Abstract
In many prokaryotes the biosynthesis of the amide aminoacyl-tRNAs, Gln-tRNAGln and Asn-tRNAAsn, proceeds by an indirect route in which mischarged Glu-tRNAGln or Asp-tRNAAsn is amidated to the correct aminoacyl-tRNA catalyzed by a tRNA-dependent amidotransferase (AdT). Two types of AdTs exist: bacteria, archaea and organelles possess heterotrimeric GatCAB, while heterodimeric GatDE occurs exclusively in archaea. Bacterial GatCAB and GatDE recognize the first base pair of the acceptor stem and the D-loop of their tRNA substrates, while archaeal GatCAB recognizes the tertiary core of the tRNA, but not the first base pair. Here, we present the crystal structure of the full-length Staphylococcus aureus GatCAB. Its GatB tail domain possesses a conserved Lys rich motif that is situated close to the variable loop in a GatCAB:tRNAGln docking model. This motif is also conserved in the tail domain of archaeal GatCAB, suggesting this basic region may recognize the tRNA variable loop to discriminate Asp-tRNAAsn from Asp-tRNAAsp in archaea. Furthermore, we identified a 310 turn in GatB that permits the bacterial GatCAB to distinguish a U1–A72 base pair from a G1–C72 pair; the absence of this element in archaeal GatCAB enables the latter enzyme to recognize aminoacyl-tRNAs with G1–C72 base pairs.
INTRODUCTION
Correct pairing of an amino acid with its cognate tRNA is an essential step to maintain the accuracy of translation. This is usually accomplished by aminoacyl-tRNA synthetases (aaRSs) that catalyze the direct attachment of an amino acid to its cognate tRNA (1). However, glutaminyl-tRNA synthetase (GlnRS) is absent in the majority of bacteria, in all known archaea, and in many organelles (2). These organisms utilize an indirect pathway for Gln-tRNAGln formation where a non-discriminating glutamyl-tRNA synthetase (ND-GluRS) synthesizes mischarged Glu-tRNAGln (3) that is then amidated to the cognate Gln-tRNAGln by glutamyl-tRNAGln amidotransferase (Glu-AdT) (2,4). Similarly, many prokaryotes lacking an asparaginyl-tRNA synthetase (AsnRS) generate Asn-tRNAAsn by the combined actions of a non-discriminating aspartyl-tRNA synthetase (ND-AspRS) and an aspartyl-tRNAAsn amidotransferase (Asp-AdT) (5,6).
Two types of AdTs exist: the heterotrimeric GatCAB (7) present in bacteria, archaea and organelles (2), and the heterodimeric GatDE found exclusively in archaea (8). GatDE specifically converts Glu-tRNAGln (8), while bacterial GatCAB acts as both a Glu-AdT and an Asp-AdT in vitro (2). The in vivo role of bacterial GatCAB is defined by the nature of ND-aaRS (ND-GluRS and/or ND-AspRS) present in the cell (2). In archaea that lack AsnRS, GatCAB is encoded (9). This enzyme (e.g. from Methanothermobacter thermautotrophicus) in vitro strongly prefers Asp-tRNAAsn over the homologous Glu-tRNAGln (10); thus archaeal GatCAB may act in vivo as an Asp-AdT.
AdTs accurately distinguish their mischarged aa-tRNA substrates (Glu-tRNAGln and/or Asp-tRNAAsn) from the cognate Glu-tRNAGlu and Asp-tRNAAsp species. Bacterial GatCAB and GatDE achieve this by recognizing the first base pair of the acceptor stem and the D-loop of their tRNA substrates (11–13). In contrast, archaeal GatCAB does not recognize the first base pair of Asp-tRNAAsn (13,14). Instead the M. thermautotrophicus GatCAB makes use of the D-loop, the nucleotide in position 49, and to a lesser extent of the length of the variable loop to distinguish Asp-tRNAAsn from Asp-tRNAAsp (14), while the Methanosarcina barkeri GatCAB appears to use primarily the length of the variable loop for the same task (13).
Previous work implicated the tail domain of the GatB and GatE in D-loop recognition of their respective aa-tRNA substrates (11,12). These tail domains share homology with the standalone YqeY proteins of unknown function (PFAM id: PF09424) that are present in a diverse array of organisms (11,12,15,16). Previous crystallographic studies did not resolve the YqeY-like domain of AdTs (11,12,17,18) or of Deinococcus radiodurans GlnRS (15). Here, we present a full-length Staphylococcus aureus GatCAB structure (containing the YqeY domain) resolved at 1.9 Å. The structure reveals a Lys rich motif in the tail domain of GatB which is absent in GatE. We also identified by biochemical studies a bacterial GatCAB-specific element that enables this enzyme to distinguish a U1–A72 base pair from a G1–C72 pair.
MATERIALS AND METHODS
Preparation of S. aureus GatCAB
The enzyme was over-produced in an Escherichia coli B834 strain and purified over a HisTrap HP column (GE Healthcare) as described (11). The sample was then diluted 5-fold with Buffer A [50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 1 mM DTT and 10% (v/v) glycerol], and applied to a HiTrap Heparin HP column (GE Healthcare) equilibrated with buffer A. The column was washed with buffer A containing 50 mM NaCl, and proteins were eluted with a linear gradient of 50–500 mM NaCl. The enzyme eluted at ∼250 mM NaCl. The enzyme fractions were then loaded onto a HiLoad 26/60 Superdex 200 pg column (GE Healthcare) equilibrated with buffer B [20 mM Tris–HCl pH 7.5, 10 mM MgCl2, 1 mM DTT and 10% (v/v) glycerol]. Pooled fractions were concentrated by ultrafiltration using Vivaspin devices (VIVASCIENCE) to a final concentration of ∼12 mg/ml. S. aureus GatCAB mutants were generated using the QuikChangeTM site-directed mutagenesis kit according to the manufacture’s protocol (Stratagene), and purified to homogeneity as described above.
Preparation of M. thermautotrophicus GatCAB and ND-AspRS
The over-production and purifications were as described (10). Methanothermobacter thermautotrophicus GatCAB mutants were generated as described above.
Preparation of S. aureus ND-GluRS
The gene encoding the ND-GluRS from S. aureus Mu50 was amplified by polymerase chain reaction (PCR). The S. aureus ND-GluRS is toxic for an E. coli, which does not possess GatCAB, therefore the gene was cloned into NcoI/XhoI site of a pTip vector, which is used to protein expression in Rhodococcus erythropolis, and then the S. aureus ND-GluRS protein was expressed in R. erythropolis as described (19). Cells were harvested (4000×g, 15 min at 4°C) and disrupted using sonication in buffer C [50 mM Tris–HCl pH 7.5, 300 mM NaCl, 5 mM MgCl2, 10% (v/v) glycerol, 0.5 mg/ml lysozyme and 0.1 mg/ml DNase I]. All the following purification processes were carried out at 4°C. Cell debris was removed by centrifugation (40 000×g, 1 h), and clarified supernatant was applied to a HisTrap HP column as described (11). Pooled fractions were loaded onto a HiLoad 26/60 Superdex 200 pg column equilibrated with buffer D [20 mM HEPES-K pH 7.6, 5 mM MgCl2, 100 mM KCl, 1 mM DTT and 10% (v/v) glycerol]. The purified S. aureus ND-GluRS was concentrated by ultrafiltration to a final concentration of 18 mg/ml, and then diluted 2-fold with 100% (v/v) glycerol, and stored at −30°C.
Preparation of S. aureus tRNAGln, and M. thermautotrophicus and C. trachomatis tRNAAsn
The tRNA isoacceptors were in vitro transcribed and purified as described (10,11).
Preparation of aminoacyl-[32P] labeled tRNA
The tRNA isoacceptors were [32P]-labeled and aminoacylated as described (10,20) with minor modification. For glutamylation of S. aureus tRNAGln, 5 µM S. aureus ND-GluRS and tRNAGln were added in the aminoacylation reaction.
Crystallization and structure determination
The high quality single crystals of S. aureus GatCAB were obtained by using the micro-seeding technique (21) from hanging drops set up in a 1 : 1 : 0.1 ratio from protein, reservoir solution [25% (w/v) PEG 600, 5 mM MgCl2, 50 mM HEPES–NaOH pH 7.2 and 3% (v/v) 2-methyl-2,4-pentanediol (MPD)] and a micro-seeds stock solution (21). The crystal of S. aureus GatCAB was rapidly soaked through the reservoir containing 50 mM MES-Na, pH 6.4, 25% (w/v) PEG 600, 5 mM MgCl2, 3% (v/v) MPD and 10% (v/v) glycerol as a cryoprotectant, and then a data set was collected to 1.9 Å resolution at SPring-8 beamline 41XU (Hyogo, Japan) under cryogenic condition (−173°C). The data set was processed and scaled using the HKL2000 package (22). The structure of S. aureus GatCAB was solved by molecular replacement using AMoRe in the CCP4 suite (23), with the refined model of the previous (11) S. aureus GatCAB structure (PDB ID: 2G5H) as a search model. The model of S. aureus GatCAB was rebuilt by automatic refinement program LAFIRE (24,25) running with CNS (26), and modified manually by using Coot (27) followed yielded the final model with the crystallographic R/Rfree factor of 19.5/21.4%. The summary of data statistics is presented in Table 1. All figures were generated by PyMol (28).
Table 1.
Data collection and refinement statisticsa
| Data collection statistics | |
| PDB ID | 3IP4 |
| Beamline | SPring-8 BL41XU |
| Wavelength | 1.00 Å |
| Space group | P212121 |
| Cell dimensions a, b, c (Å) | 71.1, 92.7, 180.4 |
| Resolution (Å) | 50.00–1.90 (1.97–1.90) |
| Rmergeb | 0.070 (0.457) |
| I/σ(I) | 23.2 (3.5) |
| Completeness (%) | 99.9 (100) |
| Redundancy | 7.0 |
| Refinement statistics | |
| Resolution (Å) | 20.0–1.90 |
| No. reflections | 94598 |
| Rworkc/Rfreed | 0.195/0.214 |
| Number of atoms | 8299/801/1 |
| (protein/water/others) | |
| Average B factors (Å2) | 33.6/35.3/26.0 |
| (protein/water/others) | |
| RMSD bond length (Å)/angles (°) | 0.006/1.3 |
| Ramachandran plot (%) | |
| Favored | 90.2 |
| Allowed | 9.6 |
| Generous | 0.2 |
aValues in parentheses are for the outermost resolution shell.
bRmerge = ΣhΣj|<I>h − Ih,j |/ΣhΣjIh,j, where <I>h is the mean intensity of symmetry-equivalent reflections.
cRwork = Σ|Fobs − Fcal|/ΣFobs, where Fobs and Fcal are observed and calculated structure factor amplitudes.
dRfree value was calculated for R factor, using only an unrefined subset of reflections data.
Small-angle X-ray scattering
For preparation of the tRNAGln-bound S. aureus GatCAB, the S. aureus GatCAB was mixed with tRNAGln in a molar ratio of 1 : 4, and then purified by a gel filtration using a Superdex 200 10/300 GL column (GE Healthcare) in buffer B.
Small-angle X-ray scattering (SAXS) measurements were carried out at SPring-8 beamline 40B2 and 45XU (Hyogo, Japan) (29). The following SAXS analysis was carried out by using the data measured at BL45XU. A wavelength of 1.0 Å was used, and the specimen-to-detector distance was 246.5 cm. The scattering vector range was 0.008 Å−1 < s < 0.334 Å−1, where s = 4πsinθ/λ, with 2θ being the scattering angle. Data were measured at 5.0 mg/ml of samples at 25°C. Both buffers and samples were exposed 100 s. The SAXS data were normalized to the intensity of the incident beam and processed to background subtraction using the standard procedures with the program package PRIMUS (30). Maximum particle dimensions Dmax were computed using the indirect transform package GNOM (31), which also gives the distance distribution functions p(r). The discrepancy between the calculated and the experimental scattering curves was minimized using the program CRYSOL (32) as described previously (33,34). The 16 ab initio models of tRNA-free and -bound S. aureus GatCAB were generated and averaged with the program DAMMIN (35), DAMAVER (36) and SUPCOMB (37) as described previously (34).
[32P]tRNA/nuclease P1 amidotransferase assay
AdT activities of wild-type and mutant GatCAB enzymes were monitored using a [32P]tRNA/nuclease P1 amidotransferase assay as previously described (38). Determination of kinetic parameters with the S. aureus GatCAB enzymes was carried out as described for the Helicobacter pylori GatCAB (20). Determination of the kinetic parameters with the M. thermautotrophicus GatCAB enzymes was carried out as described (10).
RESULTS
Overall structure of a full-length GatCAB
We crystallized S. aureus GatCAB by adding 3–10% (v/v) MPD to the previous crystallization condition (11), which dramatically improved the crystal quality (maximum resolution from 2.3 to 1.9 Å). Surprisingly, the high-resolution structure clearly shows the GatB C-terminal region encompassing amino acids 412–475 and a histidine-tag (Figure 1A and Supplementary Figure S1), whose electron density map was not visible in the previous crystal structures. Although the unit cell dimension and crystal packing of the full-length and the previous apo-form S. aureus GatCAB are very similar, in the current structure the C-terminal GatB region is sandwiched between two GatA molecules related by 2-fold screw axes. Successful resolution of the tail-domain may be due to MPD addition in the crystallization mixture making the protein domain less flexible.
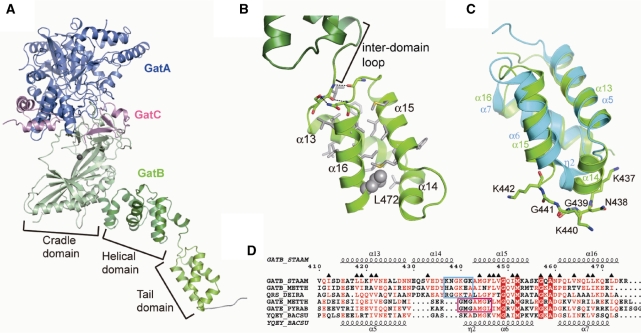
Figure 1.
(A) Overall structure of the full-length GatCAB. GatA and GatC are colored blue and magenta, respectively. GatB contains three domains: the cradle, the helical and the tail domain (light green, dark green and yellow green, respectively). Mg atom (black) in the cradle domain and C-terminal His-tag (grey) are also shown. (B) The structure of the GatB tail domain. The residues involved in the hydrophobic core are shown as a grey stick model, along with hydrogen bonds between the inter-domain loop and the tail domain (indicated as dashed lines). L472 is indicated as a grey sphere model. (C) Superposition of the S. aureus tail domain (yellow green) and YqeY-C (cyan), where the Lys rich motif is indicated as a stick model. (D) Structural based sequence alignments of the tail domain of YqeY-like proteins. Secondary structure of S. aureus tail domain is indicated at top, and that of YqeY-C is indicated at bottom (α, α-helix; β, β-strand; η, 310- turn). The number at top is corresponding to S. aureus GatB sequence. The residues constructing the hydrophobic core of S. aureus tail domain are marked with filled triangles. The Lys rich motif and the GXXAXGX motif are shown as cyan box and magenta box, respectively. The species aligned are as follows: STAAM S. aureus, METTH, M. thermautotrophicus; DEIRA, D. radiodurans; PYRAB, P. abyssi; BACSU, B. subtilis.
The GatA apo-form is essentially the same as the previously deposited S. aureus GatA structures (11). Interestingly, the high-resolution structure shows an alternative conformation for R358, which recognizes the carboxyl group of the substrate Gln (Supplementary Figure S2). Furthermore, Y310, which made a hydrogen bond with D425 in the Gln-bound S. aureus GatCAB (PDB ID: 2F2A), is flipped out from the active site of GatA, indicating that Gln binding induces only minor conformational changes in the GatA active site. GatC is also nearly identical to the deposited S. aureus GatC structures except the six C-terminal residues (95–100) were disordered and could not be modeled.
GatB is comprised of three consecutive domains: a cradle domain (1–294), a helical domain (295–407) and a tail domain (412–475). The cradle and helical domains are identical to the S. aureus GatB apo-form (PDB ID: 2G5I), and the permanent Mg2+ in the catalytic pocket was visible (11,18). The tail domain forms an anti-parallel helix bundle with three amphiphilic helices (α13, α15 and α16) that construct a hydrophobic core with α14 (Figure 1B and Supplementary Figure S1). L472 participates in this hydrophobic core and is not exposed to the molecular surface, indicating L472 is important for maintaining the structure of the tail domain rather than directly recognizing the aa-tRNA substrate as previously suggested (11). The loop between α15 and α16 interacts with I412 and S413 by two main chain and one side chain hydrogen bonds. Therefore, the S. aureus GatB tail and helical domains are linked by a ∼13 Å long inter-domain loop (408–411).
Comparison of the GatB tail domain with YqeY
The C-terminal end of GatB, comprised of the helical and tail domains (295–475), belongs to the same protein family as standalone YqeY polypeptides of unknown function present in many bacteria and in yeast (PFAM ID: PF09424) (15,16). The YqeY-like tail domain appended to the D. radiodurans GlnRS enables the enzyme to productively bind to tRNAGln (15). A similar role is proposed for this structure in GatB and GatE via recognition of the D-loop (11,12). The only YqeY structure so far resolved is from Bacillus subtilis (PDB ID: 1NG6); it can now be compared with the YqeY region in the present full-length of S. aureus GatCAB structure. The anti-parallel helix bundle of the tail domain superimposes well with the C-terminal domain of B. subtilis YqeY protein (YqeY-C: 92-146) with an r.m.s. deviation of 1.97 Å for the 51 Cα pairs compared (Figure 1C). Interestingly, the GatB tail domain has an additional α-helix (α14) and a short loop between α14 and α15 (439–442) not found in the YqeY-C. Based on the multiple sequence alignment with the tail domains and YqeY-C, this extended region contains a Lys rich motif (KXGKXK) that is highly conserved in bacterial and archaeal GatB enzymes (Figure 1D and Supplementary Figure S3). The C-terminal extension of D. radiodurans GlnRS also contains this extended region with a similar motif (RGGKTA). In contrast to GatB and D. radiodurans GlnRS, the tail domain of GatE lacks α14 and the Lys rich motif. Interestingly, the Lys rich motif of GatB is instead replaced in GatE with a GXXAXGX motif that has been implicated in GatDE distinguishing Glu-tRNAGln from Asp-tRNAAsn (16).
Docking tRNAGln into the S. aureus GatCAB structure
The GatB C-terminal tail is essential for GatCAB binding tRNAGln (11). In the co-crystal structure of the GatDE enzyme (PDB ID: 2D6F), the GatE tail domain was in the vicinity of the tRNA D-loop; however, its detailed structure could not be solved and the D-loop was fitted with the help of the B. subtilis YqeY-C structure (12). We decided to create a docking model of full-length S. aureus GatCAB with E. coli tRNAGln to understand how the enzyme uses the tail domain to bind the tRNA. The use of E. coli tRNAGln is reasonable as the sequences of the variable and D-loops of the S. aureus and E. coli tRNAGln are identical (Figure 2A), and E. coli Glu-tRNAGln serves as substrate for GatCAB in vivo (39).
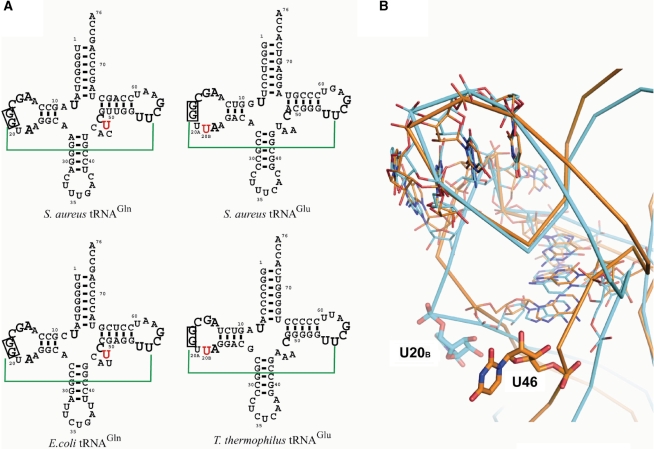
Figure 2.
Comparison with tRNAGln and tRNAGlu. (A) Secondary structures of tRNAGln tRNAGlu from S. aureus, E. coli and T. thermophilus. The G18:C56 base pair are colored green. The two guanines conserved in the tRNA D-loop are enclosed with black boxes. U8, A14–G19, A21, C48 and U54–G57, which employed on superposing of tRNAGln and tRNAGlu, are shown as black large characters. U47 of tRNAGln and U20B of tRNAGlu are colored red. (B) Superposition of the E. coli tRNAGln and T. thermophilus tRNAGlu. Orange and cyan ribbon diagram indicate E. coli tRNAGln and T. thermophilus tRNAGlu, respectively. Supplemental nucleotides of the variable loop of E. coli tRNAGln and the D-loop of T. thermophilus tRNAGlu are shown as thick stick models. The U8–A14–A21 base-triple, the conserved ‘Levitt’ pair and tRNA D-loop and TΨC -loop associating nucleotides are indicated as thin stick models.
To construct the docking model we first superposed E. coli tRNAGln (PDB ID: 1QTQ) into the GatDE:tRNAGln co-crystal structure by aligning all atoms of nucleotides U8, A14–G19, A21, C48 and U54–G57 of the tRNAs. The U8–A14–A21 base triple in the augmented D-stem is commonly found in tRNAGln while nucleotide 15 in the D-loop and nucleotide 48 in the variable loop form the conserved Levitt pair (40,41). The tertiary interaction between the D- and TΨC- loops of tRNA is also well conserved.
Next, we docked S. aureus GatCAB to tRNAGln by taking advantage of the homology between GatB and GatE (8,16), and the co-crystal structure of GatDE:tRNAGln (12), superposing GatB with M. thermautotrophicus GatE. The cradle domain of S. aureus GatB and M. thermautotrophicus GatE superpose well with an r.m.s. deviation of 1.7 Å for 244 pairs of Cα atoms compared. However, initially there were severe clashes between the helical domain and tRNAGln. Therefore, we separated the helical and tail domains into three parts (293–363, 364–381 and 382–475) and then superposed into GatE independently. Such movements are predicted based on previous AdT structures (11,12,17). Finally, the cradle and helical domains of S. aureus GatB could be superposed into that of M. thermautotrophicus GatE with an r.m.s. deviation of 1.9 Å for 321 pairs of Cα atoms compared (Supplementary Figure S4A).
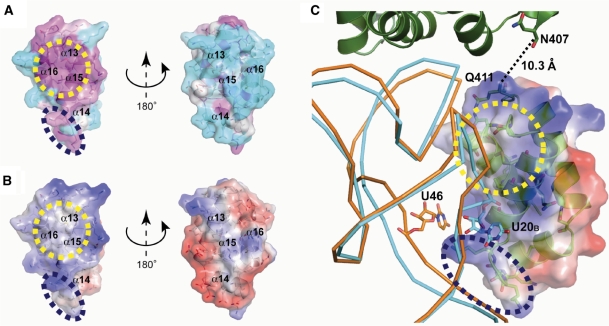
The GatB tail domain possesses a highly conserved hydrophobic pocket comprised by residues (V449, M452, G457, A459 and P461) from the α15, α16 and the loop between them (Figure 3A), surrounded by positively charged residues (Figure 3B). In contrast, the opposite surface of the tail domain is composed of non-conserved, mostly negative residues (Figure 3A and B). These observations suggest that this conserved hydrophobic pocket may recognize the shape of the tRNAGln D-loop with the surrounding positive residues interacting with the tRNA phosphate backbone (Figure 3C).
Figure 3.
(A) Surface representation with a ribbon diagram of S. aureus tail domain is shown, and is colored by the sequence conservation among bacterial GatB using the program ConSurf 3.0: from low to high (cyan to white to magenta). The conserved hydrophobic pocket and the Lys rich motif are shown as yellow and purple dashed circles, respectively. (B) Solvent-accessible surface with a ribbon diagram of S. aureus tail domain is shown in the same orientation as (A) and is colored according to the electrostatic potential calculated by the program APBS running on Pymol (blue for positively charged and red for negatively charged). (C) The S. aureus tail domain docking model. S. aureus tail domain is shown as in (B). Orange and cyan ribbon diagram indicate E. coli tRNAGln and T. thermophilus tRNAGlu, respectively. Supplemental nucleotides of the variable loop of E. coli tRNAGln and the D-loop of T. thermophilus tRNAGlu are shown as a stick model. The helical domain from the S. aureus GatB:tRNAGln docking model (described below) shown together. The inter-domain loop between the helical and the tail domain is shown as dashed lines.
Interaction of the tail domain with tRNA is expected, as deletion of the C-terminal portion of GatB gives rise to a GatCAB mutant enzyme unable to bind tRNA (11) and as mentioned the electron density map of the GatDE:tRNAGln structure places the YeqY-like tail domain in proximity of the tRNA D-loop (12). However, the initial S. aureus GatCAB:tRNAGln docking model placed the conserved hydrophobic pocket of the tail domain ∼12 Å away from the D-loop of tRNAGln (Supplementary Figure S4B). It is likely that the tail domain can move to interact with tRNAGln due to the flexible loop connecting the tail and helical domains. This domain flexibility may explain why it has been difficult to resolve the YqeY-like tail of previous AdT structures (11,12,17,18). Given the above, we manually fitted the pocket of the tail domain into the D-loop of the E. coli tRNAGln (Figure 3C). The distance between the N-terminal end of the tail domain (Q411) and the C-terminal end of the helical domain (N407) is ∼10 Å, a distance the inter-domain loop connecting the two domains can bridge (Figure 3C). Recognition of the D-loop is consistent with the fact bacterial GatCAB uses that tRNA element to distinguish transamidation substrates (Glu-tRNAGln and Asp-tRNAAsn) from Glu-tRNAGlu and Asp-tRNAAsp (11,13).
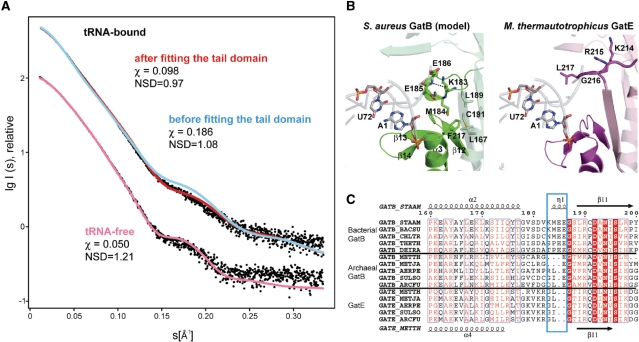
In order to verify our final S. aureus GatB:tRNAGln docking model, we recorded small-angle X-ray diffusion spectra on the tRNA-free and -bound S. aureus GatCAB purified by a size exclusion chromatography (Supplementary Figure S5). The predicted curve calculated from the crystal structure of the full-length S. aureus GatCAB with the program CRYSOL (32) fits closely to the experimental curve of tRNA-free S. aureus GatCAB, as characterized by a discrepancy value χ of 0.050 (Figure 4A). Furthermore, the crystal structure fits well to the ab initio envelope calculated with the program DAMAVER (36), of 16 dummy atom models calculated with DAMMIN (35), as reflected by a normalized spatial discrepancy (NSD) value of 1.21 (4A). The predicted scattering curve of the tRNAGln docking model fits well the experimental curve of tRNA-bound S. aureus GatCAB (χ of 0.098) and the ab initio envelope (NSD of 0.97) (Figure 4A). However, the tRNAGln docking model before fitting the tail domain shows a significantly higher discrepancy value with the experimental curve of tRNA-bound S. aureus GatCAB and the ab initio envelope (χ of 0.186 and NSD of 1.08, respectively) than using the tail domain fitting model. These results suggest that our docking model with the tail domain interacting with the tRNA is consistent with the solution structure of the S. aureus GatCAB:tRNAGln complex.
Figure 4.
(A) Experimental and computed SAXS scattering data. (Upper) the tRNA-bound GatCAB, (lower) tRNA-free GatCAB. The logarithm of the scattering intensity is plotted against the momentum transfer s=4πsinθ/λ, where 2θ is the scattering angle and λ=1.0 Å is the X-ray wavelength. The plots are displaced along the ordinate for better visualization. Red, cyan and pink curves indicated the computed scattering from tRNA-bound GatCAB model after the tail domain fitting, before the tail domain fitting, and tRNA-free GatCAB, respectively. (B) Comparison of the model of the S. aureus GatB:tRNAGln complex (left) and the crystal structure of the M. thermautotrophicus GatE:tRNAGln complex (right). S. aureus GatB, M. thermautotrophicus GatE and M. thermautotrophicus tRNAGln are shown as green, magenta and grey ribbon diagram, respectively. The A1–U72 base pair of M. thermautotrophicus tRNAGln is shown as a stick model. The 310 turn and the hydrophobic core of S. aureus GatB and the short loop of M. thermautotrophicus GatE are also shown. (C) Structural based sequence alignments of the cradle domain of bacterial GatB, archaeal GatB and GatE. The number at top is corresponding to S. aureus GatB sequence. Secondary structure of S. aureus GatB is indicated at top, and that of M. thermautotrophicus GatE is indicated at bottom. The 310 turn of bacterial GatB and the corresponding region of archaeal GatB and GatE are shown as cyan box. The species aligned are as follows: CHLTR, Chlamydia trachomatis; THETH, Thermus thermophilus HB8; METJA, Methanococcus jannaschii; AERPE, Aeropyrum pernix; SULSO, Sulfolobus solfataricus; ARCFU, Archaeoglobus fulgidus.
In our docking model, the hydrophobic pocket of the GatB tail domain nicely accommodates the curve of the tRNAGln D-loop (Figure 3C). The S. aureus enzyme distinguishes tRNAGln from tRNAGlu based on the presence of an extra base (U20B) in the D-loop of tRNAGlu (11). To gain a better understanding of how this is accomplished we superposed the structures of E. coli tRNAGln (PDB ID: 1QTQ) and Thermus thermophilus tRNAGlu (PDB ID: 1G59) in a similar manner as described above. The D-loop of T. thermophilus tRNAGlu like that of S. aureus tRNAGlu possesses a U20B supernumerary base (Figure 2A). That extra base flips out from the tRNA D-loop and TΨC-loop associating region (Figure 2B). In the docking model of the tail domain with tRNA, the flipped out U20B in the D-loop of tRNAGlu could not be accommodated; the extra base sterically clashes with the surface of the protein in the model (Figure 3C), suggesting that is the mechanism by which S. aureus GatCAB rejects tRNAGlu.
In addition to recognition of the D-loop, the GatB tail domain may also bind to the variable loop of the tRNA. In our docking model, the GatB-specific Lys rich motif of the tail domain is situated in proximity to the variable loop of tRNAGln, in particular U46 which is pushed out from the tertiary core of the tRNA (Figures 2B and 3C). Consistent with this prediction, replacement of this Lys rich motif with the GXXAXGX motif from GatE results in a mutant S. aureus GatCAB enzyme with reduced affinity for tRNA (Supplementary Figure S6).
The GatB cradle domain recognizes the first base pair of the acceptor stem
Bacterial GatCAB enzymes recognize the U1–A72 base pair of the acceptor stem of Glu-tRNAGln and Asp-tRNAAsn to discriminate them from Glu-tRNAGlu and Asp-tRNAAsp (11,13). In contrast, archaeal GatCAB does not use the first base pair of the tRNA acceptor stem to distinguish Asp-tRNAAsn from Asp-tRNAAsp, recognizing aa-tRNA species with either a U1–A72 or a G1–C72 base pair (10,13,14). However, the process of distinguishing a U1–A72 base pair from a G1–C72 base pair is not known.
In our GatCAB:tRNAGln docking model the tRNAGln acceptor stem U1–A72 base pair is located in the center of the cradle domain in a space constructed by α3, two internal loops between β12–α3 and β13–β14, and a 310 turn between α2–β11. The 310 turn is adjacent to the U1–A72 base pair (Figure 4B). This 310 turn is constructed by a hydrogen bond between the main-chain carbonyl oxygen of K183 and amide of E186. Furthermore, M184 in the 310 turn forms a hydrophobic core with L167, L189, C191 and F217, suggesting the 310 turn is fixed with α3 and β11 by a hydrophobic interaction. In the GatDE enzyme, GatE has a short loop in place of a 310 turn at the corresponding region (Figure 4B and C).
In an alignment of GatB and GatE sequences (Figure 4C), the region including the 310 turn (183–186 in S. aureus GatB) is conserved in bacterial GatB sequences, and the corresponding region in archaeal GatB is 1–3 residues shorter, suggesting archaeal GatB has a loop in place of the 310 turn like GatE. The loop region of GatE is well conserved and is rich in positive residues, however the corresponding region in archaeal GatB is not well conserved. The docking model and alignment suggest these structural differences between bacterial GatB, archaeal GatB and GatE may enable the differences in first base pair recognition between the enzymes. In particular, they imply that the 310 turn enables bacterial GatB to distinguish a U1–A72 from a G1–C72 base pair.
The 310 turn of bacterial GatB is crucial for tRNAGln acceptor helix U1–A72 base pair recognition
To evaluate the relation between the presence of the 310 turn in GatB and the specificity of first base pair recognition, we constructed a mutant S. aureus GatCAB enzyme in which the 310 turn (K183, M184 and E185) is replaced by the short putative loop from M. thermautotrophicus GatB. Based on the multiple sequence alignment (Figure 4C) we also constructed a mutant M. thermautotrophicus GatCAB where the 310 turn (residues Lys, Met and Glu) is inserted into GatB between residues Gly169 and Glu170. We then tested the transamidase activities (Tables 2 and 3) by the [32P]tRNA/nuclease P1 assay (38) of these mutant GatCAB enzymes with a variety of tRNA substrates.
Table 2.
Transamidase activity of the S. aureus (Sa) GatCAB and M. thermautotrophicus (Mt) GatCAB with different mischarged tRNA substrates
| Transamidase activity (s−1) |
||||
|---|---|---|---|---|
| aa-tRNAa |
Sa GatCABb |
Mt GatCABb |
||
| wt | 310Δ | wt | 310 ins | |
| Sa Glu-tRNAUAGln | 0.71±0.03 | 0.05±<0.01 | 0.0035±0.0003 | 0.0047±0.0002 |
| Sa Glu-tRNAGCGln | 0.05±<0.01 | 0.03±<0.01 | 0.0014±0.0002 | 0.0004±0.0002 |
| Mt Asp-tRNAGCAsn | 0.05±0.03 | 0.04±0.01 | 0.05±0.01 | 0.01±<0.01 |
| Ct Asp-tRNAUAAsn | 0.24±<0.01 | 0.02±<0.01 | 0.04±<0.01 | 0.11±<0.01 |
Measurements were from three separate experiments. Standard deviations are reported. Reactions were carried out at 37°C in the presence of ATP (4 mM), amide donor (4 mM) and aa-tRNA (1.25 µM) indicated.
aThe aa-tRNA substrates tested were the S. aureus (Sa) wild-type Glu-tRNAUAGln and mutant Glu-tRNAGCGln, the wild type of the M. thermautotrophicus (Mt) Asp-tRNAGCAsn and the C. trachomatis (Ct) Asp-tRNAUAAsn.
bIn the reactions, concentrations of the GatCAB indicated [Sa wild-type (wt) GatCAB, Sa 310 turn deleted mutant GatCAB (310Δ), Mt wt GatCAB, or Mt 310 turn insertion mutant GatCAB (310 ins)] ranged from 20 nM to 1 µM.
Table 3.
Kinetic data for the transamidase activity of the S. aureus and M. thermautotrophicus GatCAB mutants with different mischarged tRNA substrates
| Enzyme |
Sa Glu-tRNAUAGln |
Mt Asp-tRNAGCAsn |
L | ||||
|---|---|---|---|---|---|---|---|
| KM (mM) | kcat (s−1) | kcat/KM (s−1/mM) | KM (mM) | kcat (s−1) | kcat/KM (s−1/mM) | ||
| Sa wt | 1.80±0.22 | 1.7±0.1 | 930±120 | 2.05±0.48 | 0.1±<0.1 | 60±15 | 15.5 |
| Sa 310Δ | 1.85±0.59 | 0.1±<0.1 | 54±18 | 0.61±0.40 | 0.04±<0.01 | 67±44 | 0.8 |
| Mt wt | 0.70±0.26 | 0.006±0.001 | 8±3 | 0.78±0.30 | 0.10±0.01 | 125±49 | 15.6 |
| Mt 310 ins | 0.97±0.22 | 0.009±0.001 | 9±2 | 2.51±0.51 | 0.03±<0.01 | 12±3 | 1.3 |
Measurements were from three separate experiments. Standard deviations are reported. Reactions were carried out at 37°C in the presence of excess ATP (4 mM) and Gln (4 mM). L is the relative catalytic efficiency [(kcat/KM of homologous substrate)/(kcat/KM of non-homologous substrate)]. GatCAB enzymes labeled like in Table 2.
As expected (10,13,14), wild-type S. aureus GatCAB prefers tRNAs with a U1–A72 base pair over those with a G1–C72 base pair, while wild-type M. thermautotrophicus GatCAB has no strong preference (Table 2). S. aureus Glu-tRNAGln was a poor substrate for wild-type M. thermautotrophicus GatCAB (Tables 2 and 3), like B. subtilis Glu-tRNAGln for unknown reasons (10). However, the transamidase activity of wild-type M. thermautotrophicus GatCAB with wild-type Glu-tRNAGln containing a U1–A72 base pair was approximately the same as with the G1–C72 mutant Glu-tRNAGln (0.0035 and 0.0014 s−1, respectively; Table 2); this is in line with previous results archaeal GatCAB (10,13,14).
Deletion of the 310 turn from S. aureus GatCAB resulted in a mutant enzyme that was 17-fold less efficient than wild-type mostly due to a difference in kcat (Table 3), possibly due to the enzyme no longer recognizing the first base pair of the substrate aa-tRNA. However, consistent with our model, the removal of the 310 turn from the S. aureus GatB resulted in a mutant S. aureus GatCAB that no longer strongly preferred tRNA substrates with a U1–A72 base pair to those with a G1–C72 base pair (Tables 2 and 3). For example the mutant S. aureus GatCAB could use the mutant S. aureus Glu-tRNAGCGln as a substrate about as well as wild-type S. aureus Glu-tRNAUAGln (0.03 and 0.05 s−1, respectively, Table 2). In addition, the mutant S. aureus GatCAB could use M. thermautotrophicus Asp-tRNAAsn, with its G1–C72 base pair, about as well as Chlamydia trachomatis Asp-tRNAAsn, which has a U1–A72 base pair (Table 2) and was about as efficient in amidating M. thermautotrophicus Asp-tRNAGCAsn as S. aureus Glu-tRNAUAGln (Table 3).
Consistent with our model, the insertion of the 310 turn into M. thermautotrophicus GatCAB gave a mutant enzyme with preference for tRNA substrates with a U1–A72 base pair (Tables 2 and 3). The mutant M. thermautotrophicus GatCAB was about as efficient as wild-type M. thermautotrophicus GatCAB using S. aureus Glu-tRNAUAGln as a substrate (Table 3). However, compared to wild-type M. thermautotrophicus GatCAB, the mutant enzyme was 10-fold less efficient with the M. thermautotrophicus Asp-tRNAGCAsn substrate (Table 3). In addition, the mutant M. thermautotrophicus GatCAB preferred by 10-fold a substrate wild-type S. aureus Glu-tRNAUAGln substrate over the G1–C72 mutant Glu-tRNAGln (Table 2). Also, while wild-type M. thermautotrophicus GatCAB was nearly as active with C. trachomatis Asp-tRNAUAAsn as with M. thermautotrophicus Asp-tRNAGCAsn, the insertion GatCAB mutant preferred C. trachomatis Asp-tRNAAsn, which has a U1–A72 base pair (Table 2). These results indicate that insertion of the 310 turn into GatB enables M. thermautotrophicus GatCAB to distinguish aa-tRNA with a U1–A72 base pair from ones with a G1–C72 base pair. Taken with the mutant S. aureus GatCAB data, these results strongly suggest that the 310 turn in S. aureus GatCAB is crucial for distinguishing aa-tRNA with a U1–A72 from those with a G1–C72 base pair.
DISCUSSION
The bacterial GatCAB enzyme uses the first base pair of the acceptor stem and the tRNA D-loop for precise tRNA discrimination (11,13). Our results suggest that the U1–A72 base pair of tRNAAsn and tRNAGln is recognized by a 310 turn in the cradle domain of GatB, while the tail domain of GatB binds the D-loop.
The transamidosome is a ternary complex of T. thermophilus GatCAB, ND-AspRS and tRNAAsn stable during the overall catalytic process (42). Such a structure would enable Asn-tRNAAsn formation without the risk of free mischarged Asp-tRNAAsn being used in protein synthesis. The complex would also protect the Asn-tRNAAsn product from deacylation until it will be bound by EF-Tu and transported to the ribosome (42,43). Complexes between ND-GluRS, tRNAGln, and either Glu-AdT (GatCAB or GatDE) are also predicted to exist (12,42). Formation of these complexes between ND-aaRS, AdT and tRNA may explain why the AdTs recognize the specific identity elements in their tRNA substrates.
The structural models of the transamidosomes predict the ND-aaRS enzyme binds to the acceptor stem and anticodon loop of the tRNA (tRNAGln or tRNAAsn) (12,42) (Supplementary Figure S7) in the same fashion as they do in the absence of AdT (44,45). This places the tRNA 3′-end into the synthetase active site to be aminoacylated; initially the first base pair of the tRNA’s acceptor helix is not accessible to the 310 turn of bacterial GatB in its role to discriminate tRNA isoacceptors. However, the tRNA’s tertiary core (including the D-loop) is recognized (Supplementary Figure S7). Recognition of the D-loop by the AdT tail domain (11,12) may permit the amidotransferases to distinguish the ND-aaRS complexed with their tRNA transamidation substrates (tRNAGln or tRNAAsn) from other tRNA isoacceptors (tRNAGlu or tRNAAsp) (12–14). For example, GatCAB recognition of the D-loop would enable the AdT to discriminate ND-AspRS bound to tRNAAsn from the aaRS bound to tRNAAsp despite the U1–A72 base pair being initially inaccessible to GatCAB. The variable loop of the tRNA in the transamidsome models is also accessible to the AdT, in particular the Lys rich motif in the tail domain of GatB, which may explain why archaeal GatCAB uses this element to discriminate tRNAAsn from tRNAAsp (13,14).
The transamidosome models (12,42) predict that after aminoacylation the tRNA’s 3′ CCA terminus flips from active site in the ND-aaRS to the kinase active site of the AdT (42), similar to the tRNA movements seen in certain aaRSs with editing domains (46,47). In the case of the transamidosome, this movement enables the aminoacyl-moiety of the mischarged tRNA to be amidated by the AdT in the complex. Once the 3′ end of the acceptor stem flips from the aaRS active site into the transamidase active site of the AdT, the U1–A72 base pair may become accessible to the 310 turn in bacterial GatB.
Recognition of the first base pair by bacterial GatCAB and the archaeal GatDE may be a proofreading step to ensure amidation of the mischarged tRNA substrate (Glu-tRNAGln and/or Asp-tRNAAsn) and not the properly aminoacylated product of the ND-aaRS (Glu-tRNAGlu or Asp-tRNAAsp). Why this proofreading step is not required by the archaeal GatCAB is unclear, but presumably recognition of the tRNA tertiary core may be enough to ensure that GatCAB amidates Asp-tRNAAsn and not Asp-tRNAAsp (13,14).
It was speculated that the ancestor of GatB and GatE recognized the first base pair of its tRNA substrates (16). However, as the 310 turn in the cradle domain of the S. aureus GatB for recognition of the U1–A72 base pair in tRNAGln is conserved only in bacterial GatB and not in archaeal GatB and GatE, this may not be the case. Instead, the common ancestor of GatB and GatE may not have recognized the first base pair of its tRNA substrate, with bacterial GatCAB and GatDE independently evolving to recognize the first base pair of tRNA. Thus, bacterial GatCAB and GatDE both recognizing the first base pair may have been a case of convergent and not divergent evolution.
ACCESSION NUMBERS
Coordinate and structure factor have been deposited in PDB with accession number 3IP4.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
A.N. held a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists. Grants-in-Aid for Scientific Research (19370037 and 21370041 to M.Y.); Targeted Proteins Research Program (to I.T.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; Institute of General Medical Sciences and the US Department of Energy (to D.S.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to S. Chimnaronk, I. Heinemann, J. Ling and Y. Tanaka for many stimulating discussions, and to T. Tamura (National Institute of Advanced Industrial Science and Technology) for his gift of the R. erythropolis expression system. They thank the staff of SPring-8 BL41XU, 40B2 (2009A1119) and 45XU beam lines for their help during collection of the X-ray data.
REFERENCES
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Sheppard K, Yuan J, Hohn MJ, Jester B, Devine KM, Söll D. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 2008;36:1813–1825. doi: 10.1093/nar/gkn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lapointe J, Duplain L, Proulx M. A single glutamyl-tRNA synthetase aminoacylates tRNAGlu and tRNAGln in Bacillus subtilis and efficiently misacylates Escherichia coli tRNAGln1 in vitro. J. Bacteriol. 1986;165:88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilcox M, Nirenberg M. Transfer RNA as a cofactor coupling amino acid synthesis with that of protein. Proc. Natl Acad. Sci. USA. 1968;61:229–236. doi: 10.1073/pnas.61.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curnow AW, Ibba M, Söll D. tRNA-dependent asparagine formation. Nature. 1996;382:589–590. doi: 10.1038/382589b0. [DOI] [PubMed] [Google Scholar]
- 6.Becker HD, Reinbolt J, Kreutzer R, Giegé R, Kern D. Existence of two distinct aspartyl-tRNA synthetases in Thermus thermophilus. Structural and biochemical properties of the two enzymes. Biochemistry. 1997;36:8785–8797. doi: 10.1021/bi970392v. [DOI] [PubMed] [Google Scholar]
- 7.Curnow AW, Hong K, Yuan R, Kim S, Martins O, Winkler W, Henkin TM, Söll D. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl Acad. Sci. USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tumbula DL, Becker HD, Chang WZ, Söll D. Domain-specific recruitment of amide amino acids for protein synthesis. Nature. 2000;407:106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- 9.Roy H, Becker HD, Reinbolt J, Kern D. When contemporary aminoacyl-tRNA synthetases invent their cognate amino acid metabolism. Proc. Natl Acad. Sci. USA. 2003;100:9837–9842. doi: 10.1073/pnas.1632156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheppard K, Sherrer RL, Söll D. Methanothermobacter thermautotrophicus tRNAGln confines the amidotransferase GatCAB to asparaginyl-tRNAAsn formation. J. Mol. Biol. 2008;377:845–853. doi: 10.1016/j.jmb.2008.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura A, Yao M, Chimnaronk S, Sakai N, Tanaka I. Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science. 2006;312:1954–1958. doi: 10.1126/science.1127156. [DOI] [PubMed] [Google Scholar]
- 12.Oshikane H, Sheppard K, Fukai S, Nakamura Y, Ishitani R, Numata T, Sherrer RL, Feng L, Schmitt E, Panvert M, et al. Structural basis of RNA-dependent recruitment of glutamine to the genetic code. Science. 2006;312:1950–1954. doi: 10.1126/science.1128470. [DOI] [PubMed] [Google Scholar]
- 13.Bailly M, Giannouli S, Blaise M, Stathopoulos C, Kern D, Becker HD. A single tRNA base pair mediates bacterial tRNA-dependent biosynthesis of asparagine. Nucleic Acids Res. 2006;34:6083–6094. doi: 10.1093/nar/gkl622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Namgoong S, Sheppard K, Sherrer RL, Söll D. Co-evolution of the archaeal tRNA-dependent amidotransferase GatCAB with tRNAAsn. FEBS Lett. 2007;581:309–314. doi: 10.1016/j.febslet.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deniziak M, Sauter C, Becker HD, Paulus CA, Giegé R, Kern D. Deinococcus glutaminyl-tRNA synthetase is a chimer between proteins from an ancient and the modern pathways of aminoacyl-tRNA formation. Nucleic Acids Res. 2007;35:1421–1431. doi: 10.1093/nar/gkl1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheppard K, Söll D. On the Evolution of the tRNA-dependent amidotransferases, GatCAB and GatDE. J. Mol. Biol. 2008;377:831–844. doi: 10.1016/j.jmb.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitt E, Panvert M, Blanquet S, Mechulam Y. Structural basis for tRNA-dependent amidotransferase function. Structure. 2005;13:1421–1433. doi: 10.1016/j.str.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Bu W, Sheppard K, Kitabatake M, Kwon ST, Söll D, Smith JL. Insights into tRNA-dependent amidotransferase evolution and catalysis from the structure of the Aquifex aeolicus enzyme. J. Mol. Biol. 2009;391:703–716. doi: 10.1016/j.jmb.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakashima N, Tamura T. A novel system for expressing recombinant proteins over a wide temperature range from 4 to 35 degrees C. Biotechnol. Bioeng. 2004;86:136–148. doi: 10.1002/bit.20024. [DOI] [PubMed] [Google Scholar]
- 20.Sheppard K, Akochy PM, Salazar JC, Söll D. The Helicobacter pylori amidotransferase GatCAB is equally efficient in glutamine-dependent transamidation of Asp-tRNAAsn and Glu-tRNAGln. J. Biol. Chem. 2007;282:11866–11873. doi: 10.1074/jbc.M700398200. [DOI] [PubMed] [Google Scholar]
- 21.Stura EA. Seeding techniques. In: Ducroix A, Giegé R, editors. Crystallization of Nucleic Acids and Proteins: A Practical Approach, second edition. Oxford: IRL Press; 2000. pp. 177–207. [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CWJ, Sweet RM, editors. Methods in Enzymology, Volume 276: Macromolecular Crystallography, part A. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 23.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. Sect. A: Foundations Crystallogr. 1994;50:157–163. [Google Scholar]
- 24.Yao M, Zhou Y, Tanaka I. LAFIRE: software for automating the refinement process of protein-structure analysis. Acta Crystallogr. Sect. D: Biol. Crystallogr. 2006;62:189–196. doi: 10.1107/S0907444905038965. [DOI] [PubMed] [Google Scholar]
- 25.Zyou Y, Yao M, Tanaka I. New algorithm for protein model building. J. Appl. Crystallogr. 2006;39:57–63. [Google Scholar]
- 26.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D: Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. The PyMOL Molecular Graphics System. [Google Scholar]
- 29.Fujisawa T, Inoue K, Oka T, Iwamoto H, Uruga T, Kumasaka T, Inoko Y, Yagi N, Yamamoto M, Ueki T. Small-angle X-ray scattering station at the SPring-8 RIKEN beamline. J. Appl. Crystallogr. 2000;33:797–800. [Google Scholar]
- 30.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 31.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 32.Svergun DI, Barberato C, Koch MHJ. CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 33.Leulliot N, Chaillet M, Durand D, Ulryck N, Blondeau K, van Tilbeurgh H. Structure of the yeast tRNA m7G methylation complex. Structure. 2008;16:52–61. doi: 10.1016/j.str.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 34.Boczkowska M, Rebowski G, Petoukhov MV, Hayes DB, Svergun DI, Dominguez R. X-ray scattering study of activated Arp2/3 complex with bound actin-WCA. Structure. 2008;16:695–704. doi: 10.1016/j.str.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 2001;34:33–41. [Google Scholar]
- 38.Sheppard K, Akochy PM, Söll D. Assays for transfer RNA-dependent amino acid biosynthesis. Methods. 2008;44:139–145. doi: 10.1016/j.ymeth.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baick JW, Yoon JH, Namgoong S, Söll D, Kim SI, Eom SH, Hong KW. Growth inhibition of Escherichia coli during heterologous expression of Bacillus subtilis glutamyl-tRNA synthetase that catalyzes the formation of mischarged glutamyl-tRNA1Gln. J. Microbiol. 2004;42:111–116. [PubMed] [Google Scholar]
- 40.Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perona JJ, Hou YM. Indirect readout of tRNA for aminoacylation. Biochemistry. 2007;46:10419–10432. doi: 10.1021/bi7014647. [DOI] [PubMed] [Google Scholar]
- 42.Bailly M, Blaise M, Lorber B, Becker HD, Kern D. The transamidosome: a dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Mol. Cell. 2007;28:228–239. doi: 10.1016/j.molcel.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 43.Huot JL, Balg C, Jahn D, Moser J, Émond A, Blais SP, Chênevert R, Lapointe J. Mechanism of a GatCAB amidotransferase: aspartyl-tRNA synthetase increases its affinity for Asp-tRNAAsn and novel aminoacyl-tRNA analogues are competitive inhibitors. Biochemistry. 2007;46:13190–13198. doi: 10.1021/bi700602n. [DOI] [PubMed] [Google Scholar]
- 44.Ruff M, Krishnaswamy S, Boeglin M, Poterszman A, Mitschler A, Podjarny A, Rees B, Thierry JC, Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNAAsp. Science. 1991;252:1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- 45.Sekine S, Nureki O, Shimada A, Vassylyev DG, Yokoyama S. Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase. Nat. Struct. Biol. 2001;8:203–206. doi: 10.1038/84927. [DOI] [PubMed] [Google Scholar]
- 46.Nureki O, Vassylyev DG, Tateno M, Shimada A, Nakama T, Fukai S, Konno M, Hendrickson TL, Schimmel P, Yokoyama S. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- 47.Silvian LF, Wang J, Steitz TA. Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science. 1999;285:1074–1077. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.