Abstract
Sister chromatid cohesion, crucial for faithful segregation of replicated chromosomes in eukaryotes, is mediated by the multi-subunit protein complex cohesin. The Saccharomyces cerevisiae plasmid 2 micron circle mimics chromosomes in assembling cohesin at its partitioning locus. The plasmid is a multi-copy selfish DNA element that resides in the nucleus and propagates itself stably, presumably with assistance from cohesin. In metaphase cell lysates, or fractions enriched for their cohesed state by sedimentation, plasmid molecules are trapped topologically by the protein ring formed by cohesin. They can be released from cohesin’s embrace either by linearizing the DNA or by cleaving a cohesin subunit. Assays using two distinctly tagged cohesin molecules argue against the hand-cuff (an associated pair of monomeric cohesin rings) or the bracelet (a dimeric cohesin ring) model as responsible for establishing plasmid cohesion. Our cumulative results most easily fit a model in which a single monomeric cohesin ring, rather than a series of such rings, conjoins a pair of sister plasmids. These features of plasmid cohesion account for its sister-to-sister mode of segregation by cohesin disassembly during anaphase. The mechanistic similarities of cohesion between mini-chromosome sisters and 2 micron plasmid sisters suggest a potential kinship between the plasmid partitioning locus and centromeres.
INTRODUCTION
The central logic in the faithful segregation of chromosomes during mitotic division of eukaryotic cells is to keep duplicated sister chromatids together in pairs until they have been bioriented on the mitotic spindle. When pairing is annulled in anaphase, the sisters split as under, and are pulled apart by spindle forces and dynamics towards opposite cell poles. Union of sister chromatids is mediated by a multi-subunit protein complex, cohesin, and their separation by a site-specific protease, separase, that cleaves the cohesin component Mcd1 (1–3). By suitably modulating the ‘pairing-unpairing’ strategy, cohesin also promotes equal but reductional segregation of chromosomes during meiosis (4).
In the budding yeast Saccharomyces cerevisiae, cell cycle dependent assembly and disassembly of cohesin occurs not only on chromosomes but also on the 2 micron plasmid (5)—a multi-copy DNA circle that exhibits nearly chromosome-like stability in host populations. Several lines of circumstantial evidence are consistent with a functional role for cohesin in equal partitioning of the plasmid (5–8). The 2 micron circle appears to be unique among extrachromosomal elements in its ability to assimilate cohesin, and raises the prospect of an evolutionary connection between plasmid and chromosome segregation in Saccharomyces yeast. However, the mechanism by which cohesin interacts with the plasmid is poorly understood. The possibility that cohesin may play a role in plasmid physiology that is unrelated, or in addition, to segregation cannot be ruled out.
The biological function of cohesin is not restricted to sister chromatid segregation alone. Consistent with its ability to tether separate chromosomal segments, cohesin participates in DNA repair, chromosome morphogenesis and transcriptional regulation by long range activation or by blocking the spread of silencing domains (2,9–13). Several accessory factors and regulatory mechanisms specify chromatin sites for cohesin recruitment, and determine the timing of cohesin assembly, establishment of cohesion and cohesin disassembly. Mutations in cohesin components and regulatory factors have been implicated in human developmental disorders collectively termed as cohesinopathies (14).
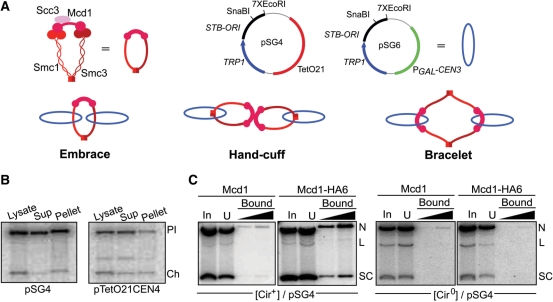
The conserved Smc1 and Smc3 subunits, characterized by a long ∼45–50 nm coiled coil connecting a hinge domain to a globular head domain, lay the foundation for cohesin’s architecture (15). Smc1 and Smc3 can form a V-shaped heterodimer through hydrophobic interactions between their hinge regions. The kleisin subunit of cohesin Mcd1 promotes noncovalent crosslinking of the Smc head domains, which potentiates the organization of two shared ATPase active sites. Mcd1 also mediates recruitment of the final component Scc3 to the complex. The ring formed by a single cohesin unit is large enough to accommodate a nascent pair of duplexes, fueling the notion that sister chromatid pairing could be established through topological embrace of DNA by cohesin rather than stable physical interaction between the two. Furthermore, variations of the basic subunit interactions in cohesin could engender alternative modes of embrace, as in the handcuff (snap) model or the bracelet model (2,15) (Figure 1A).
Figure 1.
Topological models for cohesion; trapping an STB reporter plasmid in cohesin-associated form. (A) The subunits of the yeast cohesin complex, and the ring structure they are presumed to assemble, are schematically diagrammed. Shown next to it is a simplified representation of the cohesin ring used in figures to follow. The two STB reporter plasmids, pSG4 and pSG6, employed in these studies are symbolized by blue rings. One set of control assays made use of a derivative of pSG4 lacking the TetO21 sequence (pSG5). The three models for topological interaction between cohesin and sister plasmids tested in this study are shown. The hand-cuff is drawn to be consistent with the recent finding that dimerization of human cohesin is dependent on the Scc3 (SA1/SA2) subunit (25). (B) Following high-speed centrifugation of a cell lysate, DNA from the supernatant (Sup) and ‘chromatin’ pellet fractions, digested with EcoRI, was run in agarose gels and hybridized to a radiolabeled TRP1 probe. Results from a similar fractionation of a CEN4 minichromosome pTetO21CEN4 (20) are shown for comparison. Pl, plasmid; Ch, chromosome. (C) Cleared lysates (equivalent to ‘Sup’ in B) from metaphase [cir+] or [cir0] cells harboring pSG4 were immunoprecipitated with the HA-antibody and collected on Protein A beads. DNA extracted from the different fractions (In, input; U, unbound or flow-through) was probed by a radiolabeled fragment specific to pSG4. The amount of bound DNA loaded in the right lane was five times that in the left one. This ratio was kept constant in assays shown in subsequent figures as well. SC, supercoiled plasmid; L, linear plasmid; N, nicked plasmid.
Elegant in vivo and ex vivo experiments by Nasmyth and colleagues lend credence to the embrace model for sister chromatid cohesion in S. cerevisiae (16–20). In a cohesed complex of circular minichromosome sisters, association of DNA and cohesin can be terminated by opening the DNA ring by restriction enzyme digestion or the cohesin ring by site-specific proteolysis. When the cohesin ring is covalently sealed by engineering cysteine crosslinks and a peptide linker at the protein interfaces that mediate ring closure, SDS denaturation fails to release the entrapped DNA. Based on the efficiency of crosslinking and the number of cohesin traps per pair of cohesed sisters, the embrace model (requiring monomeric cohesin rings) is favored over hand-cuff and bracelet models (requiring dimeric cohesin rings). Chromosome association with cohesin can be blocked by artificially cross-bridging the hinge domains of Smc1 and Smc3 but not by preventing the opening of the cohesin ring at the interfaces between Mcd1 and the Smc head domains. In sum, these observations suggest that transient dissociation of hinges lets a chromosome into the cohesin ring, and following replication, cohesion and spindle attachment, cleavage of Mcd1 lets sister chromatids out of the ring.
Despite the logical simplicity and parsimony of the embrace mechanism, alternative non-topological modes of cohesion have not been ruled out (21). Given the special architectural features of its subunits and the multiple cellular functions that it is involved in, distinct mechanisms for cohesin’s association with chromatin in a context dependent manner would seem plausible. Whereas cohesion at the arm regions of S. cerevisiae chromosomes promotes inter-sister pairing, cohesion at pericentric regions appears to generate DNA loops that would be consistent with intra-sister pairing (22). Transmission electron microscopy of purified minichromosome sisters with associated cohesin reveals a thick rod of cohesin, presumably containing multiple cohesin units, interacting with the replicated minichromosome copies at one end (23). It is difficult to reconcile this picture with the embrace model for cohesion. Furthermore, association of cohesin with the silent mating type locus HMR during Sir-mediated transcriptional inactivation appears to depart from the conventional topological mechanism (24). Evidence suggests that cohesin, instead of simultaneously surrounding the duplicated silent copies, encircles each of them separately. Recent data indicating potential dimerization of human cohesin in an Scc3 (SA1/SA2)-dependent manner has raised the possibility of a functional cohesin hand-cuff (25). It is argued that the greater dynamic flexibility of the hand-cuff over the more static embrace model is better suited for cohesin’s role in multiple DNA transactions. In vivo modifications such as acetylation or phosphorylation of cohesin subunits or associated factors (26–31), and the functional relevance of individual modifications to specific pathways of cohesion, may bolster the structural complexity/diversity required for modulating the mechanics and dynamics of cohesin’s interaction with DNA.
We address here the nature of cohesin’s association with the 2 micron plasmid. The motivation stems from previous observations that timely assembly and disassembly of cohesin are integral steps in the plasmid partitioning pathway (5,7). Cell biological assays suggest that cohesin recruited at the plasmid partitioning locus (STB), with assistance of the plasmid coded proteins Rep1 and Rep2, brings about cohesion between replicated plasmid copies, and subsequent dissolution of cohesion mediates plasmid segregation in a sister-to-sister fashion. The functional similarities between chromosome and plasmid segregation prompted us to examine the generality of the topological mechanism proposed for centromere-mediated replicative cohesion. Does this mechanism also apply to cohesion established at a nonchromosomal locus, namely, STB? We find that cohesin-STB interaction is topological, and best fits the embrace model, in which a pair of sister plasmids are entrapped within a single cohesin ring.
MATERIALS AND METHODS
Yeast strains and plasmids
The yeast strains used in this study are listed in Supplementary Table S1.
The plasmid pSG4 was derived from the plasmid TetO21CEN4 (20) by the following manipulations. The CEN4 sequence was replaced by an EcoRI–BamHI fragment containing the 2 micron circle replication origin and STB locus. The ARS1 sequence associated with TRP1 was removed by BamHI plus BglII digestion followed by self-ligation to obtain plasmid pSG4-1. A DNA fragment bearing six EcoRI sites was inserted into the EcoRI site of pSG4-1. The pUC19 sequences present in pGS4-1 were removed by SalI digestion and self-ligation to derive pSG4, which was recovered by transformation in yeast.
Construction of plasmid pSG5 included the following steps. The TetO21 sequence was deleted from pSG4-1 by XhoI digestion, followed sequentially by filling-in the staggered ends by Klenow polymerase, SmaI digestion and self-ligation. The resulting intermediate plasmid was named pSG5-1. After removing the pUC19 DNA by SalI digestion and self-ligation to generate pSG5, it was recovered in yeast as described for pSG4.
To obtain pSG6, the pUC19 sequences were removed from pSG5-1 by NdeI digestion and replaced by a PGAL-CEN3 fragment derived from a previously described plasmid pSG1 (6). For recovery of pSG6, the recipient yeast strain was directly transformed with the ligation mixture.
The authenticity of all plasmid constructs transformed into yeast were verified by diagnostic PCR assays and by restriction enzyme digestion and Southern analysis of total DNA prepared from the host strains containing them.
Immunoprecipitation of cohesin-associated plasmids
The procedures described by Ivanov and Nasmyth (20) were followed for immunoprecipitating cohesin or cohesin-associated plasmids. Typically, precultures of the cdc20 strains harboring the reporter plasmid were inoculated into 1 liter of Sc-Trp medium and grown to mid-log phase at 24°C. In order to arrest cells in metaphase, they were incubated at 37°C for an additional period of 2.5 h. In assays employing pTetO21CEN4 (20), 10 µg/ml nocodazole was included in the medium. Nocodazole treatment was omitted in assays with STB reporter plasmids, except when indicated otherwise. Spheroplasting of cells, cell lysis, preparation of cleared lysates, immunoprecipitations, restriction enzyme digestion and TEV protease cleavage were carried out according to published protocols (20). Immunoprecipitates were washed thrice with the lysis buffer in order to disrupt loose/non-specific associations.
Adsorption of plasmids bound by protein A-TetR to IgG beads
A subset of the assays performed in this study required the immobilization of reporter plasmids associated with Protein A-TetR on IgG beads. IgG pull-down was performed as described by Ivanov and Nasmyth (20). To minimize nonspecific binding, beads were washed three times with lysis buffer. DNA was eluted from IgG beads by two successive incubations, using a rotary shaker, in buffer containing 100 µM anhydrotetracycline (20) for 30 min each at 4°C.
DNA analysis by Southern blotting
DNA samples for Southern blot analysis were obtained by phenol extraction and ethanol precipitation. After electrophoresis, transfer to Hybond-XL membranes (GE Healthcare) and hybridization using 32P-labeled probes, bands were detected by autoradiography or phosphorimaging. Band intensities were quantitated from phosphorimages. In estimating relative amounts of a reporter plasmid co-immnoprecipitated with cohesin, DNA was digested with a restriction enzyme to completion prior to Southern blot analysis. By doing so, supercoiled, nicked and linear plasmids present in the immunoprecipitated samples were converted to a single linear form.
DNA was extracted from protein A dynabeads (Invitrogen) by incubating them twice in succession with elution buffer (50 mM Tris, pH 8.0, 10 mM EDTA and 1% SDS) for 15 min each at 65°C.
Protein analysis by western blotting
Protein samples for western blot analysis were obtained by precipitating with 10% trichloroacetic acid and redissolving the precipitate in SDS-sample buffer prior to electrophoresis. After electro-blotting onto PVDF membranes and treatment with primary antibodies, protein bands were visualized using an Amersham chemiluminescence-based detection system (GE Healthcare). The sources for primary antibodies were Covance (Princeton, NJ, USA). Peroxidase-conjugated secondary antibodies were obtained from Bio-Rad (CA, USA). The western signals were quantitated using Quantity One software (Bio-Rad).
Proteins were eluted from IgG beads by incubation in elution buffer containing 1% SDS for 10 min at 23°C and 20 min at 65°C, and were precipitated with trichloroacetic acid prior to western blot analysis.
Sucrose gradient sedimentation for separation of cohesed plasmids: EcoRI digestion and TEV protease cleavage in gradient fractions
The conditions of centrifugation were essentially according to Ivanov and Nasmyth (19), except that the gradient was from 12.5% to 37.5% sucrose. Digestions with EcoRI (New England Biolabs) and TEV protease (Invitrogen) were carried out using 30 µl gradient fractions at 4°C for 5 h. Each reaction mixture contained 20 U of EcoRI or 40 U of TEV protease. Controls were incubated for the same length of time at 4°C in reaction buffers without the addition of enzyme. Aliquots of the reactions were fractionated by electrophoresis in 1% agarose gels at 4°C for 7 h at 1.5 V/cm. The running buffer employed was TAE (pH 7.8) without ethidium bromide. When protein denaturation was required, SDS was added to samples to a final concentration of 1%, and heated at 65°C for 4 min prior to electrophoresis. Plasmid DNA was detected by Southern blotting and hybridization to radiolabeled plasmid-specific probes.
RESULTS
General experimental strategies
The general experimental strategies are briefly outlined at the outset for a better perspective of the logic of this study and limitations of the analytical methods. Their primary objective was to test whether cohesin interacts with the 2 micron plasmid topologically rather than by establishing stable physical contacts. For simplicity, the two types of interactions are distinguished as ‘topological’ or ‘physical’. For the case of topological interaction, the subsequent aim was to distinguish among three plausible models: (i) embrace, (ii) hand-cuff and (iii) bracelet (Figure 1A). The final intent was to address the stoichiometry of cohesin and DNA in the cohesed state.
The STB plasmid and epitope-tagged cohesin reporters incorporated, as explained below, relevant designs from those employed by Nasmyth and colleagues for analysis of cohesion in minichromosomes (19,20). Thus, valid comparisons could be made between results for cohesion mediated by STB and CEN4. Myc13-tagged Mcd1 and two versions of HA-tagged Mcd1 were made use of; engineered into one Mcd1-HA were three adjacent copies of the TEV protease cleavage site. Cohesin-associated plasmid molecules were immunoprecipitated from metaphase cell lysates by using antibodies to HA-6 or Myc-13 epitopes. In some experiments, the reporter plasmid harboring the TetO21 sequence was pulled down by interaction between IgG and the operator bound Protein A-TetR fusion protein. Plasmid DNA could be released by anhydrotetracycline, and the subset of cohesin-associated molecules re-trapped by a cohesin-directed antibody.
Sucrose gradient centrifugation experiments performed during this study revealed the amount of cohesed plasmids in cleared lysates from metaphase cells to be close to 10% of the total plasmids, and no >20%. Interpretations of experimental data pertain to this plasmid population. The fraction of plasmid DNA that could be immunoprecipitated by the HA- or Myc-antibody from cleared lysates ranged from 2% to 7.5% in different assays. Assuming an average of 15% cohesed (or stably cohesin-associated) plasmids, this corresponded to an efficiency of immunoprecipitation in the range of 13–50%. In experiments in which plasmid DNA (cohesed and noncohesed) was first pulled down, released and then baited with the HA- or Myc-antibody, the amount of immunoprecipitated DNA varied from 5% to 12.5%. Again, assuming 15% of the plasmid molecules to be associated with cohesin, the actual efficiency was between 33% and 83%. For assays simultaneously employing cohesin(Mcd1-HA6) and cohesin(Mcd1-Myc13), immunoprecipitation efficiency for a given antibody would be dependent on the stoichiometry of cohesin with respect to DNA. For example, if only a single monomeric cohesin complex is involved in pairing plasmid sisters, the maximum efficiency can only be 50%.
Cohesin recruitment by the 2 micron plasmid and cohesion of plasmid sisters are intricately linked to spindle integrity (6,8). Nocodazole treatment was unsuitable in our assays for instituting metaphase arrest while maintaining plasmid cohesion. Instead, metaphase cells were enriched through cdc20 arrest or by harvesting populations at appropriate times after release from G1 arrest. Only in control assays that employed a minichromosome (a CEN4-based plasmid) or aimed to disrupt cohesin assembly at STB was nocodazole employed.
Cohesion assays in 2 micron circle, unlike those in a circular minichromosome, faces the challenge of the multi-copy state of the plasmid and its clustered organization. Previous experiments showed that a fluorescence tagged single copy STB reporter plasmid undergoes cohesion in metaphase in the context of the native cluster of endogenous plasmids (6). Furthermore, two such reporters, one tagged by red fluorescence and the other by green, segregate red-to-red and green-to-green during anaphase. Pairing, even among a population of plasmid molecules, appears to be restricted to sisters. The majority of experiments reported here, though performed in the multi-copy context, implicitly assume that two sister plasmids constitute the basic unit of cohesion. This assumption was validated by a final set of experiments utilizing a stand-alone single copy STB plasmid complemented with Rep1 and Rep2 proteins in trans.
Plasmid-cohesin association is broken by linearizing DNA or cleaving Mcd1
Three plausible topological models for chromosome cohesion are diagrammed in Figure 1A. The ends of the DNA are artificially shown as closed to highlight the linkage between DNA and protein. The circular DNA form applies to the plasmids used in the studies reported here. The embrace, bracelet and hand-cuff models are consistent with the structural features of, and physical interactions among, cohesin subunits. Additional models, based on more complex variations of the cohesin ring theme, may be envisaged but are not considered here. While published results from one series of experiments support the embrace model (16–20), other lines of evidence leave open alternative possibilities (22–25). In experiments described below, we address whether the interaction of cohesin with the STB locus is accommodated by a ring or rings of cohesin encircling the DNA sisters.
The STB reporter plasmid pSG4 (Figure 1A) harboring the 2 micron plasmid replication origin, the yeast TRP1 marker and the TetO21 sequence (but no other non-yeast DNA) could be maintained with relatively high stability in a [cir+] host strain whose endogenous plasmids supplied the partitioning proteins Rep1 and Rep2. High-speed centrifugation of an extract from cdc20 arrested metaphase cells expressing Mcd1-HA6 yielded supernatant fractions (cleared lysates) containing roughly 40–50% of the pSG4 minichromatin with very little contamination from chromosomal chromatin (Figure 1B, left). For comparison, a similar procedure applied to nocodazole (10 µg/ml) treated metaphase cells partitioned nearly 80% of a CEN4 containing minichromosome (20) into the cleared lysate with slightly higher chromosomal contamination (Figure 1B, right). The pSG4 minichromatin, presumably associated with cohesin(Mcd1-HA6), could be immunoprecipitated by the HA-antibody (Figure 1C, left). Consistent with the requirement of the Rep proteins and an intact spindle for cohesin assembly at STB (5,8), immunoprecipitation of pSG4 was not detected in lysates from [cir0] host cells (Figure 1C, right) or nocodazole treated [cir+] cells (Supplementary Figure S1).
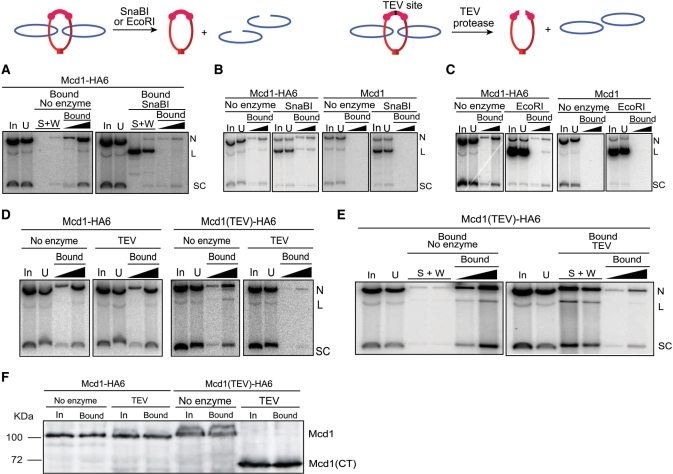
Next, we inquired into the nature of the DNA–protein interaction in cohesion-associated pSG4. When the plasmid was digested with SnaBI on Protein A beads used to pull down the HA-antibody–cohesin–plasmid complex, the linearized DNA was released nearly quantitatively into the supernatant (S + W in Figure 2A). The DNA retained on the beads was almost exclusively circular, supercoiled or nicked. That is, only the fraction that escaped SnaBI digestion remained trapped by cohesin (‘Bound’ in Figure 2A). The Mcd1 (and by inference cohesin) stayed bound to the beads under the conditions of the digestion, as indicated by western blot analysis (data not shown).
Figure 2.
Release of an STB reporter plasmid from cohesin’s grasp by linearizing DNA or cleaving Mcd1. The consequences of cutting DNA or protein on the topological association between plasmid and cohesin are schematically indicated. (A) Cohesin associated pSG4 was adsorbed on HA-antibody and immobilized on Protein A beads as described under Figure 1. After digestion with SnaBI, DNA released into the supernatant (S) and wash fractions (W) and that retained on the beads (Bound) were analyzed. (B and C) SnaBI or EcoRI digestion was performed in the cleared lysates prior to immunoprecipitation by HA-antibody. (D) Cleared lysates were treated with TEV protease, and then subjected to pull-down by HA-antibody and Protein A beads. (E) Cohesin bound plasmid from cleared lysates was treated with HA-antibody, trapped on Protein A beads, and subjected to TEV protease treatment. (F) Cleavage of Mcd1-HA6 by TEV protease in the cleared lysates (corresponding to the DNA analysis shown in D) was monitored by western blotting using HA-antibody. The amount of protein analyzed from the bead-bound fraction was four equivalents of that from the input. The identity of the weak band above Mcd1 seen in some of the lanes is not known. Its mobility would be consistent with phosphorylation of Mcd1, which occurs during the establishment of cohesion in response to DNA damage (27).
The choice of the single SnaBI site for plasmid digestion was based on its presence in a relatively nucleosome-free locale of the STB-ORI segment of the 2 micron circle, at least in the native context of the plasmid genome (32). For verification of the SnaBI result, plasmid digestion was also performed with EcoRI. Incorporation of multiple tandem EcoRI sites into the design of pSG4 was intended to increase cutting efficacy by this enzyme. SnaBI or EcoR1 treatment prior to immunoprecipitation left behind nearly all of the linear pSG4 DNA in the supernatant, and pulled down almost exclusively the undigested circular DNA (Figure 2B and C). When plasmid digestion was carried to completion in the supernatant by an even higher excess of the enzyme than that used in standard assays, no DNA was brought down by the antibody (Supplementary Figure S2). The restriction enzymes were active not only during the digestion phase but also during the immunoprecipitation phase of the assay. Continued cutting of the plasmid on the beads until the point of DNA extraction could account for the slight increase in linear DNA in the bound fraction from enzyme treated samples compared to that from non-treated samples (Figure 2B and C).
Digestion of Mcd1 in the cleared lysate or in the immunoprecipitated pSG4–cohesin complex with TEV protease annulled the association between DNA and protein (Figure 2D and E). TEV protease treatment per se did not affect the topology of the plasmid. The DNA in the lysate remained almost exclusively circular even though it was not pulled down by the HA-antibody once Mcd1 was cleaved (Figure 2D). Similarly, the DNA released from the beads as a result of TEV protease digestion was predominantly circular (Figure 2E). The cleavage of Mcd1 in the lysate by TEV protease was nearly quantitative, as determined by western blotting of the total protein fraction in the lysate or the protein fraction bound to the HA-antibody-Protein A beads (Figure 2F).
The combined results from DNA digestion and protein cleavage suggest that the mainstay of the interaction between cohesin and replicated STB plasmids in metaphase is topological. The possibility that cohesin has poor affinity for linear DNA is unlikely, as it normally acts on linear chromosomes as they are being replicated. It is difficult to imagine how cohesin, acting locally at or near the replication fork, can sense the global topology of DNA. If cohesin can slide or track along DNA, it would fall off from the ends of linear DNA regardless of whether DNA-protein association is topological or physical. However, it would seem unlikely that opening a peptide bond would terminate physical association of DNA with cohesin. Ivanov and Nasmyth (20) showed that severance of either the Mcd1 or the Smc3 subunits of cohesin would suffice to free minichromosomes from cohesin’s grasp. Furthermore, for small DNA molecules, it is circularity rather than supercoiling that is important for stable association with cohesin. The effect of nicking a single strand on cohesin’s association with DNA is much smaller compared to that of breaking both strands (20). The more or less unbiased association of supercoiled or nicked plasmid circles with cohesin, contrasted by the lack of association of linear molecules, is also evident in our assays (Figure 2A–C).
Strictly, our interpretation applies only to the fraction of plasmids that is stably associated with the cohesin complex, and can be recognized by anti-cohesin antibodies. While the data favor interlinked cohesin and DNA rings, they do not discriminate among the three models in Figure 1A. Based on previous results regarding the nature of 2 micron plasmid cohesion and segregation (6,33), we assume tentatively that two sister plasmid molecules are held together by the cohesin ring (or rings).
Cohesin encircles DNA in the form of solitary rings and not conjoined ones
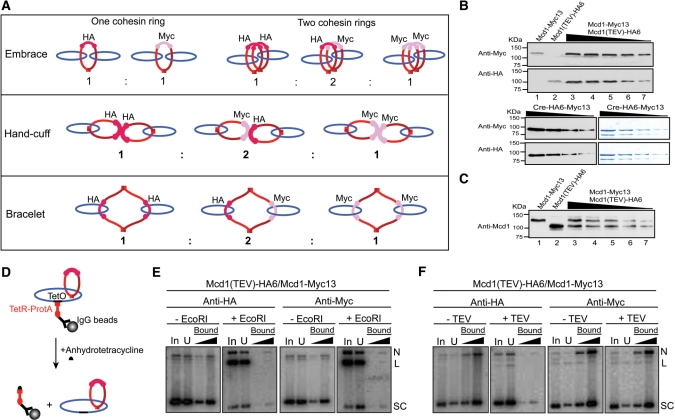
In the embrace and bracelet models for cohesion, sister chromosomes are enfolded by one cohesin ring (or multiple units of a solitary ring) (Figure 1A). The ring sizes are different in the two cases, the bracelet being a cohesin dimer. In contrast, the hand-cuff model propounds two distinct, but mutually associated, monomeric cohesin rings. The one ring versus two ring models could be distinguished by expressing two types of cohesins, differentially tagged by HA6 and Myc13, in the same cell and in roughly equal amounts (Figure 3). Mcd1-HA6 was cleavable by TEV protease; Mcd1-Myc13 was not. The types of cohesion resulting from the embrace, hand-cuff and bracelet models are schematically diagrammed in Figure 3A.
Figure 3.
Plasmid–cohesin association in metaphase cells expressing two differentially tagged cohesin moieties. (A) The types of sister plasmid cohesion expected from the embrace, hand-cuff and bracelet models in presence of cohesin(Mcd1-HA6) and cohesin(Mcd1-Myc13) in equivalent amounts are indicated (see also Table 1). Whereas Mcd1-HA6 in cohesin could be cleaved by TEV protease, Mcd1-Myc13 could not. (B) Aliquots of cell lysates were probed by western analysis using HA- or Myc-antibody to reveal relative levels of cohesin(Mcd1-HA6) or cohesin(Mcd1-Myc13). Data are shown for the haploid strains expressing either cohesin(Mcd1-HA6) (lane 1) or cohesin(Mcd1-Myc13) (lane 2) and the diploid generated from them expressing both Mcd1 variants (lanes 3–7). Signals from the two antibodies were normalized using aliquots of ∼75% pure Cre recombinase tagged at its carboxyl-terminus with HA6 as well as Myc13. The mean Myc13 to HA6 signal intensity was 1.83 ± 0.18. The dilution factor between Cre samples stained by Coomassie blue (right) and the corresponding ones analyzed by western blotting (left) was 500 to 1. (C) Aliquots of cell lysates from the haploid and diploid strains, run as in B, were probed using an antibody to native Mcd1. The mean ratio of Mcd1-Myc13 to Mcd1-HA6 was 0.96 ± 0.11. (D) pSG4 molecules from the cleared lysate were first immobilized on IgG beads, and then released from them by disrupting TetO–TetR interaction using anhydrotetracycline. (E and F) Following treatment with EcoRI or TEV protease, plasmid pull-down was attempted using HA- or Myc-antibody.
Table 1 summarizes the expectations from the three models for the loss or retention of cohesin–DNA linkage following the opening of the Mcd1-HA6 containing ring by TEV protease. The simplest prediction, and the easiest to verify, is that the HA-antibody would not be able to trap plasmid DNA following the action of TEV protease according to embrace and bracelet models. All plasmid molecules embraced by cohesin(Mcd1-HA6) rings, and even those associated with mixed cohesin(Mcd1-HA6)–cohesin(Mcd1-Myc13) bracelets, would have escaped through the opening created in the cohesin(Mcd1-HA6) ring. This prediction is independent of the number of rings surrounding a given pair of DNA sisters. However, the hand-cuff model predicts 25% of the DNA to be immunoprecipitated by this antibody, so long as the fissured cohesin(Mcd1-HA6) stays associated with the intact cohesin(Mcd1-Myc13), which is non-cleavable by TEV protease. If there is more than one hand-cuff per DNA sisters, the DNA fraction immunoprecipitated by the HA-antibody will be >25%.
Table 1.
Predictions by the three topological models on the nature of plasmid cohesion
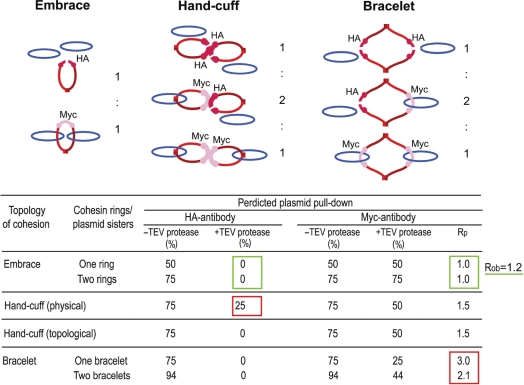 |
Schematic diagrams for plasmid cohesion established by the embrace, bracelet and hand-cuff models from an equal mixture of cohesin(Mcd1-HA6) (cleavable by TEV protease) and cohesin(Mcd1-Myc13) (resistant to TEV protease) are shown in Figure 3 (top). Treatment with TEV protease will cleave all cohesin(Mcd1-HA6) containing rings (see drawings above), opening gates for trapped plasmids to escape. Only those plasmid molecules surrounded by the closed cohesin(Mcd1-Myc13) ring(s) will be stopped. Rp is the predicted molar ratio of plasmid that can be pulled down by the Myc-antibody before and after TEV protease cleavage; Rob is the observed value. Agreement between experiment (Figure 3B and C) and prediction is indicated by the green rectangles; disagreement by red ones. The embrace model is the winner (with two green rectangles) over the hand-cuff and bracelet models (each with a red rectangle). These assays do not permit a clean distinction between the embrace and the topological hand-cuff models.
To ensure approximately equal amounts of cohesin tagged by HA6 and Myc13 in the diploid host harboring pSG4, the Mcd1 variants were expressed from the native MCD1 promoter and the native chromosomal locale. This expectation was further verified by a western blot analysis of the steady state levels of Mcd1-HA6 and Mcd1-Myc13 (Figure 3B and C). Differences in the HA6 and Myc13 signals were normalized using Cre recombinase harboring both these epitope tags at its carboxyl-terminus as a reference protein (Figure 3B). With appropriate correction, we estimated the cellular ratio of cohesin(Mcd1-Myc13) to cohesin(Mcd1-HA6) to be 1.09 ± 0.12. This result was further confirmed by quantifying the proteins using an antibody to native Mcd1, cohesin(Mcd1-Myc13): cohesin(Mcd1-HA6) = 0.96 ± 0.11 (Figure 3C). The extents of cohesin immunoprecipitation by the HA- and Myc-antibodies were more or less equal under our experimental conditions, as indicated by the Southern signals from the co-precipitated DNA (Supplementary Figure S3; Figure 4C and D).
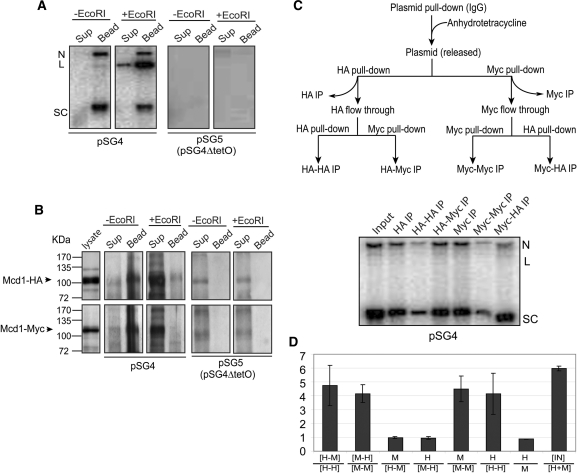
Figure 4.
Distinction between embrace and bracelet models for plasmid cohesion: cohesin stoichiometry tested by sequential immunoprecipitation. (A and B) The expected outcomes for plasmid immobilization via TetO affinity interaction followed by DNA linearization were experimentally verified. Plasmid molecules associated with Protein A-TetR bound to TetO were pulled down by IgG beads. DNA and protein remaining associated with the beads or released into the supernatant in the absence of EcoRI treatment or following EcoRI digestion were followed by Southern and western analyses, respectively. (C) The flow-chart for the two-step immunoprecipitation assays is diagrammed at the top. Plasmids were first trapped on IgG beads as in A, and then released by treatment with anhydrotetracycline. Equal amounts of the supernatant containing the freed plasmid were immunoprecipitated with the HA- or Myc-antibody. The leftover plasmid molecules in the supernatant were subjected to a second round of immunoprecipitations. (D) The histograms denote the mean ratios of Southern blot signals for immunoprecipitated DNA from three independent experiments, with the error bars showing standard deviations. Immunoprecipitations with HA- and Myc-antibodies are represented by ‘H’ and ‘M’, respectively. Sequential immunoprecipitations by these antibodies are indicated by the two letters separated by a dash. The ratio of the input (IN) plasmid DNA to that immunoprecipitated by the HA- and Myc-antibodies combined is given as IN/[H + M]. The immunoprecipitable plasmid fractions were 17.33%, 16.50%, 16.46% in individual assays.
In order to improve the sensitivity of the assay, pSG4 was enriched from metaphase cells by IgG pull-down (Figure 3D), followed by its release in the presence of anhydrotetracycline. The released plasmid could be pulled down again by either the HA- or Myc-antibody with approximately equal efficiency (Supplementary Figure S3). Attempts to immunoprecipitate DNA after EcoRI digestion of the released plasmid with the HA- or Myc-antibodies failed (Figure 3E), as expected from earlier results. Digestion with TEV protease yielded a distinct, and significant, result. While the HA-antibody failed, the Myc-antibody succeeded in reprecipitating DNA, almost entirely as intact circles (Figure 3F). This result is consistent with the embrace and bracelet models but inconsistent with the hand-cuff model (Table 1).
Quantitations suggest that the molar ratio of plasmid brought down by the Myc-antibody in the absence of, and following, TEV protease digestion was ∼1.2, or nearly equal to 1.0—(compare the ‘Bound’ lanes in the right two panels of Figure 3C). This value is close to that predicted by the embrace model, and significantly smaller than that anticipated from the single bracelet (3.0) or the double bracelet (2.14) models (Table 1). Thus, for one cohesin ring or a reasonably small number of such rings per pair of sister plasmids, these results discount the bracelet in favor of the embrace model (Table 1). Multiple cohesin rings per sister pair would increase the relative amount of DNA pulled down by the Myc antibody after TEV protease cleavage, due to increased mole fraction of cohesin(Mcd1-Myc13)/cohesin(Mcd1-Myc13) bracelets around sisters. However, the stoichiometry of cohesin to DNA will have to be quite high in order to blur the distinction between the two models.
A variation of the hand-cuff model in which the two cohesin rings are topologically, not physically, linked cannot be easily ruled out by the above experiments. In a topological hand-cuff, opening of cohesin(Mcd-HA6) by TEV protease would automatically end its association with cohesin(Mcd1-Myc13). As a result, in the pull-down test using the HA-antibody, it would be no different from the embrace model in which two cohesin rings are formed around sister plasmids (Table 1). Further experiments argue against a hand-cuff formed by a catenated pair of cohesin rings (see below).
It may be noted that, the physical and topological handcuffs will give identical results for DNA pull-down, before and after TEV protease digestion, by the Myc-antibody (Table 1). They do differ from embrace by one or two monomeric cohesin rings in causing one third reduction (from 75% to 50%), following TEV protease cleavage, in plasmid DNA associated with cohesin(Myc-13). However, as is evident from Table 1, the Myc-antibody pull down offers better distinction between the embrace and bracelet models than between embrace and hand-cuff models.
Stoichiometry of cohesin-plasmid association: two-step immunoprecipitations support embrace by a single monomeric cohesin ring
The experimental outcomes so far favor monomeric cohesin ring(s) (embrace) or dimeric cohesin ring(s) (bracelet) over a cohesin hand-cuff as the unit entity in cohesion. However, they do not distinguish between the ring and the bracelet; nor do they reveal the number of rings or bracelets assembled around paired sister plasmids. The ability to immobilize pSG4 alternatively by IgG or the HA- or Myc-antibody offers a potential tool to resolve these uncertainties. Once again, the assays were performed using metaphase cells of the host strain expressing Mcd1-HA6 and Mcd1-Myc13 in approximately equivalent amounts.
The plasmid, trapped on IgG beads through Protein A-TetR bound to TetO, is expected to bring down with it associated cohesin(s), cohesin(Mcd1-HA6) and cohesin(Mcd1-Myc13) monotypes (according to embrace or bracelet) or cohesin(Mcd1-HA6)/cohesin(Mcd1-Myc13) hybrid type (according to bracelet). Linearization of the bound pSG4 should release all associated cohesin into the supernatant, irrespective of the stoichiometry of cohesin rings to DNA. The linear form of pSG4, however, should stay bound to the beads. These expectations were satisfied (Figure 4A and B). As shown by the results in Figure 3, plasmid released from the beads by treatment with anhydrotetracycline could be immunoprecipitated using antibodies to associated cohesin(s). A two-step immunoprecipitation assay could then be performed to test whether or not cohesin(Mcd1-HA6) and cohesin(Mcd1-Myc13) are capable of simultaneous association with the plasmid.
Predictions concerning plasmid pull-down by the HA- or Myc-antibody are dependent on both the cohesion model and cohesin stoichiometry. According to the one ring embrace model, immunoprecipitation by the HA-antibody would deplete only plasmid molecules embraced by cohesin containing Mcd1-HA6, and leave those embraced by cohesin containing Mcd1-Myc13 unscathed. As a result, a second round of immunoprecipitations of the ‘depleted’ supernatant individually by HA- and Myc-antibodies would skew plasmid pull-down in favor of the latter. If the first immunodepletion is performed by the Myc-13 antibody, the bias in DNA pull-down during the second immunoprecipitation would be directed oppositely. According to the one bracelet model, the first immunoprecipitation with either the HA- or Myc-antibody would pull down two thirds of the cohesin bracelets bearing the opposite epitope specificity in the form of choesin(Mcd1-HA6)–cohesin(Mcd1-Myc13) hybrid bracelets. The two ring embrace model is also subject to a similar depletion effect due to cohesion mediated by a mixed pair of cohesin(Mcd1-HA6) and cohesin(Mcd1-Myc13) rings. Hence the relative advantage for the virgin antibody in the second immunoprecipitation step is less than that anticipated from the single ring embrace model.
Quantitations of the Southern signals of immunoprecipitated DNA from three repeats of the two-step assay (Figure 4C) are graphed in Figure 4D. The virgin antibody displayed a strong advantage in the second step (HA-Myc IP or Myc-HA IP) over the experienced antibody (HA-HA IP or Myc-Myc IP). The [HA-Myc]/[HA-HA] and [Myc-HA]/[Myc-Myc] ratios were 4.74 ± 1.46 and 4.15 ± 0.64, respectively. In contrast, the molar ratio of DNA brought down by the HA-antibody in the first step to that in the second step following immunoprecipitation by the Myc-antibody was ∼1 (0.95 ± 0.01; HA/[Myc-HA] in Figure 4D). The corresponding ratio for immunoprecipitation by the Myc antibody was also the same, 0.98 ± 0.07 (Myc/[HA-Myc] in Figure 4D). The absence of cross-depletion by either antibody during plasmid pull-down is consistent with only one cohesin ring (carrying a single epitope specificity; either HA or Myc) bridging a pair of plasmid sisters. A similar analysis with the CEN4 containing minichromosome gave concordant results (Supplementary Figure S4).
The total amount of immunoprecipitated DNA, by HA- and Myc-antibodies combined, in the two step assay added up to approximately 15-20% of the input DNA (that was released from the IgG beads), accounting for nearly the entire fraction of DNA in the cohesed state (IN/[H + M] = ∼6.0 in Figure 4D). Furthermore, the molar DNA ratio of approximately 4 for the HA/[HA-HA] or Myc/[Myc-Myc] sequence (Figure 4D) corresponds to an immunoprecipitation efficiency of ∼75% for each antibody. Note also that the DNA amounts pulled down by the HA-antibody and the Myc-antibody in the respective first step immunoprecipitations were nearly equal (HA/Myc of 0.88 ± 0.01 in Figure 4D). These values are in agreement with the one ring embrace model, which proscribes pull-down by one antibody of the other’s cognate epitope. One would then expect 75% of the plasmid DNA bearing cohesin of one epitope type, which constitutes half of all the cohesed molecules, to come down during the first step, and ∼19% [(100 − 75 = 25) × 75%] during the second step.
By the two ring embrace or the single bracelet model, 50% of the cohesed DNA molecules would display dual epitope specificity in cohesin, and be subject to immunoprecipitation by either the HA- or Myc-antibody. For 75% immunoprecipitation efficiency, the first antibody would bring down [(25 + 50 = 75) × 75%)] = ∼56% of all cohesed DNA molecules. Note that 25% of the DNA molecules displaying a single epitope specificity would also be competent for immunoprecipitation at the first step. This step would deplete (50 × 75%) = 37.5 % of the dual specificity molecules from the subsequent round of immunoprecipitation. The fraction of DNA molecules that the virgin antibody can recognize would thus be (50 − 37.5) = 12.5% with dual epitope specificity plus 25% with single specificity. The expected immunoprecipitation by this antibody is [(12.5 + 25 = 37.5) × 75%] = ∼28%. The predicted two fold reduction in DNA pull-down between the two antibodies (from 56% to 28%) for their primary encounters with cohesed plasmids is not in agreement with the experimental result, which showed no such reduction (Figure 4C and D).
The two-step immunoprecipitation data are also inconsistent with a cohesin hand-cuff, even one in which the individual rings are linked by catenation.
Isolation of STB plasmid sisters paired by cohesin and their separation by linearizing DNA or cleaving Mcd1 ex vivo
Data from Figures 1–4 support the entrapment, in metaphase cells, of an STB reporter plasmid as a DNA–protein catenane containing a single protein ring of cohesin. The assumption that two DNA rings are present within such a catenane is based on the earlier finding that cohesin assembly at STB promotes plasmid cohesion followed by sister-to-sister plasmid segregation (6). In order to directly test our assumption, we have isolated the cohesed form of a single copy STB reporter plasmid from metaphase cells by sucrose gradient sedimentation (19), and interrogated ex vivo the DNA status within it.
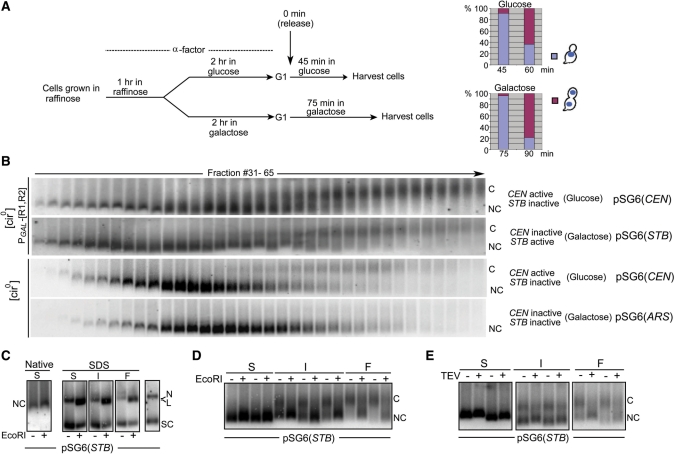
The single copy reporter plasmid pSG6, ∼4 kb long, was fashioned after the CEN-STB reporter plasmids used in previously published work (6,33). In this pSG4 derivative, the TetO21 locus was replaced by a PGAL-CEN3 DNA fragment. The CEN sequence, while it helped maintain the copy number at or close to unity, could be conditionally inactivated by galactose induced transcription. The plasmid was housed in a [cir0] strain or an isogenic [cir0] strain expressing Rep1 and Rep2 from the bidirectional GAL1–GAL10 promoter. The plasmid would behave as a true CEN plasmid (or a minichromosome) in either host strain in the presence of glucose as the carbon source. In galactose, though, it would behave as an ARS plasmid in the [cir0] host and as an STB plasmid in the [cir0]::PGAL[REP1 REP2] host. Fractionation of cleared lysates from metaphase cells by centrifugation through a 12.5–37.5% sucrose gradient resolved the plasmid into two forms: an ‘uncohesed’ monomer form and a ‘cohesed’ 2 × monomer form (Figure 5).
Figure 5.
Enrichment of plasmids in their cohesed form from metaphase cells by velocity sedimentation: test of the topological model for cohesion. (A) The experimental regimen for enriching metaphase cells from the appropriate [cir0] host strain harboring pSG6 and going through the cell cycle in glucose or galactose is schematically indicated. At 45 min for the cell cycle in glucose and at 75 min for that in galactose, the predominant population consisted of large budded cells with a single DAPI staining mass at the bud neck. (B) The sedimentation patterns of pSG6 in 12.5–37.5% sucrose gradient were followed under conditions where the plasmid-borne CEN alone or STB alone or neither of the two was active. Samples were run in agarose gels in the cold (4°C) and probed by pSG6-specific radio-labeled DNA. C, cohesed plasmids; NC, non-cohesed plasmids. (C) Representative fast (F), intermediate (I) and slow (S) sedimenting fractions from the gradient were reanalyzed by agarose gel electrophoresis with or without EcoRI digestion, followed by SDS treatment. For reference, S fractions treated or untreated with EcoRI but without subsequent SDS addition (left panel) and DNA prepared form the lysate by phenol extraction and ethanol precipitation (rightmost lane) were included in the run. (D and E) Representative fractions from the sucrose gradient (fast, slow and intermediate) were treated with EcoRI (D) or with TEV protease (E), and subjected to agarose gel electrophoresis under native conditions.
The general scheme for enriching metaphase cells from populations arrested in G1, conditioned in glucose or galactose, and then released into the cell cycle is outlined in Figure 5A. The sedimentation profiles of pSG6 in its CEN, STB and ARS incarnations (Figure 5B) shed light on the DNA stoichiometry in the cohesed form of the plasmid. Fractions were divided into three categories based on their sedimentation velocities: fast (F; cohesed?), slow (S; non-cohesed?) and intermediate (I; cohesed plus non-cohesed?). When they were analyzed by electrophoresis in native agarose gels in the cold (4°C), there was a reversal (as expected) in their relative mobility: the fast-sedimenting (heavy) fractions migrated more slowly than the slow-sedimenting (light) fractions. In a typical run, the S group comprised of fractions 31 (start point of Southern signal for DNA) to 44, the I group 45 to 54 and the F group 55 to 65. The lower mobility band of DNA (C for cohesed) was characteristic of pSG6(CEN) and pSG6(STB), and was absent or nearly so for pSG6(ARS). The sedimentation and gel migration profiles of the plasmid containing fractions were reminiscent of those reported by Ivanov and Nasmyth (19) for their minichromosome assays. We inferred therefore that the upper band comprised presumably of cohesed sister plasmids (in association with protein factors in addition to cohesin) while the lower band contained noncohesed (NC) plasmid molecules. This inference was subjected to further verification (see below). The fact that the pSG6(ARS) profile was almost entirely free of the upper band implies little contribution from catenated or covalent plasmid dimers to the ‘cohesed’ DNA fraction.
Digestion by EcoRI at 4°C did not alter the DNA mobility of the S (non-cohesed) fractions of pSG6(STB) in electrophoresis under native conditions (Figure 5C). However, electrophoresis in presence of SDS without EcoRI treatment revealed monomeric plasmid DNA, primarily as the supercoiled form (∼70–80%) and the remainder as nicked circles in S, I and F fractions. EcoRI treatment resulted in conversion of ∼60–70% of the DNA into linear molecules which migrated below (but almost overlapping with) the nicked circle under SDS-electrophoresis. Consistent with the extent of linearization of plasmid by EcoRI, native gel electrophoresis revealed a similar conversion of the cohesed form of pSG6 from the I and F fractions to the noncohesed form (Figure 5D). Conversion from the cohesed to the non-cohesed form was also promoted by TEV protease cleavage (Figure 5E). Breaching of cohesion either by linearization of pSG6(STB) or by Mcd1 cleavage was as expected for topology mediated cohesion. The outcomes of EcoRI and TEV protease digestions were quite similar between pSG6(STB) and pSG6(CEN) (Supplementary Figure S5A–C). Furthermore, the sedimentation assay revealed little cohesed dimers of pSG6 in galactose grown but nocodazole treated cells (Supplementary Figure S5D, bottom). This result attests to the complete inactivation of CEN by galactose-induced transcription, and verifies the authenticity of STB-mediated cohesion when the mitotic spindle is intact (Supplementary Figure S5D, top; Figure 5B).
The data for plasmid sedimentation followed by ex vivo cleavage assays support the presence of two plasmid monomers per cohesed unit. Results from the previous pull-down analyses are most easily explained by the occupancy of this unit by one monomeric cohesin ring. Together they are consistent with the embrace model for plasmid cohesion, in which a monomeric cohesin ring holds two sister rings of the STB reporter plasmid in a tripartite catenane.
DISCUSSION
The topology model for sister chromatid cohesion derives its support from experiments that convert DNA from circular to linear form or break a polypeptide chain in cohesin or non-covalently or covalently seal borders at critical points where cohesin subunits interface with each other (16–20). Closure of the hinge gate in cohesin apparently blocks entry of a chromosome into its interior (16); and circular minichromosomes already associated with cohesin remain trapped until an opening is introduced either in the DNA or in the protein (18–20). These observations suggest a double-gate mechanism for the establishment and annulment of chromosomal cohesion. The logic is similar to that used by DNA topoisomerase II to transport a DNA segment through two oppositely located entrance and exit gates during DNA relaxation (34). The difference in the case of cohesin is that the second gate opening event involves proteolytic cleavage of a protein subunit. Notwithstanding the evidence favoring cohesion by topological DNA–protein association, the possibility of cohesion by physical interaction remains a viable alternative (21–24,35). We have now shown that 2 micron plasmid molecules, or at least those amongst them that remain cohesed under the assay conditions, also conform to cohesion by the topological dictum.
Sister plasmid cohesion in the 2 micron circle
The stable propagation of the 2 micron plasmid is conferred by a partitioning system consisting of the plasmid coded Rep1 and Rep2 proteins and the cis-acting STB locus (36,37). Requirement of an active partitioning system is mandated by the organization of the multi-copy plasmid into a tight cluster of 3–5 foci, the cluster being the unit of segregation (38). The REP-STB system appears to couple plasmid segregation to chromosome segregation either by appropriating chromosome segregation factors or tethering the plasmid cluster to a chromosome (5). Association of cohesin with STB, assisted by the Rep proteins and an intact mitotic spindle (5,8,39), establishes cohesion between plasmid sisters that subsequently segregate one-to-one from each other (6). Our present analyses favor the embrace model for plasmid cohesion, and suggest a stoichiometry of one cohesin ring per two sister plasmids (or STBs).
The fraction of cohesin-associated STB reporter plasmid obtained in pull-down and sedimentation assays is <20%, comparable to 10–30% reported for minichromosomes (20). In contrast, cohesion of STB reporter plasmids as assayed by cell biological methods in metaphase populations is much higher, >70% (6). The loss of cohesion during biochemical manipulations could be due to a physical, and less stable, mode of cohesin–DNA interaction. However, mutually concordant outcomes from distinct affinity interaction and velocity sedimentation assays are consistent with a uniform, and topological, mode of cohesion in a finite fraction of cohesin-associated plasmids.
Molar ratio of DNA rings and cohesin within cohesed plasmid species: a 2:1 DNA–cohesin tri-link?
Chromatin immunoprecipitation assays reveal a periodic distribution of cohesin at 10–15 Kb intervals along chromosome arms in S. cerevisiae with a higher density of localization at and around centromeres (40–42). Fluorescent intensity of Smc3-GFP at kinetochores normalized to one copy of the histone H3 variant Cse4 per centromere (43), suggests the presence of one cohesin molecule for every 4 Kb of centric/pericentric DNA (22). Our finding that a pair of sister STB reporter plasmids is likely held together by one cohesin ring agrees well with this estimate. It is also consistent with in vivo FRET results indicating lack of interaction between more than one cohesin complex (44) and biochemical outcomes from induced cohesin ring closure within cohesed minichromosomes (18).
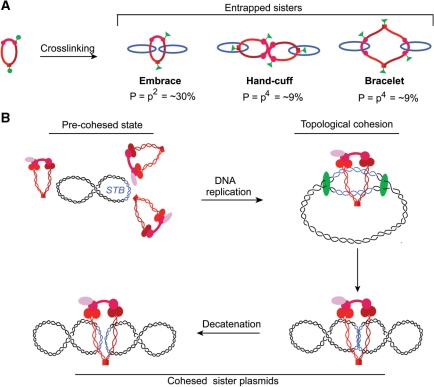
Based on the extent of cohesed minichromosomes entrapped by chemically crosslinking cohesin subunits, Haering et al. (18) argue in favor of cohesion by monomeric cohesin rings (embrace model) over dimeric cohesin rings (hand-cuff and bracelet models). In their assays, covalent protein closure in a monomeric ring required crosslinking of two pairs of cysteine neighbors resident at distinct locations (green circles) to form a pair of chemical bridges (green triangles) [Figure 6A; adapted from (18)]. For a probability ‘p’ of forming a single cross-link (p = efficiency of the crosslinking agent), the expectation for entrapment of DNA sisters by a monomeric ring is p2, since two crosslinks have to be formed to seal the ring. For a dimeric ring of the bracelet or hand-cuff type, this value is p4. For a value of ‘p’ = ∼55%, experimentally determined by Haering et al., the observed extent of entrapment ‘P’ matches p2 = 30%, and not p4 = 9%. The Haering et al. result is also contrary to two or more monomeric cohesin rings surrounding the sister DNAs (Supplementary Figure S6A). In the case of two rings, the expected P is 1−[1−p2]2 = 2p2−p4 = 51%. However, double ring models in which the DNA sisters reside within one ring cannot be ruled out (Supplementary Figure S6B). A biased hand-cuff, physical or topological, predicts P=p2=30%, since two crosslinks would suffice to trap both sisters.
Figure 6.
A single ring formed by a monomeric cohesin complex as the unit of cohesion at STB. (A) The results from Haering et al. (18) for CEN cohesion ruling out ‘double’ rings of cohesin are schematically represented. In their experimental design, covalent protein ring closure required crosslinking two neighboring pairs of cysteines at two locations (green circles) to form a pair of chemical bridges (green triangles). Cross-linking efficiency (or probability ‘p’ of circle to triangle conversion) was ∼55% in their assays. Experiments agreed with entrapment probability of DNA sisters ‘P’ equal to p2 (30%) and not p4 (9%). Note that, upon SDS treatment during the assay, a physical hand-cuff (but not a topological one) would fall apart to yield monomeric cohesin rings, each with a single trapped plasmid molecule. (B) In the pre-cohesed state of the 2 micron circle, multiple cohesin molecules may interact physically and dynamically at or near STB. Such interactions could be promoted by the cohesin loading factors Scc2 and Scc4, which are required for cohesin assembly on the plasmid (5). Transition to the stable topological association may be mediated by passage of the replisome and closure of a single cohesin ring around a pair of STB sisters.
Although multiple cohesin molecules could be involved in precohesive interactions, likely mediated through DNA-bound cohesin loading factors Scc2 and Scc4 (45). Only a subset of such interactions may be consummated to stable topological cohesion by passage of the replisome. In the case of the 2 micron plasmid, only one cohesin ring, in most cases, may be closed to encircle the nascent STB duplexes (Figure 6B).
Conserved mode of cohesion at CEN and STB: evolutionary implications
The genetically defined ‘point’ centromere of S. cerevisiae, contrasted by the epigenetically specified centromeres of fungi in general, poses a rather puzzling transitional oddity in centromere evolution (46). It has been suggested that the point centromere is a domesticated version of the partitioning locus of an ancestral 2 micron-like plasmid that functionally replaced the canonical epigenetic fungal centromere. Miniaturization of the centromere and loss of the machinery for establishing pericentric heterochromatin and RNA interference (47) appear to have been related events. It is noteworthy in this regard that the existence of 2 micron-related plasmids is limited to fungal lineages belonging to Saccharomycetaceae (46,48). The emergence of a novel centromere had necessarily to engender mechanisms for integrating it into the established chromosome segregation pathway. Principally, it had to be rendered competent in recruiting components of the kinetochore complex. Several subunits (Ndc10 and Ctf13, for example) of the CBF3 complex, which binds to the CDE III region and provides the platform for kinetochore assembly, are distinguishing hallmarks of Saccharomycetaceae among fungi (46,48). Of particular significance is the conspicuous absence of these proteins in Schizosaccharomyces pombe and Candida albicans. The Rep1 and Rep2 proteins of the extant 2 micron circle (and perhaps related plasmids) also orchestrate the assembly of a partitioning complex that couples plasmid segregation to chromosome segregation. The individual plasmid partitioning systems must have co-evolved with the respective centromere-based segregation machineries to preserve functional coupling between the two. Recruitment of cohesin at STB and the conserved topological mechanism for establishing cohesion between sister duplexes at CEN and STB may represent evolutionary vestiges of their common ancestry.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM064363); Robert F Welch Foundation award (F-1274, partial). Funding for open access charge: National Institutes of Health (GM064363).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank D. Ivanov, K. Nasmyth, A. Johnson, V. Guacci and Doug Koshland for generously sharing yeast strains, plasmids, reagents and equipment. We are grateful to C. Haering and D. Ivanov for helpful comments on the article. We acknowledge Chien-Hui Ma for excellent technical assistance. We appreciate the insightful critique from the reviewers that helped improve the article in style and content.
REFERENCES
- 1.Nasmyth K, Schleiffer A. From a single double helix to paired double helices and back. Phil. Trans. R Soc. Lond. B Biol. Sci. 2004;359:99–108. doi: 10.1098/rstb.2003.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu. Rev. Cell Dev. Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- 3.Uhlmann F. The mechanism of sister chromatid cohesion. Exp. Cell Res. 2004;296:80–85. doi: 10.1016/j.yexcr.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe Y. Sister chromatid cohesion along arms and at centromeres. Trends Genet. 2005;21:405–412. doi: 10.1016/j.tig.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Mehta S, Yang XM, Chan CS, Dobson MJ, Jayaram M, Velmurugan S. The 2 micron plasmid purloins the yeast cohesin complex: a mechanism for coupling plasmid partitioning and chromosome segregation? J. Cell Biol. 2002;158:625–637. doi: 10.1083/jcb.200204136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh SK, Hajra S, Jayaram M. Faithful segregation of the multicopy yeast plasmid through cohesin-mediated recognition of sisters. Proc. Natl Acad. Sci. USA. 2007;104:13034–13039. doi: 10.1073/pnas.0702996104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghosh SK, Hajra S, Paek A, Jayaram M. Mechanisms for chromosome and plasmid segregation. Annu. Rev. Biochem. 2006;75:211–241. doi: 10.1146/annurev.biochem.75.101304.124037. [DOI] [PubMed] [Google Scholar]
- 8.Mehta S, Yang XM, Jayaram M, Velmurugan S. A novel role for the mitotic spindle during DNA segregation in yeast: promoting 2 micron plasmid-cohesin association. Mol. Cell Biol. 2005;25:4283–4298. doi: 10.1128/MCB.25.10.4283-4298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hakimi MA, Bochar DA, Schmiesing JA, Dong Y, Barak OG, Speicher DW, Yokomori K, Shiekhattar R. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature. 2002;418:994–998. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]
- 11.McKee BD. Does cohesin regulate developmental gene expression in Drosophila? Proc. Natl Acad. Sci. USA. 2008;105:12097–12098. doi: 10.1073/pnas.0805712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol. Cell Biol. 2004;24:3100–3111. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peric-Hupkes D, van Steensel B. Linking cohesin to gene regulation. Cell. 2008;132:925–928. doi: 10.1016/j.cell.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Krantz ID. Cohesin and human disease. Annu. Rev. Genomics Hum. Genet. 2008;9:303–320. doi: 10.1146/annurev.genom.9.081307.164211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 16.Gruber S, Arumugam P, Katou Y, Kuglitsch D, Helmhart W, Shirahige K, Nasmyth K. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell. 2006;127:523–537. doi: 10.1016/j.cell.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 17.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 18.Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 19.Ivanov D, Nasmyth K. A physical assay for sister chromatid cohesion in vitro. Mol. Cell. 2007;27:300–310. doi: 10.1016/j.molcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Ivanov D, Nasmyth K. A topological interaction between cohesin rings and a circular minichromosome. Cell. 2005;122:849–860. doi: 10.1016/j.cell.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 21.Guacci V. Sister chromatid cohesion: the cohesin cleavage model does not ring true. Genes Cells. 2007;12:693–708. doi: 10.1111/j.1365-2443.2007.01093.x. [DOI] [PubMed] [Google Scholar]
- 22.Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Surcel A, Koshland D, Ma H, Simpson RT. Cohesin interaction with centromeric minichromosomes shows a multi-complex rod-shaped structure. PLoS ONE. 2008;3:e2453. doi: 10.1371/journal.pone.0002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang CR, Wu CS, Hom Y, Gartenberg MR. Targeting of cohesin by transcriptionally silent chromatin. Genes Dev. 2005;19:3031–3042. doi: 10.1101/gad.1356305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang N, Kuznetsov SG, Sharan SK, Li K, Rao PH, Pati D. A handcuff model for the cohesin complex. J. Cell Biol. 2008;183:1019–1031. doi: 10.1083/jcb.200801157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben-Shahar TR, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 27.Heidinger-Pauli JM, Unal E, Guacci V, Koshland D. The kleisin subunit of cohesin dictates damage-induced cohesion. Mol. Cell. 2008;31:47–56. doi: 10.1016/j.molcel.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Strom L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjogren C. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–245. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- 29.Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- 30.Unal E, Heidinger-Pauli JM, Koshland D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7) Science. 2007;317:245–248. doi: 10.1126/science.1140637. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Nelson RG, Fangman WL. Nucleosome organization of the yeast 2-micron DNA plasmid: a eukaryotic minichromosome. Proc. Natl Acad. Sci. USA. 1979;76:6515–6519. doi: 10.1073/pnas.76.12.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui H, Ghosh SK, Jayaram M. The selfish yeast plasmid utilizes the nuclear motor Kip1p but not Cin8p for its localization and equal segregation. J. Cell. Biol. 2009;185:251–264. doi: 10.1083/jcb.200810130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roca J, Wang JC. DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell. 1994;77:609–616. doi: 10.1016/0092-8674(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 35.Huang CE, Milutinovich M, Koshland D. Rings, bracelet or snaps: fashionable alternatives for Smc complexes. Phil. Trans. R Soc. Lond. B Biol. Sci. 2005;360:537–542. doi: 10.1098/rstb.2004.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jayaram M, Mehta S, Uzri D, Voziyanov Y, Velmurugan S. Site-specific recombination and partitioning systems in the stable high copy propagation of the 2-micron yeast plasmid. Prog. Nucleic Acid Res. Mol. Biol. 2004;77:127–172. doi: 10.1016/S0079-6603(04)77004-X. [DOI] [PubMed] [Google Scholar]
- 37.Jayaram M, Yang XM, Mehta S, Voziyanov Y, Velmurugan S. The 2 micron plasmid of Saccharomyces cerevisiae. In: Funnell BE, Phillips G, editors. Plasmid Biology. Washington DC: ASM Press; 2004. pp. 303–324. [Google Scholar]
- 38.Velmurugan S, Yang XM, Chan CS, Dobson M, Jayaram M. Partitioning of the 2-micron circle plasmid of Saccharomyces cerevisiae. Functional coordination with chromosome segregation and plasmid-encoded Rep protein distribution. J. Cell Biol. 2000;149:553–566. doi: 10.1083/jcb.149.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- 41.Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:e259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- 43.Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl Acad. Sci. USA. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mc Intyre J, Muller EG, Weitzer S, Snydsman BE, Davis TN, Uhlmann F. In vivo analysis of cohesin architecture using FRET in the budding yeast Saccharomyces cerevisiae. EMBO J. 2007;26:3783–3793. doi: 10.1038/sj.emboj.7601793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol. Cell. 2006;23:787–799. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 46.Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 47.Aravind L, Watanabe H, Lipman DJ, Koonin EV. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl Acad. Sci. USA. 2000;97:11319–11324. doi: 10.1073/pnas.200346997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blaisonneau J, Sor F, Cheret G, Yarrow D, Fukuhara H. A circular plasmid from the yeast Torulaspora delbrueckii. Plasmid. 1997;38:202–209. doi: 10.1006/plas.1997.1315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.