Abstract
The C/D box scaRNA2 is predicted to guide specific 2′-O-methylation of U2 snRNA. In contrast to other SCARNA genes, SCARNA2 appears to be independently transcribed. By transient expression of SCARNA2-reporter gene constructs, we have demonstrated that this gene is transcribed by RNA polymerase II and that the promoter elements responsible for its transcription are contained within a 161 bp region upstream of the transcription start site. In mammals, we have identified four cross species conserved promoter elements, a TATA motif, an hStaf/ZNF143 binding site and two novel elements that are required for full promoter activity. Binding of the human hStaf/ZNF143 transcription factor to its target sequence is required for promoter activity, suggesting that hStaf/ZNF143 is a fundamental regulator of the SCARNA2 gene. We also showed that RNA polymerase II continues transcription past the 3′-end of the mature RNA, irrespective of the identity of the Pol II promoter. The 3′-end processing and accumulation are governed by the sole information contained in the scaRNA2 encoding region, the maturation occurring via a particular pathway incompatible with that of mRNA or snRNA production.
INTRODUCTION
Pseudouridines and 2′-O-methylriboses are the most two common modifications in ribosomal and small nuclear RNAs (1,2). In rRNAs and U6 snRNA, they are performed in the nucleolus by small nucleolar ribonucleoprotein particles (snoRNPs) (3,4). Box C/D snoRNPs direct the 2′-O-methylation of riboses, while H/ACA snoRNPs catalyze pseudouridylation (5–8). Box C/D snoRNPs are composed of a sequence-specific guide RNA associated with a set of at least four proteins: fibrillarin, Nop56, Nop58 and 15.5 kDa (4). Each box C/D snoRNA contains the conserved C, C′, D and D′ box motifs. Typically, box H/ACA snoRNAs exhibit a common hairpin–hinge–hairpin–tail structure with the H motif ANANNA in the single-stranded hinge region, and the ACA triplet located three nucleotides upstream of the 3′-termini (4). H/ACA snoRNAs associate with the four H/ACA snoRNP proteins, the pseudouridine synthase dyskerin/Cbf5, Gar1, Nhp2 and Nop10 (9).
U6 snRNA is synthesized by RNA polymerase III (Pol III) and is modified and assembled with proteins in the nucleus. In contrast, U1, U2, U4 and U5 (and likely U11 and U12) snRNAs are synthesized by RNA polymerase II (Pol II) and possess a distinct maturation pathway (10–12). In the nucleus, the Pol II-specific snRNAs acquire a monomethyl guanosine (7mG) cap structure and are extended for at least 250 nucleotides beyond the 3′-end of the mature RNA (13). Stable pre-snRNAs intermediate bearing a short 3′-end extension are generated by a transcription-linked cleavage event that requires a cis-acting element called the 3′-box (14–16). After export to the cytoplasm, the pre-snRNA is assembled with the Sm proteins to form a pre-snRNP. Binding of the Sm proteins is needed for dimethylation of 7mG to form the 2,2,7-trimethylguanosine cap structure and for removal of the 3′ trailer. Subsequently, snRNPs are imported to the nucleoplasm and transiently accumulate in Cajal bodies (CB) where the snRNAs undergo specific 2′-O-methylation and pseudouridylation directed by scaRNAs which accumulate in CB (17–19). Vertebrate guide scaRNAs contain either one single (C/D or H/ACA) or two snoRNA boxes. The composite structures H/ACA-H/ACA, C/D-H/ACA, C/D-C/D or (C/D-C/D like box) were observed in scaRNAs harboring two associated boxes. The scaRNAs are synthesized in the nucleoplasm and a common sequence motif specifically determines the CB localization of box H/ACA scaRNAs (20). Very recently, the human protein WDR79 was shown to be associated with scaRNAs containing one single or two snoRNA boxes, the WDR79 binding being required for CB localization of a scaRNA (21). In vertebrates, sequences encoding sno- and scaRNAs are generally located in introns of host genes (22). These intronic sno/scaRNAs are processed from the spliced and debranched host introns. U3 and U8 and most likely U13 snoRNAs, however, are transcribed from independent units by RNA polymerase II. In vertebrates, the basal promoter of the U3 snoRNA genes include an essential promoter sequence element (PSE) also found in the basal promoter of snRNA genes (10,23). Associated to the basal promoter of snRNA and snoRNA genes, the distal sequence element (DSE) plays a major role in transcription efficiency (10). The DSE are essentially composed of two functional submotifs, the octamer and the Staf Binding Site (SBS), that can reside simultaneously or individually (10,24,25). The capped scaRNA2 (mgU2-25/61) and scaRNA17 (mgU12-22/U4-8) are apparently encoded in intergenic regions and the presence of conserved motifs in the upstream region of the genes suggest that scaRNA2 and scaRNA17 are synthesized from independent units. The capped scaRNA2 contains a C/D box associated to a C/D like box domain. It is predicted to guide the specific 2′-O-methylation of the U2 snRNA G25 and C61 positions (26). To understand how scaRNA2 synthesis is controlled, we have cloned the gene and its flanking regions and demonstrated that 161 bp of the 5′-flanking region are sufficient for efficient promoter activity. In this region, we have mapped four essential promoter elements and identified hStaf/ZNF143 as a critical transactivator interacting with the cognate cis-element. Furthermore, we have shown that transcription of the SCARNA2 gene is governed by RNA polymerase II and continues beyond the region encoding the mature scaRNA2. The 3′-end processing and accumulation of the scaRNA2 are directed only by sequences within the scaRNA2 encoding region, contrasting with the mRNA or snRNA production pathways.
MATERIALS AND METHODS
Reporter constructs
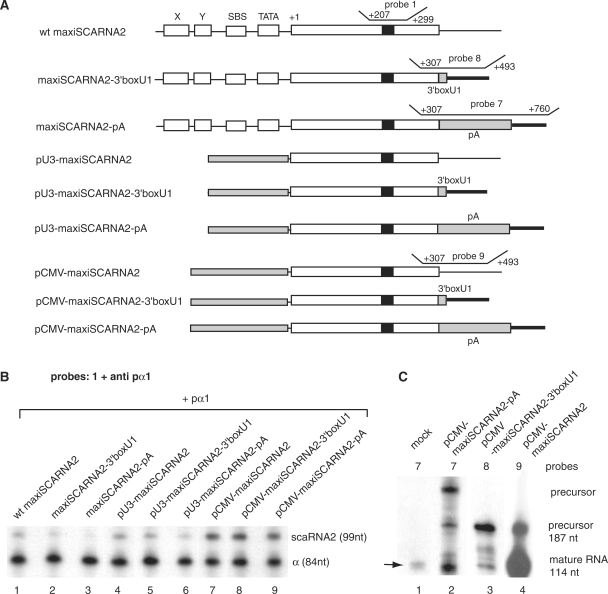
The human SCARNA2 promoter fragment −250/+532 was PCR amplified from genomic DNA and cloned clockwise into pGEMTeasy (Promega) to create SCARNA2 −250/+532. The SCARNA2 promoter fragments −250/+145, −250/+8, −161/+145, −88/+145 and −25/+145 were PCR amplified from SCARNA2 −250/+532 using direct and reverse primers incorporating SacI and BamHI sites, respectively. The PCR products were cloned 5′ to the luciferase reporter gene in the SacI/BamHI cut promoterless pFLASH1 vector (Synapsis) to generate SCARNA2 −250/+145 Luc, SCARNA2 −250/+8 Luc, SCARNA2 −161/+145 Luc, SCARNA2 −88/+145 Luc and SCARNA2 −25/+145 Luc. The SCARNA2 −250/+532 maxigene (maxiSCARNA2) was obtained by insertion of a BamH1 site at position +281 of SCARNA2 −250/+532. Substitution mutants giving rise to constructs Xsub, Ysub, SBSsub and TATA sub are listed in Figure 1B. Substitution and insertion mutants were generated with the QuickChange II XL site-directed mutagenesis kit (Stratagene). The SCARNA2 maxigene with a deletion or an inverted orientation of the +421 to +532 region (positions relative to the wt gene) were obtained as follows. A Pst1 site was introduced at position +421 in maxiSCARNA2 and the resulting mutant was digested with Pst1 (Pst1cut at +425 and in the polylinker). Self-ligation yielded the SCARNA2 −250/+421 maxigene Δ3′ (maxiSCARNA2Δ3′) and SCARNA2 −250/+421 inverted 3′ (maxiSCARNA2-inverted 3′). pCMV-maxiSCARNA2 and pU3-maxiSCARNA2 were generated as follows. A SacII site was introduced at position −8 of the maxiSCARNA2 containing a Pst1 site at position +421 and the SCARNA2 promoter was swapped with the entire CMV immediate early promoter from pcDNA3.1(+) (positions +206 to +855, Invitrogen) or the −324 to −1 fragment of the U3 snoRNA promoter amplified from human cell genomic DNA, to yield pCMV-maxiSCARNA2 and pU3-maxiSCARNA2, respectively. Swapping of the SCARNA2 3′ flanking sequence in maxiSCARNA2, pCMV-maxiSCARNA2 and pU3-maxiSCARNA2 (containing a Pst1 at position +421) with the SV40 polyadenylation signal (pA), amplified by PCR as a Pst1 fragment from pcDNA3.1(+) (positions +2947 to +3192), yielded maxiSCARNA2-pA, pCMV-maxiSCARNA2-pA and pU3-maxiSCARNA2-pA, respectively. MaxiSCARNA2-3′boxU1, pCMV-maxiSCARNA2-3′boxU1 and pU3-maxiSCARNA2-3′boxU1 were created by swapping the SCARNA2 3′ flanking sequence in maxiSCARNA2, pCMV-maxiSCARNA2 and pU3-maxiSCARNA2 (with a Pst1 at position +421) digested by Pst1 and Nde1 with the oligonucleotide ctgcagaGTTTCAAAAGTAGActgtacatatg containing the U1snRNA 3′ box (3′-box in capitals). Maxi U6 was created by inserting the 16 bp GACCTCGAGGCGGTTC sequence at position 87 of the human U6.2 gene. All constructs were verified by automated DNA sequencing.
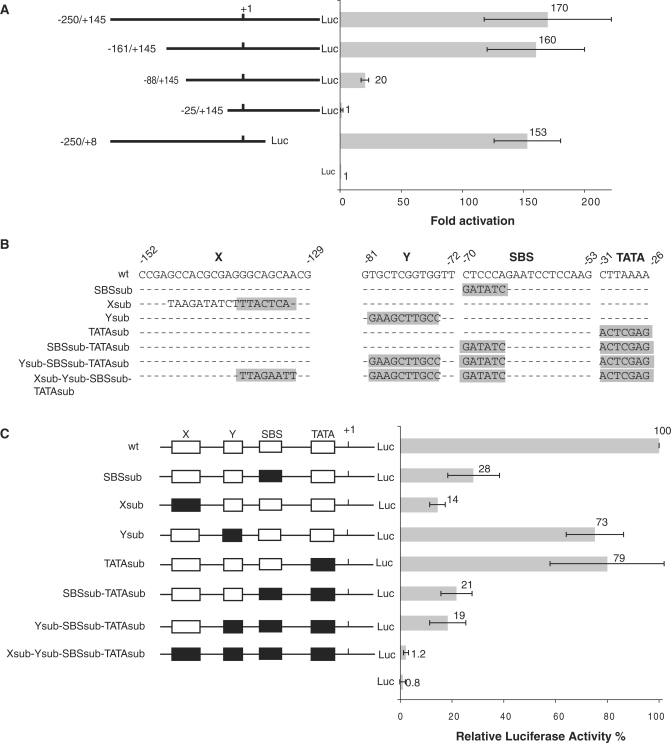
Figure 1.
Identification and functional analysis of the elements in the human SCARNA2 promoter. (A) Detection of promoter activity in the 5′-flanking sequence of the human SCARNA2 gene. Schematic representation of the SCARNA2 constructs fused to a luciferase (Luc) reporter and their activities in COS-7 cells. For each construct, the length of the sequence upstream and downstream of the +1 is shown to the left and the luciferase activity to the right. Cells were transiently transfected with the constructs and assayed for luciferase activity. The firefly luciferase activity was normalized to that of the renilla luciferase provided by co-transfection of the control vector. Data for each construct were plotted as fold-activation which represents the ratio between the normalized luciferase activity of each reporter construct to that of the empty luciferase vector (pFLASH1). Data are presented as the mean ± SD of three separate experiments. (B) Sequences of the wild-type and mutant X, Y, SBS and TATA elements of the SCARNA2 promoter. Mutations are highlighted in gray. (C) Contribution of the X, Y, SBS and TATA elements to SCARNA2 promoter activity. Left panel: schematic representation of the SCARNA2 promoter-luciferase (Luc) constructs substituted individually or simultaneously in the X, Y, SBS and TATA elements. Open and solid boxes represent wild-type and mutant elements, respectively. Right panel: COS-7 cells were transfected with the reporter constructs and the luciferase activity was normalized to that of the renilla lucifease obtained from the co-transfected control vector. The normalized luciferase activity of the wild-type construct −250/+145 was arbitrarily set to 100%. Data are presented as the mean ± SD of three separate experiments.
Transfection and dual luciferase reporter assays
COS-7 cells (8 × 104 cells) were transfected with Lipofectamine 2000 (Invitrogen). Cells received 500 ng of the SCARNA2 promoter plasmid containing the firefly luciferase reporter with 50 ng of pRL-TK (Promega) as the renilla luciferase internal control. The total amount of DNA was kept constant at 800 ng with carrier DNA. After 48 h, cells were lysed and the activities of both the firefly and renilla luciferases were determined with the dual luciferase assay kit (Promega). A minimum of three independent transfections were performed in duplicate and the firefly luciferase activity was normalized to that of the Renilla luciferase.
Transfection, RNase protection and reverse transcription assays
HeLa cells (9 × 105 cells) were transfected with Lipofectamine 2000 with 3.5 µg of promoter test plasmid and 1.5 µg of pα1x72 as internal control (27). For transcription assays in the presence of α-amanitin, the medium before transfection was changed to growth medium containing 20 µg/ml of α-amanitin and after tranfection the medium was replaced 19 h later by a fresh medium containing 20 µg/ml of α-amanitin. Cells were collected after 48 h and total RNA was extracted using the TRI-REAGENT (Euromedex). Synthesis of the internally labeled antisense RNA probes 1 to 9, complementary to various parts of the maxiscaRNA2 and variants, was performed by in vitro transcription with T7 for probes 1 to 6 (201, 193, 172, 264, 250, 226 nt long) or SP6 for probes 7 to 9 (454, 187 and 187 nt long) polymerases, as described in ref. (28). Probes 1–4 are complementary to positions +207/+299, −14/+71, +368/+441 and +368/+531 of the maxiSCARNA2 construct, respectively. Probes 5 to 9 are complementary to positions +283/+460, +368/+495, +307/+760, +307/+493 and +307/+493 of the maxiSCARNA2Δ3′, maxiSCARNA2-inverted3′, maxiSCARNA2-pA, maxiSCARNA2-3′boxU1 and pCMV-maxiSCARNA2 constructs, respectively (positions relative to the wt gene). Before utilization, each probe was purified on a 6% sequencing gel and further used for RNase protection assay (RPA) as previously described in refs (28,29). Briefly, after DNase I treatment, 10 µg of total RNA was annealed with 105 c.p.m. of probe at 85°C for 5 min and then allowed to hybridize overnight at 45°C. The unprotected RNA was next digested by incubation with a mixture of RNases A and T1 and the protected fragments were separated on a 6% sequencing gel. For reverse transcription assays, total RNA was analyzed by primer extension of a labeled oligonucleotide complementary to positions +88/+104 (wt human U6 gene numbering). The extended products were separated on a 6% sequencing gel. Results were quantitated with a Fuji Bioimage Analyzer.
hStaf/ZNF143 preparation and DNA binding assays
Full-length hStaf/ZNF143 was synthesized by in vitro coupled transcription–translation with the TnT system (Promega). Fifty microliter reactions were programmed with 1 µg of pSK(−)-ZNF143 (30). The various probes containing the wild-type and mutant versions of the SBS in the SCARNA2 promoter were obtained by PCR amplification of the −88/+145 region, using 32P-labeled primers. Gel retardation assays were performed essentially as described in refs (24,31) with 20 fmol of the labeled probe in the presence of 2.5 µl or 5 µl of programmed lysate.
ChIP assay and PCR analysis
The ChIP procedure was essentially as described in refs (32,33). Purified immunoprecipitated DNA was analyzed in 25 µl PCR reactions in the presence of 3 µCi (α-32P) dCTP (3000 Ci/mmol) with the test primer pair CCTGTGCTCGGTGGTTTC and GCAGGAGGAGAGCTTTTCAT, specific for the SCARNA2 promoter and complementary to positions −87/−69 and +228/+247 of the promoter, respectively. The tRNASec gene test primer pairs hybridized to sequences −391/−365 and −205/−181. For negative controls of the ChIP assay, the PP1 primer pair hybridizing to unique regions lying 2.4 kb upstream of the tRNASec was used. A 1/500 and 1/2000 of the immunoprecipitated DNA were used in PCR analysis. Decreasing amounts of input DNA (1/10 000, 1/25 000 and 1/100 000) were used to determine the linear range of PCR reaction for each primer pair. Cycling parameters were 95°C for 3 min, 35 cycles at 95°C for 30 s, 52–72°C (depending on each primer pair) for 30 s, 72°C for 30 s and one cycle at 72°C for 5 min.
RESULTS
Biological activities of putative cis-elements in the human SCARNA2 promoter and identification of RNA polymerase II as the acting polymerase on the SCARNA2 gene
To identify whether the 5′-flanking DNA of the SCARNA2 gene exhibits promoter activity, various sequences were fused to a firefly luciferase reporter gene. As the capped 5′-end of the 420 nt long scaRNA2 was previously mapped by primer extension (26), various constructs containing sequences upstream and downstream of the +1 of the scaRNA2 were generated. Constructs containing progressive 5′ truncated portions of the −250/+145 regions were transiently transfected into COS-7 cells. The luciferase activity of the resulting cell extracts was further measured (Figure 1A). Analysis of the firefly luciferase values, normalized to the activity of the renilla luciferase control, revealed that similar promoter activity was observed with fragments containing 161 bp or more upstream of the +1 (compare the luciferase activity of fragments −250/+145 and −161/+145 in Figure 1A). In contrast, the luciferase activity of the −88/+145 construct was about 8-fold lower than that of −161/+145. Construct −25/+145, which contains only 25 bp of the SCARNA2 gene flanking sequence, led to abrogation of the luciferase activity, showing it is not sufficient on its own for promoter activity (Figure 1A). Finally, the luciferase activity of construct −250/+8 containing only 8 nucleotides of the transcribed region was equivalent to that of construct −250/+145 (Figure 1A). Thus, the 5′-flanking sequence of the SCARNA2 gene possesses a promoter activity and the minimal promoter sequence can be defined as covering the −161/+8 region.
To identify putative elements in the promoter region of the SCARNA2 gene, we used the phylogenetic footprinting approach. A multiple alignment was performed with sequences of the chimpanzee, dog, mouse and rat genomes that are orthologous to the −250/−1 region of the human SCARNA2 gene. This revealed the presence of the four interspecies conserved elements X, Y, SBS and TATA at positions −152/−129, −81/−72, −70/−53 and −31/−26, respectively (Supplementary Figure S1A and Figure 1B). Sequence analysis of the region −250/−1 with the Matinspector software (34) established that the SBS element located at positions −70/−53 matches the consensus binding site for transcription factor hStaf/ZNF143 (Supplementary Figure S1C) (25,35). This analysis did not allow us to identify transcription factor binding sites for the conserved motifs X and Y. Furthermore, the 5′-flanking region of the SCARNA2 gene contains no obvious initiator (Inr) or downstream promoter element (DPE) in the vicinity of the transcription start site (TSS). Collectively, this computational analysis revealed the presence of four conserved motifs in the basal promoter of the mammalian SCARNA2 gene, constituting putative elements regulating its expression. Multiple sequence alignments of the Fugu, Tetraodon, Stickleback, Medaka and Xenopus tropicalis SCARNA2 promoters (Supplementary Figure S1B) revealed conservation of only the SBS and TATA element identified in the mammalian promoters, suggesting their crucial role in SCARNA2 gene expression.
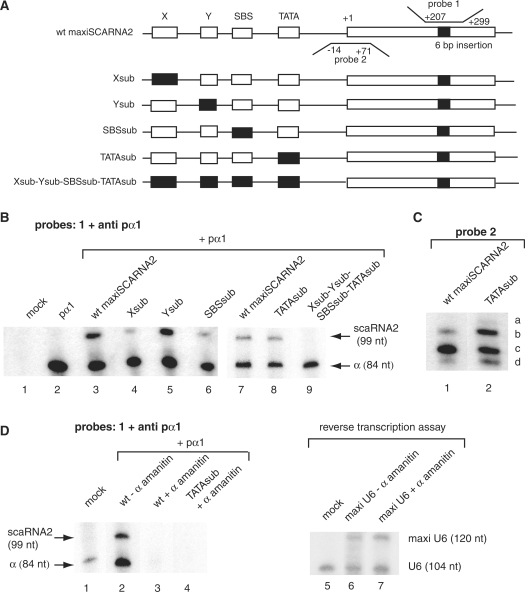
To evaluate the role of the phylogenetically conserved elements, we introduced individual mutations into the X, Y, SBS and TATA elements (Xsub, Ysub, SBSsub and TATAsub in Figure 1B). Expression of the reporter gene directed by the mutated SCARNA2 promoters was monitored by the luciferase activity after transfection into COS-7 cells. Mutation of the SBS reduced the SCARNA2 promoter activity to about 30% of the wt level (Figure 2C, compare wt and SBSsub). These results provide strong evidence for the presence of a biologically active SBS in the SCARNA2 promoter. The effect on transcription activity of substituting the Y element (Ysub, 73% of the wt level) is of the same order as that of the TATA element mutation (TATAsub, 79% of the wt level). In contrast, substitution of the X element reduced drastically expression from the promoter to 14% of the wt level (Figure 1C, compare wt and Xsub). Finally, SBSsub was modified by sequentially introducing substitutions into the TATA, Y and X elements (Figure 1B). As shown in Figure 1C, the simultaneous mutations of the SBS and TATA element reduced the luciferase activity to about 20% of the wt level (compare wt, SBSsub and SBSsub-TATAsub). The reporter activity of the triple mutant Ysub-SBSsub-TATAsub was of the same order as that of the double SBSsub-TATAsub mutant, indicating that the Y element is not essential for SCARNA2 gene expression. In contrast, introduction of a fourth mutation by substitution in the X element led to basal levels of luciferase activity (compare wt, Ysub-SBSsub-TATAsub and Xsub-Ysub-SBSsub-TATAsub in Figure 1C). Next, the RPA was used to evaluate the functionality of the identified four promoter elements in the context of the SCARNA2 gene and its 3′-flanking regions. We made use of the SCARNA2 maxigene with a 6 bp insert in the central part of the gene. The 6 bp insert offers the possibility to discriminate the endogenous scaRNA2 from the product of the transfected gene. HeLa cells were transfected with the wt maxiSCARNA2, Xsub maxiSCARNA2, Ysub maxiSCARNA2, SBSsub maxiSCARNA2, TATAsub maxiSCARNA2 and Xsub-Ysub-SBSsub-TATAsub maxiSCARNA2 constructs together with plasmid pα1 as the internal transfection control (Figure 2A). RNAs were analyzed by RPA of an antisense RNA probe complementary to region +207/+299 of the nontranscribed strand maxigene and containing the 6 bp insert (probe 1 in Figure 2A). Results were normalized to the expression of the α-globin mRNA included as an internal standard. Analysis of the mutation effects revealed an expression pattern of SCARNA2 that is globally similar to that observed with the SCARNA2 promoter mutants placed in front of a luciferase reporter gene. The predominant role of the X and SBS motifs is still observed, with Xsub and SBSsub expressed at 32% and 48% of the wt level, respectively (Figure 2B, compare lane 3 with lanes 4 and 6). Next, to investigate the possible alteration of the TSS by the different mutant promoters, we performed an RPA with probe 2 (Figure 2A) that is complementary to a region of the nontranscribed strand containing the TSS of the SCARNA2 gene. The presence of three protected fragments when the wt maxiSCARNA2 gene was used (fragments b, c and d in Figure 2C, lane 1) suggests the existence of three different TSS. The other analyses revealed that transcription initiation occurred at exactly the same position when the Xsub, Ysub and SBSsub maxiSCARNA2 mutants were used (data not shown). Surprinsingly, with the TATAsub mutant the relative alteration in the yield of fragments b, c and d, plus the detection of a fourth product (product a in Figure 2C, lane 2) revealed an alteration of the TSS when the TATA element is mutated.
Figure 2.
Transcription assays of wild-type and mutated human SCARNA2 gene promoters and polymerase transcribing status of the SCARNA2 gene. (A) Structures of the templates used in the in vivo analyses. The +1 indicates the start of transcription. Open boxes, wild-type elements; solid boxes, mutant elements. The solid box in the transcribed region represents the 6 bp insertion creating the maxigene. Probes 1 and 2 and their hybridization regions in the nontranscribed strand are indicated (wt gene numbering). (B) In vivo expression of wild-type and mutated gene promoters. HeLa cells were transfected with the constructs indicated above the lanes together with plamid pα1 as the internal control. RNA recovered 48 h after transfection were analyzed by RNase protection assay with probe 1 and probe anti pα1 complementary to the α-globin standard. scaRNA2 and α are the protected RNAs derived from the scaRNA2 and pα1 globin internal standard. Results from lanes 1 to 6 and 7 to 9 arose from separate experiments. Normalized transcription activity values: wt, 100%; Xsub, 32 ± 7%; Ysub, 90 ± 9%; SBSsub, 48 ± 10%; TATAsub, 95 ± 7%. (C) Identification of altered TSS of the wt and mutants SCARNA2 promoters. RPA with probe 2 was used to determine whether transcription of the TATAsub mutant gene initiated at the same position. HeLa cells were transfected with the constructs indicated above the lanes; a, b, c and d indicate protected fragments. (D) Pol II transcription of the SCARNA2 gene. HeLa cells were transiently transfected with the constructs indicated above the lanes and treated simultaneously with low α-amanitin concentration (+α-amanitin, lanes 3, 4 and 7) or untreated (−α-amanitin, lanes 2 and 6). Lanes 1–4, RPA performed as in (B) with probe 1 and anti pα1. The lengths of protected fragments are indicated (nt). Lanes 5–7, primer extension assay performed with RNA isolated from transfected or untransfected cells and a 32P-labeled oligonucleotide complementary to a region located downstream of the 16 bp insertion creating the maxi U6 gene. Data are representative views of experiments performed in triplicate. Results from lanes 1 to 4 and 5 to 7 arose from separate experiments.
The characteristics of the SCARNA2 promoter and the absence of a T strech at the proximity of the mature scaRNA2 3′-end suggest that the SCARNA2 gene is transcribed by Pol II. To verify this hypothesis, we performed transcription assays with the wt maxiSCARNA2 gene in the presence of plasmid pα1 as the Pol II control under α-amanitin conditions inhibiting Pol II but not Pol III transcription (Figure 2D). The extracted RNAs were analyzed by RPA with probes 1 and anti pα1; the absence of protected RNA products in the presence of α-amanitin demonstrated that the SCARNA2 gene is transcribed by RNA polymerase II (Figure 2D, compare lanes 1, 2 and 3). In an additional control experiment, we showed that expression of a Pol III maxi U6 gene was unaffected under the same α-amanitin conditions (Figure 2D, compare lanes 5, 6 and 7). Lastly, the absence of transcriptional activity of the TATAsub mutant in the presence of α-amanitin under conditions inhibiting Pol II transcription (Figure 2D, lane 4) demonstrated that Pol II is responsible for the activity of the TATA sub template (Figure 2B, lane 7).
Taken together, these results indicate that Pol II is the polymerase acting on the SCARNA2 gene and that there are four positive regulatory elements in the human SCARNA2 promoter, the function of the X and SBS motifs being predominant for full promoter activity. These results also suggest an important role of the TATA motif in TSS selection.
The hStaf/ZNF143 transcription factor binds the cognate element in the human SCARNA2 promoter
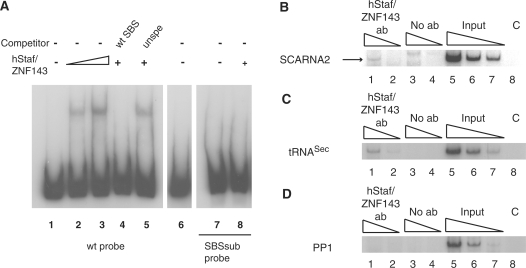
Gel mobility shift assays were used to ask whether hStaf/ZNF143 interacts with the putative SBS located at positions −70/−53 in the SCARNA2 promoter. In the binding assay, the full-length hStaf/ZNF143 was used with the 32P-labeled fragment containing the putative SBS. Figure 3A shows that hStaf/ZNF143 bound to the SCARNA2 promoter and generated one retarded complex (Figure 3A, compare lane 1 with lanes 2 and 3). In contrast, we were unable to detect a retarded complex with the unprogrammed reticulocyte lysate (compare lane 1 with lanes 3 and 6 in Figure 3A). The specificity of binding was confirmed by competition studies. As expected, competition by a 1000-fold molar excess of unlabeled oligonucleotide containing the consensus SBS (35) abolished complex formation (Figure 3A, compare lanes 2 and 4). In contrast, an unrelated oligonucleotide was unable to compete (Figure 3A, lane 5). To demonstrate that the hStaf/ZNF143 binding pattern was strictly dependent on the SBS integrity, similar binding assays were carried out with hStaf/ZNF143 and a promoter mutant having the −70 CTCCCA −65 sequence in the SBS substituted by GATATC (24,36,37). Indeed, this mutation abolished totally formation of the DNA-protein complex (Figure 3A, lane 8).
Figure 3.
Identification of an occupied hStaf/ZNF143 transcription factor binding site in the human SCARNA2 promoter. (A) Gel retardation assays were performed with 32P-labeled promoter fragments encompassing positions −88/+145 of the SCARNA2 promoter and containing the wild-type SBS sequence (wt probe, lanes 1–6), mutant SBS (SBSsub probe, lanes 7 and 8). In the SBSsub probe, the −70 CTCCCA−65 sequence was replaced by GATATC. Probes were incubated in the absence (lanes 1 and 7) or presence of hStaf/ZNF143 (2.5 µl of programmed reticulocyte lysate in lanes 2, 4, 5 and 8; 5 µl in lane 3). Reactions in lanes 4 and 5 were performed in the presence of a 1000-fold molar excess of unlabeled specific competitor (wt SBS) and unspecific competitor (unspe). Reaction in lane 6 was performed in the presence of 5 µl of unprogrammed reticulocyte lysate. Lanes 1–6 and 7–8 are from separate experiments. (B–D) Immunoprecipitation with hStaf/ZNF143 antibody of the SCARNA2 promoter from formaldehyde cross-linked chromatin prepared from HeLa cells. DNA fragments recovered from input chromatin, or ChIP assays with anti-hStaf/ZNF143 or control experiment without antibody, were analyzed by semi-quantitative PCR with specific primer pairs in the presence of (α−32P) dCTP. Lanes 1, 2 and 3, 4: serial dilutions of DNA immunoprecipitated with anti-hStaf/ZNF143 or no antibody, respectively. Lanes 5–7: serial dilutions of input material to demonstrate that the assays were within the linear range of PCR amplification. Lane 8 (C, PCR control): PCR lacking DNA template. (B) PCR was performed with specific primer pairs for the human SCARNA2 promoter or (C) for the tRNASec promoter acting as the positive control. (D) Negative control with the PP1 primer pair targeting a region located upstream of the tRNASec gene and lacking SBS.
To determine whether hStaf/ZNF143 is associated in vivo with the active SCARNA2 promoter, we performed a chromatin immunoprecipitation assay in HeLa cells. After immunoprecipitation with the hStaf/ZNF143 antibody, enrichment of the SCARNA2 promoter was monitored by semi-quantitative PCR amplification using primers amplifying the SCARNA2 promoter in the region surrounding the SBS. To confirm the specificity of binding, positive and negative controls were performed. PCR amplification of the tRNASec promoter, which binds hStaf/ZNF143 as well, was used as the positive control (33). The negative control amplified a segment located several kilobasepairs upstream from the tRNASec transcribed region, which does not harbor any hStaf/ZNF143 binding site. As a supplementary control, a similar experiment was performed without antibody. The SCARNA2 promoter was specifically immunoprecipitated by the hStaf/ZNF143 antibody (Figure 3B, compare lanes 1, 2 and 3, 4; the specific product is indicated by an arrow). As expected, the positive control with anti-hStaf/ZNF143 yielded a specific signal of intensity higher than in the no-antibody control (Figure 3C, compare lanes 1, 2 and 3, 4). In contrast, no specific PCR product could be observed with the PP1 primers amplifying SBS-lacking regions (Figure 3D, lanes 1–4). We conclude from these experiments that an SBS resides at positions −70/−53 and that hStaf/ZNF143 is recruted to the SCARNA2 promoter in living cells.
Transcription of the SCARNA2 gene continues past the 3′-end of the mature RNA
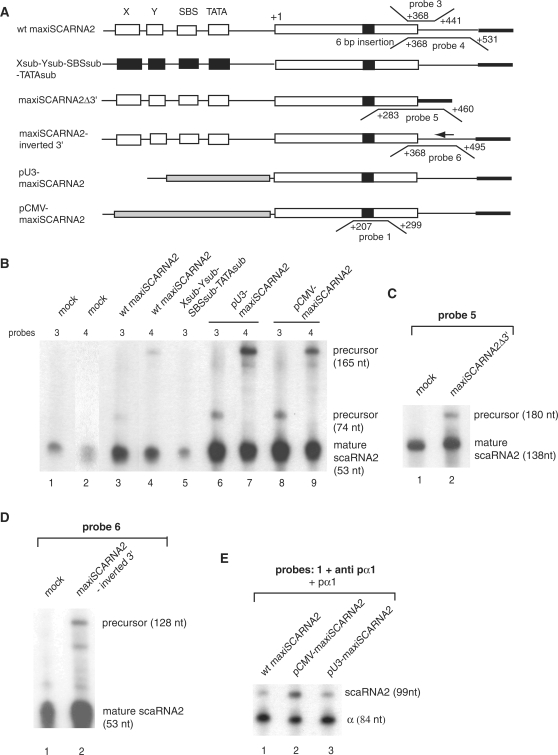
The 3′-end formation of the independently transcribed U1–U5 snRNAs, U3, U8 and U13 snoRNAs requires a cis-acting sequence known as the 3′-box, located downstream of the 3′-end of the mature RNA. Transcription of the genes encoding these RNAs continues beyond the 3′-box in vivo and produces a precursor form that is not abundant in a steady-state level. The 3′-box is not an efficient transcription terminator but may rather act as a processing element functioning in the precursor RNA (13,10,14,16). Scrutiny of the sequence located at proximity of the 3′-end of the mature scaRNA2 did not show any motif with sequence similarity to the 3′-box of Pol II snRNA genes, reading typically GTTTN0-3AAARN2AGA (38). The 3′-end of the mature scaRNA2 was previously mapped to position +420 of wt scaRNA2 gene (26). Accumulation and processing of the expressed scaRNA2 were tested here by RPA using specific antisense probes 3 and 4 that map to the 3′-end of the RNA (Figure 4A). Each probe is complementary to the same 3′-part of the mature scaRNA2 (positions +368/+420 of the wt gene) and to 21 and 111 nucleotides of downstream regions for probes 3 and 4, respectively. With both probes, protected fragments arising from the mature scaRNA2 could be detected with RNA extracted from untransfected cells (Figure 4B, lanes 1 and 2). With RNA isolated from maxiSCARNA2 gene transfected cells, probes 3 and 4 detected the mature scaRNA2 and longer products corresponding to scaRNA2 precursors (Figure 4B, compare lanes 3 and 4). Due to the low concentration of the endogenous scaRNA2, the probes detected only the transfected overexpressed RNA precursors. As expected, the RNA precursor was undetected in an RPA performed with RNA isolated from cells transfected with maxiSCARNA2 gene bearing mutations in the four promoter elements (Figure 4B, lane 5). To investigate whether the 3′-flanking region of the SCARNA2 contains elements that promote proper 3′-end processing or accumulation, we deleted the genomic sequence downstream of the mature scaRNA2 3′-end in the maxiSCARNA2 gene to create maxiSCARNA2Δ3′ (Figure 4A). We also generated construct maxiSCARNA2-inverted 3′ bearing the downstream region in an inverted orientation (Figure 4A). After cell transfection, we examined the accumulation and the presence of an RNA precursor by RPA using RNA probes complementary to the 3′-part of the mature scaRNA2 and downstream regions (Figure 4A, probes 5 for maxiSCARNA2Δ3′ and 6 for maxiSCARNA2-inverted 3′). We detected the presence of RNA precursors and accumulation of properly processed scaRNA2 upon expression from maxiSCARNA2Δ3′ (Figure 4C, lane 2, ratio signal/mock: 2.5) and maxiSCARNA2-inverted 3′ (Figure 4D, lane 2, ratio signal/mock: 5). Therefore, changes in the downstream sequence adjacent to the mature scaRNA2 3′-end had no impact on scaRNA2 accumulation driven by the wt promoter.
Figure 4.
The 3′ flanking genomic region of the SCARNA2 gene is not required for the processing step and the mature scaRNA2 can be produced from various Pol II promoters. (A) Structures of the various templates used in the analyses. Probes 1, 3–6 and their hybridization regions in the nontranscribed strand are indicated (wt gene numbering). (B) Various Pol II promoters produce correctly 3′-end processed scaRNA2. HeLa cells were transfected with the constructs indicated above the lanes. After quantitation, the signal/mock ratios in lanes 3–9 relative to the mature RNA are: 3.25 ± 0.25, 5 ± 0.3, 1 ± 0.1, 6.7 ± 0.35, 13 ± 0.5, 9.25 ± 0.4, 11 ± 0.25. Results from lane 2 arose from a longer exposure. (C) Deletion of the 3′ flanking genomic region of the SCARNA2 gene does not impede production of the mature scaRNA2. Signal/mock ratio relative to the mature RNA in lane 2: 2.5 ± 0.3 (D) The mature scaRNA2 can be obtained with a construct harboring the 3′-flanking region in an inverted orientation. Signal/mock ratio relative to the mature RNA in lane 2: 3.5 ± 0.5. (E) The U3 snoRNA and CMV promoters are compatible with scaRNA2 production. RNA extracted from cells transfected with the constructs indicated above the lanes were analyzed by RPA with the indicated probe. pα1: vector used as the internal transfection control; anti pα1: probe monitoring the RNA produced from the transcribed pα1 gene. scaRNA2 and α: protected RNAs derived from the scaRNA2 and pα1 globin internal standard. Normalized transcriptional activity values: wt, 100%; pCMV-maxiSCARNA2, 300 ± 25%; pU3-maxiSCARNA2, 145 ± 15%. The lengths of protected fragments are indicated (nt). Data presented in (B–D) are representative views of experiments performed in triplicate.
The scaRNA2 can be obtained from a variety of Pol II promoters. 3′end processing and accumulation are solely governed by information in the mature scaRNA2
Monitoring RNA expression from different gene contexts constitutes a powerful approach to obtain insight into the pathway of transcript maturation. We employed this strategy to study the scaRNA2 biosynthesis by promoter exchange or addition of motifs known to be essential for processing of other RNAs.
In a first step, we substituted the scaRNA2 promoter in construct maxiSCARNA2 with either an mRNA promoter from the CMV or an snRNA promoter from the U3 snoRNA gene (Figure 4A). The resulting constructs pCMV-maxiSCARNA2, pU3-maxiSCARNA2 and wt maxiSCARNA2 were transfected separately into HeLa cells cells in the presence of pα1 as the internal control for transfection. After 48 h, RNAs were extracted and analyzed by RPA with three different probes (probes 1, 3 and 4 in Figure 4A). RPA with probe 1 complementary to the central part of the maxiscaRNA revealed that scaRNA2 is expressed more efficiently from pCMV-maxiSCARNA2 (3.5-fold) and pU3-maxiSCARNA2 (1.45-fold) than from the wt maxiSCARNA2 (Figure 4E, compare lane 1 with lanes 2 and 3). RPA with probes 3 and 4 revealed that expression from the CMV or the U3 snRNA promoters resulted in the accumulation of scaRNA2 correctly processed at the 3′-end (Figure 4B, compare lanes 3, 4 with lanes 6, 7 and 8, 9). From these results, we conclude that scaRNA2 can be produced and properly 3′-end processed from a variety of Pol II promoters. The RPA performed with probe 5 and RNA from cells transfected with pCMV-maxiSCARNA2Δ3′ and pU3-maxiSCARNA2Δ3′ (harboring deletions downstream of the mature scaRNA2 3′-end) detected accumulation of correctly processed scaRNA2 (data not shown). These results confirm that the region downstream of the mature 3′-end is inert in 3′-end processing and that only signals in the mature scaRNA2 direct its processing and accumulation.
In a second step, we swapped the wild-type 3′-flanking sequence in the SCARNA2maxigene with the mRNA polyadenylation signal (polyA) of the SV40 or the human U1 snRNA 3′-box (3′-boxU1) downstream element (contructs maxiSCARNA2-pA and maxiSCARNA2-3′boxU1 in Figure 5A). Note that the poly A signal is essential for processing and stability of mRNAs, while this role in snRNAs is devoted to the 3′-box. The polyA signal and U1 snRNA 3′-box were also introduced into the constructs harboring the U3 snoRNA or CMV promoters in place of the wt SCARNA2 promoter (pU3-maxiSCARNA2-pA, pU3-maxiSCARNA2-3′boxU1, pCMV-maxiSCARNA2-pA and pCMV-maxiSCARNA2-3′boxU1 in Figure 5A). The accumulation and presence of an RNA precursor were evaluated by RPA using RNA probes complementary to the central part of the maxiRNA (probe 1) or to the 3′-part of the mature scaRNA2 and downstream regions (Figure 5A, probes 7, 8 and 9). First, RPA results with probe 1 and anti pα1 as the internal standard showed that addition of the 3′-box U1 or polyA signal in the context of the wt SCARNA2 promoter reduced the accumulation of the RNA to 61% and 34% of the wt level, respectively (Figure 5B, compare lane 1 with lanes 2 and 3). Similarly, in the context of the U3 snoRNA promoter, the presence of the U1 snRNA 3′-box reduced the accumulation of the RNA to 75% of the wt level, that obtained with the addition of the polyA signal being 66% (Figure 5B, compare lane 4 with lanes 5 and 6). The yield of the transcribed RNA probed with internal probe 1 is only slightly affected by the presence of the 3′-end processing elements in the context of the CMV promoter (Figure 5B, compare lane 7 with lanes 8 and 9). Probes 7, 8 and 9 will be used to follow formation of the 3′- end from constructs containing the polyA signal, the 3′-box U1 or the wt sequence (Figure 5A). In the RPA experiment with probe 7 and RNA from untransfected cells, the correctly processed endogenous 3′-end mature scaRNA2 is indicated by an arrow in Figure 5C (lane 1). The correctly processed 3′-end mature and precursor scaRNA2 are easily detectable using probe 9 with RNA isolated from cells transfected with pCMV-maxiSCARNA2 (Figure 5C, lane 4). In contrast, in the presence of the polyA signal or 3′-box U1, the transcribed RNA was uncorrectly processed and we were unable to detect accumulation of similar amounts of mature scaRNA2 (compare Figure 5C, compare lanes 2 and 3 with lane 4). Similar results were obtained when the pA signal or the 3′-box U1 were inserted in the context of the wt or U3snoRNA driven maxiSCARNA2. However, a lower yield of products was observed (data not shown). Although processing of the scaRNA2 was independent of the genomic sequence located 3′ to the mature RNA, addition of the poly A signal or snRNA 3′-box reduced significantly the accumulation of scaRNA2 correctly processed at the 3′-end.
Figure 5.
Introduction of 3′-end processing elements inhibits production of mature scaRNA2. (A) Schematic representation of the structures of the various constructs used in the assays. (B and C) In vivo expression of the various constructs depicted in (A). HeLa cells were transfected with the constructs indicated above the lanes. The recovered RNAs were analyzed by RNase protection assay with the indicated probes. pα1: vector used as the internal transfection control; anti pα1: probe monitoring the RNA produced from the transcribed pα1 gene. scaRNA2 and α: protected RNAs as in Figure 2. The lengths of protected fragments are indicated (nt). Results from lanes 1–3 and lane 4 in (C) arose from separated experiments. Data presented are representative views of experiments performed in triplicate. Normalized transcription activity values for lanes 1–9 in (B): lane 1, 100%; lane 2, 61 ± 25%; lane 3, 34 ± 15%; lane 4, 100 %; lane 5, 75 ± 17%; lane 6, 66 ± 9%; lane 7, 100%; lane 8, 90 ± 9%; lane 9, 85 ± 10%.
From these results, we conclude that in contrast to snRNA genes, the scaRNA2 can be obtained from various Pol II promoters and without dependence on 3′ elements. In addition, scaRNA2 maturation occurs via a pathway incompatible with that of mRNAs or snRNAs.
DISCUSSION
In vertebrates, sequences encoding scaRNAs are generally located in the introns of their host genes. Intronic scaRNAs are produced after the splicing step by processing of the debranched intron (19). Here, we demonstrated that the 161 bp region located upstream of the mature scaRNA2 bears sufficient information for Pol II transcription of the SCARNA2 gene. This is consistent with the high-resolution map of the genome wide distribution of RNA polymerase II which localizes it to a region covering positions −345 to +40 of the SCARNA2 gene (39). Furthermore, the present study established that transcription of the human SCARNA2 gene is governed by four cis-acting elements that are evolutionarily conserved, a TATA motif, a SBS and two novel, previously uncharacterized motifs that have been identified in this study. The position of the TATA element suggests that it represents a bona fide TATA binding protein (TBP) motif, a key basal cis regulatory element shared by Pol II promoters of protein coding genes and a few RNA polymerase III promoters. The CTTAAAAG sequence of the TATA element diverges in the 5′-part from the TATAAAAG consensus (40). However, structural studies of TBP–DNA complexes concluded that such a nucleotide sequence variability does not impair TBP recognition, the protein being able to accommodate C at position 1 and T at position 2 of the TATA element (41). Indeed, we showed that nucleotide substitutions in the TATA element had only a minor impact on transcription efficiency of the SCARNA2 gene. Besides, substitution in the TATA sequence led to new TSS, as already observed for protein-coding genes lacking TATA elements (42). It is therefore likely that the function of the TATA element is to ensure correct initiation of transcription as in other Pol II transcribed genes (42). Motifs X and Y that we characterized in this study do not correspond to any cis-acting motif previously identified. They are only conserved in mammalian SCARNA2 promoters but are surprisingly absent in Xenopus and pufferfish SCARNA2 genes. This suggests that these two elements act in a species-specific manner. The third cis-acting motif that we identified is specifically recognized by hStaf/ZNF143 which was found associated to the SCARNA2 promoter in vivo. Although the Xenopus Staf was originally identified as the transcription activator of the tRNASec gene, it also controls expression of snRNA and snRNA-type genes transcribed by RNA polymerases II and III (24,25,30). Furthermore, hStaf/ZNF143 has been reported to regulate transcription of nine protein-coding gene (33,36,37). Lastly, a genome-wide analysis led us to identify 1175 Staf/ZNF143 binding sites distributed in 938 mammalian protein gene promoters (33). This strongly suggests that hStaf/ZNF143 is involved in the transcriptional regulation of a very large number of protein coding genes. The present work extends the role of hStaf/ZNF143 to the transcription of the scaRNA2 gene which harbors a promoter organization different from that of snRNA and snRNA-type genes. Interestingly, the SCA2RNA promoter does not contain the PSE motif which is a key element in the transcription of snRNA genes (10). Instead, the SCARNA2 gene carries promoter elements that look like those of protein-coding genes, as is the case for the telomerase RNA gene (TERC) and most likely SCARNA17. However, despite this resemblance, the organization of the elements is unique within the promoters of the SCARNA2 and TERC genes (43–45). The vast majority of snRNA genes are ubiquitously expressed while, in contrast, expression of most of the mRNA coding genes is temporally and spatially restricted. Thus, the presence of an mRNA-type promoter in the scaRNA2 gene can offer the possibility to also regulate its expression.
We have not detected elements in the context of the SCARNA2 gene that can influence the specificity of scaRNA2 synthesis. Furthermore, context elements known to promote accumulation of mRNA and snRNA inhibited rather than enhanced production of mature scaRNA2. Instead of a precursor context dependent pathway, processing of the scaRNA2 very likely relies on the integrity of the mature scaRNA2 and in particular on the C/D-C/D like boxes essential for scaRNP assembly. We propose, as described for the yeast snoRNP and human telomerase, that protein association with the scaRNA2 precursor to form scaRNP is a prerequesite to 3′-end processing of the scaRNA2 (44,46,47). This will be addressed in future investigations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Schweigert for excellent technical assistance. C. Allmang was thanked for the generous gift of T7 RNA polymerase. M.A.G. was a recipient of an Allocation de Recherche from the Ministère de l’Enseignement Supérieur et de la Recherche.
REFERENCES
- 1.Limbach PA, Crain PF, McCloskey JA. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maden BE, Hughes JM. Eukaryotic ribosomal RNA: the recent excitement in the nucleotide modification problem. Chromosoma. 1997;105:391–400. doi: 10.1007/BF02510475. [DOI] [PubMed] [Google Scholar]
- 3.Smith CM, Steitz JA. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 4.Reichow SL, Hamma T, Ferre-D'A;mare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 6.Tycowski KT, Smith CM, Shu MD, Steitz JA. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc. Natl Acad. Sci. USA. 1996;93:14480–14485. doi: 10.1073/pnas.93.25.14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 8.Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 9.Meier UT. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005;114:1–14. doi: 10.1007/s00412-005-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez N. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem. 2001;276:26733–26736. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- 11.Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 12.Kiss T. Biogenesis of small nuclear RNPs. J. Cell Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- 13.Cuello P, Boyd DC, Dye MJ, Proudfoot NJ, Murphy S. Transcription of the human U2 snRNA genes continues beyond the 3′ box in vivo. EMBO J. 1999;18:2867–2877. doi: 10.1093/emboj/18.10.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Medlin JE, Uguen P, Taylor A, Bentley DL, Murphy S. The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J. 2003;22:925–934. doi: 10.1093/emboj/cdg077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uguen P, Murphy S. The 3' ends of human pre-snRNAs are produced by RNA polymerase II CTD-dependent RNA processing. EMBO J. 2003;22:4544–4554. doi: 10.1093/emboj/cdg430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 17.Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2'-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiss AM, Jady BE, Darzacq X, Verheggen C, Bertrand E, Kiss T. A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res. 2002;30:4643–4649. doi: 10.1093/nar/gkf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiss AM, Jady BE, Bertrand E, Kiss T. Human box H/ACA pseudouridylation guide RNA machinery. Mol. Cell Biol. 2004;24:5797–5807. doi: 10.1128/MCB.24.13.5797-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richard P, Darzacq X, Bertrand E, Jady BE, Verheggen C, Kiss T. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. 2003;22:4283–4293. doi: 10.1093/emboj/cdg394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol. Cell. 2009;34:47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 23.Ach RA, Weiner AM. Cooperation between CCAAT and octamer motifs in the distal sequence element of the rat U3 small nucleolar RNA promoter. Nucleic Acids Res. 1991;19:4209–4218. doi: 10.1093/nar/19.15.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuster C, Myslinski E, Krol A, Carbon P. Staf, a novel zinc finger protein that activates the RNA polymerase III promoter of the selenocysteine tRNA gene. EMBO J. 1995;14:3777–3787. doi: 10.1002/j.1460-2075.1995.tb00047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaub M, Myslinski E, Schuster C, Krol A, Carbon P. Staf, a promiscuous activator for enhanced transcription by RNA polymerases II and III. EMBO J. 1997;16:173–181. doi: 10.1093/emboj/16.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tycowski KT, Aab A, Steitz JA. Guide RNAs with 5′ caps and novel box C/D snoRNA-like domains for modification of snRNAs in metazoa. Curr. Biol. 2004;14:1985–1995. doi: 10.1016/j.cub.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Lobo SM, Hernandez N. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell. 1989;58:55–67. doi: 10.1016/0092-8674(89)90402-9. [DOI] [PubMed] [Google Scholar]
- 28.Myslinski E, Ame JC, Krol A, Carbon P. An unusually compact external promoter for RNA polymerase III transcription of the human H1RNA gene. Nucleic Acids Res. 2001;29:2502–2509. doi: 10.1093/nar/29.12.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez N, Lucito R. Elements required for transcription initiation of the human U2 snRNA gene coincide with elements required for snRNA 3' end formation. EMBO J. 1988;7:3125–3134. doi: 10.1002/j.1460-2075.1988.tb03179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myslinski E, Krol A, Carbon P. ZNF76 and ZNF143 are two human homologs of the transcriptional activator Staf. J. Biol. Chem. 1998;273:21998–22006. doi: 10.1074/jbc.273.34.21998. [DOI] [PubMed] [Google Scholar]
- 31.Myslinski E, Krol A, Carbon P. Optimal tRNA((Ser)Sec) gene activity requires an upstream SPH motif. Nucleic Acids Res. 1992;20:203–209. doi: 10.1093/nar/20.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinmann AS, Farnham PJ. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods. 2002;26:37–47. doi: 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- 33.Myslinski E, Gerard MA, Krol A, Carbon P. A genome scale location analysis of human Staf/ZNF143-binding sites suggests a widespread role for human Staf/ZNF143 in mammalian promoters. J. Biol. Chem. 2006;281:39953–39962. doi: 10.1074/jbc.M608507200. [DOI] [PubMed] [Google Scholar]
- 34.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaub M, Krol A, Carbon P. Flexible zinc finger requirement for binding of the transcriptional activator staf to U6 small nuclear RNA and tRNA(Sec) promoters. J. Biol. Chem. 1999;274:24241–24249. doi: 10.1074/jbc.274.34.24241. [DOI] [PubMed] [Google Scholar]
- 36.Gerard MA, Krol A, Carbon P. Transcription factor hStaf/ZNF143 is required for expression of the human TFAM gene. Gene. 2007;401:145–153. doi: 10.1016/j.gene.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 37.Myslinski E, Gerard MA, Krol A, Carbon P. Transcription of the human cell cycle regulated BUB1B gene requires hStaf/ZNF143. Nucleic Acids Res. 2007;35:3453–3464. doi: 10.1093/nar/gkm239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiner AM. E Pluribus Unum: 3′ end formation of polyadenylated mRNAs, histone mRNAs, and U snRNAs. Mol. Cell. 2005;20:168–170. doi: 10.1016/j.molcel.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 39.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 41.Patikoglou GA, Kim JL, Sun L, Yang SH, Kodadek T, Burley SK. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13:3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 43.Zhao JQ, Hoare SF, McFarlane R, Muir S, Parkinson EK, Black DM, Keith WN. Cloning and characterization of human and mouse telomerase RNA gene promoter sequences. Oncogene. 1998;16:1345–1350. doi: 10.1038/sj.onc.1201892. [DOI] [PubMed] [Google Scholar]
- 44.Fu D, Collins K. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol. Cell. 2003;11:1361–1372. doi: 10.1016/s1097-2765(03)00196-5. [DOI] [PubMed] [Google Scholar]
- 45.Zhao J, Bilsland A, Hoare SF, Keith WN. Involvement of NF-Y and Sp1 binding sequences in basal transcription of the human telomerase RNA gene. FEBS Lett. 2003;536:111–119. doi: 10.1016/s0014-5793(03)00038-3. [DOI] [PubMed] [Google Scholar]
- 46.Morlando M, Ballarino M, Greco P, Caffarelli E, Dichtl B, Bozzoni I. Coupling between snoRNP assembly and 3′ processing controls box C/D snoRNA biosynthesis in yeast. EMBO J. 2004;23:2392–2401. doi: 10.1038/sj.emboj.7600254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ballarino M, Morlando M, Pagano F, Fatica A, Bozzoni I. The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae. Mol. Cell Biol. 2005;25:5396–5403. doi: 10.1128/MCB.25.13.5396-5403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.