Abstract
The complexity in intracellular signaling mechanisms relevant for the conquest of many diseases resides at different levels of organization with scales ranging from the subatomic realm relevant to catalytic functions of enzymes to the mesoscopic realm relevant to the cooperative association of molecular assemblies and membrane processes. Consequently, the challenge of representing and quantifying functional or dysfunctional modules within the networks remains due to the current limitations in our understanding of mesoscopic biology, i.e., how the components assemble into functional molecular ensembles. A multiscale approach is necessary to treat a hierarchy of interactions ranging from molecular (nm, ns) to signaling (μm, ms) length and time scales, which necessitates the development and application of specialized modeling tools. Complementary to multiscale experimentation (encompassing structural biology, mechanistic enzymology, cell biology, and single molecule studies) multiscale modeling offers a powerful and quantitative alternative for the study of functional intracellular signaling modules. Here, we describe the application of a multiscale approach to signaling mediated by the ErbB1 receptor which constitutes a network hub for the cell’s proliferative, migratory, and survival programs. Through our multiscale model, we mechanistically describe how point-mutations in the ErbB1 receptor can profoundly alter signaling characteristics leading to the onset of oncogenic transformations. Specifically, we describe how the point mutations induce cascading fragility mechanisms at the molecular scale as well as at the scale of the signaling network to preferentially activate the survival factor Akt. We provide a quantitative explanation for how the hallmark of preferential Akt activation in cell-lines harboring the constitutively active mutant ErbB1 receptors causes these cell-lines to be addicted to ErbB1-mediated generation of survival signals. Consequently, inhibition of ErbB1 activity leads to a remarkable therapeutic response in the addicted cell lines.
Introduction
ErbB family receptors (named because of their homology to the erythroblastoma viral gene product, v-erbB, and consisting of the epidermal growth factor receptor or EGFR/ErbB1/HER1, ErbB2/HER2, ErbB3, and ErbB4) signal by activating crucial pathways1 in response to activation by ligands such as the epidermal growth factor (EGF) and other related peptide growth factors. Through ligand-stimulated formation of various homodimeric and heterodimeric complexes, ErbB receptors are activated, leading to the phosphorylation of multiple tyrosine residues on the C-terminal tail of the receptors as well as on other substrate proteins. Through specific interactions of the phospho-tyrosine sites to binding domains, the receptors bind to cytosolic partners that are responsible for the recruitment and activation of multiple down-stream cascades.2–7 Activation through the mitogen-activated protein kinase (MAPK) cascades of the extracellular signal-regulated kinases (ERKs) is functionally linked to proliferation. The phosphoinositide 3-kinase (PI3K) pathway leads to the activation of the serine/threonine protein kinase Akt (cellular homologue of the viral oncogene v-Akt) which is linked to survival. Other significant pathways mediated by ErbB signaling include activation and nuclear translocation of signal transducers and activators of transcription proteins (STATs)8 and clathrin mediated endocytosis. 9 Yet, the molecular context in which ErbB receptors activate and regulate signaling has not been fully recognized. More specifically, it is of great interest to investigate the molecular mechanisms that lead to the disregulation of ErbB signaling in several pathologies such as cancer, psoriasis, atherosclerosis, impaired cardiac development, and schizophrenia. 10,11 With rapid progress in high-performance computing methods and infrastructure,† we advocate that multiscale modeling offers a powerful, quantitative, and complimentary alternative for the study of functional intracellular modules. We describe the application of a hierarchical multiscale modeling procedure (summarized in the section: Models and methods) to signaling in ErbB family receptors to describe how point-mutations in the ErbB1 receptor can profoundly alter signaling characteristics leading to the onset of oncogenic transformations.
Models and methods
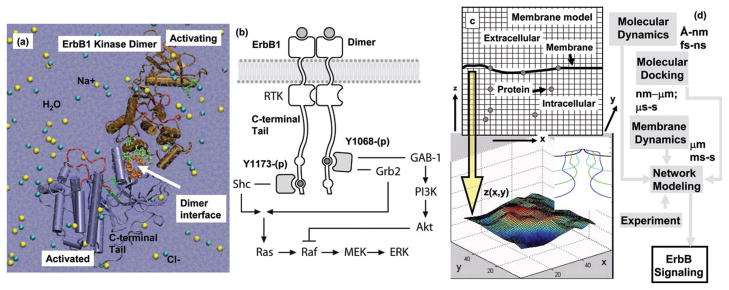
We provide a brief summary of the hierarchical multiscale modeling scheme we have employed for describing ErbB signaling (see Fig. 1a–d). The flow of information between the models (summarized below) is depicted in Fig. 1d. We model the dimer-mediated receptor activation characteristics of the ErbB1 receptor tyrosine kinase using molecular dynamics simulations.12,13 Fig. 1a depicts the atomistic model of the explicitly solvated ErbB1 kinase dimer employed in the molecular dynamics: 10–30 ns trajectories of fully atomistic, explicitly solvated systems of wildtype and mutant ErbB1 kinase monomers and dimers are obtained and analyzed for specific stabilizing interactions such as hydrogen bonds and salt-bridges, see also Fig. 2.12,13 Based on the model in Fig. 1a, we also model the interactions of ErbB1 substrate tyrosines derived from the C-terminal tail of ErbB1 with the kinase using molecular docking simulations.12 Specifically, we perform a global conformational search for bound conformations of substrate peptides to the ErbB1 kinase domain using a multiple conformation docking strategy in which the protein flexibility is taken into account implicitly. We treat receptor-mediated signaling using a deterministic network-based kinetic model,12,14,15 see Fig. 1b: the figure depicts the branched signaling network model for ErbB1-mediated signaling employed in this study. In the branched kinetic model, signaling through ErbB1 is modeled by combining three published models and augmented by our own set of reactions: phosphorylation and docking reactions are modeled according to ref. 5; the MAPK pathway reactions are modeled after ref. 3; Akt and PI3K activation are incorporated into the model as described in ref. 16. We resolve phosphorylation of the active ErbB1 dimer at tyrosine site Y1068, which when phosphorylated can bind to growth-factor- receptor bound-2 protein (Grb2) or Grb2-associated binding protein (GAB-1), or at tyrosine site Y1173, which when phosphorylated can bind to the Src-homology-2-containing (Shc) adaptor protein. Phosphorylation of downstream factors Akt and ERK are used as indicators of downstream activation (see also Table 1). Differences in ERK and Akt phosphorylation levels due to mutations in the ErbB1 receptor are accounted for through changes to the kinetic parameters of the deterministic model. The altered parameters are obtained through a combination of our molecular dynamics and molecular docking simulations as well as through experiments published in the literature.15 Altogether, our network model comprises 74 reactions and 67 species. 17 of these reactions are novel to this work and represent enhanced molecular resolution in the ErbB1 activation, phosphorylation, and docking reactions, and enable separate parameterization for ErbB1 wildtype and mutant systems.12,14,15 Recently, we have extended the signaling module to include ErbB1 receptor internalization, which we treat using a hybrid discrete/continuum stochastic dynamics protocol.12,17,18 Fig. 1c depicts the hybrid stochastic model for ErbB1 internalization: (top) grids in finite difference scheme for solving membrane dynamics (based on a continuum field model) and for modeling protein diffusion via lattice hopping; (bottom) snapshot of vesicle bud on the membrane in response to a specific spatial ordering of the curvature-inducing protein, epsin, on the membrane.
Fig. 1.
Hierarchical multiscale modeling scheme for ErbB signaling. The dimer-mediated receptor activation characteristics of ErbB1 receptor tyrosine kinase are studied using molecular dynamics simulations. The interactions of substrate tyrosine containing peptides derived from the C-terminal tail with the ErbB1 kinase are studied using molecular docking simulations. We have employed a deterministic network-based kinetic modeling scheme to study ErbB1-mediated signaling, and a hybrid discrete/continuum stochastic dynamics protocol to study the initiation of ErbB1 receptor internalization. (a) Atomistic model for ErbB1 dimer employed in the molecular dynamics and molecular docking calculations. (b) Branched network model for ErbB1-mediated signaling in which phosphorylation of the ErbB1 dimer occurs at either tyrosine Y1068, which can bind GAB-1 or Grb2, or at tyrosine Y1173, which binds Shc. Phosphorylation of the factors Akt and ERK was used as indication of downstream activation. (c) Hybrid stochastic model for ErbB1 internalization. (top) Grids in finite difference scheme for membrane dynamics and the lattice in the kinetic Monte Carlo scheme for protein diffusion; (bottom) snapshot of vesicle bud on the membrane in response to a specific spatial ordering of the curvature-inducing protein, epsin, on the membrane; inset depicts a stabilized vesicle neck. (d) Flow of information between different simulation methods.
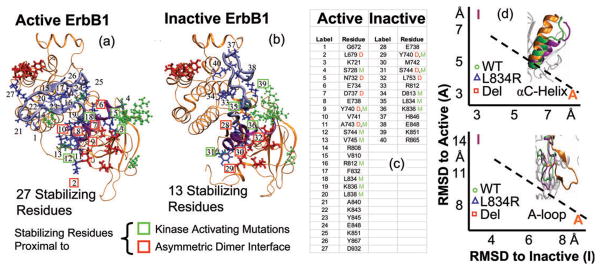
Fig. 2.
(a, b) Stabilizing network residues in ErbB1 kinase in its active and inactive conformations. The numbers in (a, b) correspond to the entries in (c). Compared to inactive, the active state presents a significantly increased number of stabilizing residues, with approximately the same number of residues affected by the asymmetric dimerization (marked D and boxed red) and mutation (marked M and boxed green); so the inactive system is much more susceptible to a conformational shift triggered by the asymmetric dimer or mutation. We identify the residues participating in stabilizing bonds that are also proximal (≤3 Å between heavy atoms) to the dimer interface. Cross-referencing this list with the stabilizing residues provides a list of potential residues possibly affected by dimerization: entries marked D in (c) and boxed red in (a, b). (d): Symbols represent trace during dimer molecular dynamics simulations of wildtype and the two mutants L834R and Del L723–P729 ins S or del: (top) root-mean-squared deviation or RMSD of αC-helix, and (bottom) RMSD of the activation loop (or A-loop). Insets depict the relative positions of the αC-helix and the A-loop at the end of the dynamics runs. The purple corresponds to the inactive conformation, the orange corresponds to the active conformation, and the green corresponds to the conformation in the wildtype dimer following 20 ns of molecular dynamics from the inactive state. The conformation of the activation loop stays close to that in the inactive conformation even upon dimerization (see bottom inset). However, the conformation of the αC-helix in the wildtype dimer undergoes considerable rearrangement and shift toward the active conformation (see top inset).
Table 1.
(a) Comparison of signaling and inhibition characteristics predicted by our network simulations for wildtype and mutant systems under different conditions; Del stands for the deletion mutant del L723–P729 ins S. Concentrations of Akt-(p) or ERK-(p) are used to indicate the downstream effects of receptor signaling. EC50 values are calculated as a measure of the degree to which signals downstream are inhibited by the ATP analogue, erlotinib. (b) Kinetic rate constants or species concentrations constituting the top three principal components for network hyper-sensitivity calculated through global sensitivity analysis: kf, turnover for phosphorylation; (•) denotes bound complex; the square brackets represent concentrations
| (a) | Wildtype | L834R | Del |
|---|---|---|---|
| ERK-(p) in nM for normal ErbB1 expression | |||
| −EGF/+EGF | 0.2/5.0 | 1.0/1.0 | 8.0/8.0 |
| Akt-(p) in nM for normal ErbB1 expression | |||
| −EGF/+EGF | 1.0/15.0 | 13.0/13.0 | 20.0/20.0 |
| ERK-(p) in nM for ErbB1 over-expression | |||
| −EGF/+EGF | 3.0/3.0 | 5.0/5.0 | 1.0/1.0 |
| Akt-(p) in nM for ErbB1 over-expression | |||
| −EGF/+EGF | 20.0/20.0 | 22.0/22.0 | 19.0/19.0 |
| Cellular EC50 in nM for inhibition of ERK-(p) and Akt-(p) for normal ErbB1 expression/ErbB1 over-expression | |||
| ERK-(p) | 700/4000 | 100/1000 | −/− |
| Akt-(p) | 1200/500 000 | 300/50 000 | −/− |
| (b) Rate and binding constants | |||
| kf: Y1068; kf: Y1173 | |||
| Ki: inhibitor; KM: ATP•RTK | |||
| KM: GAB-1•ErbB1-(p) | |||
| Initial concentrations | |||
| [Raf•Ras•GTP] | |||
| [Pase3] phosphatase for ERK-(p) | |||
| [Pase4] phosphatase for Akt-(p) | |||
| [MEK-(p)] | |||
| [PI3K inactive] | |||
| [MEK•Raf active] | |||
| [ErbB1•Shc•Grb2•SOS• RasGTP] | |||
| [inhibitor] | |||
Rationalizing and predicting the effects of molecular perturbations on receptor kinase activation
Small molecule tyrosine kinase inhibitors for ErbB1/2 tyrosine kinase such as gefitinib, erlotinib, and lapatinib, which are ATP analogues, are of significant interest as cancer drugs. While the receptor tyrosine kinase inhibition approach has shown promise in some clinical trials, success has been mixed. In particular, the occurrence of somatic mutations in the ErbB1 kinase domain (L834R: where the leucine residue in position 834 is replaced by an arginine, L837Q: leucine at 837 replaced by a glutamine, G685S, del L723–P729 ins S: deletion of residues 723–729 and an insertion of a serine) as seen in patients of non-small cell lung cancer19‡ renders the cell lines harboring such mutations more sensitive to treatment.19,20
In order to determine how such molecular perturbations can shape cellular fates, we sought to determine how the mutations affect the regulatory mechanisms operational within the kinase domain of ErbB1, ErBb2, and ErbB4. A recent structural and biochemical study involving ErbB1 by Zhang et al.21 proposed a new dimer-mediated allosteric activation mechanism according to which ErbB1 receptor tyrosine kinase dimerizes in an asymmetric head-to-tail configuration. While the monomer ErbB1 kinase is stable in an inactive conformation which interferes with ATP binding, in the asymmetric dimer configuration (see section: Models and methods), one ErbB1 kinase domain serves as an activating protein and activates the other ErbB1 kinase in the dimer, through allosteric contacts. The kinase–kinase contact at the asymmetric dimer interface allosterically stabilizes the active conformation. Recently, we hypothesized that an underlying network of stabilizing hydrogen bonds dominates the relative stabilities of the inactive and active conformations and governs the kinase activation. We then performed a hydrogen bond analysis of the molecular dynamics data, focusing on the interactions surrounding the activation-loop and the αC-helix sub-domains in the active and inactive conformations, and identified this network, see Fig. 2a and b. The total number of 27 residues participating in the stabilizing network in the active state (residues 1–27 marked in Fig. 2a) out-numbers the 13 residues participating in the stabilizing network of the inactive state (residues 28–40 marked in Fig. 2b). This stark contrast of 27 to 13 stabilizing interactions led us to hypothesize that a particular stimulus such as the asymmetric dimerization is poised to preferentially destabilize the inactive kinase conformation triggering the conformational change to the active state.
In order to consider the effect of ErbB1 kinase dimerization on the network of stabilizing interactions, we identified the protein residues participating in stabilizing bonds that are also proximal (≤3 Å between heavy atoms) to the residues involved in the formation of the asymmetric dimer interface, (which are P675, L679, L680, I682, V736, L758, V762 in the kinase undergoing activation). The rationale for the proximity analysis is that the interactions in the stabilizing network could be compromised due to the molecular level reorganization upon kinase dimerization. The proximity analysis suggests that 3 out 13 stabilizing residues in the inactive state may be perturbed or lost upon dimerization (these are marked with D in Fig. 2c), which suggests a potential molecular relay mechanism by which the kinase dimerization event activates the kinase domain. Indeed, when we performed 20 ns molecular dynamics simulations of an ErbB1 kinase dimer system (shown in Fig. 1a), we observed a significant rearrangement (change in the root-mean-squared deviation or RMSD of 3 Å ) of the αC-helix position: see Fig. 2d, the inset to Fig. 2d shows the shifting of the αC-helix (top) in the wildtype dimer but no significant rearrangement of the activation loop (bottom). The shift in the αC-helix position was accompanied by several changes in the stabilizing network consistent with the predicted bond-patterns in Fig. 2a–c. In particular, in the inactive conformation, the bonds between Y740–S744, H846–R865, K851–R812 surrounding the activation loop and the αC-helix were severed. Thus, already in the wildtype dimer, due to the re-configuration of the αC-helix, the bond pattern is found to be shifting significantly toward that observed in the active kinase. The key bonds stabilizing the wildtype inactive ErbB1 kinase are the E738–K836 and E848–R865 salt bridge interactions, as well as the L834–D813 hydrogen bond. In our dimer simulations, the E738–K836 salt bridge has considerably weakened: the fraction of the time this bond was present decreased from >90% in the monomer trajectory to ~60% in the dimer trajectory; moreover this bond has undergone considerable stretching allowing E738 to hydrogen bond to F832, which is one of the bonds seen in the active kinase. The L834–D813 interaction is at the threshold of still being considered a stabilizing hydrogen bond. The residue K851 is hydrogen bonded to E725, moving away from the inactive bond K851–R812 and towards the active salt bridge K851–E734. The E848–R865 salt bridge is not perturbed significantly due to the formation of the asymmetric dimer interface. Thus, we hypothesize that a few specific bonds act as gate-keepers for each step of the conformational change, namely the E738–K836 salt guards against the αC-helix movement and the E848–R865 salt bridge guards against the activation loop rearrangement. Our dimer simulations re-affirm our notion that the stabilizing network is susceptible to perturbation in the inactive conformation of the kinase, and that formation of the asymmetric dimer will have the effect of directly breaking the network of interactions around the αC-helix, thereby destabilizing the inactive state. The loss of these interactions and the shift of the αC-helix conformation towards the active state will provide the impetus for kinase domain activation. Intriguingly, several of the clinically identified mutations that have been reported to constitutively activate the kinase also directly perturb the stabilizing network by breaking key stabilizing bonds: these are marked by the symbol M in Fig. 2c. In addition, the deletion of residues L723 to P729 in the del L723–P729 ins S mutant re-configures the αC-helix in the inactive state to a conformation closer to the active state (data not shown). Thus, our delineation of the stabilizing hydrogen bond network provides molecular-level insight into the possible mechanisms by which activating mutations of ErbB1 kinase such as L834R and del L723–P729 ins S destabilize the inactive conformation. This preferential destabilization of the inactive conformation renders the receptor kinase constitutively active even as a monomer, producing high basal activation levels of the kinase even in the absence of a growth-factor induced dimerization.
Considering that there is an excellent correlation between the stabilizing network of interactions and the clinically identified activating mutations in ErbB1, our structural studies on kinase activation are well poised to forecast the mutation landscape associated with other ErbB members. We have extended the analysis we have presented for ErbB1 to ErbB2 and ErbB4 kinases in which we have identified similar networks of stabilizing interactions. Based on the similarities between the stabilizing interactions between ErbB1 and ErbB4 kinase domains, we can predict the effect of analogous mutations in ErbB4 on kinase activation: (1) based on the location of the mutations E690G, G700S, the mutants are expected to be activating through directly impacting dimerization (similar to the activating mutants E685G and G695S of ErbB1). (2) Del 728–G733 ins S and S749I are poised to cause a conformation shift of the αC-helix of ErbB4 and hence are predicted to be activating. (3) Mutations F740A, L839R and L842Q in ErbB4 are poised to perturb the bond network of the inactive kinase and hence are expected to be activating. We note that in support of our predictions of ErbB4, the F740A and L839R have been tested independently by Qiu et al.22 and were indeed found to be activating. Based on the subtle differences we have noted in the stabilizing bond networks of ErbB1 and ErbB4, we are also able to suggest new activating ErbB4 mutations (that do not have an obvious counter-part in the ErbB1 system). In particular, R841 in ErbB4 is featured prominently in the stabilizing network and mutating R841 to either alanine or aspartic acid is expected to promote activation. Mutation of the two residues blocking the catalytic aspartate, E743 and G838, to residues with smaller side chains, alanine or glycine, is also expected to promote activation. Thus, with experimental validation of the above predictions, we believe that our approach can be valuable for evaluating the likely effect of mutations on ErbB2 inhibition efficacies in cancer, and ErbB4 inhibition in cardiac development and schizophrenia.11
Differential ErbB1 signaling due to substrate specificity and a branched signaling model for transcribing the effects of molecular alterations into downstream signal activation
The preferential binding characteristics of different cytosolic substrates to different phospho-tyrosine locations of ErbB family kinases were reported recently.23 The variations in the phosphorylation kinetics associated with the different tyrosine sites of the cytoplasmic C-terminal tail of the ErbB1 kinase can induce differential patterns of downstream signaling leading to differences in the activation of key transcription factors. This leads us to hypothesize that the clinically identified activating mutations of ErbB1 kinase can also potentially influence cellular homeostasis by directly altering the phosphorylation kinetics of ErbB1 substrate tyrosines. Indeed the identity-specific phospho-tyrosine kinetics for Y1068 and Y1173 (as well as other phospho-tyrosine sites in ErbB1) for wildtype, L834R and del L723–P729 ins S mutant systems are supported by the kinetic experiments of Mulloy et al.24 In particular, the relative catalytic turnover (λ = kcat/KM, where kcat represents the rate of tyrosine phosphorylation in the bound complex, and KM represents the affinity of the tyrosine substrate to the ErbB1 kinase) rates of Y1068 phosphorylation and Y1173 phosphorylation measured in the experiments are as follows. For Y1068, wildtype: log10[λwildtype/λ] = 0.0; L834R: log10[λwildtype/λ] = 0.0; del L723–P729 ins S: log10[λwildtype/λ] = −1.5. For Y1173, wildtype: log10[λwildtype/λ] = 0.0; L834R: log10[λwildtype/λ] = −1.0; del L723–P729 ins S: log10[λwildtype/λ] = +0.5. The structural basis for the context- specific kinetics of the C-terminal tyrosine substrates is provided by our computational docking calculations:12 substrate peptides derived from tyrosine sites of the ErbB1 C-terminal tail— Y1068 (VPEYINQ) and Y1173 (NAEYLRV)—bound to the wildtype and the L834R mutant ErbB1 kinase revealed how the structure of the bound peptide–protein complex is altered at the catalytic site due to the arginine substitution of leucine in L834R.
In order to translate differences in substrate specificity into tangible differences in the downstream response (ERK and Akt activation), we introduced a branched signaling model for ErbB1, see Fig. 1b, that features two parallel phosphorylation pathways corresponding to Y1068 and Y1173.12,24 Based on the results of ref. 23 we developed a molecularly resolved systems model in which phosphorylated Y1068 binds only to Gab-1 and Grb2 and not Shc, and phosphorylated Y1173 binds only to Shc and not to Gab-1 and Grb2 as depicted in Fig. 1b. We were then able to reparameterize the model based on the identity-specific phospho-tyrosine kinetics of Y1068 and Y1173 for wildtype and mutant (L834R and del L723–P729 ins S) ErbB1 based on the relative catalytic turnover (λ) rates.12,24 We were also able to extend this model for ErbB1 kinase inhibition upon treatment with small molecule inhibitor erlotinib in wildtype and mutant, again based on experimentally available inhibitor/ATP affinity data.20
Using the different parameter values corresponding to wildtype, L834R, and del L723–P729 ins S mutant systems, we ran network simulations for different levels of EGF stimulation and ErbB1 expression levels. The parameters explored were: normal receptor expression (initial concentration of ErbB1 [ErbB1] = 100 nM or 30 000 receptors per cell), and over-expression of ErbB1 (initial [ErbB1] = 1000 nM or 300 000 receptors per cell), no EGF stimulation, and 8 nM (50 ng ml−1) EGF stimulation. Based on the altered λ (KM and kcat values derived form Mulloy et al.24), the L834R had a stronger preference for both Y1068 and Y1173 phosphorylation compared to the wildtype receptor, while the del L723–P729 ins S mutant showed increased Y1068 and decreased Y1173 phosphorylation. To gauge the downstream effects of differential signaling through Y1068 and Y1173 phosphorylation sites of ErbB1, we calculated the levels of ERK-(p) (phosphorylated ERK) and Akt-(p) (phosphorylated Akt) in our system simulations in response to changes in the phosphortyrosine kinetics (λ values) of Y1068 and Y1173. Table 1a summarizes our simulation results where each entry corresponds to the peak level of phosphorylation over a simulated time of 900 s. The effect of altered affinities of the Y1068 and Y1173 sites to the catalytic domain of ErbB1 is that the L834R under normal ErbB1 expression exhibits differential downstream response, i.e., a pronounced decrease in ERK activation (~5-fold) and relatively smaller decrease Akt activation (~15% decrease). The del L723–P729 ins S mutant, however, shows sustained ERK as well as Akt activation relative to wildtype. For ErbB1 over-expressed cells, both ERK and Akt activation characteristics show relative insensitivity to ErbB1 as a result of signal saturation. The trends in Table 1a also show that the mutants can continue to signal even in the absence of the growth factor. In addition, the mutant signaling can be different due to changes in the ATP affinity. However, neither of these factors introduce any differential characteristics (in terms of preferring Y1068 to Y1173); i.e., each factor impacts the overall activation levels of ERK and Akt uniformly. Our calculated responses agree with the qualitative experimental observations of Sordella et al.19 and Tracy et al.,25 i.e., the preferential activation of Akt in L834R and del L723–P729 ins S mutant cell lines predicted from our simulations is consistent with the reported experiments. 19,24–26
We also examined the sensitivity of downstream signaling molecules ERK-(p) and Akt-(p) to inhibition by a range of erlotinib (ErbB1 inhibitor) concentrations. Specifically, we compute the EC50, which is the inhibitor concentration at which 50% of the phosphorylation activity is suppressed in the cellular context, see Table 1a. Under normal expression levels of ErbB1, we predict nearly a 7-fold decrease in EC50 for ERK-(p) inhibition for L834R (EC50 = 100 nM) compared to wildtype (EC50 = 700 nM) with and without EGF ligand present (and a 4-fold decrease for over-expressed ErbB1). With respect to Akt activation, we report a 4-fold decrease in EC50 for L834R compared to wildtype (300 nM vs. 1200 nM) with and without ligand present15 (and 10-fold decrease for overexpressed ErbB1). These results are consistent with several experimental results that have reported the effect of the closely related inhibitor gefitinib on normal and non-small cell lung cancer cells,19,24–26 providing a mechanistic basis for the inhibitor efficacy in mutant cell lines. This agreement supports the notion that the branched signaling in our model can indeed represent a possible mechanism for preferential down-stream activation. In summary, we find that the mutant cell line L834R is more susceptible to inhibition through curbing downstream (ERK and Akt) activation. Considering that the absolute Akt-(p) levels are 5-fold higher than those for ERK-(p) in the wildtype and 20-fold higher in the mutant, see Table 1a, the gain in efficacy with respect to Akt inhibition may be a crucial difference between the wildtype and the mutant cell lines. We discuss this aspect below.
Clinical implications from the multiscale modeling of ErbB receptor signaling
One of the conditions for cellular homeostasis can be viewed as a balance between pro-survival signals and death-inducing signals, both being triggered and balanced by a variety of interacting intracellular pathways. Recently, using a simplified model for the effect of Akt activation on cell response,§ we showed that preferential Akt activation is conducive for the cell to rely on (be “addicted to”27,28) the most efficient Akt-(p) generating pathway for generation of pro-survival signals while requiring the generation of death-inducing signals from other pathways. Our simplified model illustrates a mechanism by which inhibiting the dominant source of the pro-survival signals shifts the cellular homeostasis to a cellular state devoid of pro-survival signals, thereby providing grounds for a remarkable inhibitor sensitivity.15
We hypothesized that the mechanisms that lead to inhibitor hypersensitivity (as well as resistance) attack points of network hypersensitivity and fragility. Since preferential Akt activation is a hallmark of the hyper-sensitive mutants and the efficacy of the inhibitors, we determine through a global sensitivity analysis14,15 the combinations of model parameter perturbations that drive enhanced production of Akt-(p) and ERK-(p). Table 1b reports the components (rate constants and initial concentrations) of the top 3 principal eigenvectors derived from the parameter variations in our network in Fig. 1b. Perturbing the components of these principal eigenvectors (i.e. the entries in Table 1(b)) produces maximum changes to the Akt-(p) and ERK-(p) levels. Interestingly, there is a striking correlation between the components of Table 1(b) resulting from our model sensitivity analysis and patterns of oncogenic mutations and mechanisms of drug resistance found in clinical studies. High frequency of mutations of PI3K, Ras (a GTPase named as an acronym for rat sarcoma), Gab-1, MEK (mitogen activated protein kinase kinase), Raf (a serine/threonine kinase which activates MEK) have all been observed in several human cancers.29–33 Moreover, it has been established in screened breast and colorectal cancer patients that the GAB-1, MEK, and Ras mutations are nonrandom and likely arise from selective evolutionary pressures that give the cancer cells a survival advantage.33 With reference to the hyper-sensitive ErbB1 mutants found in non-small cell lung cancer patients, the perturbation of the phospho-tyrosine kinetics of Y1068 and Y1173 through mutations (L834R and del L723–P729 ins S) is directly responsible for the differential signaling leading to preferential Akt activation, as we have shown in Table 1a. Other sensitive quantities reported in Table 1b also have direct relevance to hyper-sensitive signaling and drug resistance in the L834R and del L723–P729 ins S mutant cell lines. The inhibitor concentration is the most obvious and is generally depleted in the cells via multi-drug resistance mechanisms involving drug efflux pumps.34 The restoration of signaling via reduction of Ki of the inhibitor and the simultaneous enhancement of KM associated with ATP binding has also been reported through a double mutation of L834R/T766M.35,36 This double mutant increases receptor phosphorylation (Y1068 and Y1173) kinetics 100-fold24 while simultaneously decreasing inhibitor affinity.36 Another drug resistance mechanism related to Y1068 kinetics (i.e. by circumventing Y1068 involvement and restoring downstream signaling through an alternative branch) has been identified: in the presence of ErbB3, a branch of signaling analogous to that through Y1068 becomes available through ErbB1–3 heterodimerization directly resulting in PI3K recruitment on ErbB3 and subsequent Akt activation. Indeed phosphorylation of ErbB3 by the Met receptor kinase, (Met receptor tyrosine kinase is a high-affinity transmembrane receptor for the hepatocyte growth factor), due to overexpression of the Met receptor leads to drug resistance to gefitinib/erlotinib treatment (inhibition) of ErbB1.37 A drug resistance mechanism involving the change in expression of the phosphatase associated with Akt has also been identified to restore Akt-(p) levels upon inhibitor treatment.27,38 Roles for the phosphatase for ERK and the multimeric complex [ErbB1•Shc•Grb2•SOS•RasGTP] (which are our remaining predictions from Table 1b) in enhanced signaling and/or drug resistance have not yet been established experimentally.
Conclusion and future outlook
We have employed a multiscale modeling platform to study ErbB1-mediated signal transduction, to help rationalize and integrate the collective results emerging from structural, biochemical, cell biological, and clinical studies. At the molecular level, our results suggest that the clinically identified mutations of ErbB1 kinase induce network fragility in the stabilizing interactions of the inactive kinase conformation, thereby providing a persistent stimulus for kinase activation even in the absence of any growth factor. At a cellular level, parameter perturbations driving network hypersensitivity through the enhancement of phosphorylated ERK and Akt levels show a striking correlation with observed mutations of specific proteins in oncogenic cell lines as well as the observed mechanisms of drug resistance to ErbB1 inhibition. Therefore, subject to the well-appreciated limitations of computational modeling, i.e., uncertainty in network topology and parameters, neglect of molecular cross-talk, autocrine loops,39 we suggest that cascading mechanisms of network hypersensitivity/fragility at multiple scales enable molecular-level perturbations (clinical mutations) to induce oncogenic transformations and mechanisms of drug resistance. Moreover, our results describe a possible mechanism of signal branching leading to preferential activation of down-stream molecules (Akt) in the ErbB1 activating mutants. This preferential activation of a survival factor makes the cell-lines harboring the mutant receptors to be conducive to oncogenic addiction, i.e. reliance on the L834R or del L723–P729 ins S ErbB1- mediated generation Akt-(p) for survival signals. The survival pathway addiction also results in a remarkable sensitivity to ErbB1 kinase inhibition.
A more complete model description, which we are currently pursuing, will not only require resolving the differential characteristics of all of the tyrosine phosphorylation sites in ErbB1 (Y992, Y1086, Y1114, Y1148 Gab1:Y627, Shc:Y317, phosphoinositide-specific phospholipase Cγ or PLCγ-1:Y771) and their associated substrate recognition properties in ErbB1 (i.e., exploring the interactions of Gab1, Shc, and PLCγ-1 binding to phospho-tyrosines of ErbB1 (see recent work by Kholodenko et al.,40), but also the extension to other Erb family members in the context of homo- and heterodimers.41 These extensions are especially crucial because transactivation of ErbB1 occurring through ligand-induced receptor heterodimerization 42 combined with a potential for differential signaling adds a palette of finer control elements in the ErbB-family signaling network. This view is further bolstered by the over-expression of ErbB3 in drug resistance to ErbB1 targeting (as discussed above) and mutations and over-expression of ErbB2 in different cancers.43 Moreover, the internalization mechanism of receptors via endocytosis, which we have not considered in our model, features in crucial regulatory roles:44 attenuation of endocytosis 45 leading to impaired deactivation of receptor tyrosine kinases is linked to hyper-proliferative conditions such as cancer.46–48 Hendriks et al.48 proposed a mechanism of differential signaling and preferential activation of Akt in ErbB1 mutants based on reduced internalization rates of ErbB1 mutants (relative to wildtype). Hence we are developing the orchestrated vesicular assembly (OVA) model, which will serve as a multiscale, spatially-resolved model for describing the ErbB1 receptor internalization through endocytotic vesicle formation. In the OVA model, the membrane dynamics are represented by the time-dependent Ginzburg Landau formalism, and the dynamics of accessory proteins, including the curvature-inducing protein, epsin, are represented by the kinetic Monte Carlo formalism. The integration of the two formalisms (Fig. 1c) is described in ref. 12 and 17. Protein–protein interactions and protein– membrane interactions are considered using coarse-grained potentials, and a phenomenological model for the effect of a clathrin coat is employed and the treatment of the membrane to allow for extreme deformations is through the surface evolution approach (see inset in Fig. 1c) (unpublished results). Through the OVA model simulations, we propose to address the biological question of how the cell stages the endocytosis nucleation event to specific sites on the membrane, i.e. to the site of a phosphorylated receptor protein; in particular, how does the network of accessory proteins identify the site of nucleation and synchronize with the receptor phosphorylation. A comprehensive multiscale model for ErbB activation and internalization can then be achieved by integrating the OVA model, Fig. 1c, with the network model we have outlined in Fig. 1b.
At the molecular level, considering that there is an excellent correlation between the stabilizing network of interactions and the clinically identified activating mutations in ErbB1, our structural studies on kinase activation are well poised to forecast the mutation landscape associated with other ErbB members. Indeed based on our simulations of ErbB2 and ErbB4, we have identified similar networks of stabilizing residues and are already able to predict activating mutations (data not presented) in these receptors that have not yet been reported clinically, which together with the extensions proposed above can be valuable for evaluating the likely effect of mutations on ErbB2 inhibition efficacies in cancer, and ErbB4 inhibition in cardiac development and schizophrenia.11
Acknowledgments
We thank M. A. Lemmon, B. N. Kholodenko, Yingting Liu, Neeraj Agrawal, and Shannon Telesco for many insightful discussions. We also acknowledge funding from the National Science Foundation through grant CBET- 0730955, and funding from the Whitaker Foundation and the Greater Philadelphia Bioinformatics Alliance. High-performance computing resources were available in part from the National Partnerships for Advanced Computational Infrastructure through grant MRAC MCB060006.
Biographies

Andrew J. Shih (top) is a fifth year PhD candidate in the Department of Bioengineering at the University of Pennsylvania. He works on understanding the structural biology of ErbB receptor kinases through computational simulation techniques. Jeremy Purvis is a fourth year PhD student in the Genomics and Computational Biology Graduate Group at the University of Pennsylvania. He is currently working on understanding the behavior of growth factor signaling networks as they relate to cancer and developing a signaling model of the human platelet using a combination of high-throughput experiments and computational modeling.

Ravi Radhakrishnan (Assistant Professor of Bioengineering) obtained his PhD in chemical physics from the Department of Chemical Engineering at Cornell University in 2001. After two postdoctoral assignments in chemical physics and biophysics at MIT and at the New York University/Howard Hughes Medical Institute, he joined the faculty of the University of Pennsylvania in 2004. He has a background in chemical physics and molecular biophysics, and is involved in developing multiscale modeling and simulation protocols to predict single molecule properties as well as signal transduction mechanisms for mammalian systems.
Footnotes
Our applications of multiscale algorithms to ErbB signaling are enabled by high-performance computing infrastructure. As we transition from teraflops or 1014 flops (flops: floating point operations per second) computing to petaflops (1015 flops) computing in the near future, we expect that extension of our molecular and systems modeling to complete cellular pathways will become tractable.
There are 2 main types of lung cancer and they are treated differently: small cell lung cancer and non-small cell lung cancer. About 85% to 90% of all lung cancers are of the nonsmall cell type with three sub-types. The cells in these sub-types differ in size, shape, and chemical make-up: (1) squamous cell carcinoma linked to smoking, (2) adenocarcinoma usually found in the outer part of the lung, and (3) large-cell (undifferentiated) carcinoma exhibiting rapid growth and spreading.
The phenomenological model can be summarized by: Akt ⇆ Akt-(p) → S, with equilibrium constant K1 = [Akt-(p)]/[Akt] and rate constant k1; D-ind → D, with rate constant k2; Akt-(p) + D-ind ⇆ Akt-(p)•D-ind with equilibrium constant K2 = [Akt-(p)•D-ind]/[Akt-(p)][D-ind] and can be solved analytically. In this model, the cellular states S and D denote survival and death and D-ind denotes a death-inducing factor. The three reactions capture the effect of Akt on cell response: i.e., Akt activating survival pathways and simultaneously inhibiting death-inducing pathways.
References
- 1.Yarden Y, Sliwkowski MX. Untangling the ErbB Signaling Network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 2.Citri A, Yarden Y. EGF-ERBB signaling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 3.Schoeberl B, Jonsson CE, Gilles ED, Muller G. Computational modeling of the dynamics of MAPKinase cascade activated by surface and internalized receptors. Nat Biotechnol. 2002;20:370–375. doi: 10.1038/nbt0402-370. [DOI] [PubMed] [Google Scholar]
- 4.Hatakeyama M, Kimura S, Yumoto N, Ichikawa M, Kawasaki T, Naka T, Konagaya A. A computational model on the modulation of MAPK and Akt pathways in Heregulin-induced ERB signaling. Biochem J. 2003;373:451–463. doi: 10.1042/BJ20021824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kholodenko BN, Demin OV, Moehren G, Hoek JB. Quantification of short term signaling by the epidermal growth factor receptor. J Biol Chem. 1999;274:30169–30181. doi: 10.1074/jbc.274.42.30169. [DOI] [PubMed] [Google Scholar]
- 6.Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 8.Kloth MT, Laughlin KK, Biscardi JS, Boerner JL, Parsons SJ, Silva CM. STAT5b, a Mediator of Synergism between c-Src and the Epidermal Growth Factor Receptor. J Biol Chem. 2003;278:1671–1679. doi: 10.1074/jbc.M207289200. [DOI] [PubMed] [Google Scholar]
- 9.Vieira AV, Lamaze C, Schmid SL. Control of EGF Receptor signaling by clathrim-mediated endocytosis. Science. 1996;274:2066–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 10.Mendelsohn J, Baselga J. The EGF receptor family as targets for cancer therapy. Oncogene. 2000;19:6550–6565. doi: 10.1038/sj.onc.1204082. [DOI] [PubMed] [Google Scholar]
- 11.Linggi B, Carpenter G. ErbB receptors: new insights on mechanisms and biology. Trends Cell Biol. 2006;16:649–656. doi: 10.1016/j.tcb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Purvis J, Shih A, Weinstein J, Agrawal N, Radhakrishnan R. A Multiscale Computational Approach to Dissect Early Events in the Erb Family Receptor, Mediated Activation Differential Signaling, and Relevance to Oncogenic Transformations. Ann Biomed Eng. 2007;35:1012–1025. doi: 10.1007/s10439-006-9251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shih A, Choi S-H, Lemmon MA, Radhakrishnan R. Network of Stabilizing Hydrogen Bonds is Integral to the Dimer-Mediated Allosteric Activation Mechanism in the ErbB1 and ErbB4 Kinase. 2008 submitted. [Google Scholar]
- 14.Purvis J, Liu Y, Ilango V, Radhakrishnan R. Proc Foundations in Systems Biology II. IRB Verlag; Stuttgart: 2007. Efficacy of tyrosine kinase inhibitors in the mutants of the epidermal growth factor receptor: A multiscale molecular/systems model for phosphorylation and inhibition; pp. 289–294. [Google Scholar]
- 15.Purvis J, Ilango V, Radhakrishnan R. Role of Network Branching in Eliciting Differential Short-Term Signaling Responses in the Hyper-Sensitive Epidermal Growth Factor Receptor Mutants Implicated in Lung Cancer. Biotechnol Prog. 2008;24:540–553. doi: 10.1021/bp070405o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown KS, Hill CC, Calero GA, Myers CR, Lee KH, Sethna JP, Cerione RA. The statistical mechanics of complex signaling networks: nerve growth factor signaling. Phys Biol. 2004;1:184–195. doi: 10.1088/1478-3967/1/3/006. [DOI] [PubMed] [Google Scholar]
- 17.Weinstein J, Radhakrishnan R. A coarse-grained methodology for simulating interfacial dynamics in complex fluids: application to protein mediated membrane processes. Mol Phys. 2006;104:3653–3666. doi: 10.1080/00268970600997580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agrawal N, Weinstein J, Radhakrishnan R. Landscape of membrane-phase behavior under the influence of curvature- inducing proteins. 2008 submitted. [Google Scholar]
- 19.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 20.Carey KD, Garton AJ, Romero MS, Kahler J, Thomson S, Ross S, Park F, Haley JD, Gibson N, Sliwkowski MX. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 2006;66:8163–8171. doi: 10.1158/0008-5472.CAN-06-0453. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An Allosteric Mechanism for Activation of the Kinase Domain of Epidermal Growth Factor Receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 22.Qiu C, Tarrant MK, Choi SH, Sathyamurthy A, Bose R, Banjade S, Pal A, Bornmann WG, Lemmon MA, Cole PA, Leahy DJ. Mechanism of activation and inhibition of the HER4/ErbB4 kinase. Structure. 2008;16:460–467. doi: 10.1016/j.str.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol. 2005:1. doi: 10.1038/msb4100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulloy R, Ferrand A, Kim Y, Sordella R, Bell DW, Haber DA, Anderson KS, Settleman J. Epidermal growth factor receptor mutants from human lung cancers exhibit enhanced catalytic activity and increased sensitivity to gefitinib. Cancer Res. 2007;67:2325–2330. doi: 10.1158/0008-5472.CAN-06-4293. [DOI] [PubMed] [Google Scholar]
- 25.Tracy S, Mukohara T, Hansen M, Meyerson M, Johnson BE, Janne PA. Gefitinib induces apoptosis in the EGFRL858R non-small-cell lung cancer cell line H3255. Cancer Res. 2004;64:7241–7244. doi: 10.1158/0008-5472.CAN-04-1905. [DOI] [PubMed] [Google Scholar]
- 26.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 27.Sharma SV, Gajowniczek P, Way IP, Lee DY, Jiang J, Yuza Y, Classon M, Haber DA, Settleman J. A common signaling cascade may underlie “addiction” to the Src, BCR-ABL, and EGF receptor oncogenes. Cancer Cell. 2006;10:425–435. doi: 10.1016/j.ccr.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinstein IB. Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 29.Duesbery N, Vande Woude G. BRAF and MEK mutations make a late entrance. Sci STKE. 2006:pe15. doi: 10.1126/stke.3282006pe15. [DOI] [PubMed] [Google Scholar]
- 30.Bentires-Alj M, Gil SG, Chan R, Wang ZC, Wang Y, Imanaka N, Harris LN, Richardson A, Neel BG, Gu H. A role for the scaffolding adapter GAB2 in breast cancer. Nat Med. 2006;12:114–121. doi: 10.1038/nm1341. [DOI] [PubMed] [Google Scholar]
- 31.Mascaux C, Iannino N, Martin B, Paesmans M, Berghmans T, Dusart M, Haller A, Lothaire P, Meert AP, Noel S, Lafitte JJ, Sculier JP. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer. 2005;92:131–139. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JKV, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High Frequency of Mutations of the PIK3CA Gene in Human Cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 33.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JKV, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The Consensus Coding Sequences of Human Breast and Colorectal Cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 34.Venter H, Shilling RA, Velamakanni S, Balakrishnan L, Veen HWv. An ABC transporter with a secondary-active multidrug translocator domain. Nature. 2003;426:866–870. doi: 10.1038/nature02173. [DOI] [PubMed] [Google Scholar]
- 35.Blencke S, Ullrich A, Daub H. Mutation of threonine 766 in the epidermal growth factor receptor reveals a hotspot for resistance formation against selective tyrosine kinase inhibitors. J Biol Chem. 2003;278:15435–15440. doi: 10.1074/jbc.M211158200. [DOI] [PubMed] [Google Scholar]
- 36.Liu B, Bernard B, Wu JH. Impact of EGFR point mutations on the sensitivity to gefitinib: Insights from comparative structural analyses and molecular dynamics simulations. Proteins: Struct, Funct, Bioinf. 2006;65:331–346. doi: 10.1002/prot.21111. [DOI] [PubMed] [Google Scholar]
- 37.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Janne PA. MET Amplification Leads to Gefitinib Resistance in Lung Cancer by Activating ERBB3 Signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 38.Sergina NV, Rausch M, Wang DH, Blair J, Hann B, Shokat KM, Moasser MM. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445:437–441. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersen SL, Wang L, Yalcin-Chin A, Li L, Peyton M, Minna J, Harran P, Wang X. Autocrine TNFalpha signaling renders human cancer cells susceptible to Smac-mimetic-induced apoptosis. Cancer Cell. 2007;12:445–456. doi: 10.1016/j.ccr.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suenaga A, Hatakeyama M, Kiyatkin AB, Radhakrishnan R, Taiji M, Kholodenko BN. Tyr-317 Phosphorylation Reduces Shc Binding Affinity for Phosphotyrosyl Residues of Epidermal Growth Factor Receptor. 2008 doi: 10.1016/j.bpj.2008.11.018. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Birtwistle MR, Hatakeyama M, Yumoto N, Ogunnaike BA, Hoek JB, Kholodenko BN. Ligand-dependent responses of the ErbB signaling network: experimental and modeling analyses. Mol Syst Biol. 2007;3:144. doi: 10.1038/msb4100188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waterman H, Yarden Y. Molecular mechanisms underlying endocytosis and sorting of Erb receptor tyrosine kinases. FEBS Lett. 2001;490:142–152. doi: 10.1016/s0014-5793(01)02117-2. [DOI] [PubMed] [Google Scholar]
- 43.Lee JW, Soung YH, Seo SH, Kim SY, Park CH, Wang YP, Park K, Nam SW, Park WS, Kim SH, Lee JY, Yoo NJ, Lee SH. Somatic mutations of ERBB2 kinase domain in gastric, colorectal, and breast carcinomas. Clin Cancer Res. 2006;12:57–61. doi: 10.1158/1078-0432.CCR-05-0976. [DOI] [PubMed] [Google Scholar]
- 44.Sorkin A, Von Zastrow M. Signal transduction and endocytosis: close encounters of many kinds. Nat Rev Mol Cell Biol. 2002;3:600–614. doi: 10.1038/nrm883. [DOI] [PubMed] [Google Scholar]
- 45.Bache KG, Slagsvold T, Stenmark H. Defective downregulation of receptor tyrosine kinases in cancer. EMBO J. 2004;23:2707–2712. doi: 10.1038/sj.emboj.7600292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polo S, Pece S, Di Fiore PP. Endocytosis and cancer. Curr Opin Cell Biol. 2004;16:156–161. doi: 10.1016/j.ceb.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 47.Floyd S, Camilli PD. Endocytosis proteins and cancer: a potential link. Trends Cell Biol. 1998;8:299–301. doi: 10.1016/s0962-8924(98)01316-6. [DOI] [PubMed] [Google Scholar]
- 48.Hendriks BS, Griffiths GJ, Benson R, Kenyon D, Lazzara M, Swinton J, Beck S, Hickinson M, Beusmans JM, Lauffenburger D, De Graaf D. Decreased internalisation of ErbB1 mutants in lung cancer is linked with a mechanism conferring sensitivity to gefitinib. IEE Proc Syst Biol. 2006;153:457–466. doi: 10.1049/ip-syb:20050108. [DOI] [PubMed] [Google Scholar]