Abstract
Small interfering RNAs (siRNAs) are short, double-stranded RNAs that use the endogenous RNAi pathway to mediate gene silencing. Phosphorylation facilitates loading of a siRNA into the Ago2 complex and subsequent cleavage of the target mRNA. In this study, 2′, 3′ seco nucleoside modifications, which contain an acylic ribose ring and are commonly called unlocked nucleic acids (UNAs), were evaluated at all positions along the guide strand of a siRNA targeting apolipoprotein B (ApoB). UNA modifications at positions 1, 2 and 3 were detrimental to siRNA activity. UNAs at positions 1 and 2 prevented phosphorylation by Clp1 kinase, abrogated binding to Ago2, and impaired Ago2-mediated cleavage of the mRNA target. The addition of a 5′-terminal phosphate to siRNA containing a position 1 UNA restored ApoB mRNA silencing, Ago2 binding, and Ago2 mediated cleavage activity. Position 1 UNA modified siRNA containing a 5′-terminal phosphate exhibited a partial restoration of siRNA silencing activity in vivo. These data reveal the complexity of interpreting the effects of chemical modification on siRNA activity, and exemplify the importance of using multiple biochemical, cell-based and in vivo assays to rationally design chemically modified siRNA destined for therapeutic use.
INTRODUCTION
RNA interference (RNAi) is a mechanism of posttranslational gene silencing where double-stranded RNAs (dsRNAs), such as a small interfering RNAs (siRNAs), induce degradation of sequence specific homologous mRNA (1). siRNAs are RNA duplexes of 19–21 nucleotides and are widely used for functional genetic or cellular screens (2–4). RNAi-based therapy is also being developed whereby a synthetic siRNA can be used to enter the endogenous RNAi pathway and eliminate a gene of interest, making it an attractive way to therapeutically target genes outside of the small molecule druggable genome (5,6). In order for a siRNA to be effective upon entering the cell, the siRNA must become phosphorylated on the 5′-end by Clp1 kinase and be incorporated into the endogenous RNA-induced silencing complex (RISC), which consists of Ago2, Dicer, and TRBP (2,7–10). During this assembly, one of the two strands of the siRNA, known as the passenger strand, is cleaved, whereby the other strand, referred to as the guide strand, binds to the Argonaute protein, Ago2 (11–13). The guide strand in RISC associates with the complementary mRNA strand, cleaves the mRNA, and thereby regulates gene expression (14).
Phosphorylation of the 5′-end of siRNA is one of the first steps required for it to function in RISC (15,16). Recently, Clp1 was identified as a mammalian RNA kinase and shown to be responsible for the 5′-end phosphorylation of siRNAs (7). Specifically, when cell lysates were immunodepleted of Clp1, exogenous siRNA lacking a 5′-phosphate could no longer stimulate cleavage of its target mRNA. In addition, modified siRNAs that had attenuated silencing potency were found to have increased potency after synthetically applying a 5′-end phosphate to their guide strand (17–20). These studies emphasize the critical role of Clp1-mediated phosphorylation on the ability of a siRNA to function in the cell. Identifying the parameters of Clp1 substrate specificity such as positional base preference, nucleoside modification tolerance or siRNA length will be essential for optimizing siRNAs as therapeutics. Similarly, understanding the efficiency of phosphorylation of a siRNA and how that relates to Ago2 binding and mRNA degradation will also improve this optimization process.
Many of the chemical modifications that are currently tolerated in siRNAs reside in the 2′ position on the ribose ring. 2′-methoxy modifications at position 2 on the guide strand can reduce off target effects and, when on either the passenger or guide strand, reduce immune responses (21,22). Other modifications such as 2′-fluoro or 2′-deoxy can increase potency, serum stability and/or half life while reducing immunostimulatory responses (23,24). 2′-O,4′-C-methylene bicyclonucleosides, also known as locked nucleic acids (LNAs), increase structural stability and prolong serum half life by having an additional linkage between the 2′ and 4′ carbons, forcing the ribose sugar to maintain a C3′-endo pucker which is found in A-form helices such as RNA (25). However, many of these modifications have not been evaluated for their impact on 5′-phosphorylation or on siRNA strand loading into Ago2 when placed near the 5′-end of the guide strand. Understanding how modifications to a siRNA can impact these steps will be important for the rational design of chemically optimized siRNA therapeutics.
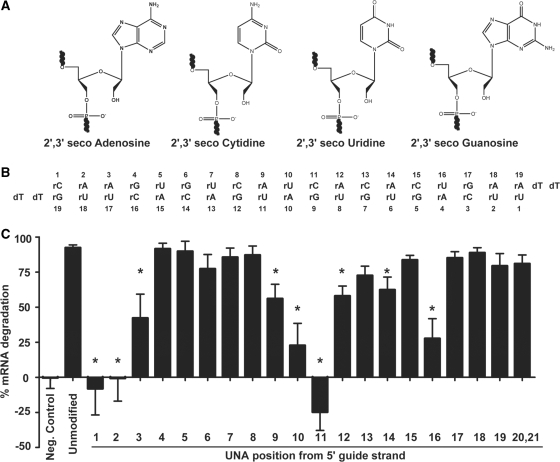
2′,3′ Seco nucleosides, also known as unlocked nucleic acids (UNA’s), are a class of modifications whereby the ribose ring becomes acyclic by removing the bond between the 2′ and 3′ carbon atoms (26–29) (Figure 1A). This change in sugar structure increases the nucleic acid analog flexibility, and, when incorporated into duplexes, can reduce duplex stability as observed by lower thermal denaturation temperatures (26,28,30–32). Recently, UNAs were incorporated into the guide strand of siRNAs designed against GFP at positions 3, 6, 12 and 18 and found to have target specific mRNA degradation (32).
Figure 1.
UNAs at position 1, 2 and 3 of the guide strand impair mRNA degradation. (A) Structure of the nucleotides used for oligonucleotide synthesis. (B) ApoB siRNA sequence used in this study. (C) Percent mRNA degradation of ApoB mRNA as determined by qPCR. Unmodified refers to the ApoB siRNA sequence containing only ribonucleotides and dTdT overhangs. The negative control is a scrambled siRNA sequence. Individual UNAs were substituted at the indicated position on the guide strand of the duplex. A paired t-test was conducted to evaluate UNA modifications to the unmodified control. The asterisk indicates a significant change in mRNA degradation from unmodified siRNA (P < 0.001).
Here, we evaluate siRNA duplexes that were individually modified at every position in the guide strand with UNAs. We found that modified positions 1–3, 9–12, 14 and 16 had decreased siRNA-mediated mRNA degradation in cells when compared to an unmodified siRNA. UNA modifications at positions 1, 2, and 3 of the guide strand were particularly detrimental to target mRNA degradation. UNA modified siRNAs at position 1 or 2 were also unable to be phosphorylated by Clp1 kinase, exhibited decreased binding to Ago2 and impaired Ago2 cleavage of the mRNA target. Cell based mRNA degradation and Ago2 binding activity of the UNA modified siRNA at position 1 were successfully recovered by the chemical addition of a 5′-phosphate on the UNA modified siRNA. When tested in mice, the 5′-terminal phosphate was also able to recover the activity of the position 1 UNA modified siRNA.
MATERIALS AND METHODS
Oligo synthesis and sequence
All UNA containing oligos were synthesized at RiboTask (Denmark). The oligo is derived from the ApoB gene at positions 10 162–10 182 (5′-UUCAGUGUUGAUGACACUUG-3′ guide strand) with dTdT overhangs on the 3′-ends of both the guide and passenger strands. Negative control siRNA for the in vitro studies is (5′-GUAUGACCGACUACGCGUA-3′-passenger strand).
Transfection and qPCR
Hepa1-6 cells were cultured in Dulbecco’s Modified Eagle Medium (Mediatech Cellgro) containing 10% serum. Cells were plated in a 96-well plate (3500 cells/well) and were transfected 24 h after plating in Opti-MEM I Reduced Serum Media (Gibco) and Lipofectamine RNAiMax reagent (Invitrogen) for a final concentration of 10 nM siRNA for the initial screening. For IC50 curves the final concentration ranged from 0.15 pM to 160 nM along a 12-point titration curve. Approximately 24 h after transfection, cells were washed with phosphate-buffered saline and lysed in Cells-to-CT Lysis Buffer (Ambion) with rDNase I (RNase-Free) added. Stop Solution (Ambion) was used to halt the reaction. RT–PCR was performed using 7 µl of cell lysate in 2X RT Buffer with 20X RT Enzyme Mix (Ambion) added. Conditions were as follows: 37°C for 60 min and 95°C for 5 min. ApoB mRNA levels were detected by quantitative PCR with ApoB-specific probes from Applied Biosystems. All cDNA samples were added to a 10 µl reaction volume with the following cycling conditions: 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. qPCR was assayed using an ABI Prism 7900 sequence detector using 2x Taqman Gene Expression Master Mix (Applied Biosystems). GAPDH mRNA levels were used for data normalization.
Melting temperature determination
Melting temperature (Tm) of siRNA duplexes was measured using SYBR Green I (Invitrogen) and a 7500 Real-time PCR system with SDS software version 2.0.1 (Applied Biosystems). A 25 µl reaction containing 5 µM siRNA, 5X SYBR Green stain and 50% PBS was run using the 7500 instrument. The siRNA-SYBR mixture was denatured at 99.9°C for 30 s then subjected to a 10% ramp to 30°C for 8 min. Samples were then heated in 0.4°C steps with 30 s holds from 30°C to a final temperature of 99.9°C. The SDS software performed the melting curve analysis and Tm.
Ago2 binding
HA-FLAG-hAgo2 was over-expressed in the Flp-In T-Rex 293 cell line from Invitrogen. Cells were grown as specified by the manufacturer. Protein expression was induced for 24 h with 1 µg/ml tetracycline. An equivalent amount of cells were grown similarly but without induction of protein expression. After 24 h of protein expression, the media was aspirated and the cells were incubated with lysis buffer consisting of 50 mM KCl, 20 mM HEPES pH 7.5, 0.5 mM Tris(2-carboxyethyl)phosphine (TCEP), 0.05% NP-40 and 1X protease inhibitor cocktail (Sigma-Aldrich), for 30 min at room temperature with gentle agitation. Lysate was pooled and spun for 15 min at 25 000g. The supernatant was immediately flash frozen on dry ice and stored at −80°C until needed. 10 µl of 0.1 μM siRNA was added to 130 µl of thawed cell lysate (2 mg/ml) with 1.5 µl of RNase inhibitor (Applied Biosystems). Samples were incubated at 37°C for 4 h. A total of 100 µl of the siRNA/cell lysate was transferred to an Anti-FLAG High Sensitivity, M2 coated 96-well plate from Sigma-Aldrich. The plate was incubated overnight with gentle agitation at 4°C. The next day the wells are washed with Tris-buffered saline (TBS) and the siRNA eluted with 30 µl of Cells-to-Ct lysis buffer. The resulting siRNA was quantified by Taqman PCR. The amount bound was calculated as the Ct resulting from the experimental sample subtracted from that of the negative control.
Ago2 cleavage
A total of 0.4 µl of 10 µM single strand siRNA was added to a reaction containing 1.5 μM purified hAgo2, 1X cleavage buffer (20 mM HEPES pH 7.5, 60 mM NaCl and 5 mM MgCl2) and 20 U of RNase inhibitor (Ambion) in a final volume of 20 μl. The reaction was incubated at 37°C for 30 min. The target mRNA (Cy3-AUUUCUCCUUUAAAUCAAGUGUCAUCACACUGAAUACCAAUGCUGAACUU) was heated to 90°C for 1 min then quick cooled on ice for three minutes in order to eliminate any secondary structure. A 0.6 µl of the prepared target was added to the reaction after the 30 min incubation and allowed to incubate at 37°C for an additional 90 min. The reaction was quenched with 20 µl of cleavage loading dye (40 mM EDTA, 0.2% SDS, 2 mg/ml ProteinaseK and 1.78X TBE–urea sample buffer (Invitrogen). The samples were then heated to 90°C for 90 s and quickly cooled on iced to eliminate any secondary structure. 20 µl of each sample was loaded onto a 10-well, 15% TBE-urea gel (Invitrogen) and run at 180V for ∼1 h. The gel was rinsed in water and visualized on a Typhoon 8600 (GE Healthcare) with the Cy3 fluorescence settings. The bands were quantitated using ImageQuant software and the graph expresses the percent cleavage of the total.
Clp1 purification
The glutathione S-transferase (GST) tagged Clp1 (NM_006831) fusion plasmid was generated by recombining full length Clp1 into pDEST15 (Invitrogen), which was then transformed into the Rossetta strain of Escherichia coli. (Novagen) following the manufacturers instructions. A modified procedure was used to induce the production of the GST-Clp1 fusion protein (7). In brief, 500 ml of plasmid transformed E. coli in Luria broth containing ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) were induced with 0.4 mM isopropyl β-d-thiogalactosidase (IPTG) at 37°C for 2 h. The bacteria induced to express Clp1 were then collected by centrifugation at 10 000 r.p.m. for 20 min at 4°C.
The cell pellet was resuspended in 35 ml of extraction buffer [PBS at pH 7.4, containing 137 mM NaCl, 27 mM KCl, 5 mM MgCl2, 2.5 mM DDT, 0.1 mM 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), and 0.1% Triton X-100]. The cell suspension was kept on ice and disrupted using the Sonic Dismembrator (Fisher, model 500) by 6 consecutive 15-s pulses with 1 min pause. The cell lysate was centrifuged at 4°C for 20 min at 12 000 r.p.m. and filtered through 0.22 µm filter. Affinity purification of Clp1 was performed by chromatograph using GSTrap column on AKTApurifier system (GE) following the manufacturers instructions with some modification. Briefly, a GSTrap 5 ml column containing Glutathione-Sepharose Fast Flow beads were equilibrated with 25 ml of binding buffer (extraction buffer without Triton X-100). The 35 ml of filtered cell lysate was loaded on a column followed by washing with 25 ml binding buffer. The column was then eluted with 50 mM Tris–HCl (pH 8.5) buffer containing 20 mM reduced glutathione, 100 mM NaCl, 5 mM MgCl2, 0.1 mM AEBSF and 2.5 mM DTT. Fractions containing Clp1 were pooled and concentrated using 10 KDa Centriprep centrifugal filter units (Milipore). The concentrated protein was then exchanged into kinase assay buffer (50 mM Tris–HCl, pH 8.0 containing 50 mM KCl, 5 mM MgCl2, 5 mM DTT). GST-Clp1 recombinant protein was analyzed by SDS-PAGE gel. The identity of Clp1 was further confirmed by gel band protein ID on a Mass Spectrometer (Proteomics). Protein sample concentration was determined by UV on a NanoDrop-8000.
Clp1 assay
The siRNA-kinase activity assay was performed in 40 µl of 50 mM Tris–HCl (pH 8.0) reaction mix, containing 10 µg of recombinant GST–Clp1, 50 mM KCl, 5 mM MgCl2, 5 mM DTT, 1 mM ATP, 0.2 mM GTP and 1 μM siRNA, followed by incubation at 30°C for 1 h. The reaction was stopped by heating at 95°C for 5 min. The samples were then run directly on the mass spectrometer. The percent phosphorylation was derived from the ratio of phosphorylated to non-phosphorylated of the passenger or guide oligonucleotide strand.
Mass spectrometry
All samples were run on a LC/MS system comprised of an Applied Biosystems MDS Sciex API 4000 mass spectrometer (Concord, Ontario, Canada) equipped with a Turbo Ion Spray source and operating in the negative-ion mode, two Shimadzu LC-20AD UFLC pumps (Colombia, MD, USA), a Leap Technologies HTS PAL autosampler (Carrboro, NC, USA) and a column heater (Phenomenex, Tustin, CA, USA) operated at 70°C. Data acquisition was performed with ABI/Sciex Analyst software, version 1.5 and zero-charge state molecular weights were determined by deconvolution using the BioAnalyst software module. Ion-pair reversed phase HPLC chromatographic separations were performed on an XBridge OST C18 column (Waters, Milford, MA, USA) 50 mm × 2.1 mm, 2.5 µm particle size at a flow rate of 200 µl/min. Mobile phase A consisted of 1.7 mM TEA and 100 mM HFIP (pH 7.5) in water, and mobile phase B was methanol. A 20 µl injection of each sample was loaded onto the column and separated using the following elution gradient: 3% B for 1 min, 3% to 25% B over 3 min, 95% B for 1 min followed by initial conditions for 2 min for column re-equilibration.
Modeling methodology
The 3.0-Å structure of Argonaute from Thermus thermophilus with bound DNA guide strand and RNA passenger strand (PDB code 3F73) was aligned with the human Ago2 sequence (GenBank accession number NP_036286.2). Alignment was performed with MOE software (Chemical Computing Group, Montreal, Canada) and modified manually on observed and predicted secondary structure. A homology model was built using MOE, with the duplex oligonucleotides present during model construction. The structure of the human PIWI domain (1SI3) was grafted onto the model by aligning the linking residues Ile346-Val347 and Pro229-Val230, excising the model PIWI domain, and attaching the PIWI domain from 1SI3. The crude chimeric structure was cleaned up by rebuilding a full-length Ago2 homology model using the chimera as template. Next, the DNA guide strand was converted into RNA, and the complex was minimized with the protein heavy atoms fixed, the oligonucleotide heavy atoms constrained with a flat-bottomed potential of 0.5 A width, and all hydrogen atoms unrestrained.
5′RACE
5′-RACE was performed using a GeneRacer Kit (Invitrogen). Total RNA was obtained from livers of mice injected with 6 mg/kg siRNA and sacrificed at 24 h. GeneRacer RNA Oligo was ligated to the 5′-ends of 3–5 µg of total RNA. The RNA was then precipitated and reverse transcribed into cDNA using a gene specific primer, 5′-CTTGTGATCAATGCCTCCTGTTGC-3′, according to the GeneRacer Kit protocol. Two microliters of cDNA was used for PCR with GoTaq Colorless Master Mix (Promega), 1 µl of the gene-specific primer, and 3 µl of GeneRacer 5′ Primer. The following cycling parameters were used: 2 min at 95°C, 35 cycles of 30 s at 95°C, 1 min at 55°C, 1 min at 72°C, and a final 7 min at 72°C. A second nested PCR was performed to aid in the identifying of real RACE products. 2 µl of the pervious PCR reaction was used with GoTaq Colorless Master Mix (Promega), 1 µl of a second gene-specific primer, 5′-GAGAGACAGCTGTGGCTAGTTTCAAT-3′, and 1 µl of GeneRacer 5′ Nested Primer. Cycling parameters were kept the same but only 25 cycles were performed. Samples were run on a 3% agarose gel with ethidium bromide and imaged.
siRNA formulation
Lipid nanoparticles (LNPs) were made using the cationic lipid CLinDMA (2-{4-[(3b)-cholest-5-en-3-yloxy]-butoxy}-N,N-dimethyl-3-[(9Z,12Z)-octadeca-9,12-dien-1-yloxy]propan-1-amine), cholesterol, and PEG-DMG (monomethoxy(polyethyleneglycol)-1,2-dimyristoylglycerol) in 50.3:44.3:5.4 molar ratio as previously described (33). Briefly, siRNAs were incorporated in the LNPs with high encapsulation efficiency by mixing siRNA in buffer into an ethanolic solution of the lipid mixture, followed by a stepwise diafiltration process. Cholesterol was purchased from Northern Lipids, PEG-DMG was purchased from NOF Corporation and CLinDMA was synthesized by Merck and Co. The encapsulation efficiency was determined using a SYBR Gold fluorescence assay and the particle size measurements were performed using a Wyatt DynaPro plate reader. The siRNA and lipid concentrations in the LNP were quantified by a HPLC using a PDA detector.
In vivo
C57BL/6 male mice 20–23 g were purchased from Taconic Farms, Inc. After receiving, the mice rested for a week, divided in groups of eight and injected intravenously with 200 µl containing 6 mg/kg siRNA formulated in a lipid nanoparticle. Five mice per group were sacrificed at 24 h following siRNA injection. Livers were harvested and processed to assess ApoB mRNA levels by qPCR as described above.
RESULTS
Gene silencing activity of siRNAs containing UNAs in their guide strand
A synthetic, double-stranded 21-mer corresponding to positions 10 162–10 182 of the mRNA from the ApoB gene was used. Positions 1–19 of both strands were ribonucleotides, and the overhangs at positions 20 and 21 contained 2′-deoxyribonucleotide thymidines (Figure 1B). This unmodified siRNA was the template for systematic evaluation of modified siRNAs containing a single UNA at every position along the guide strand. Mass spectrometry was used to confirm that the modified siRNA strands were intact and the most adundant species (Supplementary Figure S1).
Functional ApoB siRNAs demonstrate target specific mRNA degradation in cell based assays that can be detected by quantitative PCR (qPCR). Hepa 1–6 cells were transfected with either unmodified ApoB siRNA, UNA-modified ApoB siRNA, or negative control siRNA. Negative control siRNA did not induce ApoB mRNA degradation. Unmodified siRNA resulted in degradation of 92% of the total ApoB mRNA (Figure 1C). For UNA modified siRNA, mRNA degradation activity varied depending on the position of the substitution on the guide strand. UNA substitutions at position 1 or 2 did not exhibit any mRNA degradation activity. UNA substitutions at position 3 exhibited only 42% of activity as compared to unmodified siRNA. UNA substitutions at positions 4–8 had similar activity to unmodified siRNA, ranging from 77% to 92%. Substitutions at positions 13, 15, 17–19 or on the overhang positions 20 and 21 had activity similar to unmodified siRNA (79–88%). UNA substitutions at positions 9–12, 14 or 16 had attenuated mRNA degradation and were significantly different from unmodified (paired t-test, P < 0.001), with values ranging from −25% to 62% compared to the unmodified siRNA, which was 92%. This is similar to a previously published result where UNA modifications closer to the 5′-end (position 3) had reduced silencing activity and modifications farther away from the 5′-end at positions 6 and 18 had similar activity to unmodified (32).
Dose response analysis was conducted for each UNA modified siRNA (Table 1). An unmodified ribonucleotide containing siRNA had an IC50 of 0.123 nanomolar (nM). For UNA modifications at positions 4–8, which have similar mRNA degradation activity compared to the unmodified siRNA, the IC50s were also similar. While UNA modifications at positions 17–19 showed mRNA degradation at 10 nM similar to unmodified siRNA, the IC50s were 10-fold higher (1.29, 1.02 and 1.20 nM, respectively). The IC50s were also increased for UNA modifications at positions 3, 9, and 12–14, which also showed attenuated mRNA activity.
Table 1.
IC50s and melting temperatures of UNA modified siRNAs
| siRNA | Guide sequence (5′→3′) | IC50 (nM) | Tm (°C) | ΔTm (°C) |
|---|---|---|---|---|
| unmodified | UUCAGUGUGAUGACACUUGdTdT | 0.123 ± 0.023 | 67.4 | |
| UNA1 | unaUUCAGUGUGAUGACACUUGdTdT | ND | 66.4 | −1.0 |
| UNA2 | UunaUCAGUGUGAUGACACUUGdTdT | ND | 65.0 | −2.4 |
| UNA3 | UUunaCAGUGUGAUGACACUUGdTdT | 3.83 ± 2.28 | 63.0 | −4.4 |
| UNA4 | UUCunaAGUGUGAUGACACUUGdTdT | 0.13 ± 0.08 | 61.2 | −6.2 |
| UNA5 | UUCAunaGUGUGAUGACACUUGdTdT | 0.21 ± 0.05 | 58.2 | −9.1 |
| UNA6 | UUCAGunaUGUGAUGACACUUGdTdT | 0.23 ± 0.17 | 58.2 | −9.1 |
| UNA7 | UUCAGUunaGUGAUGACACUUGdTdT | 0.39 ± 0.12 | 58.8 | −8.6 |
| UNA8 | UUCAGUGunaUGAUGACACUUGdTdT | 0.15 ± 0.06 | 58.2 | −9.1 |
| UNA9 | UUCAGUGUunaGAUGACACUUGdTdT | 2.31 ± 0.73 | 60.4 | −7.0 |
| UNA10 | UUCAGUGUGunaAUGACACUUGdTdT | ND | 59.2 | −8.2 |
| UNA11 | UUCAGUGUGAunaUGACACUUGdTdT | ND | 59.8 | −7.6 |
| UNA12 | UUCAGUGUGAUunaGACACUUGdTdT | 6.78 ± 5.21 | 61.0 | −6.4 |
| UNA13 | UUCAGUGUGAUGunaACACUUGdTdT | 2.17 ± 0.46 | 59.8 | −7.6 |
| UNA14 | UUCAGUGUGAUGAunaCACUUGdTdT | 3.09 ± 0.85 | 56.8 | −10.5 |
| UNA15 | UUCAGUGUGAUGACunaACUUGdTdT | 0.76 ± 0.17 | 60.4 | −7.0 |
| UNA16 | UUCAGUGUGAUGACAunaCUUGdTdT | ND | 61 | −6.4 |
| UNA17 | UUCAGUGUGAUGACACunaUUGdTdT | 1.29 ± 0.28 | 64 | −3.4 |
| UNA18 | UUCAGUGUGAUGACACUunaUGdTdT | 1.02 ± 0.24 | 65 | −2.4 |
| UNA19 | UUCAGUGUGAUGACACUUunaGdTdT | 1.20 ± 0.74 | 64.8 | −2.6 |
| UNA20,21 | UUCAGUGUGAUGACACUUGunaUunaU | 0.56 ± 0.20 | 65.6 | −1.8 |
UNA, unlocked nucleic acid; ACGU, RNA; dTdT, deoxynucleotide overhang; ΔTm, difference between test and unmodified siRNA Tm; ND, could not be determined.
Clp1 siRNA kinase cannot phosphorylate siRNAs with UNAs at position 1 or 2
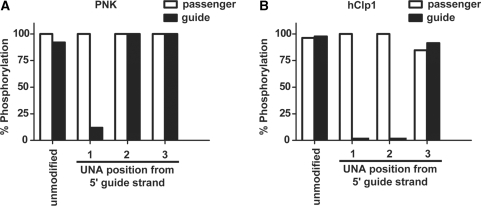
Human Clp1 was expressed and purified from E. coli and kinase activity assays were performed using duplexed, unmodified siRNA. Mass spectrometry was used in order to detect phosphorylation of both the passenger and guide strands of the siRNA. Polynucleotide kinase (PNK), a nucleotide kinase found in bacteria, was used as a positive control to confirm the integrity of the siRNAs (Figure 2). PNK was able to phosphorylate both RNA and DNA and work in a variety of buffer conditions. Both strands of the unmodified siRNA were completely phosphorylated by Clp1 following a 1 h incubation under optimized kinase conditions. Duplexed modified siRNA containing UNAs at positions 1, 2 or 3 of the guide strand (UNA1, UNA2, UNA3) were incubated with Clp1. All passenger strands, which contain ribonucleotides and no UNA modifications, were phosphorylated to completion. However, guide oligos UNA1 and UNA2 failed to be phosphorylated by Clp1. In contrast, UNA3 was phosphorylated to completion by Clp1. Interestingly, PNK was also not able to phosphorylate UNA1 above 20% after 1 h of incubation, but was able to phosphorylate UNA2 and UNA3 to completion. Mass spectrometry of UNA1 confirmed its integrity (Supplementary Figure 1). These data suggest that Clp1 fails to recognize position 1 or 2 UNA-modified siRNAs as nucleic acid substrates for 5′-terminal phosphorylation.
Figure 2.
Clp1 kinase failed to phosphorylate the 5′-termini of siRNAs containing UNAs at position 1 or 2. (A) PNK was used as a control kinase to evaluate phosphorylation of the siRNAs and to ensure the integrity of the synthesized siRNAs. siRNAs were incubated with PNK and samples were run on the mass spectrometer as described in Materials and Methods section. Mass spectrometric analysis of the siRNAs indicated that the siRNAs were intact. (B) Clp1 siRNA kinase assay was performed to evaluate the role of Clp1 in phosphorylation of UNA modified siRNA as described in (A). Unmodified refers to the ApoB siRNA sequence containing only ribonucleotides and dTdT overhangs.
5′-terminal phosphorylation restores gene silencing when the UNA is at position 1 but not at positions 2 or 3
Synthetic 5′-terminal phosphorylation has been shown to recover the mRNA degradation activity of siRNAs (17–20,34). Since UNA1 and UNA2 are unable to be phosphorylated on the 5′-terminus by Clp1, we asked whether these siRNAs could recover their ability to induce degradation of target mRNA when prepared in a phosphorylated form (7).
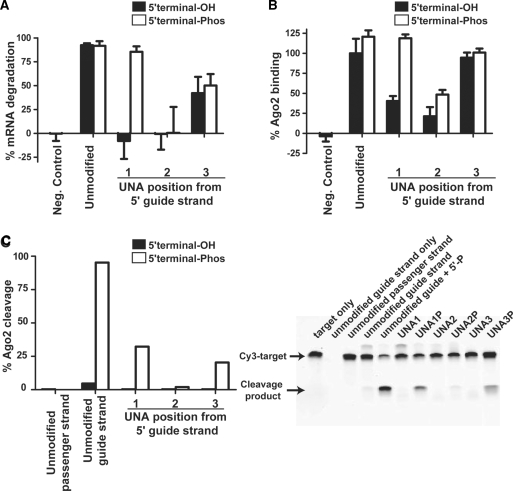
UNA1, 2 and 3 with a 5′-terminal phosphate on the guide strand were synthesized (UNA1P, UNA2P and UNA3P, respectively). UNA1P had nearly full recovery of mRNA degradation activity, with 85% mRNA degradation, compared to its unmodified counterpart. UNA1 had no detectable target mRNA degradation when administered at 10 nM (Figure 3A). A dose–response evaluation of UNA1P showed an IC50 of 0.26 nM, which is slightly less potent than the unmodified siRNA (Table 2). UNA2P had no improvement in mRNA degradation when compared to UNA2. UNA3P also showed no improvement in mRNA degradation. Unmodified siRNA with a 5′-terminal phosphate on the guide strand exhibited no change in mRNA degradation consistent with previous results that siRNAs are phosphorylated in cells (16). These data suggest that the loss of activity of UNA1 substituted siRNA in cell based assays is predominantly due to its inability to be phosphorylated by Clp1, whereas the loss of activity for UNA2 and UNA3 substituted siRNA occurs for other reasons.
Figure 3.
Chemical phosphorylation of siRNAs containing UNAs at position 1, but not position 2 or 3, restores biological activity in cell based assays. (A) 5′-terminal phosphorylation of the guide strand restores activity when UNAs are at position 1. Unmodified refers to the ApoB sequence containing only ribonucleotides and dTdT overhangs. The negative control is a scrambled sequence. Individual UNAs are placed at the indicated position on the guide strand. Oligonucleotides with a 5′-terminal hydroxyl group on the guide strand are shown in black and oligonucleotides with a 5′-terminal phosphate on the guide strand are shown in white (all passenger strands contained a 5′-terminal hydroxyl group). (B) Binding of oligonucleotides to epitope-tagged Ago2 in cell lysates. Ago2 binding studies were performed as described in Materials and Methods section. The percent of the UNA modified oligonucleotides bound to Ago2 was normalized to unmodified siRNA. The negative control refers to a sequence that is mismatched to the stem loop PCR primers. (C) Cleavage by purified Ago2 of a Cy3 target oligo that mimics the cellular mRNA target. The gel is on the right and quantification of the cleavage is on the left. Ago2 cleavage studies were performed as described in ‘Materials and Methods’ section.
Table 2.
IC50 and melting temperature of UNA modified siRNAs with a 5′-terminal phosphate on the guide strand
| siRNA | Guide sequence (5′→3′) | IC50 (nM) | Tm (°C) | ΔTm (°C) |
|---|---|---|---|---|
| unmodified | UUCAGUGUGAUGACACUUGdTdT | 0.123 ± 0.023 | 67.4 | – |
| UNA1 | unaUUCAGUGUGAUGACACUUGdTdT | ND | 66.4 | −1.0 |
| UNA1P | P-unaUUCAGUGUGAUGACACUUGdTdT | 0.26 ± 0.18 | 66.6 | −0.8 |
| UNA2 | UunaUCAGUGUGAUGACACUUGdTdT | ND | 65.0 | −2.4 |
| UNA2P | P-UunaUCAGUGUGAUGACACUUGdTdT | ND | 66.4 | −1.0 |
| UNA3 | UUunaCAGUGUGAUGACACUUGdTdT | 3.83 ± 2.28 | 63.0 | −4.4 |
| UNA3P | P-UUunaCAGUGUGAUGACACUUGdTdT | 4.95 ± 2.39 | 63.4 | −4.0 |
P, phosphate; ΔTm, difference between test and unmodified siRNA Tm; ND, could not be determined.
The thermodynamic stability of siRNA does not change with phosphorylation
Melting temperature experiments using SYBR Green intercalation were conducted to assess the effect of UNA incorporation on the thermodynamic stability of the siRNA duplexes. The SYBR Green melting temperature assay has been validated as an acceptable way of measuring oligo melting temperatures (35). We also validated the assay using differential scanning calorimetry (DSC) and UV melting temperature and saw a strong linear correlation between SYBR Green and DSC methods (R2 = 0.9066, data not shown). All UNA modified positions along the guide strand showed a decrease in melting temperature, as compared to unmodified siRNA (Table 1). These results are comparable to previous literature reports that evaluated the destabilizing effect of UNA substitutions in oligonucleotides (28,30,32). The smallest changes in melting temperature were seen at the 5′- and 3′-positions including UNA1-3 and UNA18-19 where the largest were in the central region of the siRNA. There were also small changes in melting temperature for the overhang positions UNA20, 21. These results for UNA20,21 are to be expected as the overhangs do not make contact with any complementary bases and should not affect the overall thermodynamic stability of the siRNA. Replacement of the 5′-terminal hydroxyl with a phosphate in the UNA modified oligonucleotides did not have any impact on the melting temperature of UNA1P, UNA2P or UNA3P indicating that the addition of a phosphate would not have a stabilizing effect (Table 2).
Inhibition of Ago2 binding to the UNA1 siRNA can be reversed by the addition of a 5′-terminal phosphate
The crystal structure of T. thermophilus Ago2 revealed that the 5′-terminal phosphate and the position 1 nucleotide of the guide strand are not associated with the complementary mRNA target strand (37,38). The 5′-terminal phosphate does have strong interactions with four invariant residues in the Mid domain of Ago2. In contrast to position 1, the nucleotides in the seed region of the guide strand (positions 2–8) are base paired to the complementary strand in an A-form helix.
We investigated if the binding affinity for Ago2 was influenced by UNA modifications, and the impact of a 5′-terminal phosphate on this affinity. Duplexed siRNAs containing various UNA substitutions, with or without a 5′-terminal phosphate, were incubated in cell lysate containing overexpressed and epitope tagged Ago2. The cell lysate based Ago2 binding assay supports the introduction of duplex siRNAs that interact with the RISC machinery and allow guide strand loading into Ago2. Following immunoprecipitation of Ago2, stem-loop qPCR was performed to determine the amount of guide strand bound. Stem loop PCR is a real-time Reverse Transcriptase (RT)–PCR method in which a stem loop RT primer is hybridized to a siRNA strand, then reverse transcribed. The RT products are then quantified using conventional Taqman PCR (36). All data were normalized to the binding observed for the unmodified siRNA (Figure 3B). An immunoblot blot was also performed to verify equal amounts of Ago2 had been immunoprecipitated (Supplementary Figure S2). When the unmodified siRNA was synthetically phosphorylated on the 5′-terminus, Ago2 binding increased slightly (an increase of 20% compared to the unmodified siRNA). For UNA1 and UNA2, Ago2 binding was 40% and 21% of the binding efficiency observed for unmodified siRNA, respectively. When UNA1P was incubated with Ago2, the binding increased to the same binding efficiency of unmodified siRNA with a 5′-phosphate. This represents a 3-fold preference of UNA1P for Ago2 over UNA1. Similarly, UNA2P bound with a 2-fold higher efficiency than UNA2, although the total binding for UNA2P was still only 40% of that seen with the unmodified siRNA with a 5′-phosphate. In contrast, UNA3 exhibited nearly equal levels of binding to that seen for the unmodified siRNA; thus, phosphorylation did not increase the binding efficiency of UNA3. These results demonstrate that position 1 UNA modifications have the unique ability to have both mRNA degradation activity and Ago2 binding rescued by a 5′-end phosphate.
The cell-based mRNA degradation results (Figure 3A) were also confirmed with an Ago2 cleavage assay (Figure 3C). This assay monitors cleavage of a Cy3 labeled oligo that is complementary to the siRNA guide strand. The Cy3 target oligo is cleaved by Ago2 RNAse activity when incubated with the siRNA guide strand and Ago2. Neither the unphosphorylated all-ribose guide nor the passenger strand gave any detectable cleavage of the Cy3 target oligo. However, when the unmodified guide strand siRNA was synthetically phosphorylated on the 5′-terminus, 95% cleavage of the Cy3 target oligo was detected. The same results were observed for UNA1P and UNA3P, whereby the unphosphorylated siRNAs gave no detectable cleavage of the Cy3 target oligo but when a synthetic phosphate was applied to the 5′-terminus of the siRNA cleavage was observed of the Cy3 target oligo. The Cy3 target oligo was cleaved less when incubated with UNA3P than with UNA1P, having only 20% compared to 33%, respectively. There was no detectable cleavage of the Cy3 target oligo when incubated with UNA2 or UNA2P. These results correlate with the in vitro mRNA degradation and Ago2 binding results (Figure 3A and B). They also demonstrate that 5′-end phosphorylation is necessary for cleavage of mRNA target by these siRNAs and that these siRNAs proceed through a RNAi-mediated mechanism to induce mRNA degradation.
5′-terminal phosphorylation restores gene silencing in vivo for UNA1
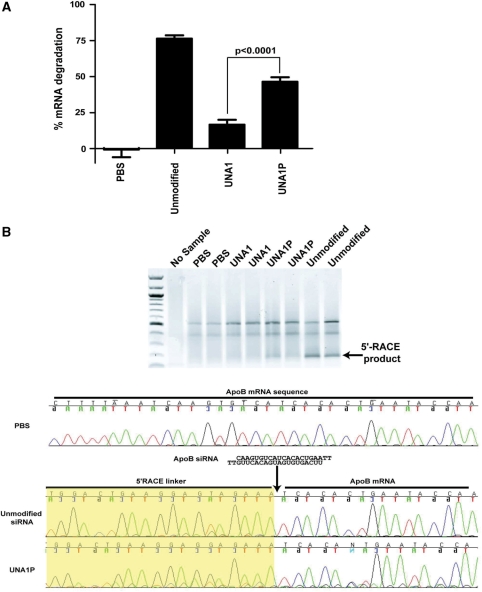
In order to assess the if 5′-terminal phosphorylation of UNA1 also improves potency in vivo, ApoB siRNAs corresponding to unmodified or UNA modified with or without a 5′-terminal phosphate were formulated into lipid nanoparticles and delivered by intravenous (IV) tail injection into mice. PBS was delivered as a control. Lipid nanoparticles localize to the liver, where ApoB is highly expressed. Livers were harvested 24 h after injection, and ApoB mRNA levels were assessed by qPCR (Figure 4). Mice treated with unmodified siRNA showed significant ApoB mRNA degradation compared to the PBS control, with 76% mRNA degradation 24 h after injection. For UNA1, 16% mRNA degradation was observed after 24 h and was not significantly different from the PBS control (paired t-test, P < 0.05). For UNA1P, 46% of the ApoB transcript was degraded following IV administration. The ability of UNA1P to silence ApoB mRNA following injection was significantly different from UNA1 although phosphorylation alone was not able to rescue all the potency observed with the unmodified siRNA (Figure 4). This decrease of in vivo potency in UNA1P compared to the unmodified siRNA is consistent with the IC50 data (Table 2), where UNA1P had a two-fold decrease in IC50 value from the unmodified siRNA.
Figure 4.
UNAs demonstrate in vivo activity at 24 h post-IV injection. (A) Unmodified siRNA, siRNA with an UNA at position 1 (UNA1), and siRNA containing a 5′-terminal phosphate with UNA at position 1 (UNA1P) were formulated into lipid nanoparticles and injected in mice at 6 mg/kg. The livers were harvested and ApoB mRNA levels were evaluated by qPCR 24 h after injection (student’s t-test, P < 0.0001). Bars represent the mean and standard deviation of five mice per group. (B) UNA1 and UNA1P function via an Ago2 mechanism. 5′RACE was used to detect the ApoB mRNA cleavage site. Mouse liver RNA samples were evaluated for each sample group. 5′RACE products are 150 bp. The precise Ago2-mediate cleavage site on ApoB was mapped by sequencing and is designated by an arrow. The yellow box indicates the 5′-RACE linker sequence.
5′RACE was performed on the in vivo samples to demonstrate that mRNA cleavage and degradation occurred through an Ago2 specific mechanism. 5'R;ACE involves ligation of a tag to the 5′-end of the cleavage site (if cleavage occurs) and then amplification using a ligation-specific primer and a gene specific downstream primer. If Ago2-mediated cleavage occurs, the amplification of this region would be 150 bp. If no cleavage occurs there would not be an amplification product since the length of the ApoB from its endogenous 5′ terminus to the cleavage site would be 10 162 bp and the PCR extension time is optimized for 150 bp. 5′-RACE does not provide quantification of the amount of cleavage that occurs, only that cleavage occurred via an Ago2 mechanism. 5′RACE was performed on the samples and run on an agarose gel. Bands corresponding to 150 bp appear for the unmodified all ribose siRNA and UNA1P siRNA. No band was detected for UNA1 which may be due to the low level of cleavage that could not be detected by 5′RACE. These bands were sequenced and cleavage of the mRNA corresponded to between position 10 and 11 of the guide strand of the siRNA, thus confirming UNA1P mRNA degradation in vivo proceeds through an Ago2-mediated mechanism.
DISCUSSION
In this study, we systematically evaluated UNA substitutions throughout the guide strand of a siRNA targeting the ApoB transcript. Substitution of an UNA at position 1 or 2 of the siRNA had a detrimental effect on mRNA degradation, Clp1 phosphorylation, Ago2 binding, and Ago2 cleavage. Chemical addition of a 5′-terminal phosphate allowed for the recovery of mRNA degradation activity and Ago2 binding and cleavage to UNA1, but not UNA2. 5′-phosphorylation of UNA1 also rescued siRNA silencing activity in vivo and was consistent with the determined IC50s for the unmodified siRNA and UNA1P. These data indicate the importance of conducting dose response analysis as well as maximal mRNA degradation measurements in order to fully characterize the impact of a chemical modification on the activity of a siRNA.
Clp1 is the kinase responsible for exogenous siRNA phosphorylation. The ideal parameters under which Clp1 operates remain to be defined (7). In this study, mass spectrometry was used to identify phosphorylation of the passenger and guide strand by Clp1. These data suggest for the first time that Clp1 may recognize chemical features of position 1 and 2 of the siRNA in order to phosphorylate the 5′-terminus. If either of these positions are modified with a UNA, Clp1 can no longer phosphorylate the 5′-terminus. This failure to be phosphorylated is the most likely reason UNA1 has decreased silencing activity since synthetic addition of a 5′-phosphate rescues mRNA degradation activity in cells and in mice. The loss of silencing activity for UNA2 and UNA3 is more likely due to the structural complexity induced by the UNA modifications rather than the lack of phosphorylation since chemical addition of a 5′-terminal phosphate to these oligonucleotides does not rescue this activity.
The molecular events responsible for 5′-phosphate recognition during RISC assembly remain to be defined. Position 1 is unique in that it does not seem to have a role in mRNA target identification or in maintaining the structural stability of the siRNA, but instead has a role in RISC loading and guidance of the rest of the siRNA into Ago2 (9,37). In the Ago2 crystal structure, the 5′–phosphorylated nucleotide at position 1 fits into a basic pocket with the 5′ base stacking on the aromatic ring of an invariant tyrosine, further stabilizing this binding (38,39). Interestingly, the UNA modification at position 1 does not impair Ago2 binding when a 5′-terminal phosphate is present, indicating that it is the lack of phosphorylation, not the modification that prevents efficient binding. In the Ago2 crystal structure at position 1 there is a flexible protein loop that should accommodate additional movements that could be caused by the UNA modifications (Supplementary Figure S3). In contrast, the opposite is true for UNAs in positions 2 and 3, where the contacts involve the sugar phosphate backbone of the nucleotides and the bases are positioned next to each other to direct bonding to the target mRNA. For position 2 the base of the nucleoside would be between the base of position 3, and the helix of Ago2 between residues 556 and 571. This makes the position 2 base less able to accommodate the movements in the sugar and the base that could be introduced by the UNA. The base at position 3 would be between position 2 and 4 and suffer from the same steric hindrances. The lack of the 2′C-3′C bond in the ribose sugar increases the structural instability, diminishes the strength or number of contacts of the backbone with Ago2 and the target mRNA at positions 2 and 3, and leads to a lack of degradation of target mRNAs due to incorrect positioning of the position 2 or 3 UNA modified siRNA. In addition, UNAs at positions 1–2 also cause duplex instability, as reflected in the data obtained from thermal denaturation studies (Table 1). This would translate into decreased stability of the duplex formed between the UNA containing guide strand with the mRNA and the decreased mRNA degradation.
The UNA1P is less potent than the unmodified siRNA in vitro and in vivo. One reason for this may be due to 5′-terminal phosphate instability. If the 5′-phosphate is removed in the cytoplasm the UNA could not be rephosphorylated by Clp1 since UNAs are not substrates for Clp1 at position 1 (Figure 2). Another possibility could be due to decreased Ago2 mediated cleavage by UNA1P as compared to the unmodified siRNA. Both UNA1P and the unmodified siRNA bind equally to Ago2. However, less Ago2 mediated mRNA cleavage is observed from UNA1P as compared to the unmodified siRNA. The lack of activity seen from UNA1P would therefore be driven by its inability to cleave its mRNA target with the same efficiency as unmodified. Further pharmacokinetic studies examining 5′-terminal phosphate stability and mRNA cleavage in vivo will elucidate the mechanism for the UNA1 decrease in potency.
Chemical modifications of siRNAs are critical for optimizing siRNA as human therapeutics. In this study, we systematically evaluated UNA substitutions throughout the guide strand of a siRNA targeting the ApoB transcript. Dose response analysis of the UNA substituted siRNAs in positions 2–8 showed that UNAs were tolerated at positions 4-8 but were detrimental when substituted at positions 1, 2 or 3 of the guide strand. Furthermore, we demonstrate an UNA substitution at position 1 or 2, but not position 3, blocked human Clp1 phosphorylation, subsequent Ago2 binding and Ago2 mediated target cleavage. Taken together these results suggest that human Clp1 utilizes specific nucleic acid recognition elements at both positions 1 and 2 to allow siRNA phosphorylation and RISC loading. Based on these findings, UNA modifications, as well as other novel nucleoside modifications that function similarly in the guide strand with respect to Clp1 phosphorylation, may be best suited for position 1 or 2 substitution of the passenger strand to block phosphorylation and abrogate RISC loading. Such a strategy would improve potency due to lack of RISC loading of the passenger. Further studies incorporating these novel nucleosides will help identify the additional beneficial properties that they can confer to siRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We would like to thank Steven Bartz, Minh Thoa Lai, Michelle Caguyong, Walter Strapps, Victoria Pickering and Ahn Le for their help with the coordination of this study and helpful suggestions. We would also like to thank Dipali Ruhela, Ray Fucini, Yi Pei, Alan Wei, Elmer Payson, Jeff Jones, Jing Wang and the Formulations and Pharmacology groups for their help.
REFERENCES
- 1.Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 4.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 5.Corey DR. RNA learns from antisense. Nat. Chem. Biol. 2007;3:8–11. doi: 10.1038/nchembio0107-8. [DOI] [PubMed] [Google Scholar]
- 6.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weitzer S, Martinez J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature. 2007;447:222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- 8.Sen GL, Blau HM. A brief history of RNAi: the silence of the genes. FASEB J. 2006;20:1293–1299. doi: 10.1096/fj.06-6014rev. [DOI] [PubMed] [Google Scholar]
- 9.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 10.Doi N, Zenno S, Ueda R, Ohki-Hamazaki H, Ui-Tei K, Saigo K. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr. Biol. 2003;13:41–46. doi: 10.1016/s0960-9822(02)01394-5. [DOI] [PubMed] [Google Scholar]
- 11.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 12.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 13.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Ye X, Jiang F, Liang C, Chen D, Peng J, Kinch LN, Grishin NV, Liu Q. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325:750–753. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boutla A, Delidakis C, Livadaras I, Tsagris M, Tabler M. Short 5′-phosphorylated double-stranded RNAs induce RNA interference in Drosophila. Curr. Biol. 2001;11:1776–1780. doi: 10.1016/s0960-9822(01)00541-3. [DOI] [PubMed] [Google Scholar]
- 16.Schwarz DS, Hutvagner G, Haley B, Zamore PD. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell. 2002;10:537–548. doi: 10.1016/s1097-2765(02)00651-2. [DOI] [PubMed] [Google Scholar]
- 17.Watts JK, Choubdar N, Sadalapure K, Robert F, Wahba AS, Pelletier J, Pinto BM, Damha MJ. 2′-fluoro-4′-thioarabino-modified oligonucleotides: conformational switches linked to siRNA activity. Nucleic Acids Res. 2007;35:1441–1451. doi: 10.1093/nar/gkl1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher M, Abramov M, Van Aerschot A, Xu D, Juliano RL, Herdewijn P. Inhibition of MDR1 expression with altritol-modified siRNAs. Nucleic Acids Res. 2007;35:1064–1074. doi: 10.1093/nar/gkl1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, Wanders L, Griffey RH, Swayze EE, Bhat B. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 20.Lima WF, Wu H, Nichols JG, Sun H, Murray HM, Crooke ST. Binding and cleavage specificities of human Argonaute2. J. Biol. Chem. 2009;284:26017–26028. doi: 10.1074/jbc.M109.010835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robbins M, Judge A, Liang L, McClintock K, Yaworski E, MacLachlan I. 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol. Ther. 2007;15:1663–1669. doi: 10.1038/sj.mt.6300240. [DOI] [PubMed] [Google Scholar]
- 23.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elmen J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, Wahren B, Liang Z, Orum H, Koch T, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangos MM, Min KL, Viazovkina E, Galarneau A, Elzagheid MI, Parniak MA, Damha MJ. Efficient RNase H-directed cleavage of RNA promoted by antisense DNA or 2′F-ANA constructs containing acyclic nucleotide inserts. J. Am. Chem. Soc. 2003;125:654–661. doi: 10.1021/ja025557o. [DOI] [PubMed] [Google Scholar]
- 27.Vandendriessche F, Augustyns K, Van Aerschot A, Busson R, Hoogmartens J, Herdewijn P. Acyclic oligonucleotide: possibilities and limitations. Tetrahedron. 1993;49:7223–7238. [Google Scholar]
- 28.Jensen TB, Langkjaer N, Wengel J. Unlocked nucleic acid (UNA) and UNA derivatives: thermal denaturation studies. Nucleic Acids Symp. Ser. 2008;52:133–134. doi: 10.1093/nass/nrn068. [DOI] [PubMed] [Google Scholar]
- 29.Kvaerno L, Kumar R, Dahl BM, Olsen CE, Wengel J. Synthesis of abasic locked nucleic acid and two seco-LNA derivatives and evaluation of their hybridization properties compared with their more flexible DNA counterparts. J. Org. Chem. 2000;65:5167–5176. doi: 10.1021/jo000275x. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen P, Dreioe LH, Wengel J. Synthesis and evaluation of oligodeoxynucleotides containing acyclic nucleosides: introduction of three novel analogues and a summary. Bioorg. Med. Chem. 1995;3:19–28. doi: 10.1016/0968-0896(94)00143-q. [DOI] [PubMed] [Google Scholar]
- 31.Sipa K, Sochacka E, Kazmierczak-Baranska J, Maszewska M, Janicka M, Nowak G, Nawrot B. Effect of base modifications on structure, thermodynamic stability, and gene silencing activity of short interfering RNA. RNA. 2007;13:1301–1316. doi: 10.1261/rna.538907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N, Babu BR, Hojland T, Abramov M, Van Aerschot A, et al. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burchard J, Jackson AL, Malkov V, Needham RH, Tan Y, Bartz SR, Dai H, Sachs AB, Linsley PS. MicroRNA-like off-target transcript regulation by siRNAs is species specific. RNA. 2009;15:308–315. doi: 10.1261/rna.1326809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen PY, Weinmann L, Gaidatzis D, Pei Y, Zavolan M, Tuschl T, Meister G. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA. 2008;14:263–274. doi: 10.1261/rna.789808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gudnason H, Dufva M, Bang DD, Wolff A. Comparison of multiple DNA dyes for real-time PCR: effects of dye concentration and sequence composition on DNA amplification and melting temperature. Nucleic Acids Res. 2007;35:e127. doi: 10.1093/nar/gkm671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 38.Yuan YR, Pei Y, Chen HY, Tuschl T, Patel DJ. A potential protein-RNA recognition event along the RISC-loading pathway from the structure of A. aeolicus Argonaute with externally bound siRNA. Structure. 2006;14:1557–1565. doi: 10.1016/j.str.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.