Abstract
The cohesin protein complex holds sister chromatids together after synthesis until mitosis. It also contributes to post-replicative DNA repair in yeast and higher eukaryotes and accumulates at sites of laser-induced damage in human cells. Our goal was to determine whether the cohesin subunits SMC1 and Rad21 contribute to DNA double-strand break repair in X-irradiated human cells in the G2 phase of the cell cycle. RNA interference-mediated depletion of SMC1 sensitized HeLa cells to X-rays. Repair of radiation-induced DNA double-strand breaks, measured by γH2AX/53BP1 foci analysis, was slower in SMC1- or Rad21-depleted cells than in controls in G2 but not in G1. Inhibition of the DNA damage kinase DNA-PK, but not ATM, further inhibited foci loss in cohesin-depleted cells in G2. SMC1 depletion had no effect on DNA single-strand break repair in either G1 or late S/G2. Rad21 and SMC1 were recruited to sites of X-ray-induced DNA damage in G2-phase cells, but not in G1, and only when DNA damage was concentrated in subnuclear stripes, generated by partially shielded ultrasoft X-rays. Our results suggest that the cohesin complex contributes to cell survival by promoting the repair of radiation-induced DNA double-strand breaks in G2-phase cells in an ATM-dependent pathway.
INTRODUCTION
DNA double-strand breaks (DSBs) are a major threat to the genomic integrity of a cell. They can result in cell death if left un-repaired, or, if incorrectly repaired, can produce chromosomal aberrations and are thought to induce cancer (1,2). DSBs are induced by ionizing radiation, a range of chemotherapeutic drugs and are formed endogenously during DNA replication or as initiators of programmed genetic rearrangement processes that occur during lymphocyte differentiation and meiosis. In order to repair DSBs, higher eukaryotic cells primarily utilize two conceptually different pathways, non-homologous end-joining and homologous recombination. Non-homologous end-joining repairs DSBs with no requirement for sequence homology at the break ends and operates throughout the mammalian cell cycle. Homologous recombination, which utilizes an undamaged template of a homologous sequence for faithfully restoring the sequence at the break site, preferentially contributes to DSB repair in late S/G2 when a sister chromatid is available to serve as template (3–5).
During replication, the newly synthesized sister chromatids are tied together by the cohesin complex that forms a ring around chromatids (6). It consists of Smc1, Smc3, Scc1/Mcd1/Rad21 and Scc3/SA1/SA2 (7,8). The cohesin complex plays an important role in the fidelity of sister chromatid separation and chromosome segregation during anaphase (9) but is also involved in other aspects of chromosome metabolism. Cohesin is believed to facilitate DNA repair by tethering sister chromatids. In yeast and human cells, proteins needed to load cohesin onto chromosomes and generate cohesion during the S phase (Scc2, Eco1, sororin) are also shown to be required for repair (10,11). Furthermore, cohesin is recruited to chromatin regions surrounding an enzymatically induced DSB in a γH2AX-dependent manner in Saccharomyces cerevisiae (12,13). Interestingly, recent findings suggest that one DSB induced enzymatically in one S. cerevisiae chromosome results in increased sister chromatid cohesion of all chromosomes (14,15). Cohesin is recruited to regions of laser scissor-induced nuclear damage in mammalian cells (16), but only at very high power settings (17).
There is evidence that the securin–separase complex has a DNA damage repair role in interphase by cleavage of the Rad21 subunit (18,19). It is not yet clear if this occurs after damage to promote repair or whether it happens after repair to release the additional loaded cohesin subunits.
A number of studies have presented evidence for the involvement of cohesin in DSB repair in yeast and vertebrate cells. However, many of the previous studies were done with enzymatically induced breaks that differ significantly in their chemical structure from radiation-induced ones. Also, these approaches tend to monitor very specific repair pathways and events at a specific genomic site. To date, two less selective approaches have been used to study the role of cohesin in DNA repair. The first was based on pulsed field gel electrophoresis of asynchronous S. pombe cells (20) or S. cerevisiae cells that had been chemically arrested in prometaphase (10,12–14). In the second approach chromosome aberrations were analysed in mitotic vertebrate cells following chemical synchronization in G1/S and gamma-irradiation in the late S phase (21). To avoid any interference from the G2/M checkpoint in an otherwise very similar approach, Schmitz et al. (11) treated cells with caffeine, a cell cycle checkpoint inhibitor that can block the key DNA damage response kinases ATM, ATR and DNA-PKcs.
In contrast to previous studies, we wished to determine, in defined cell cycle stages without the use of chemical inhibitors, the role of cohesin in the repair of bulk DNA damage induced by an environmentally and clinically relevant agent, ionizing radiation. Using a number of independent approaches, we have attempted to analyse DSB repair in SMC1 or Rad21-depleted human G1- and G2-phase cells whilst minimizing interference from checkpoint functions or chemical agents.
MATERIALS AND METHODS
Antibodies
Rabbit polyclonal antibodies against SMC1, SA1, SA2 and SMC3 were purchased from Bethyl Laboratories (Universal Biologicals, Cambridge, UK), the mouse anti-human Cyclin A (clone 6E6) was from Vector Laboratories (Peterborough, UK) and another rabbit polyclonal SMC1 antibody from Autogen Bioclear (ABF021 190; used only in Figure 6C and D). Anti-actin antibodies and horseradish peroxidase-conjugated secondary antibodies were obtained from Sigma (Gillingham, UK). Mouse monoclonal anti-Rad21 and anti-γH2AX (clone JBW301) were from Millipore (Dundee, UK). Rat anti-bromodeoxyuridine (BrdU) was from Cancer Research UK. The anti-CENP-F antibody (ab5) and anti-53BP1 antibody were obtained from Abcam (Cambridge, UK). AlexaFluor 488 and 532-conjugated secondary antibodies were purchased from Invitrogen (Paisley, UK) and all other secondary antibodies for immunofluorescence microscopy were from Jackson ImmunoResearch (Stratech, Suffolk, UK). \
Figure 6.
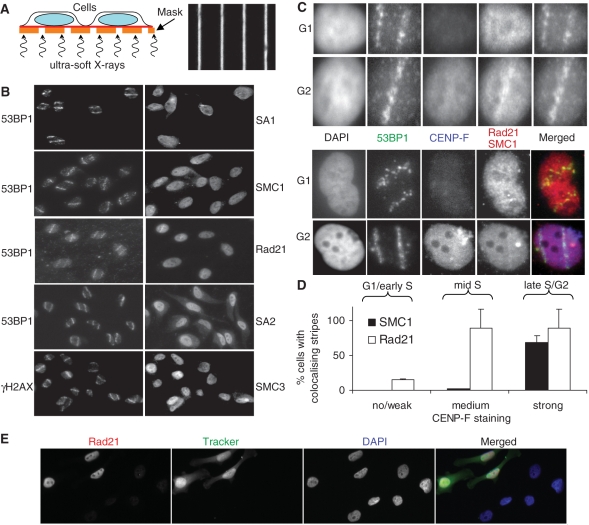
Recruitment of cohesin to ionizing radiation-induced DNA double strand breaks. (A) Schematic representation of micro-irradiation setup (left) and bright field image of the grid with 1 μm wide gaps/9 μm wide shielding used for generating defined patterns of DNA damage within the nucleus (right). (B) HeLa cells were grown on Mylar foil and irradiated with soft X-rays. After 1 h incubation, cells were pre-extracted with 0.1% Triton X-100, fixed and stained with the indicated antibodies. Each image is 125 μm wide. (C) As in (B) but cells were furthermore stained with CENP-F to determine the cell cycle phase. Each image is 15 μm wide. (D) Quantitative analysis of Rad21 and SMC1 stripe formation. Cells were divided into three groups of no or weak, medium and strong CENP-F staining. Microbeam tracks were predicted using the Rad21 or SMC1 signal, respectively, and then tested for co-localization with the 53BP1 signal. (E) Immunofluorescence microscopic analysis of Rad21 depletion. Control siRNA-transfected cells labelled with CellTrackerTM Green CMFDA (Invitrogen) were mixed with Rad21 siRNA-treated cells and stained for Rad21.
Cell culture
HeLa cells were obtained from Cancer Research UK Research Services. Cells were grown in Eagles minimum essential medium with 10% foetal calf serum, 2 mm l-glutamine, 1 mm sodium pyruvate, 0.1x nonessential amino acids, 0.135% sodium bicarbonate, 100 U/ml penicillin, 0.1 mg/ml streptomycin with 90%N2, 5%O2, 5% CO2 in a well-humidified incubator at 37°C. Exponentially growing cultures with a fraction of ∼65% G1 and ∼35% S/G2 cells were used for all experiments.
RNA interference
SiRNA transfections were performed using DharmaFECT 1 (Dharmacon) and Oligofectamine (Invitrogen), according to the suppliers’ instructions. SMC1-1 (5′-GCA AUG CCC UUG UCU GUG AUU-3′) and SMC1-3 (5′-GAA CAA AGA CAC AUG AAG AUU-3′) siRNAs (Dharmacon) target SMC1. RAD21 (5′-AUA CCU UCU UGC AGA CUG U-3′) siRNA (MWG Biotech) targets the human RAD21 homologue (22). SiCo1 (5′-UAG CGA CUA AAC ACA UCA A-3′), siCo2 (5′-UAA GGC UAU GAA GAG AUA CUU-3′) and lacZ (5′-CGU ACG CGG AAU ACU UCG A-3′) are control siRNAs (Dharmacon). All siRNAs were used at a final concentration of 100 nM unless specified otherwise. Cells were assayed 20–24 h after transfection.
Drug treatment
The KuDOS Pharmaceuticals agents ATMi/KU55 933 (23) and DNA-PKi/NU7026 (24,25) are potent inhibitors of the DNA damage response kinases ataxia telangiectasia mutated (ATM) and DNA-dependent protein kinase (DNA-PK), respectively. Inhibitors were added to the cells 1 h before irradiation to give a final concentration of 0.01 mM. Inhibitors were present during and after irradiation. For one set of experiments, cells were incubated in 0.02 mM bromodeoxyuridine (Sigma Aldrich) immediately before, during and after irradiation.
Irradiation
Conventional X-ray exposures were performed at room temperature with a 240 kV (constant potential) X-ray set (Pantak) at a dose rate of ∼1 Gy/min. For ultrasoft X-ray irradiations, cells were grown in a glass-walled dish with a 0.9 μm Mylar base, through which the cells were irradiated. The ultrasoft X-rays were produced using a cold cathode discharge tube (26) with an aluminium transmission target to produce predominantly Al-K characteristic X-rays (1.48 keV) with a discharge of 5 mA at a potential of 5 kV. Masks for partial shielding were made from 1 μm thick gold deposited on a 0.5 μm silicon nitride membrane. The gold was deposited in 9 μm wide stripes separated by 1 μm. The average radiation dose per cell was about 2.5 Gy with the majority of the energy and therefore damage being deposited in 1 μm stripes across the cell separated at 10 μm intervals. Following irradiation, cells were incubated in a well-humidified incubator at 37°C with 5% CO2.
Colony assay
Cells were trypsinized, counted and serial dilutions in triplicates incubated for colony formation following X-ray treatment. Flasks were stained with crystal violet and only colonies consisting of more than 50 cells were scored.
Micronucleus assay
Cells were grown and siRNA-transfected in LabTek four-well chamber slides. Thirty minutes post-irradiation, cells were treated with 2 μg/ml cytochalasin B for 4 h at 37°C, washed with phosphate-buffered saline and fixed for 10 min in ice-cold methanol. Dried slides were stained with Giemsa, washed in deionized water and analysed by bright field light microscopy using ×400 magnification.
Immunofluorescence microscopy
Cells grown in LAB-TEK four-well chamber slides were washed in phosphate-buffered saline (PBS) and fixed for 10 min in either 100% methanol at −20°C or in 3.7% formaldehyde at room temperature and permeabilized for 10 min with 0.25% Triton X-100. For ultrasoft X-ray experiments cells grown in 0.9 μm Mylar glass dishes were treated with 0.1% TritonX-100 in PBS for 2 min at room temperature to extract loosely bound proteins and then fixed with formaldehyde as above. Bromodeoxyuridine-treated cells were microwaved at 850 W for 5 min in 10 mM citric acid (pH 6.0) to denature DNA. Cells were blocked in PBS with 5% BSA for a minimum of 30 min at room temperature. All samples were incubated with primary antibodies for 1 h at room temperature, washed in PBS, 2% BSA for three times 5–10 min and incubated with fluorophore-conjugated secondary antibodies for 1 h at room temperature. Cells were counterstained with 4,6 diamidino-2-phenylindole (DAPI), washed in PBS and mounted. Fluorescence images were captured using a Nikon Eclipse TE200 or 90i fluorescence microscope equipped with cooled charge-coupled device camera and acquisition software. Identical illumination and camera settings were used within each data set. For quantitative analysis, intensities of SMC1 in siRNA-treated cells were determined using the ImageJ software. 53BP1 and γH2AX foci were counted by eye during the imaging process using a 100× objective. In at least two independent experiments for each data point, 40–100 cells were analysed. Samples were coded to prevent any bias.
Western blotting
Cell pellets were lysed in the ice-cold lysis buffer (1 X TBS, 1% Triton X-100) containing protease inhibitor cocktail (Sigma, Gillingham, UK) and then sonicated for 1 min. Samples were fixed in the Laemmli sample buffer and heated for 3 min at 100°C. Protein samples were separated on NuPage Novex 3–8% Tris-Acetate gels (Invitrogen) according to the manufacturer’s protocol. Proteins were then transferred onto Immobilon FL membranes in a transfer buffer for 60 min at 100 V. Membranes were blocked in Tris-buffered saline/0.1% Tween20 (TBST) containing 0.5% milk and 1% BSA for 1 h and then washed and incubated in TBST with 2.5% milk and primary antibody overnight at 4°C. They were then washed and incubated with secondary antibodies [Qdot655-conjugated goat anti-chicken (Invitrogen) and horseradish peroxidase-conjugated goat anti-rabbit (Dako Cytomation)] for 1 h at room temperature, washed several times and incubated with ECL Advance detection reagent (Amersham BioSciences) for 5 min prior to scanning. Fluorescence and chemiluminescence imaging was carried out using a Kodak ImageStation 4000MM system, and quantification was performed using Kodak MI software. The level of RNAi knockdown was determined using the level of actin as a standard.
Comet assay
Cells were X-irradiated on ice, incubated for 0, 1 or 2 h at 37°C, trypsinized and 12 500 cells were embedded in agarose on glass slides. Lysis in 1.2 M NaCl, 0.1% N-lauroyl sarcosine, 0.26 M NaOH, 100 mM Na2EDTA, pH > 12.5 (all Sigma, Poole, UK) was performed for 18–20 h at 4°C. Slides were rinsed three times in the 0.03 M NaOH/2 mM Na2EDTA electrophoresis buffer (pH > 12.5) before starting alkaline electrophoresis at 0.6 V/cm for 30 min. After electrophoresis, slides were placed immediately in cold 70% ethanol followed by 90% and 100% ethanol for 10 min each. Slides were air dried, stained with Sybr–Gold solution diluted 1: 10 000 and covered with a coverslip using the DABCO antifade mounting medium. The edges were sealed with nail varnish. Comet imaging and analysis was performed using an automated microscope system and analysis software developed by Vojnovic et al. (unpublished data). It calculates automatically various parameters including the percentage of DNA in the comet tail, tail length and tail moment for each cell analysed and gives the staining intensity of each individual cell that allows attribution of the cell cycle phase based on the DNA content. 200–500 G1-phase cells and 100–300 S/G2-phase cells were analysed for each data point.
Statistical analysis
Standard errors of the mean were calculated by dividing the standard deviation of the mean of more than two experiments by the square root of the number of experiments. The significance of any observed effects was tested individually for each siRNA using the Student’s t-test. In some cases, differences were only significant for pooled analysis of both SMC1-targeting siRNAs. This is stated in the text, where applicable.
RESULTS
SMC1 knockdown cells are radiosensitive
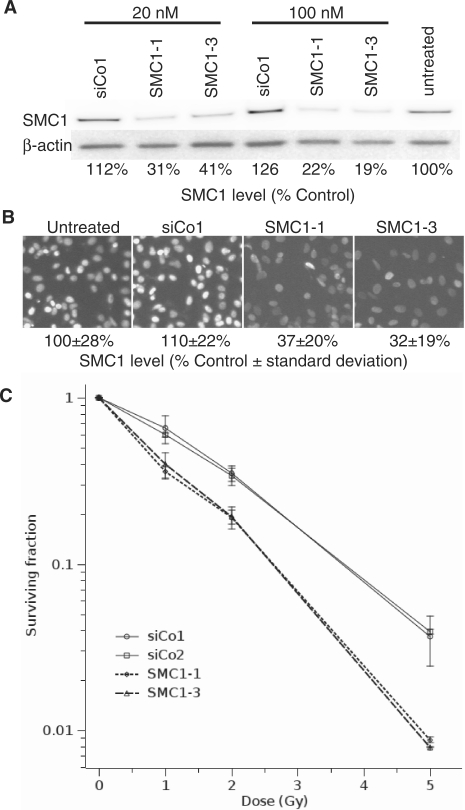
If the phosphorylation of SMC1 after DNA damage is suppressed, cells show decreased survival, increased chromosomal aberrations and a defect in the S-phase checkpoint (27,28). Here we wanted to investigate whether the SMC1 protein itself is contributing to cell survival and DNA damage repair. To do so we established small interfering RNA (siRNA) to specifically target the SMC1 subunit of the cohesin complex. SMC1 expression was depleted to ∼20% levels in HeLa cells by these two targeted siRNAs (SMC1-1 and SMC1-3) but was not reduced in samples treated with control siRNA (Figure 1A). Depletion of SMC1 was homogenous in the cell population (Figure 1B) and radiosensitized HeLa cells 2-fold at the 37% survival level, as measured by colony-formation (Figure 1C) but had no significant effect on the plating efficiency of un-irradiated cells (43 ± 6% for SMC1 siRNA versus 44 ± 2% for control siRNA). These data show that the SMC1 protein is important for cell survival after X-irradiation.
Figure 1.
SMC1 depletion radiosensitizes HeLa cells. (A) Depletion of SMC1 in HeLa cells using RNA interference. Cells were transfected with two different siRNAs against SMC1 (SMC1-1 and SMC1-3), a non-targeting siRNA (siCo1) or untransfected. Protein levels were analysed by western blotting. The percentage of SMC1 levels with respect to the untransfected control is shown. (B) Immunofluorescence microscopic analysis of SMC1 depletion. Details as in (A). (C) Colony formation assay for control and SMC1-depleted cells. Cells were transfected with two different siRNAs against SMC1 (SMC1-1 and SMC1-3) and two different non-targeting siRNAs (siCo1 and siCo2). Error bars are standard errors of the mean from three experiments.
SMC1 promotes the repair of radiation-induced DSBs in the G2 phase of the cell cycle
To determine whether SMC1 depletion affects the repair of ionizing radiation-induced DSBs, RNAi-treated exponentially growing HeLa cells were X-irradiated, incubated for repair, fixed and immunostained for the DSB marker γH2AX (Figure 2A). To distinguish cells in G1, S and G2 phase, cells were co-immunostained for CENP-F, a protein whose expression is tightly cell cycle-regulated (29,30). Radiation-induced γH2AX foci were scored in CENP-F-negative cells that were classified as G1-phase cells and in cells with a high CENP-F staining intensity and interphase-like DAPI staining at the time of analysis that were classified as late S/G2-phase cells. Cells with intermediate CENP-F staining were in early to mid-S phase at the time of analysis and were ignored as their cell cycle stage at the time of irradiation could have been either G1 or S.
Figure 2.
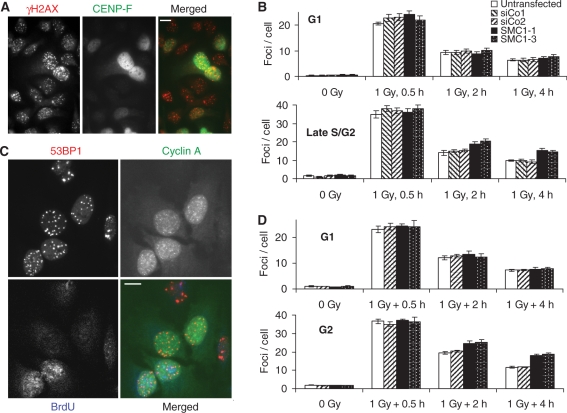
SMC1 depletion impairs the repair of DSBs in G2-phase cells. (A) γH2AX and CENP-F staining in SMC1-depleted cells 2 h after 1 Gy X-irradiation. (B) Time course for the loss of 1 Gy X-ray-induced γH2AX foci in G1- (CENP-F negative) and late S/G2-phase cells (CENP-F strongly positive). (C) 53BP1, Cyclin A and BrdU staining in SMC1-depleted cells 2 h after 1 Gy X-irradiation. (D) Time course for the loss of 1 Gy X-ray-induced 53BP1 foci in G1 (BrdU-negative cyclin A-negative) and G2-phase cells (BrdU-negative cyclin A positive). Error bars are standard errors of the mean from two to three experiments.
Depletion of SMC1 by RNAi had no significant effect on the number of foci present in un-irradiated cells; however, late S/G2-phase cells contained about three times more foci per cell than G1-phase cells (1.47 ± 0.14 versus 0.47 ± 0.05), irrespective of the RNAi treatment (Figure 2B). Also, 30 min after treatment with 1 Gy X-rays, foci levels were higher in late S/G2-phase cells (36.8 ± 0.8 per cell) than in G1-phase cells (22.5 ± 0.5 per cell) but again very similar across the different RNAi treatments. This difference likely reflects the higher DNA content of late S/G2-phase cells compared to G1-phase cells. At the same radiation dose, a cell with twice the amount of DNA would be expected to encounter twice the number of DSBs. Importantly, 2 and 4 h following X-irradiation significantly higher levels of residual γH2AX foci were present in SMC1-depleted cells than in controls in the late S/G2 phase (P < 0.005) while no difference was observed for G1-phase cells (Figure 2B). It should be noted that highly CENP-F-positive cells at the time of analysis could have been at any stage between the mid-S phase and G2 at the time of irradiation, at least for the 4 h repair samples.
To confirm this result using a different combination of DNA damage and cell cycle markers and to determine whether SMC1 promotes DSB repair even in non-replicating cells irradiated in G2, cells were labelled with bromodeoxyuridine 30 min before X-irradiation and throughout the post-irradiation incubation period. Cells were immunostained for bromodeoxyuridine, the cell cycle-regulated protein Cyclin A and the DSB marker 53BP1 (Figure 2C). To exclude any S-phase effects, only bromodeoxyuridine-negative cells were analysed and classified into G1 (Cyclin A negative) and G2 (Cyclin A positive, interphase-like DAPI staining) phase cells. Importantly, using this approach, cells could be selectively analysed that were in G2 at the time of irradiation and remained there throughout repair incubation. The results (Figure 2D) demonstrate the presence of a significantly higher level of residual 53BP1 foci in SMC1-depleted G2-phase cells 2 (P < 0.03) and 4 h (P < 0.001) after 1 Gy X-irradiation and thus confirm the above finding and show that cohesin contributes to the repair of DSBs that were induced after replication. A similar effect was observed when Rad21 was depleted (Figure 5), indicating that the cohesin complex is required for efficient repair of DSBs induced by ionizing radiation in replicated chromatin.
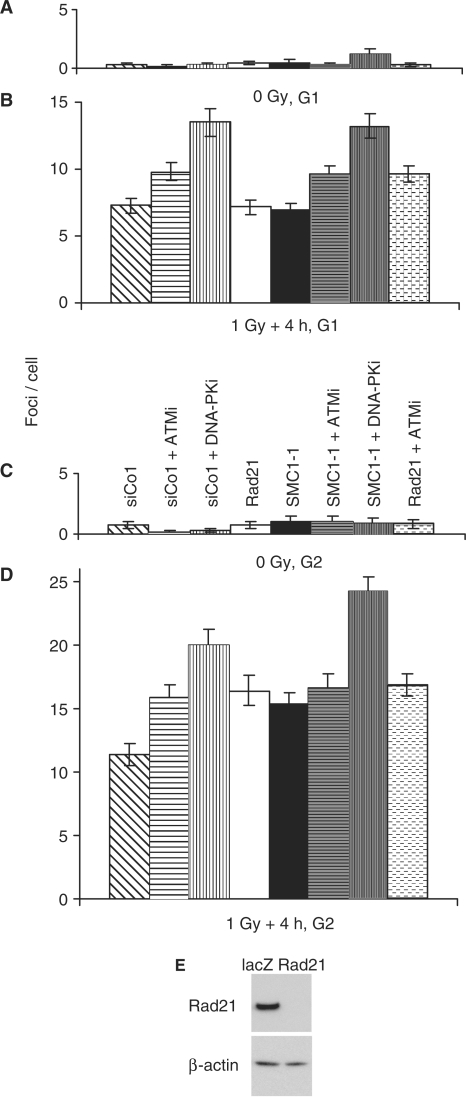
Figure 5.
Contribution of DNA damage kinases and/or the cohesin factors SMC1 and Rad21 to 53BP1 foci loss in G1 and late S/G2-phase cells. 1 Gy X-ray-induced 53BP1 foci were scored in G1- (CENP-F negative) and late S/G2-phase cells (CENP-F strongly positive) 4 h after 1 Gy X-irradiation. (A and B) Un-irradiated and irradiated G1-phase cells. (C and D) Un-irradiated and irradiated late S/G2-phase cells. Error bars are standard deviations from two experiments. (E) Depletion of Rad21 using RNA interference. Cells were transfected with siRNAs against Rad21 or non-targeting siRNA (lacZ). Protein levels were analysed by western blotting. Rad21-depleted cells contained less than 2% of the levels found in control cells.
While 53BP1 and γH2AX are well established as surrogate markers for DSBs (31–33), they do not directly measure the physical presence of a break. A physical assay for DSB repair, like pulsed-field gel electrophoresis, would be useful to confirm that the observed increase in residual foci levels in G2-phase cohesin knock-down cells does indeed reflect the presence of more residual breaks. To this end, we attempted a combination of siRNA transfection and double-thymidine block synchronization followed by irradiation and pulsed-field gel electrophoresis. However, we were unable to obtain a sufficiently pure G2 cell population. By the time most cells had left the S phase, a considerable fraction had progressed to G1.
SMC1 depletion affects progression through mitosis and causes elevated chromosome damage following irradiation
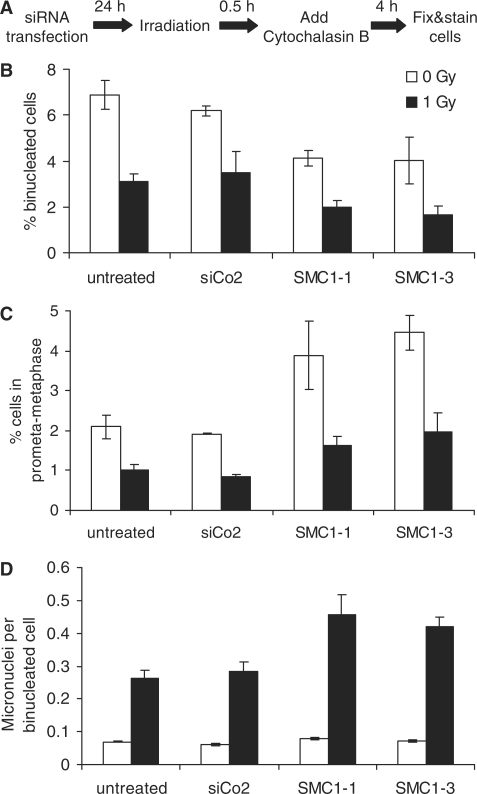
Un-repaired or mis-repaired DSBs are the most important precursor lesion for chromosome damage. To determine whether the observed deficient repair of radiation-induced DNA breaks in SMC1-depleted G2-phase cells is associated with increased chromosome damage following irradiation, siRNA-transfected cells were analysed for micronuclei (Figure 3A). The spindle poison cytochalasin B arrests cells in cytokinesis in a binuclear stage and thus enables the microscopic identification of recent mitotic cells. It was added 30 min after irradiation to allow clearance of cells that had been in mitosis at the time of irradiation. Cells were incubated in cytochalasin B for only 4 h to restrict the analysis to cells that were in late S/G2 at the time of irradiation. Accordingly, the fraction of binucleated cells was low in un-irradiated cultures and dropped further following 1 Gy X-irradiation (Figure 3B), likely due to radiation-induced activation of the G2/M checkpoint. Interestingly, SMC1 depletion resulted in an even lower fraction of binucleated cells in un-irradiated and irradiated cultures (P < 0.003 for pooled analysis of SMC1-1 and SMC1-3), indicating that SMC1 is required for efficient cell cycle progression from G2 through to cytokinesis. The mitotic index was higher in unirradiated SMC1-depleted cells than in controls, which is consistent with the notion that SMC1-depleted cells spend more time in mitosis (Figure 3C). Four hours after irradiation with 1 Gy, however, a similar 50–60% reduction in the mitotic index was observed in both controls and SMC1-depleted cells, suggesting that loss of SMC1 does not compromise the DNA damage-induced G2/M checkpoint arrest. SMC1 depletion had no effect on spontaneous levels of micronuclei in binucleated cells (Figure 3D). Following irradiation, significantly higher levels of micronuclei per binucleated cell were observed in SMC1 knock-down cells than in un-transfected or control siRNA-transfected cells (P < 0.04), indicating that SMC1 promotes chromosome integrity.
Figure 3.
SMC1 depletion affects progression through mitosis and enhances micronuclei induction. (A) Schematic representation of the experimental protocol. (B) Percentage of binucleated cells, (C) mitotic index and (D) numbers of micronuclei per binucleated cell obtained for different siRNA treatments following 0 or 1 Gy X-irradiation. Error bars are standard deviations from two experiments. The following numbers of cells were analysed in total for each data point (from left to right): 3366, 4290, 3654, 4566, 4877, 5211, 4718 and 5926.
The formation and repair of DNA single-strand breaks in irradiated cells is not affected by SMC1 depletion
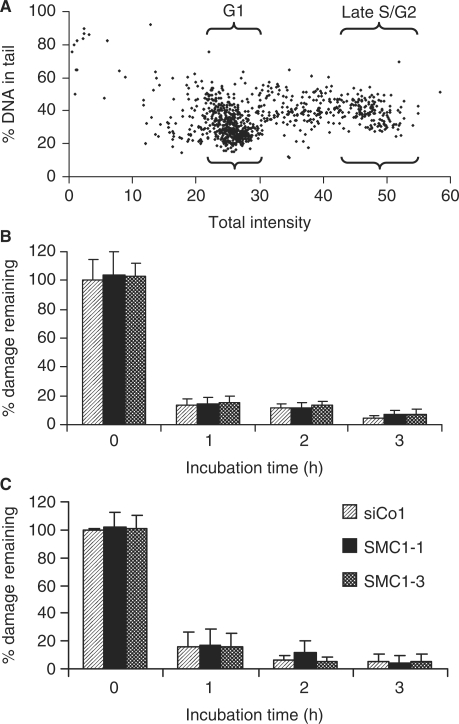
SMC1 is required for the ionizing radiation-induced S-phase checkpoint (27,28,34) and cohesin has previously been implicated in sister chromatid-dependent repair of DSBs arising because of replication failures, for example, when replication encounters nicks (35). To test whether depletion of SMC1 results in a general increase in DNA damage levels following irradiation, 4 Gy X-irradiated SMC1 knock-down cells were analysed with the alkaline comet assay that detects both single- and double-strand breaks but, given the 40 : 1 prevalence of the former over the latter for sparsely ionizing radiation, mainly measures the induction and repair of single-strand breaks. Automated image acquisition and quantitative analysis enabled several hundred cells to be analysed per sample, and G1 and late S/G2 populations were ‘gated’ based on the DNA content and analysed separately (Figure 4A). Similar levels of initial breaks induced by 4 Gy X-rays and a rapid decrease over time with similar residual levels in control and SMC1-depleted cells were observed in both G1 (Figure 4B) and late S/G2-phase populations (Figure 4C).
Figure 4.
Comet analysis of DNA strand break induction and repair. (A) Percentage of DNA in tail versus total DNA signal in a 4 Gy X-irradiated sample. The areas gated for the analysis of G1 and late S/G2-phase cells are indicated. (B and C) Time course for the percentage of damage remaining in G1 and late S/G2-phase cells following 4 Gy X-irradiation. Error bars are standard deviations from two experiments.
Effect of ATM and DNA-PK inhibition on radiation-induced foci loss in cohesin-depleted cells
Following irradiation, SMC1 is phosphorylated on Ser 966 and Ser 957 by the DNA damage kinase ATM but not by DNA-PK (27,28). Furthermore, SMC3 is phosphorylated at Ser 1083 by ATM [Bauerschmidt et al. unpublished data and (36)]. To test whether the observed role of SMC1 in the repair of radiation-induced DSBs in replicated chromatin requires ATM or DNA-PK, we treated siRNA-transfected cells with the ATM inhibitor Ku55 933 (23) or the DNA-PK inhibitor Nu7026 (24), irradiated cells with 1 Gy X-rays and determined residual 53BP1 foci in G1 (CENP-F negative) and late S/G2-phase cells (CENP-F strong positive, interphase DAPI staining) by immunofluorescence microscopy (Figure 5). In addition, siRNA targeting the cohesin subunit Rad21 was used to test whether the effects observed when knocking down SMC1 apply to the cohesin complex in general. Rad21 expression was efficiently knocked down to very low residual levels (Figure 5E) using a previously published siRNA (9,22). Like SMC1 knock-down cells, Rad21-depleted cells contained higher levels of unresolved foci compared to control siRNA-treated cells when irradiated in late S/G2 (P < 0.001) but not after irradiation in G1 (Figure 5B and D).
Inhibition of ATM or DNA-PK significantly increased the level of residual foci in siCo-treated cells (P < 0.003) as well as in SMC1- (P < 0.0002) or Rad21-depleted cells (P < 0.003) following irradiation in G1. In cells irradiated in G2, however, ATM inhibition caused higher levels of residual foci only in control siRNA-treated cells (P < 0.001) but did not have any additional effect on foci loss in SMC1- or Rad21-depleted cells. These findings indicate that cohesin and ATM may function together in the same pathway(s) that facilitate the efficient repair of radiation-induced DSBs in the G2 phase of the cell cycle. Treatment with the DNA-PK inhibitor, on the other hand, resulted in higher residual foci levels in both control (P < 0.00001) and SMC1 siRNA-transfected cells (P < 0.00001) (Figure 5B and D). These data suggest that DNA-PK-independent repair pathways benefit from the presence of cohesin in G2-phase cells.
Recruitment of cohesin to radiation-induced DNA double strand breaks
As no recruitment of cohesin subunits to sites of isolated DSBs induced by conventional X-rays was observed (data not shown), we used partially shielded soft X-rays (37) to generate microbeam tracks and thereby induce localized DNA damage in 1 μm wide ‘stripes’ (Figure 6A). After extraction of unbound proteins immunofluorescence microscopy was performed for all components of the cohesin complex. SMC1, SMC3, SA1 and SA2 did not form microscopically visible stripes (Figure 6B). However, a possible recruitment of Rad21 to X-ray-induced DNA damage was observed in a subset of cells that showed colocalization of Rad21 and 53BP1 stripes. Analysis of cells triple-stained for Rad21, CENP-F and 53BP1 revealed that Rad21 stripes were only present and colocalized with 53BP1 in CENP-F positive, i.e. late S/G2-phase cells but not in CENP-F-negative G1-phase cells (Figure 6C and D). This indicates that Rad21 is preferentially recruited to sites of X-ray-induced DNA damage in late S/G2-phase cells. Similar results were obtained for SMC1, using a different antibody (Autogen Bioclear) from the one used in Figure 6B (Bethyl Laboratories), albeit with weaker stripe formation than seen for Rad21 (Figure 6C and D). To determine whether the immunofluorescence signal of the Rad21 antibody is specific, control siRNA-transfected cells labelled with CellTrackerTM Green CMFDA were mixed with Rad21 siRNA-transfected cells and stained for Rad21. No Rad21 staining was observed in Rad21 siRNA-treated Tracker-negative cells while Tracker-positive control siRNA-treated cells showed strong Rad21 staining (Figure 6E). This result confirms that the Rad21 ‘stripes’ shown in panels B and C are specific.
DISCUSSION
This study shows for the first time that cohesin promotes the repair of DSBs induced by a non-lethal radiation dose in human G2-phase cells but not in G1. A function for cohesin in promoting the repair of ionizing radiation-induced DSBs in replicated chromatin provides an attractive explanation for the observation in a number of cell systems and using a variety of assays that the repair of radiation-induced DSBs, especially in end-joining mutants, is enhanced in G2 (3,38–41). Studies in S. cerevisiae using HO endonuclease and in human cells using I-SceI endonuclease to enzymatically induce a DSB at a defined site have shown that cohesin is required for efficient utilization of homologous recombination for sister chromatid-dependent DNA repair but not for intrachromosomal gene conversion (13,42). Whether this role of cohesin is also important for the repair of ionizing radiation-induced DNA breaks in humans, where homologous recombination plays a less dominant role, is not clear. Homologous recombination would intuitively be regarded as the obvious candidate, and the reported S/G2-specific radiosensitivity, increased yields of radiation-induced chromosome aberrations and DSB repair deficiency of cell lines deficient in homologous recombination (3,4) support this notion. However, a contribution of homologous recombination to the repair of ionizing radiation-induced DSBs was not detected in a number of studies (40,41,43,44). Notably, these studies used pulsed-field gel electrophoresis to quantify DSBs and, because of the limited sensitivity of this technique, had to be performed at supra-lethal doses of at least 10 Gy. The contradictory results regarding the importance of homologous recombination for the repair of ionizing radiation-induced DSBs can potentially be reconciled by the assumption that homologous recombination cannot efficiently process hundreds of DSBs induced by high doses, but contributes significantly to DSB repair at more physiological levels of DNA damage.
Figure 1 suggests that SMC1 depletion only moderately radiosensitizes HeLa cells. However, two points have to be considered in this context. (i) The knock-down efficiency of the siRNA transfections performed here was only about 80%. One could therefore assume that considerable levels of SMC1 may have been present in a subset of siRNA-treated cells that would thus have had a survival advantage and may have determined the shape of the survival curve at high doses. To address this point, SMC1 intensities of individual control and SMC1-depleted cells were quantified in immunofluorescence images (Figure 1B). The standard deviations obtained were very similar for all samples, suggesting that there was no large variation in residual SMC1 protein levels in the knock-down samples. Overall depletion appeared to be even less than 80% but quantification of immunofluorescence images may well have been skewed by background fluorescence and some non-specific binding of the SMC1 antibody. (ii) One would expect that SMC1 depletion only radiosensitizes cells in the S and G2 phase of the cell cycle. The presence of more than 60% G1-phase cells in the exponentially growing cultures used for the colony assays may well partly mask the radiosensitizing effect of SMC1 loss in S/G2. Comparison of colony formation in cells irradiated in G1 versus S versus G2 phase would not only address this point but would also help dissect SMC1’s functions in the intra-S-phase checkpoint (27,36) and DNA repair in G2. We have tried to use a fluorescence-activated cell sorter to selectively plate S- and G2-phase cells for colony formation but failed to obtain any statistically sound results.
The results presented here suggest that constitutive or DNA damage-induced sister chromatid cohesion, facilitated by the cohesin complex, promotes the repair of ionizing radiation-induced DSBs. Our observation of Rad21 recruitment to sub-nuclear regions damaged by intense X-ray microbeams in G2 but not G1-phase cells (Figure 6C and D) supports the notion of an active role of the cohesin complex in the DNA damage response that is unique to the G2 phase of the cell cycle. It is consistent with previous findings using a high power laser. Importantly, however, it rules out potential artefacts caused by the immense heat produced by dissection lasers that causes denaturation of DNA and proteins and leaves microscopically visible burn-marks in cells (16). In contrast, the ‘stripes’ induced here contained the same types and amounts of damage concentrated in a 1 μm wide stripe that are induced in a cell during homogenous exposure to 2.5 Gy ultra-soft X-rays. While these findings clearly implicate damage-induced cohesin recruitment in the radiation response, it is not yet established whether efficient repair of radiation-induced DSBs in replicated chromatin does indeed require additional damage-induced cohesion to improve local pairing of damaged and intact sister chromatid or whether it operates already more efficiently at a constitutive level of cohesion that is established during the S-phase. It is also currently not clear why only recruitment of Rad21 and SMC1, but not of SMC3 or SA1/2, was observed in this study. Given that Rad21 and SMC1 recruitment was only just detectable, despite using highly specific antibodies (Figure 6E), co-localization of the other cohesin factors may not have been observed because of a lower specificity of the antibodies used. Use of fluorescent protein fusion constructs, as employed by Bekker-Jensen et al. (2006) for recruitment to laser damage, may help resolve this issue.
As SMC1 is involved in the damage-induced intra-S-phase checkpoint and cohesin contributes to the repair of replication-associated DSBs arising at single-stranded DNA lesions, it was important to consider and, ideally, exclude any S-phase effects in order to confirm a replication-independent function of cohesin in radiation-induced DSB repair. To this end, the alkaline comet assay was used to verify that DNA single-strand break induction and repair was unaffected by SMC1 knock-down (Figure 4). Any deficiency in DSB repair would remain undetected with this assay because DSBs constitute less than 3% of all breaks induced by X-rays. Also, the finding of cohesin-dependent enhancement of DSB repair was confirmed using an assay that excludes cells in the S-phase during or after irradiation from the analysis. This was achieved by irradiation and incubation of cells for repair in the presence of bromodeoxyuridine and subsequent elimination of all bromodeoxyuridine-positive cells from the analysis (Figure 2C and D). SMC1 has not only been found to be part of cohesin but is also a subunit of recombination complex 1 (RC-1) (45). Our finding that Rad21 depletion caused phenotypes similar to the ones seen after SMC1 depletion suggests that the observed effects are due to cohesin inactivation.
The observation in previous studies that homologous recombination-deficient cells as well as non-homologous end-joining-deficient cells show enhanced DSB rejoining in G2 even following exposure to high radiation doses, where homologous recombination would likely be unable to contribute efficiently to DSB repair, may indicate that both DNA-PK-dependent end-joining and a previously described DNA-PK-independent backup end-joining pathway (46) may also benefit from damage-induced cohesion. One could speculate that either end-joining process would require the alignment of broken DSB ends that may be easier to facilitate if both ends are tightly bound by the cohesin complex to the intact sister chromatid that would thus act as a scaffold to indirectly promote repair. Figure 5 suggests that cohesin-dependent enhancement of repair in the G2 phase occurs with and without DNA-PK inhibition but it appears to depend on functional ATM. This finding suggests that cohesin may promote DSB repair via at least one additional pathway to DNA-PK-dependent end-joining but, given that both ATM and DNA-PK influence both homologous recombination and end-joining pathways (47), these data do not clarify which one or both of the discussed pathways are involved. Also, more detailed analysis of DSB repair kinetics would be required to better clarify the interaction of cohesin with ATM and DNA-PK and its contribution to specific repair pathways. The ATM and DNA-PK inhibitors are highly selective and effective at the concentrations used (23,24). They are being widely used and no non-specific effects have been reported to date. Given that, in addition, Rad21 knock-down was very efficient (Figure 5E) and similar results were obtained for Rad21 and SMC1, it is therefore unlikely that the observed effects are due to combinations of partial inhibitions and/or off-target effects.
Notably, the moderate size of the effect of cohesin depletion on DSB repair observed here in G2-phase cells is similar to that observed for a number of factors, including ATM, Artemis, NBS1, H2AX, 53BP1 and others observed in G1-phase cells (48). Our finding that cohesin-dependent enhancement of DSB repair in G2 requires ATM (Figure 5) indicates that they may act in the same pathway in this cell cycle phase. Additional work is required, however, to determine whether there is indeed a functional link between these observations.
FUNDING
Cancer Research UK (C14 504/A6116 to K.R.); Gray Laboratory Cancer Research Trust; Medical Research Council. Funding for open access charge: Core departmental budget.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr Boris Vojnovic’s group (Gray Institute for Radiation Oncology and Biology, Oxford, UK) for developing the automated comet assay system and Dr Graeme Smith (KuDos Pharmaceuticals, Cambridge, UK) for materials.
REFERENCES
- 1.Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687–696. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- 2.Rothkamm K. Different means to an end: DNA double-strand break repair. In: Kiefer J, editor. Life Science and Radiation: Accomplishments and Future Directions. Berlin Heidelberg: Springer; 2004. pp. 179–186. [Google Scholar]
- 3.Rothkamm K, Krüger I, Thompson LH, Löbrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinz JM, Yamada NA, Salazar EP, Tebbs RS, Thompson LH. Influence of double-strand-break repair pathways on radiosensitivity throughout the cell cycle in CHO cells. DNA Repair. 2005;4:782–792. doi: 10.1016/j.dnarep.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Saleh-Gohari N, Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 7.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 8.Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 9.Diaz-Martinez LA, Gimenez-Abian JF, Clarke DJ. Cohesin is dispensable for centromere cohesion in human cells. PLoS ONE. 2007;2:e318. doi: 10.1371/journal.pone.0000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sjogren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 2001;11:991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 11.Schmitz J, Watrin E, Lenart P, Mechtler K, Peters JM. Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase. Curr. Biol. 2007;17:630–636. doi: 10.1016/j.cub.2007.02.029. [DOI] [PubMed] [Google Scholar]
- 12.Strom L, Lindroos HB, Shirahige K, Sjogren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 13.Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 14.Strom L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjogren C. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–245. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- 15.Unal E, Heidinger-Pauli JM, Koshland D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7) Science. 2007;317:245–248. doi: 10.1126/science.1140637. [DOI] [PubMed] [Google Scholar]
- 16.Kim JS, Krasieva TB, LaMorte V, Taylor AM, Yokomori K. Specific recruitment of human cohesin to laser-induced DNA damage. J. Biol. Chem. 2002;277:45149–45153. doi: 10.1074/jbc.M209123200. [DOI] [PubMed] [Google Scholar]
- 17.Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagao K, Adachi Y, Yanagida M. Separase-mediated cleavage of cohesin at interphase is required for DNA repair. Nature. 2004;430:1044–1048. doi: 10.1038/nature02803. [DOI] [PubMed] [Google Scholar]
- 19.Romero F, Gil-Bernabe AM, Saez C, Japon MA, Pintor-Toro JA, Tortolero M. Securin is a target of the UV response pathway in mammalian cells. Mol. Cell. Biol. 2004;24:2720–2733. doi: 10.1128/MCB.24.7.2720-2733.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birkenbihl RP, Subramani S. Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 1992;20:6605–6611. doi: 10.1093/nar/20.24.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonoda E, Matsusaka T, Morrison C, Vagnarelli P, Hoshi O, Ushiki T, Nojima K, Fukagawa T, Waizenegger CI, Peters JM, et al. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev. Cell. 2001;1:759–770. doi: 10.1016/s1534-5807(01)00088-0. [DOI] [PubMed] [Google Scholar]
- 22.Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J. Cell Sci. 2005;118:2133–2141. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- 23.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 24.Veuger SJ, Curtin NJ, Richardson CJ, Smith GC, Durkacz BW. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 2003;63:6008–6015. [PubMed] [Google Scholar]
- 25.Willmore E, de Caux S, Sunter NJ, Tilby MJ, Jackson GH, Austin CA, Durkacz BW. A novel DNA-dependent protein kinase inhibitor, NU7026, potentiates the cytotoxicity of topoisomerase II poisons used in the treatment of leukemia. Blood. 2004;103:4659–4665. doi: 10.1182/blood-2003-07-2527. [DOI] [PubMed] [Google Scholar]
- 26.Goodhead DT, Thacker J. Inactivation and mutation of cultured mammalian cells by aluminium characteristic ultrasoft X-rays: I. Properties of aluminium X-rays and preliminary experiments with Chinese hamster cells. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1977;31:541–559. doi: 10.1080/09553007714550651. [DOI] [PubMed] [Google Scholar]
- 27.Kim ST, Xu B, Kastan MB. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 2002;16:560–570. doi: 10.1101/gad.970602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 2004;18:1423–1438. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao H, Winkfein RJ, Mack G, Rattner JB, Yen TJ. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J. Cell. Biol. 1995;130:507–518. doi: 10.1083/jcb.130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burdak-Rothkamm S, Short SC, Folkard M, Rothkamm K, Prise KM. ATR-dependent radiation-induced gamma H2AX foci in bystander primary human astrocytes and glioma cells. Oncogene. 2007;26:993–1002. doi: 10.1038/sj.onc.1209863. [DOI] [PubMed] [Google Scholar]
- 31.Rothkamm K, Löbrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc. Natl Acad. Sci. USA. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sedelnikova OA, Rogakou EP, Panyutin IG, Bonner WM. Quantitative detection of (125)IdU-induced DNA double-strand breaks with gamma-H2AX antibody. Radiat. Res. 2002;158:486–492. doi: 10.1667/0033-7587(2002)158[0486:qdoiid]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 33.Rothkamm K, Balroop S, Shekhdar J, Fernie P, Goh V. Leukocyte DNA damage after multi-detector row CT: a quantitative biomarker of low-level radiation exposure. Radiology. 2007;242:244–251. doi: 10.1148/radiol.2421060171. [DOI] [PubMed] [Google Scholar]
- 34.Yazdi PT, Wang Y, Zhao S, Patel N, Lee EY, Qin J. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 2002;16:571–582. doi: 10.1101/gad.970702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cortes-Ledesma F, Aguilera A. Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange. EMBO Rep. 2006;7:919–926. doi: 10.1038/sj.embor.7400774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo H, Li Y, Mu JJ, Zhang J, Tonaka T, Hamamori Y, Jung SY, Wang Y, Qin J. Regulation of intra-S phase checkpoint by ionizing radiation (IR)-dependent and IR-independent phosphorylation of SMC3. J. Biol. Chem. 2008;283:19176–19183. doi: 10.1074/jbc.M802299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill MA, Stevens DL, Kadhim M, Blake-James M, Mill AJ, Goodhead DT. Experimental techniques for studying bystander effects in vitro by high and low-LET ionising radiation. Radiat. Prot. Dosimetry. 2006;122:260–265. doi: 10.1093/rpd/ncl429. [DOI] [PubMed] [Google Scholar]
- 38.Mateos S, Slijepcevic P, MacLeod RA, Bryant PE. DNA double-strand break rejoining in xrs5 cells is more rapid in the G2 than in the G1 phase of the cell cycle. Mutat. Res. 1994;315:181–187. doi: 10.1016/0921-8777(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 39.Lee SE, Mitchell RA, Cheng A, Hendrickson EA. Evidence for DNA-PK-dependent and -independent DNA double-strand break repair pathways in mammalian cells as a function of the cell cycle. Mol. Cell. Biol. 1997;17:1425–1433. doi: 10.1128/mcb.17.3.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kruger I, Rothkamm K, Lobrich M. Enhanced fidelity for rejoining radiation-induced DNA double-strand breaks in the G2 phase of Chinese hamster ovary cells. Nucleic Acids Res. 2004;32:2677–2684. doi: 10.1093/nar/gkh586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu W, Wang M, Wu W, Singh SK, Mussfeldt T, Iliakis G. Repair of radiation induced DNA double strand breaks by backup NHEJ is enhanced in G2. DNA Repair. 2008;7:329–338. doi: 10.1016/j.dnarep.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 42.Potts PR, Porteus MH, Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25:3377–3388. doi: 10.1038/sj.emboj.7601218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, Zeng ZC, Bui TA, Sonoda E, Takata M, Takeda S, Iliakis G. Efficient rejoining of radiation-induced DNA double-strand breaks in vertebrate cells deficient in genes of the RAD52 epistasis group. Oncogene. 2001;20:2212–2224. doi: 10.1038/sj.onc.1204350. [DOI] [PubMed] [Google Scholar]
- 44.Rothkamm K, Kühne M, Jeggo PA, Löbrich M. Radiation-induced genomic rearrangements formed by nonhomologous end-joining of DNA double-strand breaks. Cancer Res. 2001;61:3886–3893. [PubMed] [Google Scholar]
- 45.Jessberger R, Riwar B, Baechtold H, Akhmedov AT. SMC proteins constitute two subunits of the mammalian recombination complex RC-1. EMBO J. 1996;15:4061–4068. [PMC free article] [PubMed] [Google Scholar]
- 46.Iliakis G, Wang H, Perrault AR, Boecker W, Rosidi B, Windhofer F, Wu W, Guan J, Terzoudi G, Pantelias G. Mechanisms of DNA double strand break repair and chromosome aberration formation. Cytogenet. Genome Res. 2004;104:14–20. doi: 10.1159/000077461. [DOI] [PubMed] [Google Scholar]
- 47.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 48.Riballo E, Kühne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]