Abstract
The 7SK snRNP represents a major reservoir of activity where P-TEFb, a general transcription factor key for RNA polymerase II elongation, can be withdrawn to promote gene expression, cell growth and development. Within this complex, 7SK snRNA is a central scaffold that coordinates key protein–protein interactions and maintains P-TEFb in an inactive state. Although the stability of 7SK directly affects the amount of active P-TEFb in vivo, relatively little is known about how it is maintained and how the 7SK methylphosphate capping enzyme MePCE and LARP7, a La-related protein associated with the 3′-poly(U) of 7SK, contribute to this process. Here, we show that 7SK is capped by the LARP7-free MePCE and in probably a co-transcriptional manner prior to its sequestration into 7SK snRNP. However, upon interacting with LARP7 within 7SK snRNP, MePCE loses its capping activity, probably due to the occlusion of its catalytic center by LARP7. Despite its lack of capping activity in 7SK snRNP, MePCE displays a capping-independent function to promote the LARP7–7SK interaction, which in turn stabilizes 7SK and facilitates the assembly of a stable MePCE–LARP7–7SK subcomplex. Our data indicate that MePCE and LARP7 act cooperatively to stabilize 7SK and maintain the integrity of 7SK snRNP.
INTRODUCTION
Recent global analyses indicate that the elongation phase of RNA polymerase (Pol) II transcription plays a much more important role in controlling metazoan gene expression than previously thought (1). Composed of Cdk9 and its regulatory subunit Cyclin T1 (CycT1) or three other C-type cyclins (CycT2a/b and CycK), the positive transcription elongation factor b (P-TEFb) plays a key role during transcriptional elongation. P-TEFb stimulates the processivity of Pol II through phosphorylating the C-terminal domain of the largest subunit of Pol II and a pair of negative elongation factors. This leads to the synthesis of full-length RNA transcripts and the coupling of transcription with other pre-mRNA processing events (2,3). In human cells, not only is P-TEFb critical for the expression of a vast array of cellular genes, but is also an indispensable host cofactor for efficient transcription of the HIV-1 genome (2,3).
Recent evidence indicates that most of cellular P-TEFb exist in two mutually exclusive complexes that are characterized by their different P-TEFb-associated factors and Cdk9 kinase activity (2). A catalytically inactive complex termed 7SK snRNP sequesters a major fraction of nuclear P-TEFb and also contains the 7SK snRNA and three nuclear proteins, HEXIM1 (or the minor HEXIM2 protein), LARP7 (also termed PIP7S) and MePCE (also known as BCDIN3) (4–11). Within this complex, 7SK, an abundant 331-nt long transcript produced by RNA Pol III and highly conserved in vertebrates, functions as a molecular scaffold to coordinate the interactions among key protein components and maintain the integrity of 7SK snRNP (2). While HEXIM1 inhibits the Cdk9 kinase activity in a 7SK-dependent manner (6,12), LARP7 and MePCE are known to ensure the stability of 7SK (9,10,13). LARP7 is a La-related protein that binds to the 3′-UUUU-OH sequence of 7SK and protects it against cleavage by exonucleases (10,11,13). In the nucleus, nearly all the 7SK molecules are bound by LARP7, which explains the observation that the depletion of LARP7 by specific short hairpin (sh)RNA caused an almost complete co-depletion of 7SK (10,11,13). MePCE, on the other hand, contains a methyltransferase domain and is responsible for adding a unique γ-monomethyl phosphate cap structure onto the 5′-end of 7SK (9). It has been shown that the siRNA-mediated silencing of MePCE reduced the cellular 7SK level by about half (9).
In contrast to the P-TEFb sequestered in the inactive 7SK snRNP, another major portion of P-TEFb exists in the transcriptionally active state through association with the bromodomain protein Brd4, which recruits P-TEFb to chromatin templates through interacting with acetylated histones and/or the mediator complex (14,15). Recent data indicate that the recruitment occurs at late mitosis and is essential for promoting early G1 gene expression and cell cycle progression (16). Thus, through alternatively interacting with its positive and negative regulators, P-TEFb is kept in a functional equilibrium (2). Accumulating evidence indicates that this equilibrium can be dynamically controlled by extracellular signals to modulate the overall levels of active P-TEFb in the cell for optimal gene expression, growth and development (2,17,18). Notably, disrupting 7SK snRNP and shifting the P-TEFb functional equilibrium toward the Brd4-bound, active state can contribute to cardiac hypertrophy or mammary epithelial transformation (13,19).
Despite the fact that 7SK snRNP represents a major reservoir of activity from which P-TEFb can be withdrawn to support elevated gene transcription and accelerated cell growth, there still exist many knowledge gaps about this complex and its components. For example, among all the subunits of 7SK snRNP, MePCE remains the least studied protein, of which little is known about whether it is the only 7SK capping enzyme in vivo, how it interacts with the rest of 7SK snRNP, how its capping activity might be impacted by its presence within this complex, and finally how it may contribute to the assembly of 7SK snRNP. Here, we show that 7SK is capped by the LARP7-free MePCE and in probably a co-transcriptional manner prior to its sequestration into 7SK snRNP. However, upon the interaction with LARP7 within 7SK snRNP, MePCE loses its capping activity, probably due to the occlusion of its catalytic center by LARP7. Despite MePCE’s lack of capping activity in 7SK snRNP, it displays a capping independent function to promote the interaction of LARP7 with 7SK, which in turn stabilizes 7SK snRNA and facilitates the assembly of a stable MePCE–LARP7–7SK subcomplex within 7SK snRNP. Our data indicate that MePCE and LARP7 act cooperatively to stabilize 7SK snRNA and maintain the integrity of 7SK snRNP.
MATERIALS AND METHODS
Immunological reagents
Rabbit polyclonal anti-MePCE antibodies were generated against the C-terminal sequence (RPVYLFHKARSPSH; aa 676–689) of MePCE and affinity purified. Anti-HEXIM1, anti-LARP7 antibodies have been described earlier (6,13). All other antibodies were purchased from Santa Cruz Biotechnology.
shRNA-mediated depletion of MePCE and LARP7
To generate the HeLa-based MePCE knockdown (KD) cell line (H99-18), two DNA oligonucleotides (5′-GATCAAGCCAGAGCAGTTCAGTTCCTTCAAGAGAGGAACTGAACTGCTCTGGCTTTTTA-3′ and 5′-AGCTTAAAAAGCCAGAGCAGTTCAGTTCCTCTCTTGAAGGAACTGAACTGCTCTGGCTT-3′) for expressing shMePCE were annealed and then cloned into pSuper-retro-puro (Oligoengine, WA, USA). Retroviruses were produced in the GP2-293 packaging cell line (Clontech, CA, USA) and used to infect HeLa cells. Infected cells were selected with 0.5 μg/ml puromycin for 2 weeks to obtain individual clones. The sequence of shLARP7 used to deplete LARP7 expression has been described (13).
Generation of a stable cell line expressing F-MePCE
To generate W10, a HeLa-based cell line stably expressing Flag-tagged MePCE, the MePCE KD cell line H99-18 was stably transfected with pcDNA3-Flag-MePCE, in which the nucleotides C, A and G at positions 1941, 1942 and 1943 within the MePCE coding sequence were changed to T, T and C, respectively, in order to render the corresponding mRNA resistant to shMePCE. After selection in the presence of G418, clone W10 was chosen because F-MePCE was expressed at a similar level as endogenous MePCE expressed in parental HeLa cells.
Affinity purification of MePCE and MePCE-containing complexes
Nuclear extracts (NEs) were prepared from HeLa cells transfected with plasmids expressing F-MePCE or HeLa-based W10 cells stably expressing F-MePCE. F-MePCE and its associated factors were affinity purified from NEs by incubating at 4°C for 2 h with anti-Flag agarose beads (Sigma), followed by extensive washes with buffer D [20 mM HEPES-KOH (pH 7.9), 15% glycerol, 0.2 mM EDTA, 0.2% NP-40, 1 mM dithiothreitol and 1 mM phenylmethylsulfonyl fluride] containing 0.3 M KCl (D0.3 M). The immunoprecipitated proteins were then eluted with the Flag peptide dissolved in D0.1M. The eluted materials were analyzed by western blotting (WB) and northern blotting (NB). For the purification of F-MePCE proteins (WT and VLD-AAA) that were free of any associated 7SK snRNP components, NEs were treated with micrococcal nuclease (MNase) at room temperature for 30 min in the presence of 1 mM CaCl2 before incubation with anti-Flag agarose beads. The immunoprecipitates were washed extensively with buffer D containing 0.8 M KCl (D0.8 M) before elution with the Flag peptide and the purity confirmed by WB and NB.
Chromatin immunoprecipitation assay
The Chromatin immunoprecipitation (ChIP) assay was performed essentially as described (14) with minor modifications. Briefly, immunoprecipitation was performed with anti-Flag mAb (Sigma) from chromatin fragments derived from W10 cells that stably express F-MePCE. After DNA purification, PCR reactions containing α-[32P]dCTP (800 Ci/mmol) were carried out for 24 cycles. Input and immunoprecipitated chromatin were analyzed first in pilot experiments to ensure that PCR reactions occurred in the linear range of amplification. The primers used in ChIP assay are 5′-GACATCTGTCACCCCATTGA-3′ and 5′-AGACCGGTCCTCCTCTATCG-3′ for 7SK; 5′-CCAAGAAGAAGCGGCATT-3′ and 5′-GAGGAACTGCGTGGTGTTA-3′ for HEXIM1; 5′-ACCTGAACAAACTGGCCGAG-3′ and 5′-CAGAGAAAGGCTCCAGGTTG-3′ for TGM-2; 5′-TACAAAGAGCAGCCGCTCA-3′ and 5′-TTACCGTGAATCGAGCTCCAG-3′ for PTHLH; 5′-CAGCTTGTACCTGCAGGATC-3′ and 5′-GTCGAGGAGAGCAGAGAATC-3′ for c-MYC.
Gel mobility shift assay
Flag-tagged LARP7 was affinity purified and its purity confirmed as described earlier (13). His-tagged MePCE purified from recombinant Escherichia coli was a gift from the laboratory of Tom Alber at UC Berkeley. Purity of the proteins was confirmed by silver staining, and the concentrations were estimated by comparing with the BSA standards. 32P-labeled wild-type 7SK and 7SK(Δ4U’s) were synthesized in vitro by T7 RNA polymerase from PCR-amplified DNA templates. For the gel mobility shift assay, 0.27 pmols His-MePCE were incubated with the 7SK probe (7.3 pg; 2000 c.p.m.) in D0.05M plus 1 µg BSA, 500 ng poly(rG) at 30°C for 1h, which was followed by the incubation with or without 0.15 pmols F-LARP7 at room temperature for 20 min. The final reaction volume was 20 ul. The RNA–protein complexes were resolved in a 5% non-denaturing polyacrylamide (19:1 acrylamide:bisacrylamide) gel in 0.5× Tris–glycine electrophoresis buffer at 4°C for 4 h at 200 V.
In vitro capping assay
In vitro capping assay was performed essentially as described (9) with minor modifications. Briefly, NEs or affinity purified F-MePCE were incubated with in vitro transcribed 7SK in buffer M (20 mM Tris–HCl pH 8.0, 0.5 mM DTT, 2 mM EDTA, 50 mM KCl, 5% Glycerol, 24 U of RNase inhibitor, 10 µCi of [3H]SAM). After incubation at 30°C for 1 h, the reaction was terminated by adding 200 µl of stop solution (0.1 M NaOAc pH 5.2, 0.5% SDS, 2 mM EDTA). RNAs were isolated from the reaction by phenol/chloroform extraction and ethanol precipitation and analyzed in a 6% polyacrylamide–urea gel. The gel was fixed in 45% methanol + 10% acetic acid for 30 min, treated with the amplifier (NAMP100V, GE Healthcare) for 30 min and then exposed to an X-ray film at −80°C.
RESULTS AND DISCUSSION
MePCE is the highly predominant, if not the sole, source of 7SK capping activity in vivo
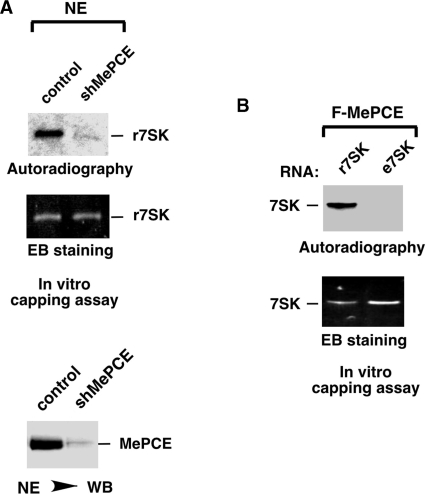
It has been shown earlier that MePCE is capable of capping 7SK through methylating the 5′-tri-phosphate at the γ-position (9). To determine whether 7SK is capped exclusively by MePCE or there may exist other unknown sources of capping activity in the cell, we incubated recombinant 7SK (r7SK) with NEs prepared from either normal HeLa cells or HeLa cells stably expressing a specific shRNA targeting MePCE (shMePCE) (Figure 1A) The capping reaction also contained the 3H-labeled methyl group donor [3H]SAM (S-adenosyl methionine). Analysis by WB and serial dilutions indicates that shMePCE depleted ∼85–90% of MePCE in HeLa cells (Figure 1A, bottom panel and data not shown). Importantly, NE from the MePCE-depleted cells shows a similar degree of reduction in its ability to cap 7SK (Figure 1A, upper panels). These data strongly indicate MePCE as the highly predominant, if not the sole, source of 7SK capping activity in vivo.
Figure 1.
7SK RNA in 7SK snRNP is likely already capped by MePCE, which is the highly predominant, if not the only, source of 7SK capping activity in vivo. (A) NE prepared from either normal HeLa cells (control) or HeLa cells stably expressing a specific shRNA targeting MePCE (shMePCE) was incubated with in vitro transcribed 7SK (r7SK) in capping reactions that also contained [3H]SAM. After the reaction, the RNA products, which were isolated and resolved in a polyacrylamide–urea gel, were subsequently analyzed by autoradiography (top panel) and ethidium bromide (EB) staining (middle panel). The levels of MePCE in NEs of control and MePCE KD cells were examined by anti-MePCE WB in the bottom panel. (B) r7SK or e7SK, which was isolated from purified 7SK snRNP, was incubated in capping reactions with affinity purified F-MePCE and [3H]SAM. The reaction products were analyzed by EB staining (bottom) and autoradiography (top) as in A.
7SK snRNA derived from 7SK snRNP is likely already capped
The experiment above used the in vitro transcribed, recombinant 7SK (r7SK) as the MePCE substrate. To determine whether endogenous 7SK (e7SK) derived from the 7SK snRNP could also serve as a substrate, we isolated e7SK by proteinase K digestion followed by phenol/chloroform extraction from 7SK snRNP that was affinity purified from NE of HH8 cells, a HeLa-based cell line stably expressing Flag-tagged HEXIM1 (6). Interestingly, compared with r7SK that was efficiently capped by Flag-tagged MePCE (F-MePCE), which was purified from transiently transfected HeLa cells (see ‘Materials and Methods’ section) and free of any associated 7SK snRNP components (see Figure 3C), e7SK was not capped at all despite the fact that about twice more of the RNA was used in the reaction (Figure 1B). Thus, it appears that 7SK present in the 7SK snRNP is already capped at the 5′-end.
Figure 3.
LARP7 prevents MePCE from capping 7SK in vivo and in vitro. (A) NE from either normal (control) or HeLa cells stably expressing shLARP7 was incubated with r7SK and [3H]SAM in capping reactions. The RNA products were isolated, resolved in a polyacrylamide–urea gel, and analyzed by autoradiography (top left) and EB staining (bottom left). The levels of the indicated proteins in NEs of control and LARP7 KD cells were examined by WB in the right panel. (B) Left panels: Purified F-LARP7 (1.2 pmols) and/or MePCE (1.3 pmols) proteins were incubated with r7SK and [3H]SAM in capping reactions. The RNA products were analyzed as in A. (Right panel) F-LAPR7 and MePCE used in the capping assay were examined on a silver-stained SDS–gel. An asterisk indicates a minor contaminating band in the F-LARP7 preparation. (C) The affinity purified F-MePCE free of any associated 7SK snRNP components and the F-LARP7-bound MePCE, which was isolated through anti-Flag immunoprecipitation and then either untreated (−) or treated (+) with MCN were analyzed by WB and NB for the presence of the indicated factors (left panel). These MePCE samples were subsequently analyzed in 7SK capping reactions as in A.
MePCE, LARP7 and 7SK form a stable subcomplex within 7SK snRNP
In light of the recent data implicating MePCE as a component of the 7SK snRNP (9,10,17), it is important to determine how the sequestration into this complex may impact MePCE’s capping activity. Toward this goal, we first performed co-immunoprecipitation experiments to examine how MePCE interacts with the rest of 7SK snRNP.
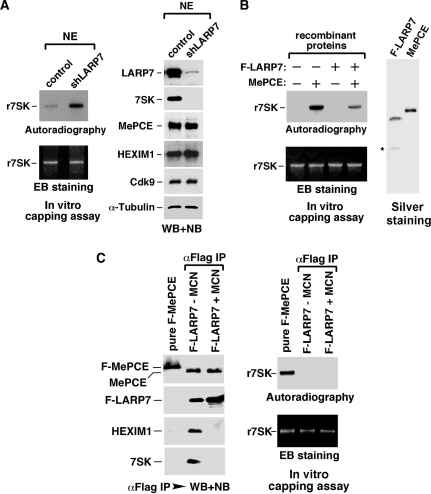
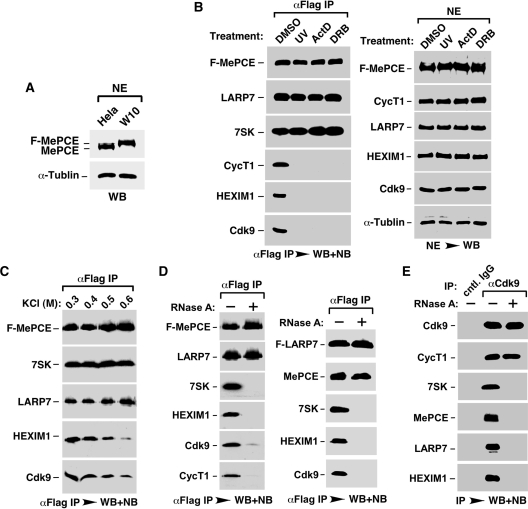
A HeLa-based cell line (W10), in which the expression of endogenous MePCE was suppressed by a specific shRNA (shMePCE) and in its place F-MePCE was stably expressed at about the same level from an shMePCE-resistant cDNA construct (Figure 2A; also see ‘Materials and Methods’ section), was established. Previous studies by others and by us have shown that treating cells with stress-inducing agents such as UV, actinomycin D and DRB, all of which can globally impact cell growth, causes the disruption of 7SK snRNP and release of P-TEFb for stress-induced gene expression (4–6). Consistent with these observations, when W10 cells were treated with UV, actinomycin D or DRB, Cdk9, CycT1 and HEXIM1 were found to dissociate efficiently from the immunoprecipitated F-MePCE (Figure 2B, left panel). However, both LARP7 and 7SK remained tightly bound to F-MePCE under these conditions, indicating the existence of a stress-resistant subcomplex consisting of MePCE, LARP7 and 7SK within 7SK snRNP. This subcomplex may serve as a platform to nucleate the re-assembly of 7SK snRNP and sequestration/inactivation of P-TEFb once the stress signals are eliminated as observed before (20,21).
Figure 2.
The 7SK-independent interaction between MePCE and LARP7 nucleates the formation of a stable MePCE–LARP7–7SK subcomplex within 7SK snRNP. (A) NEs prepared from W10, a HeLa-based cell line stably expressing F-MePCE, and the parental HeLa cells were analyzed by WB for the indicated proteins. (B) W10 cells were treated with the indicated agents. NEs prepared from the treated cells were analyzed by WB with the indicated antibodies (right panel) and subjected to anti-Flag immunoprecipitation. Upon elution with the Flag peptide, the compositions of the immunoprecipitates (αFlag IP) were analyzed by WB and NB as indicated (left panel). (C) Prior to the elution with the Flag peptide, the immobilized αFlag IP were washed with a buffer containing the indicated KCl concentrations, and their compositions were subsequently analyzed as in B. (D) NEs of W10 (left panel) and FPS86 cells, a HeLa-based cell line stably expressing F-LARP7 (right panel), were incubated with (+) or without (−) RNase A prior to anti-Flag immunoprecipitation. αFlag IP were analyzed as in B. (E) NEs of HeLa cells were pretreated with (+) or without (−) RNase A prior to immunoprecipitations with the indicated antibodies. The immunoprecipitates were analyzed as in B.
Notably, the MePCE–LARP7–7SK subcomplex was also stable when the anti-F-MePCE immune-complex was subjected to washing with increasing salt concentrations (Figure 2C). For example, at 0.6 M KCl, most of Cdk9 and HEXIM1 were already washed off, whereas LARP7 and 7SK still remained tightly bound to the immobilized F-MePCE.
7SK-independent interaction between MePCE and LARP7
To investigate the role of 7SK snRNA in the MePCE–LARP7–7SK subcomplex, we incubated the NE of W10 cells with RNase A to degrade 7SK prior to the anti-F-MePCE immunoprecipitation. As shown in Figure 2D, left panel degradation of 7SK efficiently dissociated HEXIM1, CycT1 and most of Cdk9 from F-MePCE, demonstrating their RNA-dependent interactions with MePCE. In contrast, the F-MePCE–LARP7 interaction was completely unaffected by the loss of 7SK, revealing a stable, 7SK-independent interaction between the two.
Notably, the 7SK-independent interaction was also detected between endogenous MePCE and F-LARP7 that was expressed from a stably transfected cDNA construct (Figure 2D, right panel). Finally, following a similar procedure that involves RNase treatment prior to anti-Cdk9 immunoprecipitation, a 7SK-independent interaction was also detected between Cdk9 and CycT1 (Figure 2E).
LARP7 prevents MePCE from capping 7SK in vivo and in vitro
Given the existence of a stable, 7SK-independent interaction between MePCE and LARP7 within 7SK snRNP, we next investigated whether the ability of MePCE to cap 7SK is affected by LARP7. First, NE was prepared from either control or LARP7-depleted HeLa cells and tested in capping reactions with recombinant 7SK as the substrate. Although the stable KD of LARP7 expression by an shRNA (shLARP7) that specifically targets LARP7 had little effect on the nuclear level of MePCE (Figure 3A, right panel), NE derived from the KD cells displayed a significant increase in 7SK capping activity (left panels), suggesting an inhibitory effect of LARP7 on MePCE’s catalytic activity.
To confirm the inhibition caused by LARP7, F-LARP7, which was highly purified from transiently transfected HeLa cells under conditions of high salt and micrococcal nuclease (MCN) treatment (13), and MePCE, which was purified to homogeneity from recombinant E. coli, were analyzed by silver staining (Figure 3B, right panel) and tested in capping reactions either alone or in combination. Data in Figure 3B, left panels are consistent with the notion that LARP7 can directly inhibit the capping activity of MePCE toward 7SK.
A further indication of an LARP7-mediated inhibition of MePCE capping activity came from a direct comparison between free and the LARP7-bound MePCE proteins in capping reactions. As expected, free F-MePCE, which was affinity purified from transiently transfected HeLa cells and treated with both MCN and high salt to strip away any associated 7SK snRNP components as confirmed by WB and NB (Figure 3C, left panel), was fully active in capping 7SK (Figure 3C, right panel). In contrast, the LARP7-bound MePCE, which was isolated through anti-F-LARP7 immunoprecipitation and then either untreated or treated with MCN to remove the associated 7SK RNA and HEXIM1, was completely inactive in the reaction (Figure 3C). Notably, the MCN treatment has ruled out the association of endogenous 7SK with the LARP7–MePCE complex as the reason for the failure to cap r7SK by the LARP7-bound MePCE. Taken together, these data strongly indicate LARP7 as an inhibitor of MePCE’s capping of 7SK RNA.
LARP7 may occupy the catalytic center of MePCE to inhibit the latter’s capping activity
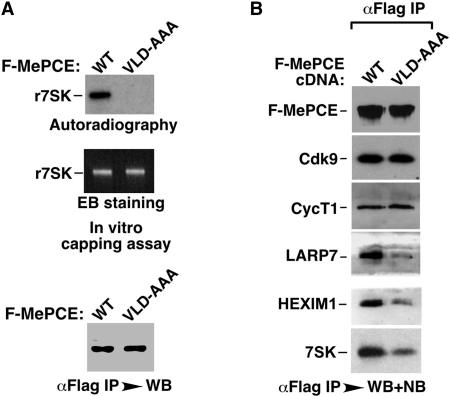
A possible explanation for how the 7SK-independent interaction between LARP7 and MePCE may lead to the inhibition of the latter’s capping activity has come from our subsequent mutagenesis analysis. When Val–Leu–Asp (aa 447–449), three highly conserved amino acids located at the catalytic center of the MePCE methyltransferase domain and likely to be involved in binding to SAM based on sequence alignment with other known methyltransferases [e.g. PRMT1, (22,23)], were changed into Ala–Ala–Ala, the resultant mutant (VLD-AAA) was completely defective in 7SK capping as expected (Figure 4A). Interestingly, this mutation also significantly reduced the interaction of MePCE with LARP7, but had no obvious effect on the interactions with Cdk9 and CycT1 (Figure 4B). The observation that VLD-AAA bound to P-TEFb with full capacity suggests that the point mutation does not simply destroy the overall structure of the protein. Given that MePCE and LARP7 have a direct, 7SK-independent interaction (Figure 2D), it is possible that the mutation at the MePCE catalytic center disrupts a critical contact point between the two proteins. In other words, the MePCE catalytic center may be occupied by residues of LARP7 when the two form a close complex. This will certainly explain the inhibition of MePCE’s capping activity by LARP7. Obviously, this notion must be put to test through future structural analyses. Finally, since the VLD-AAA mutation also reduced the interactions of MePCE with 7SK and HEXIM1 (Figure 4B), it appears that the interaction of the MePCE mutant with P-TEFb could proceed without any assistance from 7SK and HEXIM1.
Figure 4.
LARP7 may occupy the catalytic center of MePCE to inhibit the latter’s capping activity. (A) Wild-type F-MePCE and the VLD-AAA mutant were affinity purified with anti-Flag beads from NEs of transfected HeLa cells under highly stringent conditions (see ‘Materials and Methods’ section) to remove any associated 7SK snRNP components. Upon their normalization to approximately the same level by anti-Flag WB (bottom panel), these two proteins were tested in 7SK capping reactions. The RNA products were isolated and analyzed by autoradiography (top panel) and EB staining (middle panel). (B) Wild-type and VLD-AAA mutant F-MePCE were affinity purified from NEs of transfected HeLa cells under relatively mild conditions to retain their associated factors, which were subsequently detected by WB and NB as indicated.
MePCE not bound to LARP7 are responsible for all the 7SK capping activity in the nucleus
The data presented so far are consistent with the notion that 7SK is already capped when sequestered in the 7SK snRNP, in which MePCE, the likely sole source of cellular 7SK capping activity, is strongly inhibited by LARP7. These observations raise the following two questions. (i) When does 7SK become capped prior to its incorporation into 7SK snRNP? and (ii) does MePCE have additional role(s) in 7SK snRNP unrelated to its capping of 7SK?
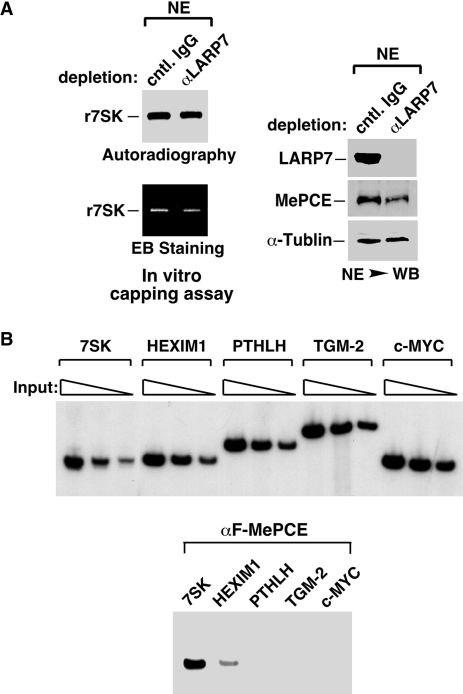
Regarding the first question, in order to explain 7SK capping that occurs before the assembly of 7SK snRNP, one has to assume that outside the snRNP there must exist MePCE proteins that are not inhibited by LARP7. To test this hypothesis, we first performed immunodepletion with immobilized anti-LARP7 antibodies and examined the level of MePCE in the depleted NE. The depletion is deemed highly efficient and specific judging from the levels of LARP7 and the internal control protein α-tubulin in NEs that were depleted with either the anti-LARP7 antibodies or unrelated control IgG (Figure 5A, right panel). Notably, the removal of LARP7 also caused the co-depletion of approximately half of MePCE in NE, indicating that not all MePCE are associated with LARP7 in the cell. When tested in 7SK capping reactions, the NE immunodepleted of LARP7 displayed the same level of capping activity as the NE mock-depleted with control IgG (Figure 5A, left panel). This result is consistent with the notion that the LARP7-bound MePCE does not contribute to 7SK capping and that the capping is entirely mediated by MePCE proteins that exist outside the MePCE–LARP7 complex. It is important to note that this result is different from the observed increase in 7SK capping activity displayed by the NE of LARP7 KD cells (Figure 3A), in which MePCE were produced in the absence of LARP7 and thus more MePCE were present in the active form to methylate 7SK.
Figure 5.
MePCE not bound to LARP7 are responsible for all the 7SK capping activity in the nucleus and preferentially associated with the 7SK gene promoter. (A) HeLa NEs were subjected to immunodepletion with the indicated antibodies and then analyzed by WB for the presence of the indicated proteins (right panel). The depleted NEs were then tested in 7SK capping reactions, with the RNA products analyzed by autoradiography (top left) and EB staining (bottom left). (B) The levels of input chromatin derived from the HeLa-based W10 cells stably expressing F-MePCE were carefully adjusted (in 2-fold increments) and then PCR amplified with primer sets corresponding to the indicated genes (top panel). Once the input chromatin corresponding to the five genes were normalized to approximately the same level, ChIP with anti-Flag mAb was performed. After DNA purification, PCR reactions containing α-[32P]dCTP were carried out and the products analyzed by gel electrophoresis and autoradiography (bottom panel).
Preferential association of MePCE with the 7SK gene promoter
For pre-mRNAs transcribed by RNA Pol II, capping is known to take place co-transcriptionally when nascent transcripts reach the length of ∼25–30 nt (24). Given the above demonstration that the methyl-phosphate cap is likely added onto 7SK prior to its sequestration into 7SK snRNP, it is reasonable to speculate that like pre-mRNAs, 7SK is also capped co-transcriptionally. To test a key aspect of this hypothesis, we performed ChIP assay to determine whether MePCE can be detected at the 7SK gene promoter. As an important control, the genes encoding HEXIM1, PTHLH, TGM-2 and c-MYC, all of which are transcribed by RNA Pol II in a P-TEFb-dependent manner (13,16,18), are also analyzed. To eliminate possible variations caused by potential differences in the copy number and PCR amplification efficiency among the five genes to be tested in the ChIP assay, the input chromatin levels corresponding to these genes were carefully titrated and normalized to approximately the same level (Figure 5B, upper panel). Under such conditions, the 7SK gene promoter was found to associate with significantly more MePCE than the other four Pol II genes (Figure 5B, lower panel), with PTHLH, TGM-2 and c-MYC displaying no signal at all. It is also interesting to note that a high concentration of MePCE was also found to associate with the promoter region of U6 (data not shown), which is known to be the only other Pol III-transcribed snRNA in human cells with the unique γ-monomethyl phosphate cap structure and can be methylated by MePCE (9,25).
Although the ChIP result in Figure 5B does not represent a definitive proof of co-transcriptional capping of 7SK, the preferential occupancy of the 7SK gene promoter by MePCE is certainly consistent with such a possibility. Moreover, the detection of no or very little MePCE at the Pol II gene promoters may also explain the fact that pre-mRNAs do not have the methylphosphate cap structure despite the existence of γ-phosphate at their 5′-end when freshly generated. It is known that for pre-mRNAs transcribed by Pol II, co-transcriptional capping is achieved through the physical association of the capping enzyme guanylyltransferase with the phosphorylated form of Pol II CTD (24). Future studies are necessary to determine whether MePCE may also associate with the Pol III transcriptional machinery at the 7SK promoter in order to accomplish co-transcriptional capping of 7SK.
Capping-independent stimulation of the LARP7–7SK binding by MePCE
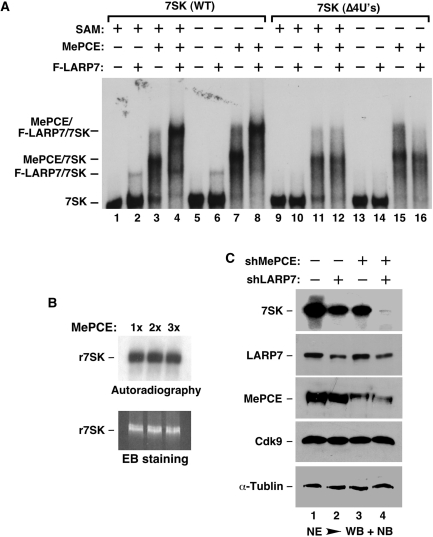
Given the above demonstrations that 7SK is capped by the LARP7-free MePCE and in probably a co-transcriptional manner and that MePCE displays no capping activity within 7SK snRNP, where 7SK is likely already capped, we next investigated whether MePCE may have another role in 7SK snRNP that is independent of its capping of 7SK. Toward this goal, we performed a gel mobility shift assay with 32P-labeled wild-type or a mutant 7SK (4U’s) lacking the 3′-UUUU-OH sequence, which is recognized by LARP7 and essential for the LARP7–7SK binding (13). The data in Figure 6A show that while recombinant MePCE (for its purity, see Figure 3B) interacted well with wild-type 7SK, F-LARP7 (see Figure 3B) formed only a weak complex with the RNA when present at a similar concentration (Figure 6A, lanes 2 and 3). However, the addition of both MePCE and LARP7 into the reaction resulted in the assembly of a robust, slower migrating complex. Antibody supershift experiments confirm that this complex contained MePCE, LARP7 and 7SK as expected (data not shown). Notably, MePCE and LARP7 were able to form this trimeric complex with wild-type 7SK but not 4U’s (compare lanes 4 and 12), indicating that the interaction of LARP7 with the 3′-poly(U) sequence of 7SK is essential for the complex formation. Thus, the binding of MePCE to the cap has been replaced by the interaction with LARP7 and then through LARP7 to the 3′-poly(U). In other words, the presence of MePCE has converted an otherwise weak LARP7–7SK binding into a much stronger one, which in turn promotes the formation of a stable MePCE–LARP7–7SK subcomplex.
Figure 6.
Capping-independent stimulation of the LARP7–7SK binding by MePCE, which leads to the stabilization of 7SK by the cooperative actions of MePCE and LARP7. (A) Gel mobility shift assay was performed with 32P-labeled wild-type 7SK or 7SK (Δ4U’s) and in the presence (+) or absence (−) of the indicated proteins or SAM. The positions of free 7SK as well as the various 7SK-containing complexes are indicated on the left. (B) 7SK capping reactions containing the same concentration (1×) as or twice (2×) or thrice (3×) more MePCE than that used in gel shift reactions in A were performed in the presence of SAM and absence of LARP7. The RNA products were isolated and analyzed by autoradiography (top panel) and EB staining (bottom panel). (C) HeLa cells were transfected with constructs expressing the indicated shRNAs to KD the expression of MePCE or/and LARP7. The levels of 7SK and the other indicated proteins in NEs of transfected cells were analyzed by WB and NB.
It is important to point out that this stimulatory effect of MePCE is unrelated to its capping activity, as the omission of SAM, the methyl group donor, in the reactions produced no effect on the MePCE-mediated MePCE–LARP7–7SK complex formation (Figure 6A, compare lanes 1–4 with lanes 5–8). The control experiment in Figure 6B indicates that in the presence of SAM and absence of F-LARP7, the amount of MePCE used in the gel shift reactions was able to cause a likely complete methylation of 7SK as no further methylation was observed with the addition of 2- or 3-times more MePCE. Taken together, these data indicate that despite MePCE’s lack of capping activity in 7SK snRNP, its ability to stimulate the LARP7–7SK binding and cooperate with LARP7 to promote the MePCE–LARP7–7SK subcomplex formation well justifies its existence in 7SK snRNP.
MePCE and LARP7 cooperate to stabilize 7SK snRNA
To examine the functional significance of MePCE’s stimulation of the LARP7–7SK binding and cooperation with LARP7 to form the MePCE–LARP7–7SK subcomplex, we tested whether shRNA depletion of MePCE could affect the ability of LARP7 to maintain the stability of 7SK. Previously, we have shown that the expression of shLARP7 caused a nearly complete depletion of cellular LARP7 and co-depletion of >95% of 7SK (13). However, when shMePCE was expressed in HeLa cells, ∼85% of MePCE and only ∼45% of 7SK were co-depleted (Figure 6C and data from titration analysis not shown). Thus, MePCE contributes to the metabolic stability of 7SK in HeLa cells, which is consistent with the previous observations obtained in both human 293 cells and zebrafish embryos (9,17). However, compared with LARP7, MePCE appears to be less efficient in maintaining the steady-state level of 7SK RNA.
To determine whether MePCE and LARP7 act cooperatively to maintain 7SK stability, we expressed either alone or together with shMePCE half the normal amount of shLARP7, which reduced the LARP7 level by about half in HeLa cells (Figure 6C). While shLARP7 or shMePCE alone reduced the nuclear levels of 7SK by ∼50 and 45%, respectively, the co-expression of the two together led to >99% depletion of 7SK in cells that still retained ∼50 and 15% of their original levels of LARP7 and MePCE, respectively (Figure 6C). We have previously shown that when there was little LARP7 in the cell caused by maximal shLARP7 expression, 7SK was efficiently degraded whether or not MePCE was around (13). However, when the cellular level of LARP7 was reduced by half through reduced shLARP7 expression as shown in Figure 6C, the remaining LARP7 became severely dependent on MePCE to bind to 7SK and protect it from exonucleolytic cleavage. When MePCE were also depleted under such conditions, the remaining ∼50% of LARP7 were simply unable to keep 7SK stable (Figure 6C, lane 4). Thus, the stability of 7SK depends on the cooperation between LARP7 and MePCE.
CONCLUDING REMARKS
The 7SK snRNP represents a major reservoir of activity from which functional P-TEFb can be recruited to respond to increased cellular demand for P-TEFb-dependent gene expression and cell growth (2). Recently, it has been shown that the integrity of 7SK snRNP and consequently the amount of P-TEFb sequestered in this complex are also critical for vertebrate development and the proper splicing of pre-mRNA transcripts (17). Within this multi-subunit snRNP, 7SK RNA functions as a central scaffold that holds all the protein components together and allows HEXIM1 to suppress Cdk9’s kinase activity (6,12). Thus, the stability of 7SK is of supreme importance as it determines the amount of active P-TEFb in the cell. Whereas the 7mGpppN cap structure of mRNAs is a well-known protector against 5′-exonucleolytic cleavage, it remains to be determined whether the methylated γ-phosphate cap structure of 7SK can provide a similar function. Assuming that the cap can indeed protect 7SK, the continued presence of MePCE in 7SK snRNP, which already harbors 7SK in its methylated state, can only be justified by the demonstration that MePCE has a capping-independent function of significantly stabilizing the LARP7–7SK binding and that the stability of 7SK depends on the cooperation between MePCE and LARP7 (Figure 6). In this manner, MePCE lends a helping hand to LARP7 through releasing the 5′-end of 7SK, and the MePCE–LARP7 complex then efficiently binds to the 3′-poly(U) tail to prevent cleavage by 3′-exonucleases (Figure 6A). Although the significance of LARP7 inhibition of MePCE’s capping activity remains to be determined, the data presented here have nevertheless revealed an important mechanism by which these two proteins cooperate to maintain the stability of 7SK snRNA and integrity of 7SK snRNP.
FUNDING
National Natural Science Foundation of China (30470371 and 30670408 to R.C.); Natural Science Foundation of Fujian (C0210005 to R.C.); US National Institutes of Health (AI41757 to Q.Z.); University of California Cancer Research Coordinating Committee (to Q.Z.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Seemay Chou and Tom Alber for providing us with recombinant MePCE protein.
REFERENCES
- 1.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Q, Yik JH. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol Mol. Biol. Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 4.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 6.Yik JHN, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 7.Yik JH, Chen R, Pezda AC, Zhou Q. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive positive transcription elongation factor b complexes for control of transcription. J. Biol. Chem. 2005;280:16368–16376. doi: 10.1074/jbc.M500912200. [DOI] [PubMed] [Google Scholar]
- 8.Michels AA, Nguyen VT, Fraldi A, Labas V, Edwards M, Bonnet F, Lania L, Bensaude O. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol. Cell. Biol. 2003;23:4859–4869. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Markert A, Grimm M, Martinez J, Wiesner J, Meyerhans A, Meyuhas O, Sickmann A, Fischer U. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008;9:569–575. doi: 10.1038/embor.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, Sedore SC, Price JP, Price DH, Lania L, et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23:2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, Luo K, Zhou Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell. 2008;29:588–599. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 15.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 16.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol. Cell. Biol. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barboric M, Lenasi T, Chen H, Johansen EB, Guo S, Peterlin BM. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc. Natl. Acad. Sci. USA. 2009;106:7798–7803. doi: 10.1073/pnas.0903188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He N, Pezda AC, Zhou Q. Modulation of a P-TEFb functional equilibrium for the global control of cell growth and differentiation. Mol. Cell. Biol. 2006;26:7068–7076. doi: 10.1128/MCB.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sano M, Abdellatif M, Oh H, Xie M, Bagella L, Giordano A, Michael LH, DeMayo FJ, Schneider MD. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- 20.Barrandon C, Bonnet F, Nguyen VT, Labas V, Bensaude O. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol. Cell. Biol. 2007;27:6996–7006. doi: 10.1128/MCB.00975-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Herreweghe E, Egloff S, Goiffon I, Jady BE, Froment C, Monsarrat B, Kiss T. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 2007;26:3570–3580. doi: 10.1038/sj.emboj.7601783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wada K, Inoue K, Hagiwara M. Identification of methylated proteins by protein arginine N-methyltransferase 1, PRMT1, with a new expression cloning strategy. Biochim. Biophys. Acta. 2002;1591:1–10. doi: 10.1016/s0167-4889(02)00202-1. [DOI] [PubMed] [Google Scholar]
- 23.Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 24.Sims RJ, III, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 25.Singh R, Reddy R. Gamma-monomethyl phosphate: a cap structure in spliceosomal U6 small nuclear RNA. Proc. Natl. Acad. Sci. USA. 1989;86:8280–8283. doi: 10.1073/pnas.86.21.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]