8.1 (−7.8) |
| eRF1:eRF3 | GTP | 18 (20) | 0.6 (0.5) | 3.0 (−2.2) | 11.5 (6.4) | −8.5 (−8.6) |
| eRF3f | PABP | 14 | 0.7 | −30.3 | −21.9 | −8.4 |
| eRF1:eRF3 | PABP | 14 | 0.7 | −26.3 | −17.9 | −8.4 |
| eRF3:PABP | eRF1 | 32 | 0.3 | −4.6 | 4.3 | −8.9 |
| eRF1:eRF3:PABP | GTP | 16 | 0.6 | 3.2 | 11.7 | −8.5 |
| eRF3:PABP | GDP | 15 | 0.6 | −5.9 | 2.5 | −8.4 |
| eRF1:eRF3:PABP | GDP | 14 | 0.6 | −4.6 | 3.8 | −8.4 |
| eRF3(KAKA)f | PABP | 11 | 0.9 | −9.2 | −0.9 | −8.3 |
| eRF3(AAK) | PABP | ndg | ||||
| eRF3(AAKAA) | PABP | nd |
aIn the brackets are the data for eRF3Cp, according to ref. (5). All measurements were performed in phosphate buffer (25 mM K2HPO4, 10% glycerol, 1 mM DTT and 0.1 M KCl).
bThe standard deviation did not exceed ±20%.
cCalculated as 1/Kа.
dThe standard deviation did not exceed ±8%.
eΔH was calculated taking into account the effect of protonation (see ‘Materials and Methods’ section).
fStoichiometry of the interaction is 1 PABP to 2 eRF3 molecules, and in all other cases it is close to unity.
gnd—not detected.
Table 2.
Thermodynamic parameters of eRF3 binding to eRF1, GDP and GTP at different temperatures, pH 7.5 determined by isothermal titration calorimetrya
| Sample | Ligand | T (°C) | Kа × 10−5 (M−1) | Kd (µM) | ΔH (kcal/mol) | TΔS (kcal/mol) | ΔG (kcal/mol) |
|---|---|---|---|---|---|---|---|
| eRF3 | GDP | 4 | 23 | 0.44 | −4.9 | 3.16 | −8.06 |
| 10 | 22 | 0.46 | −5.2 | 3.01 | −8.21 | ||
| 15 | 23 | 0.44 | −5.46 | 2.92 | −8.38 | ||
| 25 | 20 | 0.50 | −6.03 | 2.57 | −8.59 | ||
| 30 | 19 | 0.53 | −6.3 | 2.40 | −8.70 | ||
| 37 | 20 | 0.50 | −6.72 | 2.22 | −8.94 | ||
| eRF3 | eRF1 | 4 | 21 | 0.48 | −0.97 | 7.04 | −8.01 |
| 10 | 23 | 0.44 | −2.9 | 5.34 | −8.24 | ||
| 15 | 27 | 0.37 | −3.92 | 4.56 | −8.48 | ||
| 25 | 44 | 0.23 | −7.1 | 1.96 | −9.06 | ||
| 30 | 51 | 0.20 | −8.1 | 1.20 | −9.30 | ||
| 37 | 93 | 0.11 | −10.4 | −0.52 | −9.88 | ||
| eRF1:eRF3 | GDP | 4 | 30 | 0.33 | −3.96 | 4.24 | −8.20 |
| 10 | 20 | 0.50 | −4.46 | 3.70 | −8.16 | ||
| 15 | 12 | 0.83 | −4.97 | 3.05 | −8.02 | ||
| 25 | 9.3 | 1.1 | −5.83 | 2.31 | −8.14 | ||
| 30 | 9.1 | 1.1 | −6.44 | 1.82 | −8.26 | ||
| 37 | 7.4 | 1.4 | −7.10 | 1.22 | −8.32 | ||
| eRF1:eRF3 | GTP | 4 | 5.1 | 2.0 | 31.3 | 38.53 | −7.23 |
| 10 | 9.8 | 1.0 | 24.4 | 32.16 | −7.76 | ||
| 15 | 20 | 0.5 | 17.38 | 25.68 | −8.30 | ||
| 25 | 19 | 0.52 | 3 | 11.56 | −8.56 | ||
| 30 | 20 | 0.5 | −7.16 | 1.58 | 8.74 | ||
| 37 | 19 | 0.52 | −16 | −7.09 | −8.91 |
aSymbols, abbreviations and all other details are given in the Table 1 footnote.
Figure 2.
ITC titration curves (upper panel) and binding isotherms (lower panel) for eRF3 interaction with eRF1 (A), GDP (B) and PABP (D) and eRF3:eRF1 complex interaction with GTP (C) at 25°C in phosphate buffer (pH 7.5).
Enthalpic and entropic partitions of the interaction energetics were similar for full length eRF3 and the Cp variant only in the case of eRF1 binding, and differed considerably in the case of GTP and GDP binding to the eRF1:eRF3 complex (Table 1). In this case, enthalpic contribution to overall free-energy diminishes while the enthropic contribution increases (Table 1), compensating for the enthalpic change and resulting in virtually unperturbed ΔG (or equilibrium constant). Thus, despite the fact that the N-terminal region of eRF3 is not essential for G-nucleotide binding, it dramatically changes the thermodynamic profile of the interaction. Changes in the binding enthalpy and entropy in opposite directions upon system perturbation, such as deletion of a domain in the case of eRF3, are common for biological systems and are known as ‘enthalpy–entropy compensation’ (33).
Effect of temperature on eRF3 binding to GDP, GTP and eRF1
The binding of GDP, GTP and eRF1 to eRF3 was studied at six different temperatures from 4 to 37°C, and the results are summarized in Tables 2 and 3.
Table 3.
Heat capacity changes and solvent-accessible surface area for eRF3 binding to eRF1, GDP and GTPa
| Sample | Ligand |
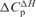 (cal mol−1 K−1) (cal mol−1 K−1) |
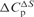 (cal mol−1 K−1) (cal mol−1 K−1) |
SAAminb (Å2) | SAAmaxb (Å2) |
|---|---|---|---|---|---|
| eRF3 | GDP | −55 | −39 | 137.5 | 203 |
| eRF3 | eRF1 | −281 | −237 | 702.5 | 1040 |
| eRF3Cp | eRF1 | −270 | −235 | 675 | 1000 |
| eRF1:eRF3 | GDP | −96 | −100 | 240 | 355.5 |
| eRF1:eRF3 | GTP | −1470 | −1480 | 3675 | 5444 |
| eRF1:eRF3Cp | GTP | −850 | −805 | 2125 | 3148 |
aHeat capacity changes were obtained as d(ΔH)/dT and d(ΔS)/d(lnT).
bChanges in SAA were estimated using the following formula: ΔCp = 0.27ΔAaromatic + 0.4ΔAnon-aromatic, where ΔAaromatic and ΔAnon-aromatic are the protected areas due to aromatic and non-aromatic amino acids in Å2, respectively (36). SAAmin and SAAmax are calculated assuming that all the changes are conferred by non-aromatic and aromatic residues, respectively.
There is almost no effect of the temperature on the Kd-value for GDP binding to eRF3, similarly to what we observed earlier for another trGTPase, EF-G (34). On the other hand, affinity of eRF3 to GDP in the eRF1:eRF3 complex decreased 4-fold when the temperature increased from 4°C (Kd = 0.33 µM) to 37°C (Kd = 1.4 µM). The Kd-value for GTP binding to the eRF1:·eRF3 complex, in contrast, decreased 4-fold when the temperature increased from 4°C (Kd = 2 µM) to 15°C (Kd = 0.5 µM), after which the change in the affinity with temperature was small (Table 2). The Kd-value for eRF1 binding to eRF3 increased 4.5-fold from 0.48 µM (4°C) to 0.11 µM (37°C).
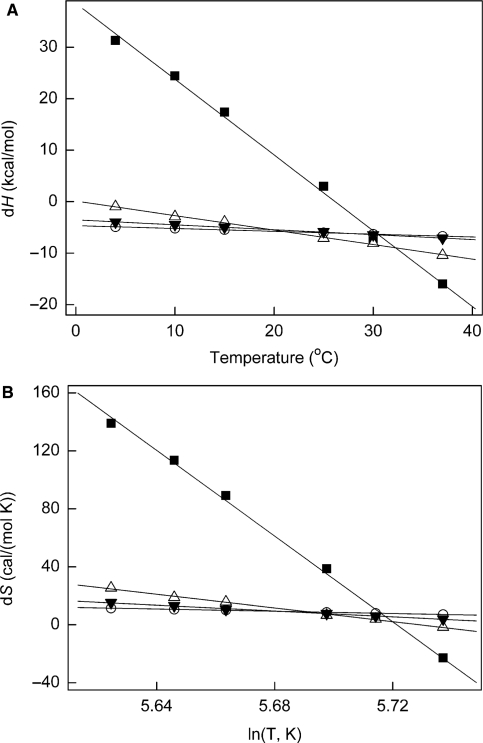
Plotting the enthalpy of the interaction versus temperature, or entropy versus the logarithm of temperature, we obtained heat capacity change (ΔCp) values for eRF3 interactions with G nucleotides and eRF1 both for the full-length eRF3 and for the N-terminally truncated Cp variant (Figure 3, Table 3). The results are similar for the two eRF3 versions: binding of eRF1 was characterized by ΔCp of −281 and −270 cal × mol−1 K−1 for eRF3 and eRF3Cp, respectively, and GTP binding to eRF1:eRF3 complex was characterized by an immense ΔCp of −1470 and −850 cal × mol−1 K−1 for eRF3 and eRF3Cp, respectively. GDP binding both to eRF3 alone and eRF1:eRF3 complex had low ΔCp of −55 and −96 cal × mol−1 K−1, respectively.
Figure 3.
Temperature dependence of entropy and enthalpy of eRF3 interactions with eRF1 (triangles) and GDP (circles) and eRF3:eRF1 complex with GDP (inverted triangle) and GTP (square). (A) Enthalpy of binding as a function of the temperature (°C) at pH 7.5. (B) Entropy of binding as functions of the logarithm of the temperature (K) at pH 7.5. Experimental errors bars were not exceeding the size of the symbols, thus are not presented on the graph.
Changes in the heat capacity are believed to reflect the change in the solvent accessible area (SAA) during the process (35), and a wealth of available experimental data has been analyzed providing an empirical formula connecting the change in ΔCp and the change in solvent accessible area: ΔCp = 0.27ΔAaromatic + 0.4ΔAnon-aromatic, where ΔAaromatic and ΔAnon-aromatic are the protected areas due to aromatic and non-aromatic amino acids in Å2, respectively (36). This formula was used to translate the ΔCp values into the SAA change estimates (Table 3).
Bioinformatic analysis of the eRF3 N-domain
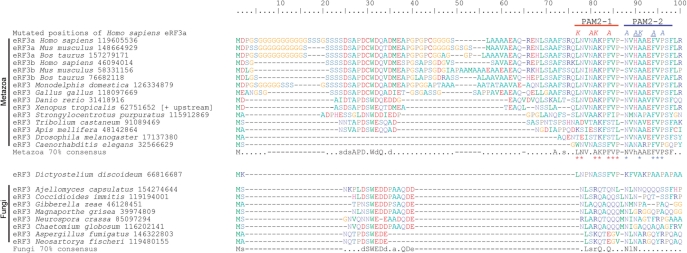
The eRF3 N-domain was excluded from previous comparative sequence analyses due to its high variability in sequence and length in both eRF3 and its close relative Hbs1p (17,19,20). However, a survey of the distribution of the Pfam HMM domain PAM2 (37) in the Swiss-Prot/TrEMBL databases suggests that within the largely unconserved N-domain, this short (15 amino acids) minidomain is conserved in eRF3 from a wide distribution of metazoa (24).
eRF3 has been found to contain two overlapping PAM2 minidomains (PAM2-1 and PAM2-2) in humans (23). However, it is not known how widespread these dual minidomains are. Therefore, we have investigated the distribution of both PAM2 minidomains of eRF3 across a broad taxonomic range. The two C-terminal domains of eRF3 (C and G) are closely related to those of other eukaryotic trGTPases Hbs1p, eEF1A and Ski7p. However, PAM2-like motifs are only found in eRF3. To investigate conservation of the PAM2 minidomains in eRF3, sequences from a wide sample of eukaryotes were aligned and reduced to a representative data set that showed homology in the PAM2 region of the N-terminal domain. Seventy percent consensus sequences were generated for two subsets of this data set: a metazoan subset and a fungal subset (Figure 4).
Figure 4.
Consensus and example sequence alignment of the extreme N-terminus of eRF3. Organism names are followed by NCBI GI numbers. The sequence from Xenopus tropicalis was extended at the N-terminus by 15 amino acids, relative to the predicted protein product in Genbank following the discovery of an alternate upstream start codon (mRNA GI number 62751651). The location of the PAM2-1 and PAM2-2 adjacent motifs are indicated above the alignment. Seventy percent conservation consensus sequences were calculated using the Python script ConsensusFinder (GCA). Uppercase letters indicate amino acids conserved in >70% of all examined sequences, and lowercase letters indicate a common amino acid substitution group conserved in >70% of the sequences. A ‘.’ denotes a position that is not conserved in sequence, and gaps are denoted by ‘-’. Asterisks below the Metazoa consensus sequence show residues that match the PAM2 motif, as represented in Pfam. The sites that were mutated are indicated above the human sequence, with the letters indicating the amino acids present in the mutant forms. Red mutated postions: the PAM2-1 KAKA mutant; Blue positions: the PAM2-2 AAKAA mutant; Blue underlined positions: the PAM2-2 AKA mutant.
Within eRF3, we have found two adjacent conserved PAM2 motifs across all sampled metazoans, with the possible exception of insects, where PAM2-1 appears unconserved (Tribolium castaneum, Apis mellifera and Drosophila melanogaster). The metazoan consensus motifs for PAM2-1 and PAM2-2 are repectively, ‘LNV.AKPFVP’ and ‘NVhAAEFVPSF’, where uppercase letters indicate amino acid conservation of over 70% (Figure 3). At deeper levels than Metazoa, conservation becomes ambiguous. The predicted eRF3 protein of the choanoflagellate Monosiga brevicollis lacks the N-domain, however, conceptual translation of the genomic sequence (NCBI GI number 163776760) identifies a miss-annotated start codon that extends the predicted protein by 80 residues (not shown). This extension shows no clear homology to other eRF3s, however it does contain a ‘LNV’ tripeptide, which may correspond to the ‘LNV’ of the PAM2-1 consensus. PAM2-1 also appears to be present in the amoebozoan Dictyostelium discoidium, with six of the nine conserved PAM2-1 residues present in the corresponding region in this organism (Figure 4). While homology in the PAM2 region appears to be present in some fungi, only the asparagines of PAM2-1/2 residues are specifically conserved. This region of possible homology could not be identified in Saccharomyces cerevisiae, suggesting that in this organism PABP binding to eRF3 is mediated by an alternative molecular interface.
The alignment has revealed other interesting conserved features of the extreme N-terminus of eRF3. Runs of Glycine and Serine are common in mammalian eRF3 (Figure 3). In organisms with both eRF3a and eRF3b, the extreme N-terminal Glycine runs are predominately in the eRF3a form. Additionally, a conserved patch with consensus ‘sdsAPD.WdQ.d’ is present in many metazoans, with a possibly homologous region in fungi with consensus ‘dSWEDd.a.QDe’ (Figure 4). As with PAM2, this conserved patch could not be identified in S. cerevisiae. The specific functions of these additional N-terminal regions are currently unknown.
Interactions of eRF3 with PABP
Using the full-length eRF3 variant we quantitatively characterized eRF3 interactions with PABP using ITC (Figure 2D, Table 1). Our data suggest that complex formation between eRF3 and PABP does not affect eRF3 interactions with G nucleotides and eRF1 (Table 1), both in the case of PABP binding to eRF3 and to the eRF1:eRF3 complex. The Cp variant lacking the N-domain was unable to bind PABP (data not shown). Current results are in contradiction with our previous report (3), where we suggested possible inhibitory effect of PABP on GTP binding by eRF3. However, in the previous work no systematic analysis was done, which necessitated the current study. This data suggest that eRF3Cp can serve as a valid model for investigation of the eRF3:eRF1:GTP interactions in the framework of termination mechanisms, but is not valid for studying effects associated with PABP binding.
In order to map the eRF3:PABP interaction, we performed mutagenesis of the conserved residues in PAM2-1 and PAM2-2 motifs. We constructed three mutants, namely a KAKA mutant disrupting the PAM2-1 motif and AKA and AAKAA mutants disrupting the PAM2-2 motif (Figure 4). The KAKA mutant was created earlier and was claimed to disrupt the eRF3:PABP interaction as judged by western blot (28). Analysis of the mutant variants in the current study demonstrates that the interaction between PABP and both PAM2-2 mutants (AKA and AAKAA) was non-detectable, suggesting absolute necessity of the AKA motif for the interaction.
However, the PAM2-1 KAKA mutant retained somewhat compromised affinity to PABP (Table 1). The equilibrium dissociation constant remained almost unaltered (0.7 µM for eRF3 wt versus 0.9 µM for the KAKA mutant), but the energetics of the interaction were altered. In the case of the wild type, protein interaction is characterized by enthalpy of −30 kcal/mol, whereas the KAKA mutant has three times lower enthalpy of −9 kcal/mol. This more labile interaction might account for the failure to detect it using western blotting in (28).
DISCUSSION
Structural rearrangements in eukaryotic termination
Formation of the ternary complex eRF1:eRF3:GTP and its interactions with an array of binding partners such as the pretermination complex and protein factors Upf1 and PABP is a pivotal event in the convergence of translation termination and NMD pathways. Our understanding of the framework of interactions among the above-mentioned components is critically dependent on determination of the network of possible mutually exclusive complexes and interconvertion pathways among these, backed up by structural information regarding the complexes.
Currently we benefit from structural insights into both eRF1 and eRF3 from X-ray (25,38) and SAXS (39,40) studies. The X-ray structure of full-length eRF1 complexed with eRF3 lacking the G-domain has also been determined (40). Taken together, the structural data suggest that in the eRF3:eRF1 complex, eRF1 assumes a bent conformation. However, available data on the complex are either of low resolution (SAXS) or obtained for the eRF3 lacking the G-domain, and it is the G-domain (and GTP binding) that is crucial for the function of the complex.
Biochemical data also suggest functionally important structural flexibility of eRF1 in the eRF1:eRF3 complex. First, the existence of two different conformations was suggested by observed activation of peptide release upon GTP hydrolysis on ribosome-bound eRF1:eRF3:GTP (9). Differences in peptide release activation by GTP hydrolysis on eRF3 with mutant eRF1 variants on different stop codons indicate that the abovementioned conformational changes might vary with each codon (41). Also, the existence of alternative eRF1 conformations was suggested in ref. (4), where interactions between separate domains of eRF1 with eRF3 were studied by ITC.
Our thermodynamical investigations of eRF3/eRF1/G nucleotide interactions are also indicative of structural rearrangements upon formation of the ternary complex (see Figure 5 for a proposed model). One of the parameters of the interactions we measured is change in heat capacity, ΔCp, which is believed to reflect the degree of structural rearrangement (35). Heat capacity changes upon GDP binding both to eRF3 and eRF1:eRF3 are relatively small (−55 and −96 cal × mol−1 K−1, respectively), similar to what was observed for another trGTPase we studied recently, EF-G (−20 cal × mol−1 K−1) (7). This is expected, since GDP binding should not promote structural rearrangements in the GTPase (42). Formation of the eRF1:eRF3 complex has moderate ΔCp (−281 cal × mol−1 K−1), although this is significantly higher than what was observed for GDP binding. According to X-ray data, the complex formation between truncated eRF3 and eRF1 buries 974 Å2 (40), which is close to our estimates of 702–1040 Å2 corresponding to ΔCp of −281 cal × mol−1 K−1 (Table 3). Complex formation between N-terminally deleted eRF3Cp and eRF1 has similar ΔCp of −270 cal ×mol−1 K−1, suggesting that the N-domain does not interfere with the formation of the complex, as was suggested in ref. (26).
Figure 5.
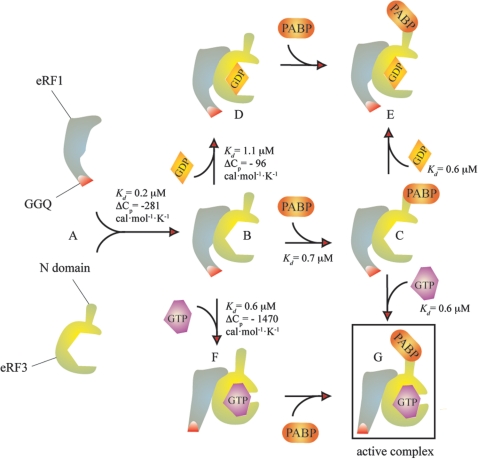
Proposed model for eRF1:eRF3:PABP:G nucleotides interactions. eRF1 and eRF3 form a tight heterocomplex [(A and B), Kd = 0.2 µM]. Formation of the complex is characterized by moderate ΔCp change which gives a SAA estimate close to the experimental value [974 Å2 (40), versus estimated 702–1040 Å2]. Binding of GDP to eRF1:eRF3 is characterized by a low ΔCp value, suggesting minor structural rearrangements (B–D), whereas binding of GTP (B–F) is characterized by high ΔCp, suggesting major rearrangements (Table 3, Figure 3). Binding of PABP to the eRF1:eRF3 complex does not interfere with GTP and GDP binding (C–E, C–G versus B–D, B–F, Table 1).
In the case of GTP binding to eRF1:eRF3, the ΔCp change is colossal (−1480 cal × mol−1 K−1, Table 3), suggesting major structural rearrangements with estimated changes is SAA ranging from 3675 to 5444 Å2 which indicates an active role for GTP. This role has previously been demonstrated in eRF1:eRF3-mediated termination (9) and structural rearrangements in the eRF1:eRF3 complex have been proposed to take place during the transition termination event (4,41).
The available X-ray reconstructions of eRF1:eRF3 were done with the complex lacking the G-domain and SAXS investigations were never carried only in the presence of GTP (40). Thus, more detailed structural investigations are required for clarification of the mechanistic details of the GTP-induced rearmaments in eRF1:eRF3 complex. Importantly, large-scale rearrangements are likely to be hindered by the crystal packing, thus the soak-in approach is likely to result in an artifactual structure not representative of the solution conformation of the complex. Therefore investigation by solution methods is necessary.
PABP interactions with termination factors
Binding of PABP to both eRF3 and the eRF1:eRF3 complex had no effect on GTP binding, suggesting that in the presence of PABP formation of the eRF1:eRF3:GTP complex is unperturbed (see Figure 5 for a proposed model). GTP binding and hydrolysis by eRF3 is necessary for efficient translation termination (9). PABP has also been reported to have an activating role in termination (11,16), and our results demonstrate that both of the activation signals (GTP and PABP binding) are compatible.
The interaction between eRF3 and PABP is characterized by a low stoichiometry of PABP binding to eRF3 (0.50 ± 0.06 PABP per eRF3). In the presence of eRF1, the stoichiometry increases reaching 0.71 ± 0.05 PABP per eRF3, suggesting possibility that the simultaneous presence of eRF1, eRF3 and PABP drives the system towards formation of the eRF1:eRF3:PABP complex. This effect of PABP, i.e. stimulation of formation of the termination-competent eRF1:eRF3:PABP:GTP complex could be the corollary of PABP’s activating role in translation termination (11,16). However, one can not rule out the possibility that this change in the stoichiometry could be insignificant and reflect alterations in the experimental system.
Despite having a conserved role in binding PABP and other proteins, the N-domain is not universally conserved in sequence across eukaryotes (17). However, among groups of closely related organisms, regions of homology are apparent. This includes the PAM2 minidomain, which we show is present across a wide distribution of metazoa, where it is comprised of 2 PAM2 motifs (Figure 4). The motifs are short and well-studied, with x-ray structure (43), as well as calorimetric analysis of PAM2 oligopeptide binding to PABP (23) available. At 25°C, the peptide mimicking the two PAM2 motifs of eRF3 binds to PABP with Kd 1.6 µM, ΔH − 16 kcal/mol and TΔS 8.73 kcal/mol, which is similar to our results for the full-length eRF3 (Kd 0.7 µM, ΔH − 30.3 kcal/mol and TΔS − 21.9 kcal/mol).
In silico analysis of the PAM2 minidomain identified this motif in many proteins, including eRF3 from several metazoan species (24). Our approach focused on the architecture of this minidomain in eRF3, considering a much larger representation of eukaryotes. In mammals, there are two genes for eRF3, which differ in their N-terminal domains (44). These are referred to as eRF3a (or GSPT1) and eRF3b (GSPT2). It is not clear how the cellular roles of these versions differ. Human eRF3a and eRF3b are both functional in translation termination (45), and mouse eRF3b can fully compensate for the effects of silenced eRF3a on termination (46). However, the two versions differ in tissue distribution and expression, with eRF3a seemingly being the major factor in most cells as it has the most widespread and abundant expression (46). We find that both eRF3a and eRF3b carry the PAM2-1 and PAM2-2 minidomains and therefore both appear capable of PABP binding (Figure 4).
We identified conserved residues in the adjacent PAM2-1 and PAM2-2 motifs in eRF3 (Figure 4), and tested the importance of conserved residues by mutagenesis. We created the PAM2-1 KAKA mutant which was earlier reported to abolish the eRF3 binding to PABP (28), as well as two mutants in the PAM2-2 motif. Unexpectedly, the PAM2-1 KAKA mutant of eRF3 was able to bind PABP, albeit with somewhat lower affinity and dramatically different binding energetics (Table 1), while both PAM2-2 mutants were completely deficient in PABP binding. This suggests that in human eRF3, PAM2-2 is the primary site of PABP interaction. However, it is possible that the primary binding site is different in different organisms. In insects, only PAM2-2 is conserved (Figure 4), while only PAM2-1 appears to be present in the amoebozoan D. discoidium.
The interaction of eRF3 and PABP is the second of two decision-making interactions that destine mRNA for NMD, the first being the eRF3 interaction with the Exon-Junction Complex [for review see refs. (47,48)]. Our detection of a motif in eRF3 indispensable for PABP interaction (the AKA site of PAM2-2) provides a tool for dissection of the connection between eRF3 activities in translation termination and PABP-mediated NMD pathways, using a point mutation in the PAM2-2 motif to enable discrimination between the two NMD pathways: the one mediated by EJC and the one mediated by eRF3:PABP interactions.
FUNDING
Presidium of the Russian Academy of Sciences (Program Molecular and Cell Biology to A.A.M. and L.Yu.F.); the Russian Foundation for Basic Research (08-04-00375a to L.Yu.F.); Estonian Science Foundation grants (6768 to T.T. and 7616 to V.H.); Grant of the President of the Russian Federation for young scientists (MK-162.2009.4 to V.A.M.); and European Regional Development Fund through the Center of Excellence in Chemical Biology (to V.H. and T.T.). Funding for open access charge: Personal grant from European Regional Development Fund through the Center of Excellence in Chemical Biology (to V.H.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to Dr Hueseyin Besir, Head of Protein Expression and Purification Core Facility, EMBL, Heidelberg, Germany for providing pETM-20 vector and to Lev Ovchinnikov for the generous gift of PAPB overexpressing construct.
REFERENCES
- 1.Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M, et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- 2.Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hauryliuk V, Zavialov A, Kisselev L, Ehrenberg M. Class-1 release factor eRF1 promotes GTP binding by class-2 release factor eRF3. Biochimie. 2006;88:747–757. doi: 10.1016/j.biochi.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Kononenko AV, Mitkevich VA, Dubovaya VI, Kolosov PM, Makarov AA, Kisselev LL. Role of the individual domains of translation termination factor eRF1 in GTP binding to eRF3. Proteins. 2008;70:388–393. doi: 10.1002/prot.21544. [DOI] [PubMed] [Google Scholar]
- 5.Mitkevich VA, Kononenko AV, Petrushanko IY, Yanvarev DV, Makarov AA, Kisselev LL. Termination of translation in eukaryotes is mediated by the quaternary eRF1*eRF3*GTP*Mg2+ complex. The biological roles of eRF3 and prokaryotic RF3 are profoundly distinct. Nucleic Acids Res. 2006;34:3947–3954. doi: 10.1093/nar/gkl549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pisareva VP, Pisarev AV, Hellen CU, Rodnina MV, Pestova TV. Kinetic analysis of interaction of eukaryotic release factor 3 with guanine nucleotides. J. Biol. Chem. 2006;281:40224–40235. doi: 10.1074/jbc.M607461200. [DOI] [PubMed] [Google Scholar]
- 7.Hauryliuk V, Hansson S, Ehrenberg M. Co-factor dependent conformational switching of GTPases. Biophys. J. 2008;95:1704–1715. doi: 10.1529/biophysj.107.127290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2:334–341. [PMC free article] [PubMed] [Google Scholar]
- 9.Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 10.Cosson B, Couturier A, Chabelskaya S, Kiktev D, Inge-Vechtomov S, Philippe M, Zhouravleva G. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol. Cell Biol. 2002;22:3301–3315. doi: 10.1128/MCB.22.10.3301-3315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J. Biol. Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- 12.Inge-Vechtomov S, Zhouravleva G, Philippe M. Eukaryotic release factors (eRFs) history. Biol. Cell. 2003;95:195–209. doi: 10.1016/s0248-4900(03)00035-2. [DOI] [PubMed] [Google Scholar]
- 13.Uchida N, Hoshino S, Imataka H, Sonenberg N, Katada T. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J. Biol. Chem. 2002;277:50286–50292. doi: 10.1074/jbc.M203029200. [DOI] [PubMed] [Google Scholar]
- 14.Hosoda N, Kobayashi T, Uchida N, Funakoshi Y, Kikuchi Y, Hoshino S, Katada T. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 2003;278:38287–38291. doi: 10.1074/jbc.C300300200. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi T, Funakoshi Y, Hoshino S, Katada T. The GTP-binding release factor eRF3 as a key mediator coupling translation termination to mRNA decay. J. Biol. Chem. 2004;279:45693–45700. doi: 10.1074/jbc.M405163200. [DOI] [PubMed] [Google Scholar]
- 16.Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atkinson GC, Baldauf SL, Hauryliuk V. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol. Biol. 2008;8:290. doi: 10.1186/1471-2148-8-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, Inge-Vechtomov SG, Smirnov VN. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol. Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 19.Inagaki Y, Blouin C, Susko E, Roger AJ. Assessing functional divergence in EF-1alpha and its paralogs in eukaryotes and archaebacteria. Nucleic Acids Res. 2003;31:4227–4237. doi: 10.1093/nar/gkg440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inagaki Y, Doolittle WF. Evolution of the eukaryotic translation termination system: origins of release factors. Mol. Biol. Evol. 2000;17:882–889. doi: 10.1093/oxfordjournals.molbev.a026368. [DOI] [PubMed] [Google Scholar]
- 21.Bailleul PA, Newnam GP, Steenbergen JN, Chernoff YO. Genetic study of interactions between the cytoskeletal assembly protein Sla1 and prion-forming domain of the release factor Sup35 (eRF3) in Saccharomyces cerevisiae. Genetics. 1999;153:81–94. doi: 10.1093/genetics/153.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urakov VN, Valouev IA, Lewitin EI, Paushkin SV, Kosorukov VS, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Itt1p, a novel protein inhibiting translation termination in Saccharomyces cerevisiae. BMC Mol. Biol. 2001;2:9. doi: 10.1186/1471-2199-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozlov G, De Crescenzo G, Lim NS, Siddiqui N, Fantus D, Kahvejian A, Trempe JF, Elias D, Ekiel I, Sonenberg N, et al. Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase. EMBO J. 2004;23:272–281. doi: 10.1038/sj.emboj.7600048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albrecht M, Lengauer T. Survey on the PABC recognition motif PAM2. Biochem. Biophys. Res. Commun. 2004;316:129–138. doi: 10.1016/j.bbrc.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 25.Kong C, Ito K, Walsh MA, Wada M, Liu Y, Kumar S, Barford D, Nakamura Y, Song H. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol. Cell. 2004;14:233–245. doi: 10.1016/s1097-2765(04)00206-0. [DOI] [PubMed] [Google Scholar]
- 26.Kodama H, Ito K, Nakamura Y. The role of N-terminal domain of translational release factor eRF3 for the control of functionality and stability in S. cerevisiae. Genes Cells. 2007;12:639–650. doi: 10.1111/j.1365-2443.2007.01082.x. [DOI] [PubMed] [Google Scholar]
- 27.Frolova LY, Simonsen JL, Merkulova TI, Litvinov DY, Martensen PM, Rechinsky VO, Camonis JH, Kisselev LL, Justesen J. Functional expression of eukaryotic polypeptide chain release factors 1 and 3 by means of baculovirus/insect cells and complex formation between the factors. Eur. J. Biochem. 1998;256:36–44. doi: 10.1046/j.1432-1327.1998.2560036.x. [DOI] [PubMed] [Google Scholar]
- 28.Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. Plos Biol. 2008;6:860–871. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen B-Y, Janes HW, editors. PCR cloning protocols. 2nd edn. Totowa, NJ: Humana Press; 2002. [Google Scholar]
- 30.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall T. BioEdit. Biological sequence alignment editor for Windows. 1998. http://www.mbio.ncsu.edu/BioEdit/bioedit.html.
- 32.Dummler A, Lawrence AM, de Marco A. Simplified screening for the detection of soluble fusion constructs expressed in E. coli using a modular set of vectors. Microb. Cell Fact. 2005;4:34. doi: 10.1186/1475-2859-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eftink MR, Anusiem AC, Biltonen RL. Enthalpy-entropy compensation and heat capacity changes for protein-ligand interactions: general thermodynamic models and data for the binding of nucleotides to ribonuclease A. Biochemistry. 1983;22:3884–3896. doi: 10.1021/bi00285a025. [DOI] [PubMed] [Google Scholar]
- 34.Hauryliuk V, Mitkevich VA, Eliseeva NA, Petrushanko IY, Ehrenberg M, Makarov AA. The pretranslocation ribosome is targeted by GTP-bound EF-G in partially activated form. Proc. Natl Acad. Sci. USA. 2008;105:15678–15683. doi: 10.1073/pnas.0807912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jelesarov I, Bosshard HR. Isothermal titration calorimetry and differential scanning calorimetry as complementary tools to investigate the energetics of biomolecular recognition. J. Mol. Recogn. 1999;12:3–18. doi: 10.1002/(SICI)1099-1352(199901/02)12:1<3::AID-JMR441>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 36.Connelly PR, Thomson JA. Heat capacity changes and hydrophobic interactions in the binding of FK506 and rapamycin to the FK506 binding protein. Proc. Natl Acad. Sci. USA. 1992;89:4781–4785. doi: 10.1073/pnas.89.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 39.Kononenko AV, Dembo KA, Kiselev LL, Volkov VV. Molecular morphology of eukaryotic class I translation termination factor eRF1 in solution. Mol. Biol. (Mosk) 2004;38:303–311. [PubMed] [Google Scholar]
- 40.Cheng Z, Saito K, Pisarev AV, Wada M, Pisareva VP, Pestova TV, Gajda M, Round A, Kong C, Lim M, et al. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 2009;23:1106–1118. doi: 10.1101/gad.1770109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan-Minogue H, Du M, Pisarev AV, Kallmeyer AK, Salas-Marco J, Keeling KM, Thompson SR, Pestova TV, Bedwell DM. Distinct eRF3 requirements suggest alternate eRF1 conformations mediate peptide release during eukaryotic translation termination. Mol. Cell. 2008;30:599–609. doi: 10.1016/j.molcel.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sprang SR. G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 43.Kozlov G, Trempe JF, Khaleghpour K, Kahvejian A, Ekiel I, Gehring K. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc. Natl Acad. Sci. USA. 2001;98:4409–4413. doi: 10.1073/pnas.071024998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoshino S, Imai M, Mizutani M, Kikuchi Y, Hanaoka F, Ui M, Katada T. Molecular cloning of a novel member of the eukaryotic polypeptide chain-releasing factors (eRF). Its identification as eRF3 interacting with eRF1. J. Biol. Chem. 1998;273:22254–22259. doi: 10.1074/jbc.273.35.22254. [DOI] [PubMed] [Google Scholar]
- 45.Jakobsen CG, Segaard TM, Jean-Jean O, Frolova L, Justesen J. Identification of a novel termination release factor eRF3b expressing the eRF3 activity in vitro and in vivo. Mol. Biol. 2001;35:672–681. [PubMed] [Google Scholar]
- 46.Chauvin C, Salhi S, Le Goff C, Viranaicken W, Diop D, Jean-Jean O. Involvement of human release factors eRF3a and eRF3b in translation termination and regulation of the termination complex formation. Mol. Cell Biol. 2005;25:5801–5811. doi: 10.1128/MCB.25.14.5801-5811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muhlemann O. Recognition of nonsense mRNA: towards a unified model. Biochem. Soc. Trans. 2008;36:497–501. doi: 10.1042/BST0360497. [DOI] [PubMed] [Google Scholar]
- 48.Muhlemann O, Eberle AB, Stalder L, Zamudio Orozco R. Recognition and elimination of nonsense mRNA. Biochim. Biophys. Acta. 2008;1779:538–549. doi: 10.1016/j.bbagrm.2008.06.012. [DOI] [PubMed] [Google Scholar]