Abstract
Parafibromin, a component of the RNA polymerase II-associated PAF1 complex, is a tumor suppressor linked to hyperparathyroidism-jaw tumor syndrome and sporadic parathyroid carcinoma. Parafibromin induces cell cycle arrest by repressing cyclin D1 via an unknown mechanism. Here, we show that parafibromin interacts with the histone methyltransferase, SUV39H1, and functions as a transcriptional repressor. The central region (128–227 amino acids) of parafibromin is important for both the interaction with SUV39H1 and transcriptional repression. Parafibromin associated with the promoter and coding regions of cyclin D1 and was required for the recruitment of SUV39H1 and the induction of H3 K9 methylation but not H3 K4 methylation. RNA interference analysis showed that SUV39H1 was critical for cyclin D1 repression. These data suggest that parafibromin plays an unexpected role as a repressor in addition to its widely known activity associated with transcriptional activation. Parafibromin as a part of the PAF1 complex might downregulate cyclin D1 expression by integrating repressive H3 K9 methylation during transcription.
INTRODUCTION
Parafibromin is a 531-amino-acid tumor suppressor protein encoded by the HRPT2 gene. It is the human homolog of yeast Cdc73 within the polymerase-associated factor 1 (PAF1) complex (1–3). Germ line and somatic mutations of HRPT2 are found in HPT-JT (hyperparathyroidism-jaw tumor) and sporadic parathyroid carcinoma patients (4–6).
The PAF1 complex, originally identified in yeast as associated with RNA polymerase II, is comprised of Paf1, Leo1, Ctr9, Rtf1 and Cdc73 (7,8). The yeast PAF1 (yPAF1) complex regulates transcription, including transcriptional initiation and elongation (9,10), histone H2B ubiquitination, histone H3 lysine (K) 4 and K79 methylation (11) and control of poly(A) length (12). A subset of genes involved in metabolism and cell cycle control is regulated by PAF1 (13,14). Drosophila parafibromin interacts with β-catenin to mediate a functional link between Wnt signaling and gene expression (15). However, the molecular mechanisms underlying the tumor suppressor activities of parafibromin remain unclear.
Posttranslational modifications of histone tails within nucleosomes, including acetylation, phosphorylation, ubiquitination and methylation, modulate transcription and chromatin structure (16,17). In general, histone H3 methylation on K4 is associated with transcriptional activation (18,19), while methylation of H3 K9 is associated with transcriptional repression (20–22). Several histone methyltransferases (HMTs) with evolutionarily conserved SET domains, such as SUV39H1 and G9a, have H3 K9 methylation activity and are responsible for mono-, di- and tri-methylation of H3 K9 (17). The biological consequences differ depending on the number of methyl groups on histone residues.
The collapse of cell cycle control is common in human cancer, where over-expression of cyclin D1 is one of the most commonly observed alterations. Cyclin D1 was identified critical in parathyroid tumors as indicated by its alternative name, PRAD1 (parathyroid adenomatosis 1) oncogene (23). Interestingly, over-expression of parafibromin leads to repression of cyclin D1 and inhibition of proliferation, thereby highlighting a potential link of parafibromin to cell cycle control through cyclin D1 as a tumor suppressor (24–26). However, the detailed molecular mechanism by which parafibromin regulates cyclin D1 is still unclear. Here, we show that parafibromin recruits SUV39H1 to the human cyclin D1 gene and promotes histone H3 K9 methylation, suggesting that parafibromin, with SUV39H1, is a transcriptional repressor targeting epigenetic control of cell cycle. It also proposes an unsuspected role for the PAF1 complex beyond its classical role in active transcription.
MATERIALS AND METHODS
Plasmid construction
DNA constructs (pcDNA3-AU5-parafibromin, pEGFP-G9a, pBS-4xnH3 and the cyclin D1 promoter-luciferase reporter construct) were kindly provided as mentioned in the ‘Acknowledgements’ section. A DNA fragment containing four tandem repeats of H3 tail (1–40) was subcloned from pBS-4xnH3 into the GST vector by PCR. Parafibromin wild-type and deletion mutants were generated by PCR and cloned into pM (Clontech), pcDNA3.1 Myc/His, or pcDNA3-Flag vectors (Invitrogen). Cloning was confirmed by sequencing.
Cell culture and transfection
293T and HeLa cells were grown with DMEM containing 10% fetal bovine serum and 50 U/ml penicillin/streptomycin (WELGENE). Cells were transiently transfected with Transfectin, according to the manufacturer’s protocol (Bio-Rad). For RNA interference assays, cells were transfected with small interfering RNAs (siRNAs) against HRPT2 (Dhamacon or Invitrogen), G9a (Santa Cruz), SUV39H1 (Santa Cruz) or control siRNA (AccuTarget™ Negative control siRNA, Bioneer) using Lipofectamine 2000 (Invitrogen).
Luciferase assays
The 293T cells were transfected with the luciferase reporters and DNA constructs and incubated for 24 h before harvest. Luciferase activity was measured according to the manufacturer’s instructions (Promega). Each transfection was performed in duplicate and repeated three to five times.
Immunoprecipitation and immunoblotting
Transiently transfected 293T cells were harvested and lysed in the lysis buffer [20 mM Tris–HCl (pH 7.4), 150 mM NaCl, 0.5 % NP-40, and inhibitors of proteases and phosphatases]. Cell lysates were immunoprecipitated with suitable antibodies along with protein-A (Amersham Pharmacia Biotech.) or protein-G beads (Santa Cruz). Immunoprecipitates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE), transferred to a nitrocellulose membrane and immunoblotted with appropriate antibodies. Antibodies were purchased from Bethyl Laboratories (parafibromin, Paf1 and Leo1), Santa Cruz (Cyclin D1, GAPDH and GFP), LabFrontier (α-tubulin), Roche [c-Myc (9E10)], Upstate (G9a and SUV39H1) and Sigma [G9a and Flag (M2)].
Preparation of GST-4xnH3
GST-4xnH3 (amino acids 1–40) was expressed in Escherichia coli by a standard protocol. Cells were disrupted by sonication in phosphate-buffered saline (PBS) (plus 1 mM DTT, 1 mM PMSF), and then Triton X-100 was added at a final concentration of 1%. After centrifugation, soluble fractions were used for incubation with glutathione agarose beads (Sigma) for 30 min at room temperature. GST-H3 was eluted from the resin with elution buffer [50 mM Tris–HCl (pH 8.0) and 5 mM reduced glutathione]. The purity and the protein concentrations were verified by Coomassie staining of SDS–PAGE gels.
In vitro histone methyltransferase assay
Immunoprecipitates were incubated with 20 µg of GST-4xnH3 (amino acids 1–40) in HMTase buffer [50 mM Tris–HCl (pH 8.5), 20 mM KCl, 10 mM MgCl2 and 50 µM S-adenosyl-l-methionine] for 1 h at 30°C. Sample buffer [2% SDS, 10% glycerol, 100 mM DTT, 60 mM Tris (pH 6.8) and 0.001% bromophenol blue] was added to stop the reaction. Proteins were separated by SDS–PAGE and further analyzed by immunoblotting using appropriate antibodies.
Quantitative real-time polymerase chain reaction analysis
Cultured HeLa cells were transfected with control, HRPT2, G9a or SUV39H1 siRNAs for 24 or 48 h. Total RNA was isolated using Trizol (Invitrogen), and complementary DNA (cDNA) synthesis was performed with the Reverse Transcription System (Promega). Quantitative real-time polymerase chain reaction (PCR) was performed with specific primers for human cyclin D1 and GAPDH using the KAPATM SYBR® FAST qPCR KIT (KAPABIOSYSTEMS) and Chromo4TM real-time PCR detector (Bio-Rad). Relative levels of mRNA were normalized to the values of GAPDH mRNA for each reaction. The following primers were used: cyclin D1 (#898 and #899) (5′-CTGTGCTGCGAAGTGGAAACC-3′ and 5′-GTCCAGGTAGTTCATGGCCAGC-3′), HRPT2 (#512 and #513) (5′-ACTGAACAGATTAGGTC-3′ and 5′-ACATCTACCTCAGCATCC-3′), SUV39H1 (#1191 and #1192) (5′-GATATGACCTCTGCATCTTCCGC-3′ and 5′-GTACACGTCCTCCACGTAGTCCAG-3′), G9a (#1193 and #1194) (5′-GTTTCCACCCTCGGCAGTTG-3′ and 5′-CTGCATTTATGTTGGCTCCAGC-3′), GAPDH (#1189 and #1190) (5′-TCAATGGAAATCCCATCACCATC-3′ and 5′-CTTCTCATGGTTCACACCCATGAC-3′).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed according to the manufacturer’s protocol (Upstate Biotechnology). Sonicated lysates were used for ChIP with antibodies against parafibromin (Bethyl Laboratory), SUV39H1(Upstate), G9a (Abcam), dimethyl H3 K4 (Upstate), dimethyl H3 K9 (Abcam), trimethyl H3 K9 (Upstate) or normal serum as a control. Immunoprecipitated DNA and input DNA were analyzed by quantitative real-time PCR using specific primers to cyclin D1: promoter region (#704 and #705) (5′-TGAAAATGAAAGAAGATGCAGTCG-3′ and 5′-CTGTAGTCCGGTTTTCATAGAAATGC-3′), coding region (#738 and #739) (5′-GTCCTACTTCAAATGTGTGCAGAAGG-3′ and 5′-CTCCCACGAAACGCTACTTCTAGC-3′), 5 kb upstream region (#744 and #745) (5′-CCCAGTTACTGTCGTTATCTCTCATC-3′ and 5′-ATCCCTTTTGTAGCATCCCAAGAG-3′).
RESULTS
Parafibromin functions as a transcriptional repressor
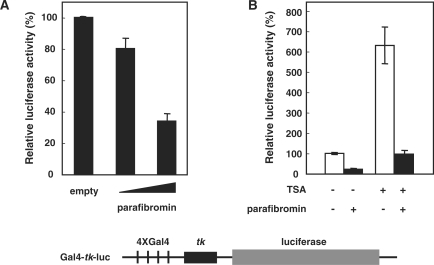
We first examined the transcriptional activity of parafibromin. Gal4-fused parafibromin was expressed in HEK293T cells along with a Gal4-tk luciferase reporter that contains four Gal4-binding sites upstream of the thymidine kinase (tk) promoter. Gal4-parafibromin dose-dependently repressed luciferase gene activity (Figure 1A). trichostatin A (TSA), a general inhibitor of histone deacetylases (HDAC), elevates general transcription activity. However, parafibromin was able to repress the reporter activity irrespective of whether TSA was added or not (∼6.5-fold in the presence and ∼4.3-fold in the absence of TSA), suggesting that parafibromin represses genes by a mechanism largely independent of HDAC (Figure 1B).
Figure 1.
Parafibromin represses transcription. (A) Gal4-parafibromin represses tk-driven luciferase reporter gene activity. The 293T cells were transfected with different amounts of Gal4DB-fused parafibromin (0.1 µg, 0.5 µg) along with the 4xGal4-tk-luciferase reporter. Reporter gene activities were normalized to protein concentration. (B) Transcriptional repression by parafibromin is not affected by an inhibitor of histone deacetylase. The 293T cells were transfected with the Gal4-tk-luciferase reporter and GAL4-parafibromin. TSA was added, as indicated, 24 h after transfection. Luciferase activities were measured 24 h after TSA treatment.
Parafibromin specifically interacts with HMT
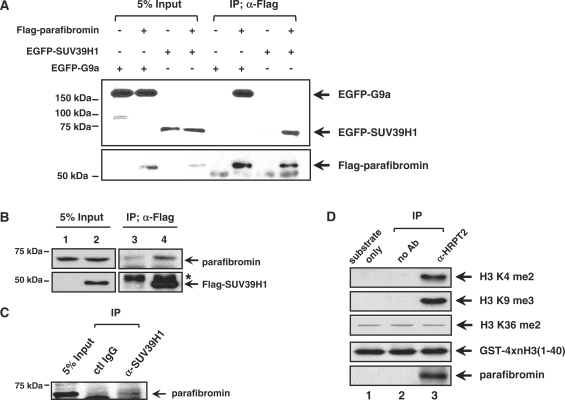
Complexes with parafibromin show HMT activity that targets H3 K4, which is implicated in transcriptional activation (2). However, in general, transcriptional repression is associated with methylation of H3 K9. Therefore, we tested whether parafibromin induces H3 methylation activity targeting K9 by first examining the physical association of parafibromin and SUV39H1 or G9a, the representative H3 K9 methyltransferases. Co-immunoprecipitation experiments indicated that parafibromin could interact with both EGFP-SUV39H1 and EGFP-G9a (Figure 2A). In addition, endogenous parafibromin interacted with Flag-SUV39H1 and endogenous SUV39H1 in HeLa cells (Figure 2B and C). Although we observed a strong interaction between parafibromin and GFP-G9a (Figure 2A), the interaction between endogenous proteins was barely detected under the same conditions as SUV39H1 (data not shown). Taken together, our data indicate that parafibromin associated with at least SUV39H1 and has a potential to integrate H3 K9 methylation for gene repression.
Figure 2.
Parafibromin specifically associates with the H3 K9 histone methyltransferase, SUV39H1. (A) Parafibromin associates with EGFP-SUV39H1 and EGFP-G9a in vivo. The 293T cells were transiently transfected with EGFP-SUV39H1 or EGFP-G9a and Flag-parafibromin. Cell lysates were immunoprecipitated with anti-Flag antibody and then immunoblotted with anti-GFP (upper panel) or anti-Flag (lower panel) antibodies, respectively. (B) Association of parafibromin with SUV39H1. The 293T cells were transfected with control (lanes 1 and 3) or Flag-SUV39H1 expressing construct (lanes 2 and 4). Flag-SUV39H1 immunoprecipitates were immunoblotted with anti-parafibromin antibody. Asterisk indicates immunoglobulin heavy chains. (C) Co-immunoprecipitation assay was used to detect interaction of endogenous parafibromin with SUV39H1 in HeLa cells. (D) Endogenous parafibromin was immunoprecipitated and HMT activity associated with precipitates was directly analyzed using GST-4xnH3 (amino acids 1–40). GST-4xnH3 consists of GST and four tandem repeats of amino acids 1–40 of H3. Parafibromin dependent histone modification was analyzed by immunoblotting using H3 K9 dimethyl, H3 K9 trimethyl, H3 K36 dimethyl and parafibromin antibodies. Probing H3 K36 methylation showed only nonspecific background. Substrate used for in vitro HMT reaction was revealed by anti GST antibody.
Given the association of parafibromin with SUV39H1, we tested whether the parafibromin-containing complex had HMT activity targeting H3 K9. Parafibromin was partially purified by immunoprecipitation using an anti-parafibromin antibody and subjected to an in vitro HMT assay using four tandem repeats of amino acids 1–40 of H3 as a substrate. Modifications on different residues of H3 were then analyzed by an immunoblotting assay with specific antibodies. The parafibromin complex immunoprecipitated from cell lysate directed H3 methylation on K9 as well as K4 (Figure 2D, lane 3). As a control, IP without specific IgG showed no HMT activity (lane 2). The substrate itself purified from bacteria gave no signals (lane 1). Importantly, parafibromin-associated H3 methylation activity was specific to K9 and K4 because H3 K36 signal was not dependent on parafibromin and seemed to be nonspecific background. In fact, Rozenblatt-Rosen et al. (2) have detected HMT activity associated with parafibromin, targeting H3 K9, which was much weaker than H3 K4 methylation in their experimental condition, although it was not described further. Overall, our data indicate that parafibromin might function through SUV39H1, and propose a combined role of parafibromin, SUV39H1 and H3 K9 methylation in regulating gene transcription.
The central region of parafibromin is important for SUV39H1 interaction and repression activity
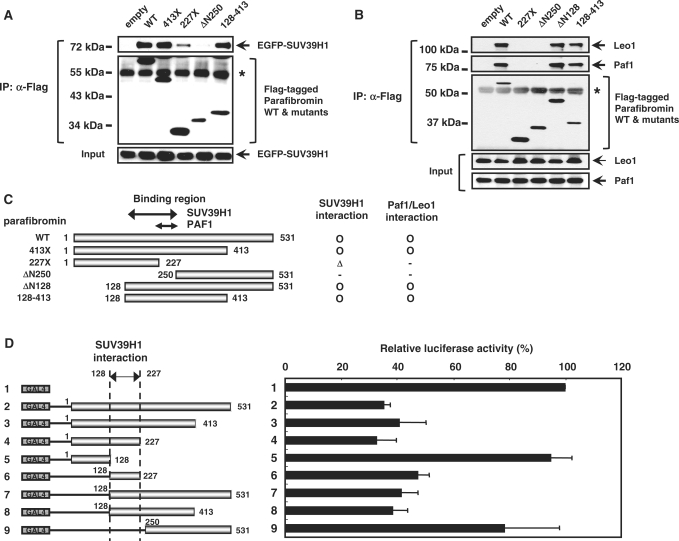
To identify the region of parafibromin responsible for the SUV39H1 interaction, we performed co-immunoprecipitation experiments with a series of Flag-parafibromin derivatives and EGFP-SUV39H1 in 293T cells. Parafibromin WT and mutants lacking far-end C-terminal regions (413X) and a mutant with only the central region (128–413) retained strong interactions with SUV39H1 (Figure 3A). The N-terminal half (227X) maintained a weak interaction with SUV39H1, but a larger deletion of the N-terminal region (ΔN250) completely disrupted the interaction with SUV39H1, indicating that the central region of parafibromin (128–250) is important and the region of 128–227 is minimally necessary for the SUV39H1 interaction (Figure 3A and C). Interestingly, 227X and ΔN250 failed to interact with the PAF1 complex (Paf1 or Leo1), while parafibromins encompassing 128–227 maintained the interaction (Figure 3B and C), suggesting that parafibromin interacts with SUV39H1 directly independently of other components of the PAF1 complex. Our domain analysis narrowed down the interaction surface of parafibromin for Paf1/Leo1 to a region of 227–250 amino acids as well (Figure 3C).
Figure 3.
SUV39H1 binds to the central region of parafibromin, which is important for repression. (A) SUV39H1 binds to the central region of parafibromin. The 293T cells were transfected with EGFP-SUV39H1 with various Flag-tagged parafibromin mutants. For interactions, cell lysates were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-GFP or Flag antibodies. (B) Paf1 and Leo1 bind to the central region of parafibromin. The 293T cells were transfected with various Flag-tagged parafibromin mutants. For interactions, cell lysates were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-Paf1, Leo1 or Flag antibodies. In (A) and (B), asterisks indicate the immunoglobulin heavy chains. (C) Diagram of deletion mutants of parafibromin and its interaction with SUV39H1 and PAF1 components. (D) The SUV39H1-binding domain in parafibromin is sufficient to repress transcription. The 293T cells were transfected with Gal4DB-fused parafibromin mutants along with the 4xGal4-tk-luciferse reporter. The values are averages from five independent experiments.
To further address whether the SUV39H1 interaction domain is important for the repression of transcription, we analyzed the ability of truncated forms of Gal4-parafibromin to repress transcription in 293T cells. All constructs were expressed at a similar level (Supplementary Figure S1). Parafibromin mutants lacking the SUV39H1 interaction domain failed to repress reporter gene activity, but the interaction domain (128–227) alone was sufficient to exert repression (Figure 3D). The weak interaction on the small fragment (128–227) seemed sufficient to support repression within the transcription complex. Taken together, these data suggest that the SUV39H1-binding region is important for transcriptional repression by parafibromin.
Parafibromin regulates cyclin D1 transcription
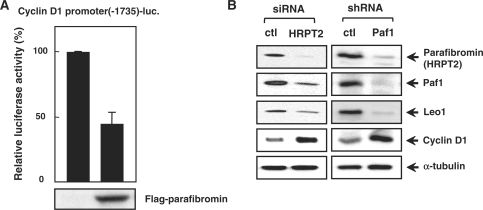
Over-expression of parafibromin decreases cyclin D1 expression, but the molecular mechanism is still unclear. Parafibromin-dependent genes, such as cyclin D1, could be a target of SUV39H1-mediated transcriptional repression. We first examined the transcriptional activity of parafibromin at the cyclin D1 promoter by a reporter assay. As expected, parafibromin repressed cyclin D1 promoter-driven luciferase activity (Figure 4A). We then depleted parafibromin by siRNA specifically targeting HRPT2 in HeLa cells. This siRNA reduced the levels of parafibromin and other components of the PAF1 complex (Paf1 and Leo1) as well, consistent with other reports (27,28), but increased cyclin D1 levels (Figure 4B, left panel). Similarly, HeLa cells stably expressing shRNA targeting a Paf1 subunit depleted Paf1, parafibromin and Leo1, and elevated cyclin D1 (Figure 4B, right panel). Thus, parafibromin, as a component of the PAF1 complex, downregulates the expression of cyclin D1.
Figure 4.
The effect of RNAi targeting HRPT2 or Paf1 on cellular cyclin D1 levels. (A) Parafibromin represses cyclin D1 promoter activity. The 293T cells were transfected with Flag-parafibromin along with a cyclin D1 promoter (–1735) reporter. The means ± SD of duplicate determinations from three separate experiments are shown. The expression level of transfected Flag-parafibromin was shown at the bottom. (B) Increased cyclin D1 levels following parafibromin or Paf1 knockdown. Immunoblotting of HeLa cells transfected with siRNAs against HRPT2 (parafibromin) or control siRNA (left panel) and stably expressed scrambled or Paf1 shRNA (right panel).
Parafibromin mediates cyclin D1 repression with SUV39H1
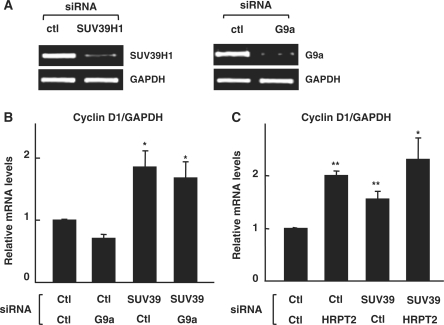
The direct role of SUV39H1 on cyclin D1 expression was analyzed by treating cells with siRNAs to inhibit the expression of SUV39H1 or G9a for comparison. These siRNAs efficiently decreased the expression level of their target HMTs (Figure 5A). Notably, depletion of SUV39H1, but not G9a, significantly increased the RNA level of cyclin D1, as observed in parafibromin-knockdown cells (Figure 5B). Importantly, depletion of both HMTs did not further affect the cyclin D1 level, indicating that SUV39H1, but not G9a, is the functional component of cyclin D1 regulation. Furthermore, although depletion of HRPT2 and SUV39H1 increased the expression of cyclin D1 separately, simultaneous depletion of both proteins did not have an additive effect on cyclin D1 level (Figure 5C). The siRNA knockdown of parafibromin had no effect on the expression of SUV39H1 or G9a (Supplementary Figure S2). Thus, our data show that SUV39H1 and HRPT2 function together at the protein level and contribute significantly to the repression of cyclin D1, putatively because of the recruitment of SUV39H1 via parafibromin and the subsequent methylation of H3 K9 onto the cyclin D1 gene.
Figure 5.
Parafibromin and SUV39H1 contribute to the cyclin D1 repression. (A) Validation of SUV39H1 (left) and G9a (right) knockdown. HeLa cells were transfected with SUV39H1, G9a siRNAs or control siRNA, and expression of SUV39H1 and G9a were checked by quantitative RT–PCR using specific primer sets. GAPDH was used as an internal control (at bottom). (B) The level of cyclin D1 mRNA was analyzed by quantitative RT–PCR with samples obtained one day after treatment of siRNAs as indicated. The PCR values have been normalized for GAPDH and presented as a relative value by considering Cyclin D1/GAPDH of control (Ctl) as 1 with the means ± SD from at least three independent experiments. (C) Quantitative analysis of cyclin D1 mRNA obtained 1 day after treatment of siRNAs as indicated at the bottom. The PCR values were normalized to GAPDH expression as described in (B). The significance of the differences was evaluated by Student’s t-test [*P < 0.001; **P < 0.05 with respect to control siRNA (Ctl)].
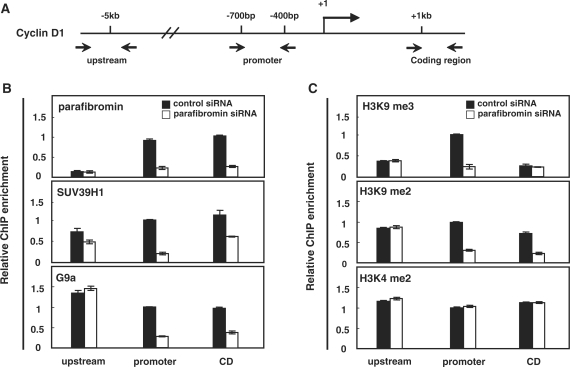
Next, we performed ChIP with primers probing the upstream (∼5 kb), promoter (–700 to –400 bp), and coding regions (∼1 kb) of the cyclin D1 gene to investigate whether the cyclin D1 regulation by parafibromin and SUV39H1 is accompanied by changes in chromatin (Figure 6A). Anti-parafibromin ChIP indicated that parafibromin was present at the promoter and coding regions, but absent at about 5 kb upstream from the cyclin D1 transcription start site (Supplementary Figure S3). The occupancy of parafibromin at both regions diminished significantly upon parafibromin depletion by siRNA (Figure 6B, top panel). We then tested the possibility that SUV39H1 was recruited to the cyclin D1 gene through the interaction with parafibromin by comparing its occupancy in the presence and absence of parafibromin. To do this, cross-linked chromatin fragments from HeLa cells pretreated with control or parafibromin-siRNA were immunoprecipitated with SUV39H1 antibody. ChIP with anti-SUV39H1 showed that SUV39H1 associated in a broad region encompassing cyclin D1 (Figure 6B, middle panel). However, this association was profoundly decreased in the parafibromin knockdown cells around the promoter and the coding regions. Although the impact of G9a depletion on cyclin D1 mRNA level was negligible (Figure 5B), G9a was associated with the cyclin D1 gene in a parafibromin-dependent manner, as its occupancy was also largely diminished by parafibromin knockdown in the promoter and coding regions (Figure 6B, bottom panel). Importantly, the level of H3 K9 trimethylation, predominantly enriched in the promoter region, was greatly reduced in parafibromin-knockdown cells (Figure 6C, top panel). Moreover, the level of H3 K9 dimethylation was also prominently affected in the promoter and coding regions but not in the upstream region (Figure 6C, middle panel). However, histone H3 K4 dimethylation was not affected by parafibromin at all (Figure 6C, bottom panel), similarly to the effect of Ctr9 knockdown reported by others (27). These results suggest that SUV39H1 (and G9a as well) was recruited to the cyclin D1 promoter and coding regions via interactions with parafibromin to induce H3 K9 methylation throughout the gene, maintaining repressive chromatin structure and downregulating cyclin D1 expression.
Figure 6.
Parafibromin recruits SUV39H1 to promote H3 K9 methylation around cyclin D1 gene. (A) Cyclin D1 locus showing regions subjected to ChIP analysis. (B, C) HeLa cells were treated with siRNA targeting HRPT2 or with an untargeted control siRNA for 24 h. Cells were then analyzed by ChIP using control antibody (rabbit IgG) and antibodies against indicated proteins or H3 modifications as described in ‘Materials and methods’ section. Immunoprecipitated DNA was analyzed in duplicates by quantitative RT–PCR to measure the relative recruitment of parafibromin, SUV39H1, or G9a (B) or the relative levels of H3K4 or H3K9 methylation along the cyclin D1 locus (C) ChIP with control IgG was subtracted from all ChIP values (IP/Input, the percentage of ChIP). Data represent the percentage of ChIP signal normalized to the value of promoter region of the control siRNA sample to compare the relative enrichment of signals along the region. (The ChIP value obtained by amplification of promoter region of the control siRNA sample was arbitrarily set to 1 in each panel.) Data represent the average of two or three independent experiments. Error bars indicate the standard deviation between experiments.
DISCUSSION
Parafibromin recruits SUV39H1 (and G9a) to the promoter and coding regions of cyclin D1 to regulate its expression by integration of H3 K9 methylation. Our study suggests a unique mechanism for transcriptional repression by parafibromin in addition to its role in active transcription. In general, PAF1 components associate with elongating RNA polymerases and transcriptionally active genes. In contrast to other typical repressors, parafibromin, as a component of the PAF1 complex, associates with actively transcribed regions and downregulates their expression while transcription is ongoing.
PAF1 is a transcription elongating complex (9–12,27,29). Importantly, it couples transcription to histone modification by promoting H3 K4 and K79 methylation. In the current study, we found that the parafibromin subunit of PAF1 mediates H3 K9 methylation in the cyclin D1 gene more actively than K4 methylation. We propose that SUV39H1 tethered by parafibromin and subsequent methylation of H3 K9 help maintain repressive chromatin structure to counteract active transcription. In budding yeasts, where H3 K9 methylation is absent, H3 K36 methylation within the body of actively transcribed genes couples transcriptional elongation to histone deacetylation for construction of repressive chromatin structure (30–32). In higher eukaryotes, H3 K9 methylation and HP1γ (Heterochromatin Protein 1γ) are unexpectedly associated with active genes in a polymerase II-dependent manner, yet it is not known why H3 K9 methylation is present at actively transcribed genes (33). Similar to H3 K36 methylation in yeast, H3 K9 methylation may be required to maintain repressive chromatin structure for keeping transcription at submaximal levels. In this regard, parafibromin and Set2 are similar. Set2, the HMT responsible for transcription elongation-coupled H3 K36 methylation, also has repressor activity when artificially tethered to a heterologous promoter, indicating its role in the downregulation of gene transcription (34).
In fact, loss of each component of PAF1 results in either an increase or decrease of transcription (35), reflecting the possibility that PAF1 has both positive (H3 K4/K79 methylation) and negative (H3 K9 methylation) effects on transcription. The sensitivity to positive and negative regulation by PAF1 probably results from the molecular environment such as interacting protein networks and upstream signals. Cyclin D1 may be dominated by PAF1’s negative effect through H3 K9 methylation via parafibromin. Of note, Takeuchi independently reported that the jumonji protein, Jmj, contributes to H3 K9 methylation by recruiting GLP (H3 K9 HMT) and G9a to repress the cyclin D1 promoter (36). As in our study, inactivation of GLP, but not of G9a, dramatically affects cyclin D1 expression, implying that GLP is critical while G9a plays a supportive role. In both studies, H3 K9 methylation seems to be an important signature for cyclin D1 regulation. Parafibromin and the PAF1 complex repress the c-myc proto-oncogene as well (28). If so, it will be of interest to determine whether H3 K9 methyltransferase activity is also involved in c-myc gene regulation.
Parafibromin functions as a tumor suppressor and plays a pivotal role in HPT-JT and sporadic parathyroid tumorigenesis. However, recently, parafibromin was reported to mediate nuclear transduction of Wnt signaling by interacting with β-catenin, in striking contrast to its role as a tumor suppressor (15). β-Catenin drives the expression of proliferative genes, such as cyclin D1 and c-myc. It is probable that parafibromin/PAF1 may play a dual role even in the same gene by alternating H3 K4/K79 (27,37) and H3 K9 methylation (this study). According to our hypothesis, an activated Wnt signal in human cancers could shift the balance toward H3 K4/K79 methylation to drive the cell cycle. In this regard, it is noteworthy that other core subunit, Paf1, is often amplified and overexpressed in many cancers (6). As control of the cell cycle is critical and widely disrupted in cancer, further work on parafibromin, the PAF1 complex, histone methylation, and their integration with other regulatory networks such as Wnt will clarify their precise role in transcriptional regulation and cancer pathogenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National R&D Program for Cancer Control (0520010-2); and the Korea Healthcare Technology R&D Project (A080181); Ministry of Health, Welfare & Family Affairs, Republic of Korea. Funding for open access charge: Ministry of Health, Welfare & Family Affairs, Korea.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr William F Simonds for providing pcDNA3-AU5-parafibromin; Dr An Woojin for pBS-4xnH3; Dr Martin J. Walsh for pEGFP-G9a; and Dr Samit Chattopadhyay and Dr R. G. Pestell for the cyclin D1 promoter-luciferase reporter construct.
REFERENCES
- 1.Carpten JD, Robbins CM, Villablanca A, Forsberg L, Presciuttini S, Bailey-Wilson J, Simonds WF, Gillanders EM, Kennedy AM, Chen JD, et al. HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nat. Genet. 2002;32:676–680. doi: 10.1038/ng1048. [DOI] [PubMed] [Google Scholar]
- 2.Rozenblatt-Rosen O, Hughes CM, Nannepaga SJ, Shanmugam KS, Copeland TD, Guszczynski T, Resau JH, Meyerson M. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol. Cell Biol. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yart A, Gstaiger M, Wirbelauer C, Pecnik M, Anastasiou D, Hess D, Krek W. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol. Cell Biol. 2005;25:5052–5060. doi: 10.1128/MCB.25.12.5052-5060.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howell VM, Haven CJ, Kahnoski K, Khoo SK, Petillo D, Chen J, Fleuren GJ, Robinson BG, Delbridge LW, Philips J, et al. HRPT2 mutations are associated with malignancy in sporadic parathyroid tumours. J. Med. Genet. 2003;40:657–663. doi: 10.1136/jmg.40.9.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shattuck TM, Valimaki S, Obara T, Gaz RD, Clark OH, Shoback D, Wierman ME, Tojo K, Robbins CM, Carpten JD, et al. Somatic and germ-line mutations of the HRPT2 gene in sporadic parathyroid carcinoma. N. Engl. J. Med. 2003;349:1722–1729. doi: 10.1056/NEJMoa031237. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhary K, Deb S, Moniaux N, Ponnusamy MP, Batra SK. Human RNA polymerase II-associated factor complex: dysregulation in cancer. Oncogene. 2007;26:7499–7507. doi: 10.1038/sj.onc.1210582. [DOI] [PubMed] [Google Scholar]
- 7.Shi X, Chang M, Wolf AJ, Chang CH, Frazer-Abel AA, Wade PA, Burton ZF, Jaehning JA. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol. Cell Biol. 1997;17:1160–1169. doi: 10.1128/mcb.17.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mueller CL, Jaehning JA. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell Biol. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 10.Rondón AG, Gallardo M, García-Rubio M, Aguilera A. Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep. 2004;5:47–53. doi: 10.1038/sj.embor.7400045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 12.Mueller CL, Porter SE, Hoffman MG, Jaehning JA. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell. 2004;14:447–456. doi: 10.1016/s1097-2765(04)00257-6. [DOI] [PubMed] [Google Scholar]
- 13.Betz JL, Chang M, Washburn TM, Porter SE, Mueller CL, Jaehning JA. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Genet. Genomics. 2002;268:272–285. doi: 10.1007/s00438-002-0752-8. [DOI] [PubMed] [Google Scholar]
- 14.Porter SE, Washburn TM, Chang M, Jaehning JA. The yeast pafl-RNA polymerase II complex is required for full expression of a subset of cell cycle-regulated genes. Eukaryot. Cell. 2002;1:830–842. doi: 10.1128/EC.1.5.830-842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mosimann C, Hausmann G, Basler K. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell. 2006;125:327–341. doi: 10.1016/j.cell.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 16.Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- 17.Rea S, Eisenhaber F, O'C;arroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl Acad. Sci. USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noma K, Grewal SI. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc. Natl Acad. Sci. USA. 2002;99:16438–16445. doi: 10.1073/pnas.182436399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 21.Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 22.Peters AH, Mermoud JE, O'C;arroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 2002;30:77–80. doi: 10.1038/ng789. [DOI] [PubMed] [Google Scholar]
- 23.Arnold A, Motokura T, Bloom T, Kronenberg H, Ruderman J, Juppner H, Kim HG. The putative oncogene PRAD1 encodes a novel cyclin. Cold Spring Harb. Symp. Quant. Biol. 1991;56:93–97. doi: 10.1101/sqb.1991.056.01.013. [DOI] [PubMed] [Google Scholar]
- 24.Woodard GE, Lin L, Zhang JH, Agarwal SK, Marx SJ, Simonds WF. Parafibromin, product of the hyperparathyroidism-jaw tumor syndrome gene HRPT2, regulates cyclin D1/PRAD1 expression. Oncogene. 2005;24:1272–1276. doi: 10.1038/sj.onc.1208274. [DOI] [PubMed] [Google Scholar]
- 25.Zhang C, Kong D, Tan MH, Pappas DL, Jr, Wang PF, Chen J, Farber L, Zhang N, Koo HM, Weinreich M, et al. Parafibromin inhibits cancer cell growth and causes G1 phase arrest. Biochem. Biophys. Res. Commun. 2006;350:17–24. doi: 10.1016/j.bbrc.2006.08.169. [DOI] [PubMed] [Google Scholar]
- 26.Zhao J, Yart A, Frigerio S, Perren A, Schraml P, Weisstanner C, Stallmach T, Krek W, Moch H. Sporadic human renal tumors display frequent allelic imbalances and novel mutations of the HRPT2 gene. Oncogene. 2007;26:3440–3449. doi: 10.1038/sj.onc.1210131. [DOI] [PubMed] [Google Scholar]
- 27.Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, Tempst P, Reinberg D. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin L, Zhang JH, Panicker LM, Simonds WF. The parafibromin tumor suppressor protein inhibits cell proliferation by repression of the c-myc proto-oncogene. Proc. Natl Acad. Sci. USA. 2008;105:17420–17425. doi: 10.1073/pnas.0710725105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of set1 histone methylase by elongating pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 30.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 31.Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 32.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 33.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 34.Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol. Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 36.Shirato H, Ogawa S, Nakajima K, Inagawa M, Kojima M, Tachibana M, Shinkai Y, Takeuchi T. A Jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. J. Biol. Chem. 2009;284:733–739. doi: 10.1074/jbc.M804994200. [DOI] [PubMed] [Google Scholar]
- 37.Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of humamn histone H2B: The factors involved and their roles in HOX gene regulation. Mol. Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.