Abstract
Physical and genetic mapping data have become as important to network biology as they once were to the Human Genome Project. Integrating physical and genetic networks currently faces several challenges: increasing the coverage of each type of network; establishing methods to assemble individual interaction measurements into contiguous pathway models; and annotating these pathways with detailed functional information. A particular challenge involves reconciling the wide variety of interaction types that are currently available. For this purpose, recent studies have sought to classify genetic and physical interactions along several complementary dimensions, such as ordered versus unordered, alleviating versus aggravating, and first versus second degree.
The successful completion of the Human Genome Project depended crucially on the integration of genetic and physical maps1. Genetic maps, also known as gene linkage maps2, were constructed by measuring the meiotic recombination frequencies between different pairs of genetic markers. On the basis of many pairwise genetic distances, markers could be placed on a number line with short distances corresponding to low recombination frequencies. Conversely, physical maps were constructed by identifying the position of markers along the chromosome. Physical distances between markers were determined by techniques such as radiation hybrid mapping3,4, fluorescence in situ hybridization (FISH)5 or, ultimately, automated DNA sequencing6. Genome assembly involved a multi-step procedure in which DNA fragments were cloned, sequenced and, on the basis of the markers they were found to contain, ordered relative to each other and to the genetic map7,8. Obtaining full coverage of the genome involved generating enough physical and genetic data so that the two maps could be reconciled. Following assembly, the physical and genetic maps were annotated and continuously updated with detailed information about functional elements9. For the physical sequence map, the primary annotation task was the identification of genes; for the genetic map, it was linking genes or their surrogate genetic markers with diseases of interest.
Remarkably, the mapping cellular regulatory and signalling networks is now proceeding in much the same way10,11 (FIG. 1). As for genomics, large-scale genetic and physical interaction mapping projects release enormous amounts of raw data that must be filtered and interpreted biologically (BOX 1). Integration of these two types of maps is important because they provide views that are highly complementary with regard to cellular structure and function: physical interactions dictate the architecture of the cell in terms of how direct associations between molecules constitute protein complexes, signal transduction pathways and other cellular machinery. Genetic interactions define functional relationships between genes, giving insight into how this physical architecture translates into phenotype. A complete picture of the cell must necessarily integrate both aspects.
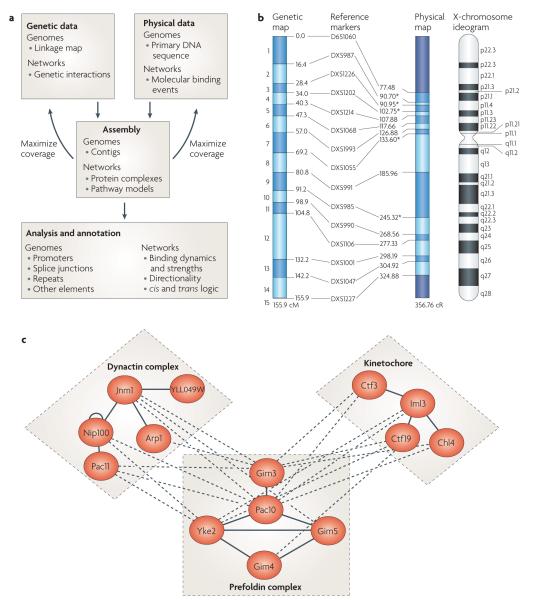
Figure 1. Genetic and physical mapping for networks and genomes.
a ∣ The assembly and analysis of genetic and physical interaction networks runs parallel to the procedures that were previously developed for assembly and analysis of DNA sequences. b ∣ An integrated map of human chromosome X. Markers are listed in the centre column, with genetic distances given on the left in centimorgans (cM) and physical distances given on the right in centirays (cR). c ∣ An integrated map of genetic and physical interactions for the yeast cytoskeleton. Solid lines represent physical protein–protein interactions, and dashed lines represent synthetic-lethal genetic interactions. The physical network defines three complexes: prefoldin, dynactin and the kinetochore, whereas the genetic network defines functional dependencies between prefoldin and dynactin or the kinetochore, respectively. Part b reproduced with permission from the Cancer Genome Anatomy Project © (2007) National Cancer Institute (USA). Part c modified with permission from Nature Biotechnology REF. 37 © (2005) Macmillan Publishers Ltd.
Box 1. Technologies for measuring physical and genetic interactions.
At least two types of physical interaction are currently measurable at high throughput: protein–protein and protein–DNA. Networks of protein–protein interactions are being built using yeast two-hybrid (Y2H) technology 11,80-86 or tandem affinity purification coupled with mass spectrometry (TAP–MS)49,87,88. Similarly, networks of protein–DNA interactions leverage the techniques of chromatin immunoprecipitation coupled with DNA microchips (ChIP–chip)51,89,90 or sequencing (ChIP–PET)91,92, DNA adenine methylase identification (DamID)93, or yeast one-hybrid assays94,95. Physical interactions can also be measured in vitro using DNA or protein arrays, which have been used to identify transcription factor binding sites96 and the substrates of yeast kinases36.
In contrast to physical interactions, genetic interactions represent functional relationships between genes, in which the phenotypic effect of one gene is modified by another29,42. Genetic interactions are identified by comparing the effect of mutating each gene individually to the effect of the double mutant. For example, ‘synthetic sickness’ (or in the extreme ‘synthetic lethality’) is a genetic interaction in which the measured phenotype is growth, and mutating both genes results in slower growth than expected from either mutation alone. In yeast, large networks of genetic interactions are being measured through the techniques of synthetic genetic arrays (SGA) and diploid-based synthetic-lethality analysis on microarrays (dSLAM)97. All of these methods allow the phenotypic consequences of double-mutant combinations to be assayed in high-throughput formats15. In worms and higher eukaryotes, genetic interactions are explored through the technique of combinatorial RNAi98 and other RNAi-based screening approaches99,100. Other types of genetic interactions are non-symmetrical and establish a ‘cause-and-effect’ ordering (BOX 2). Such ordered networks are being constructed using high-throughput growth assays41 or complex read-outs such as the ability of yeast to invade agar media40.
Increasingly, cause-and-effect genetic orderings are also being established using gene expression as the primary phenotype. In yeast, several groups52,56-59 have gathered expression profiles for panels of gene-deletion strains, in which each deletion mutant (the cause) is linked to downstream genes, the expression levels of which are affected by the deletion (the effects). This interaction differs from the classical genetic orderings that are described above in that a mutation in only one gene is involved, not two. However, as the phenotype is directly related to a second gene (that is, its expression level), a causal link from mutation to phenotype defines an ordered interaction between two genes. Similar cause-and-effect orderings are provided by expression quantitative trait loci (eQTL) analysis, which has been applied not only in yeast101 but also in higher organisms102-105. In eQTL analysis, large numbers of individuals are genotyped across a panel of polymorphic markers and simultaneously phenotyped using microarray expression profiling. Statistical methods are then used to determine linkages between markers and gene expression levels; that is, particular markers for which the pattern of genotypes over all individuals is correlated with the pattern of expression of a particular gene.
The complementarity between physical and genetic interactions has been strikingly demonstrated in yeast, for which less than 1% of synthetic-lethal genetic interactions can also be observed physically12. This complementarity has also been exploited numerous times in classical genetics and biochemistry, in which a great many pathways have been understood only through integration of both physical and genetic interactions (the LIN-12–Notch signalling pathway13 and the actin cytoskeleton14 are excellent examples).
More recently, the advent of whole-genome technologies has expanded the task of data integration dramatically. Many new types of physical and genetic measurements have appeared (BOX 1), and the use of reverse-genetic screening has increased the total number of recorded interactions from a few hundred to a few hundred thousand. This growth in data has made it possible to consider genetic or physical interactions not only on an individual level, but as building blocks of larger networks of gene and protein interactions. Unlike for individual interactions, interaction networks cannot be integrated by eye. Rather, automated methods for data collection require automated methods for bioinformatic analysis.
At present, numerous bioinformatic approaches are under development for physical and genetic network quality assessment, integration, assembly and annotation — steps that were also central to completion of the Human Genome Project (FIG. 1). In the remainder of this Review, we summarize the progress that is being made at each of these steps, as applied to interaction mapping; details on obtaining the individual interaction measurements are reviewed elsewhere11,15,16. The modes by which genetic and physical interactions complement one another have not been fully elucidated; however, a growing body of work has begun to reveal a complex but concrete set of principles governing their relationships. These studies show how interactions of different types can be combined to assemble a more comprehensive picture of biological systems.
Interaction quality assessment
An important first step in sequencing the human genome was to assign quality scores to each DNA base pair that was identified from the fluorescent traces produced by automated sequencing machines17. Not surprisingly, data quality is also a foremost concern in physical and genetic interaction measurements, and must be addressed before any biological interpretation can take place.
Dealing with false positives
The first quality-control issue concerns false-positive measurements, and the method of choice for dealing with false positives is data integration18-25. Although all large-scale studies are subject to noise, the rationale for data integration is that observations of true interactions will reinforce or complement one another when combined across different studies and/or experimental techniques. For example, the independent observation of a protein–protein interaction by both yeast two-hybrid (Y2H) and tandem affinity purification coupled with mass spectrometry (TAP–MS) methods, or by two independent TAP–MS studies, renders this interaction more likely to be true26.
Along these lines, numerous types of evidence have been integrated to bolster confidence that two genes or proteins interact. For example, if the two genes have correlated expression profiles or similar patterns of occurrence across many genomes, these findings lend further support to the raw interaction measurement27,28. Of all of the different lines of evidence that can be integrated, combining physical and genetic data can be particularly useful, because the false negatives and false positives that influence these two types of interaction measurement are generally different in character. For instance, genetic interactions mapped by synthetic genetic array (SGA) analysis are influenced by artefacts caused by gene deletion12,29, such as mis-targeted deletion constructs and deletions that alter the metabolism of the drug that is used for mutant selection. Physical interactions mapped by Y2H or TAP–MS are influenced by artefacts that result from gene tagging, which can influence the functioning of the protein that is produced30,31.
Modern network analyses use regression or likelihood functions to learn which of the multiple types of evidence for genetic or protein interactions are most predictive of a known set of interactions, and to weight them accordingly. These methods rely heavily on a set of ‘gold-standard’ (highly accurate) interactions that are used to evaluate the predictive utility of different types of evidence26,32. The result is a statistical measure that quantifies the likelihood that any given pair of biomolecules interact with each other (for example, two proteins or a protein–DNA pair)18-23. Thus, interactions are not described in a binary manner (whereby an interaction can be only either absent or present), but quantitatively20. Strictly, these quantitative confidence scores describe the probability or reproducibility of the interaction, not the interaction strength. Nonetheless, there is some evidence that stronger interactions should be more reproducible, leading to higher scores33.
Dealing with false negatives
False negatives constitute a second data quality concern in interaction mapping projects; that is, the potential to miss interactions, leading to insufficient coverage of the network. Again, data integration is key, as interactions that are missing from one study can be detected using high-confidence interactions from another. Note that integrating more information can simultaneously reduce both the false-negative and the false-positive rates: as the number of ways of detecting an interaction increases, the higher the chance that a true interaction would be detected by several of these methods, and the lower the chance that a false interaction would be.
Accounting for the dynamic nature of networks
In terms of coverage and accuracy, an important difference between genomes and interaction networks is the dynamics and plasticity of the latter: whereas the genome is largely static, interaction networks are context-dependent. Interactions might be active only in certain cell types, during particular developmental stages or under specific external conditions. This variability complicates the concept of coverage because, ideally, all possible conditions and cell types should be tested, and certain in vitro measurement techniques such as Y2H do not provide information about condition specificity. A possible solution to this problem is to develop algorithms to predict interactions on the basis of all the available data from all conditions. If those algorithms also predict the conditions under which the interactions are active, such analyses could streamline the experimental verification. For instance, Gunsalus et al. mapped mRNA expression data and early embyrogenesis RNAi phenotypic profiles onto a static Caenorhabditis elegans protein interaction map in order to extract subnetworks that function at specific stages of development34. In this case, condition-specific data aided the interpretation of a static interaction map and also predicted the conditions in which the subnetworks were most likely to be active. In terms of biological verification, context-independent interaction measurements ask the question, can they interact? By contrast, condition-specific and in vivo measurements ask the question, do they interact? This is an important distinction when navigating the burgeoning sea of available interaction data.
Assembly I: categories of interactions
In the Human Genome Project, ‘genome assembly’ was the process of putting individual ~600-bp sequence reads together to form longer sequences called contigs. In the context of molecular interactions, we refer to assembly as the integration of individual interactions into larger network structures that represent pathways, protein complexes and other components of the global cellular machinery. Given the numerous and seemingly disparate types of physical and genetic interaction measurement (BOX 1), a central question regards how all of these different types precisely interrelate. To address this question, recent studies have attempted to categorize interactions beyond the initial division into genetic and physical. Several new terminologies are emerging from these studies — some concrete and some ambiguous, some distinct and some overlapping. Which of these classifications will ultimately be most useful is still an open question; however, what is clear is that some form of interaction classification will be necessary if the various interaction measurements are to be assembled into unified models.
Ordered versus unordered measurements
An ordered interaction measurement is one that, on the basis of the underlying measurement technology, has a clear interpretation with regard to biological directionality (cause and effect). As detailed in TABLE 1, some measurement techniques imply that there is such an ordering, whereas others do not. For instance, a transcription factor–promoter binding interaction that is measured with chromatin immunoprecipitation (ChIP) is an ordered interaction, because the implication is that the transcription factor is regulating the downstream gene, not the reverse situation. A kinase–substrate interaction that is measured with a protein array is also ordered, in that it is clear which protein is the modifier and which protein is being modified. Genetic interactions also fall into ordered and unordered classes. An example of an ordered genetic interaction is epistatic masking, in which the phenotype of one mutant masks the phenotype of the other, indicating that the genes function in a regulatory hierarchy35 (BOX 2). A different instance of an ordered genetic interaction is an expression QTL (eQTL), which indicates that a polymorphism at one locus has an effect on gene expression at another (BOX 1). The litmus test for directed interaction measurements is this: does the interaction A–B have a different biological meaning from the interaction B–A? If the answer is a clear yes, then the interaction is ordered.
Table 1.
Classes of interactions and methods of identification
| Physical | Genetic | |
|---|---|---|
| Ordered | Protein–gene (ChIP–chip51,52, ChIP–PET91,92); protein–RNA (RIP–chip112); protein–protein (kinase–substrate arrays36, LUMIER113); protein–compound114; microRNA–target115 |
Epistatic orderings (fitness profiling39,41,42); knockdown expression profiles (RNAi, deletion mutants58,59); expression QTLs101,116 |
| Unordered | Protein–protein (TAP–MS49,87,88, Y2H80-83); gene–gene (co- regulon117); DNA–DNA (3C118, 5C119) |
Synthetic lethality (SGA12, dSLAM65,97, chemogenomic profiling120, combinatorial RNAi99,100) |
ChIP, chromatin immunoprecipitation; chip, microarray; coIP, co-immunoprecipitation; dSLAM, diploid-based synthetic lethality analysis on microarrays; PET, pair-end tag; RIP, ribonucleoprotein immunoprecipitation; SGA, synthetic genetic arrays; wt, wild type; Y2H, Yeast two-hybrid assay.
Box 2. Cause-and-effect ordering in genetic interactions.
Genetic interactions define logical, rather than physical, relationships between genes35. Formally, genetic interactions are determined and classified by comparing the observed phenotypes of the single mutants (Pa and Pb) to each other, to the double mutant (Pab) and to the wild type (Pwt), as shown in panel a. A genetic interaction is present if Pab deviates significantly from that which is expected on the basis of the combination of independent single mutants. Under a multiplicative model106,107, the expected phenotype is Eab = Pa * Pb. Given that a genetic interaction exists, it is classified as ‘aggravating’ if the observed phenotype of the double mutant is more severe than expected (Eab > Pab); if the opposite is true, then the interaction is considered ‘alleviating’ (Pab > Eab). ‘Synthetic lethality’ is an aggravating interaction in which Pab = 0 (no growth); alleviating interactions include suppression (Pab = Pa > Pb), masking (Pab = Pa < Pb), and co-equality (Pab = Pa, Pb). Finally, alleviating interactions can be defined as ordered or unordered depending on the relative severity of the two single-mutant phenotypes. If Pa > Pb or Pb > Pa, then the interaction is ordered. Otherwise, when Pa = Pb, no ordering of genes is implied. An ordered genetic interaction suggests that the two genes mediate different steps within a biochemical or signalling pathway35.
Panel b shows a schematic metabolic network with simple examples of alleviating, aggravating and non-interacting gene pairs (represented by green, red and black arcs, respectively). Typically, alleviating versus aggravating interactions are considered to function in the same versus related pathways, respectively. Non-interacting genes, although perhaps distantly related in the context of overall cell viability or a far-downstream metabolite (indicated by broken reaction lines), do not function in the same pathway or related pathways, and thus do not deviate from expected growth. Figure modified with permission from Nature Genetics REF. 108 © (2005) Macmillan Publishers Ltd.
Conversely, unordered interaction measurements do not imply any clear cause-and-effect directionality among the interacting genes or proteins. One example is a protein–protein interaction measured by Y2H or TAP–MS. Although these measurements distinguish which protein is the bait and which is the prey, these terms are technical and signify nothing with respect to a cause-and-effect biological ordering. Likewise, a synthetic-lethal interaction is an example of a genetic interaction that does not order the two interacting genes. Classifying an interaction as unordered does not mean that, in terms of cellular function, the interaction transmits information in both directions — only that the measurement technology is ambiguous with regard to the direction of biological signalling.
Transient versus stable interactions
Another means of classification relates to interaction dynamics — that is, distinguishing interactions that are transient in nature from those that form more stable linkages between proteins. Examples of transient interactions include kinase–substrate phosphorylation or condition-specific binding of a transcriptional regulator to DNA, whereas interactions among the cytoskeleton or the nuclear pore complex might be relatively more stable. It is likely that some interaction-measurement technologies are better at detecting transient rather than stable interactions, whereas the opposite might be true for other technologies: for instance, in vitro kinase assays are, by their nature, better suited to detect transient interactions, whereas coIP pull-downs using TAP–MS are better at detecting stable protein complexes. For many other measurement technologies, such as Y2H and chromatin immunoprecipitation combined with microarrays (ChIP–chip), it remains unclear whether the interactions that are identified are predominantly transient or stable. Various studies have suggested anecdotally that Y2H might be less able to detect transient interactions26,36, but to date no definitive evidence has been put forth. Another study reported marginal success in separating transient versus stable Y2H interactions by cross referencing with TAP–MS studies19.
Between- versus within-pathway interactions
Several analyses of genetic interactions12,37,38 have sought to distinguish those interactions that fall within the same protein complex or pathway from those that connect two related complexes or pathways. Genetic interactions that connect different pathways are generally thought to bridge genes with redundant or complementary functions, where the deletion of either gene is expected to abrogate the function of one, but not both, pathways. Genetic interactions within pathways are thought to be caused mainly by the additive effects of deletions within the pathway or the absence of an effect upon additional deletions within the same pathway. At least to some degree, aggravating genetic interactions (see BOX 2 and the following section) have been shown to occur between pathways, whereas alleviating genetic interactions occur within pathways39. The between versus within classification has also been applied to physical interactions19, in which interactions measured by both Y2H and TAP–MS were classified as within-complex interactions. In this work, Y2H interactions that were not supported by TAP–MS data were considered to consist mainly of between-complex interactions, although this assignment relies heavily on the TAP–MS data being comprehensive.
Aggravating versus alleviating interactions
Several groups29,40,41 have described genetic interactions as either ‘aggravating’, in which the double-mutant phenotype is more severe than expected given that of the single mutants, or ‘alleviating’, in which the double mutant is less severe than expected (BOX 2). The classical interpretation of an aggravating genetic interaction, such as synthetic lethality, is that it connects non-essential genes that function in parallel or redundant pathways (note the overlap with the between- versus within-pathway categories above). Conversely, alleviating interactions have been thought to indicate genes within the same pathway42. In these cases, the rationale is that a single gene mutation is sufficient to deactivate the pathway, whereas mutating a second gene from the same pathway does not further affect the phenotype and, in some cases, can even reverse it. Alleviating interactions can be further classified into ordered (for example, suppression) and unordered (for example, co-equal) subtypes (BOX 2). Although interactions that fall into these categories have long been used by classical geneticists43,44, they are now being generated at increasingly larger scales, as in the case of a recent study of 26 genes involved in the response to DNA damage41.
First- and second-degree interactions
A final and extremely useful classification has been proposed45,46 which we describe as interactions of the first versus the second degree (alternatively, the first versus the second order; here the term degree is used to avoid conflict with the ordered–unordered terminology above). In contrast to the first-degree physical or genetic interactions that have been described thus far, in which there is direct evidence for an interaction, a second-degree interaction between molecules A and B is defined as one in which A interacts with many interactors of B. That is, the network neighbours of A and B significantly overlap, whether or not A and B are themselves first-degree neighbours.
These relationships are shown schematically in FIG. 2, for different combinations of the above interaction types. A second-degree protein–protein interaction (FIG. 2a) is the hallmark of a large protein complex25,47-49 in which any two proteins have many binding partners in common. This second-degree relationship has been used to predict new components and interactions within complexes48, and to assess the confidence of an interaction given the clustering of its interaction partners50. An example of a second-degree ordered physical interaction is that which would occur between regulatory proteins that share downstream targets (FIG. 2b). Many groups51-55 have reported combinations of transcription factors that, on the basis of physical data generated by techniques such as ChIP–chip, bind to a common set of gene promoters (for example, Swi6 and Mbp1 in FIG. 3a, which form the MBF transcriptional complex53). When applied systematically, similarity between ChIP profiles has been used to enumerate entire sets of transcriptional modules containing regulators that act together55.
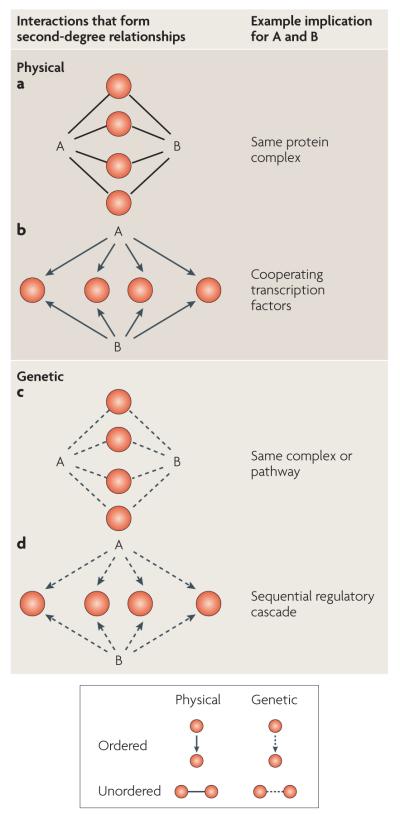
Figure 2. Second-degree interactions imply first-degree relationships.
The four example networks (panels a–d) illustrate ways in which second-degree interactions between two proteins, A and B, can imply new first-degree relationships that are complementary to the original experimental data. For protein–protein interactions (panel a), a second-degree interaction implies that A and B are in the same complex. For transcription factor–DNA interactions (panel b), A and B are possibly heterodimeric transcription factors that regulate a common set of genes. For aggravating genetic interactions (panel c), a second-degree interaction between A and B occurs if these proteins have common genetic interaction partners, implying that they act in the same pathway. For ordered genetic interactions (panel d), a second-degree interaction exists if mutations to A and B affect a common set of downstream genes or phenotypes, also suggesting that A and B act sequentially in a pathway.
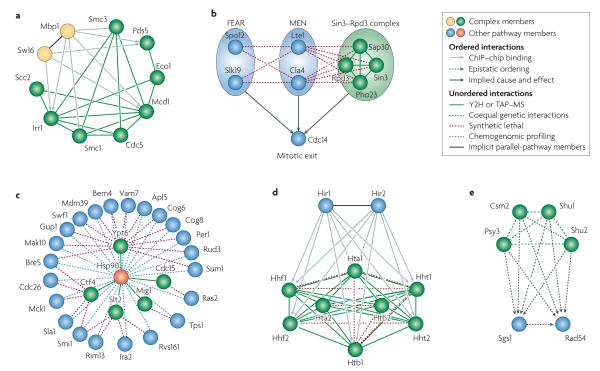
Figure 3. Examples of assembly across different interaction categories.
a ∣ Members of the cohesin complex are regulated by Swi6 and Mbp1 transcription factors, which themselves are parallel members of the MBF transcriptional complex. b ∣ Genetic interactions generated by synthetic-lethality screens identify parallel components of the Cdc14 release pathway, including members of the FEAR pathway, the MEN pathway and the Sin3–Rpd3 complex. c ∣ Identification of putative Hsp90 substrates through combined yeast two-hybrid (Y2H) and tandem affinity purification coupled with mass spectrometry (TAP–MS) screening, and synthetic genetic array (SGA) and chemogenomic profiling using an Hsp90 inhibitor. d ∣ Data that were obtained using chromatin immunoprecipitation combined with microarrays (ChIP–chip) were used to show that the transcription factors Hir1 and Hir2 regulate multiple members of a chromatin-related complex. e ∣ Members of a SHU complex (Shu1, Shu2, Csm2 and Psy3) are interconnected by coequal genetic interactions and show epistatic ordering with both Sgs1 and Rad54 in the DNA recombination/repair pathway41. Part a modified with permission from REF. 109 © (2007) National Academy of Sciences (USA). Part b modified with permission from Molecular Systems Biology REF. 46 © (2005) Macmillan Publishers Ltd. Part c modified with permission from REF. 63 © (2005) Elsevier Sciences. Part d modified with permission from REF. 66 © (2005) Biomed Central. Part e reproduced with permission from Nature Genetics REF. 41 © (2007) Macmillan Publishers Ltd.
Second-degree aggravating genetic interactions (FIG. 2c) were first identified by Tong et al.45, and were later described as showing ‘genetic congruence’46. Because first-degree aggravating genetic interactions typically run between two redundant or synergistic pathways (BOX 2, FIG. 1e), a second-degree interaction of this type (genetic congruence) places the genes within the same pathway (for example, common synthetic-lethal partners of members of the MEN pathway in yeast; FIG. 3b). Another example of a second-degree genetic interaction (FIG. 2d) is the case of two genetic perturbations that, when profiled individually using expression arrays, lead to the same set of differentially expressed genes (that is, ordered genetic interactions). This principle has been used to place genes within the galactose-utilization pathway on the basis of the similarity of their gene-knockout expression profiles56.
The importance of classification
Although it is not yet clear which of the above classifications will ultimately be most useful, given the size, scope and variety of the interaction data it seems clear that some classification system will be necessary. Classification systems invoke terminology in order to reveal the relationships between objects and to achieve precision. Consider the following two ways to describe an interaction to colleagues: the classification “a second-degree interaction of the ordered physical type”; or the more colloquial “an interaction between regulatory proteins that share downstream targets”. The second description certainly seems more intuitive; however, the first classification makes it clear that an entire set of interaction types have similar properties, in much the same way as the classification Drosophila melanogaster reveals the relationship between this and other species.
In the case of interaction networks, an important benefit of classification is that it will enable an intelligent system to compute across diverse networks to derive models. An algorithm can trivially parse the first option given above, whereas no modern computer is able to parse the structure in the second option. “A second degree interaction of the ordered physical type” tells us precisely how to treat the interaction and that it should be handled similarly to other interactions in this class. A more colloquial description would require the computer to have extensive knowledge of the English language. It would mean specifying a different set of rules for every kind of interaction and rewriting these rules whenever a new measurement technology was presented.
Assembly II: integration across categories
The above categories simplify the task of interaction assembly, by reducing the information gained from the various technologies to a few types, each with a distinct set of rules for integration. A number of studies have begun to combine interactions across several of these categories to derive integrated biological models (FIG. 4).
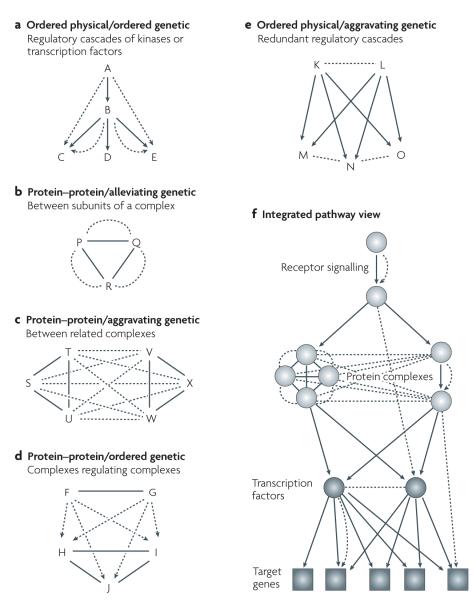
Figure 4. Network motifs assembled from different combinations of interaction measurements.
Physical interactions are shown as solid lines and arrows, and genetic interactions are shown as dashed lines and arrows. Part a shows an example of integrating ordered physical versus ordered genetic interactions, in which knockout of A or B results in changes in the activity of C, D and E (ordered genetic interactions), which are brought about because of changes in transcriptional activity or kinase–substrate binding (ordered physical interactions)52,57,110,111. Members of protein complexes (protein–protein interactions) can be connected by genetic interactions either within complexes (shown in part b) or between complexes (shown in part c) 37-39,42,46,65. In part d, members of a complex made up of F–G (protein–protein interactions) operate upstream of or epistatically to (ordered genetic) the complex H–I–J42,109. In part e, regulatory factors K and L cooperate to activate targets M, N and O (ordered physical) which function in parallel pathways (alleviating genetic46,51,66). Part f shows how the motifs of previous panels might combine within a still larger network, starting at a receptor protein and ending at transcription factors modulating the expression of target genes. Note that the motifs in each panel are summarized from the literature (see references provided) and are not intended as an exhaustive catalogue of all ways of integrating interactions.
Integration of ordered interactions
One major direction has been to integrate ordered measurements of both genetic and physical interactions (FIG. 4a). Yeang et al. attempted to explain cause-and-effect genetic relationships with regulatory pathways inferred from databases of protein–DNA (that is, transcription factor–promoter) and protein–protein physical interactions57. The genetic relationships were drawn from a panel of ~300 expression profiles measured in response to single-gene deletion experiments in yeast58. For each experiment, the modelling procedure identified the most probable regulatory pathways of physical interactions that connect the deleted gene (the cause of perturbation) to genes that are differentially expressed in response to the deletion (the effects of perturbation).
As another example, several studies52,59 have analysed expression data from transcription factor knockouts to derive sets of genes, the expression of which is affected by each knockout (ordered genetic interactions). These studies were able to derive regulatory cascades of transcription factors on the basis of integration of the genetic interactions with physical protein–DNA interactions, or by detecting second-degree genetic effects (for example, two transcription factor knockouts that affect the same set of target genes). Deplancke et al. have also applied such approaches in worms, in which expression changes in response to RNAi knockdowns were used to functionally validate protein–DNA interactions measured by yeast one-hybrid assays60.
A related approach has been used for interpreting eQTL data61. As discussed in BOX 1, eQTLs and knockout expression profiles provide similar information in that they both generate cause-and-effect ordered genetic linkages. Tu and co-workers integrated a co-expression-based network with eQTLs in order to detect pathways that link a locus to a given target gene. Assuming that only one gene at each locus is the true regulator of the target, the algorithm’s task was to identify this true regulator among the multiple candidates within a locus. The method works by connecting potential candidate genes to the target by traversing a physical network. The candidate gene with the highest-scoring physical path is predicted to be the true regulator.
As they mature, such methods may be able to address several well-known challenges of eQTL analysis. First, owing to linkage disequilibrium and sparse markers, a single locus typically contains many genes, any of which is potentially the true source of perturbation (the ‘fine-mapping’ problem62). A physical network helps resolve this problem because candidate genes that are only a few interactions away from their targets, or that cluster together in the same region of the network, are more likely to represent the true causal factors. A second challenge relates to statistical power. Classical linkage screens typically include upwards of 105 genetic markers, in which case the threshold for association is usually set high to counter the effect of multiple testing. In an integrated network analysis, one might expect that statistical power would increase, because significance depends on two independent lines of evidence: observation of high eQTL scores and correspondence with the physical network.
Integration of unordered interaction measurements
A second major group of integrative studies has focused on interrelating unordered types of physical and genetic interactions. As an example, Zhao et al. used a combination of Y2H and TAP–MS (detecting physical protein–protein interactions) as well as SGA and chemical genetic screening (detecting aggravating genetic interactions) to explore the physical and genetic neighbourhood of Hsp90 (REF. 63) (FIG. 3c). Similarly, vidal and colleagues integrated Y2H data with RNAi screens to refine a map of transforming growth factor-β (TGFb) signalling in C. elegans and to propose several DAF-7–TGFb modulators64. In both cases, the focus was on identification of proteins that act within the same or related pathways.
Following on from this work, Kelley and Ideker showed how an integrated analysis could discover such pathways automatically by identifying recognizable patterns in the physical and genetic data37. Probabilistic models were developed to capture both a within-pathway and between-pathway explanation for genetic interactions (see FIG. 4b,c and the section above on between- versus within-pathway interactions). Both models detect pathways as clusters of proteins that physically interact with each other much more often than would be expected by chance. The within-pathway model predicts that these clusters directly overlap with clusters of genetic interactions. The between-pathway model predicts that genetic interactions run orthogonal to the physical clusters, and looks for dense bipartite clusters of genetic interactions that span between clusters (FIG. 1e). In aggregate, synthetic-lethal interactions were significantly more likely to link two redundant physical complexes than they were to occur within a single pathway, as anticipated by conventional genetic wisdom.
Because synthetic lethal interactions are likely to span two pathways, genes in the same pathway should have overlapping genetic interaction partners — a second-degree genetic interaction45. Ye et al. showed that genes that are connected by such a relationship were indeed more likely to participate in the same protein complex or pathway, and to share similar protein functions46. Pan et al.65 leveraged this idea further to identify functional modules involved in genome maintenance using synthetic-lethal interactions.
Other combinations of interaction types
Other combinations of interaction types have not been as actively researched; for example, integrating ordered genetic interactions with unordered physical measurements, or the reverse. However, in one prominent example, Krogan and colleagues used the E-MAP system (epistatic miniarray profiling, a quantitative version of SGA introduced by Collins et al.29) to identify epistatic interactions (ordered genetic) among protein complexes (unordered protein–protein interaction measurements) involved in the yeast secretory42 or chromosomal organization39 pathways. The study of secretory pathways linked members of the yeast GET complex to functions within the endoplasmic reticulum–Golgi trafficking system. As an example of integrating ordered physical with aggravating genetic interactions (FIG. 4e), Zhang et al. defined a network motif that superimposes transcription factor–promoter binding interactions (ordered physical) with synthetic lethality66. An example of this motif is provided by the Hir1 and Hir2 transcription factors, which bind genes functioning in chromatin maintenance that are also interconnected by many synthetic-lethal interactions (FIG. 3d).
Annotating further details on the scaffold
Network assembly reveals the connectivity of the network, producing a static wiring diagram of the molecular interactions that make up the molecular machinery in a cell. Numerous questions remain. For instance, in the case of ordered interactions, what is the direction of information flow? Is each upstream component an activator or repressor of the component that lies immediately downstream? It is also desirable to understand the kinetics and dynamics of signal transduction; that is, to resolve the timescale of regulatory processes. The level of detail that is required depends on the question at hand. For instance, for identification of potential drug targets, it might initially be sufficient to know the position of a protein in a static pathway map. Alternatively, if the system should be optimized to increase the yield of some biochemical product, more quantitative information, including dynamics, might be necessary.
In the case of interactions for which the ordering is ambiguous, one next step is to try to infer the directionality of the interaction (if one exists) and its effect (activating versus repressing). St Onge et al. were able to order some previously unordered genetic interactions using theory from classical genetics on the basis of quantitative single- and double-mutant growth rates41. Alleviating genetic interactions were subdivided into masking and suppression interactions (BOX 2), which were then used to order various genes involved in the DNA-damage response on the basis of a positive regulatory model35, including the suppression of SGS1 and RAD54 deletions by members of a Shu1, Shu2, Csm2 and Psy3 complex (FIG. 3e). Nguyen and D’Haeseleer derived the inhibitory or activating activity of transcriptional regulators through analysis of expression data and transcription factor binding motifs67. For each transcriptional regulator, a gene-specific cis-regulatory code was identified that determines the (positive or negative) interaction between the regulator and its target. Yeang et al. also attempted to infer additional details of protein–protein and protein–DNA interactions (directionality, repressing versus enhancing) through integration with expression data57,68.
Finally, the most detailed models quantify the interaction dynamics. Studies of interaction dynamics are numerous, and lie outside the scope of this Review; for an excellent treatment, see REF. 69. As one of many examples, the yeast osmotic-shock signalling pathway has been modelled kinetically70 using a system of differential equations to describe in detail the dynamic activation and deactivation of the pathway components, including feedback control. Importantly, the model predicted unexpected features of the pathway that could later be experimentally verified, such as the role of phosphatase transcription in negative-feedback control. However, although differential equations certainly provide a high degree of detail, they often suffer from a lack of knowledge of kinetic constants. The most successful approaches so far have coordinated the measurement of kinetic constants iteratively, between modellers and experimental biologists, as the models can identify which parameters have the greatest influence over the simulation outcome69.
Conclusions
Given the hindsight of the Human Genome Project, what lessons can be learned and applied at the network level? Perhaps most trivially, both genetic and physical data are absolutely essential to our understanding of biological systems. Genetic data explain the ‘what’ of biological systems: what is the function of a gene, what is its phenotype and what is its target? Physical data explain the ‘how’: how does a gene or protein execute its function? Given that the final stages of the Human Genome Project were driven by physical shotgun sequencing (which revealed genome structure), it is easy to forget that genetic linkage studies were used to map most of the known disease genes (revealing gene function). A second important lesson is that full coverage is not needed for deriving meaningful biological information from the physical or genetic data. In case of the Human Genome Project, decades passed from the sequencing of the first human gene to completion of the full genome sequence71. In the case of interaction networks, the examples covered in this Review suggest that the available interaction maps already have sufficient coverage to reveal the structure and function of hundreds of pathways. However, physical and genetic interaction networks provide a higher level of complexity (in two or three dimensions) compared with the one-dimensional genome sequence. Therefore, extra effort and care should be taken when analysing these data, especially considering that different technologies provide data on different types of interactions, and that networks are dynamic structures in contrast to the genome, which is relatively static.
A particularly important sub-class of genetic interactions are combinations of SNPs that are causative for complex diseases72. In such cases, no single SNP can cause the disease alone, but in combination they form a condition under which the disease may develop. Such genetic relationships are close relatives of the synthetic-lethal interactions that are measured in model organisms. Mapping these interactions onto physical networks will be an important way to elucidate the mechanistic foundation of disease72,73. Such integration will be particularly important to the current wave of genome-wide association studies, in which, often, little is known a priori about the functions of the significant loci74,75. To achieve this level of integration, greater coverage will be needed of the human protein network as well as that of the mouse, in which many QTL and eQTL studies are carried out.
All of this is not to say that physical and genetic networks are the ultimate representation of cellular function. There are a host of other philosophical frameworks for understanding cells, some undoubtedly remaining to be discovered. For instance, the cell has been variously depicted as a compartmentalized bag of enzymes76, a collection of enzyme complexes77, a hydrogel78 or a broadcast transmission or Petri network79. However, it is clear that the current paradigm in molecular biological research has shifted focus from protein sequences to protein networks. And there is much work to be done to fully realize this world view, before we graduate to the next one.
DATABASES
Entrez Gene: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=geneRAD54|SGS1
OMIM: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM
UniProtKB: http://ca.expasy.org/sprotHir1|Hir2|Mbp1|Notch|Swi6
FURTHER INFORMATION
The Idaker laboratory homepage: http://chianti.ucsd.edu/idekerlab
Cancer Genome Anatomy Project: http://gai.nci.nih.gov/cgi-bin/histo.cgi?c=23&o=h
CellCircuits DB: http://cellcircuits.org
Database of Interacting Proteins (DIP): http://dip.doe-mbi.ucla.edu
General Respository for Interaction Datasets (GRID): http://www.thebiogrid.org
Human Protein Interaction Database (HPID): http://wilab.inha.ac.kr/hpid
Human Protein Reference Database (HPRD): http://www.hprd.org
IntAct: http://www.ebi.ac.uk/intact/site/index.jsf
MIPS Mammalian Protein–Protein Interaction Database: http://mips.gsf.de/proj/ppi
Molecular INTeractions Database (MINT): http://mint.bio.uniroma2.it/mint/Welcome.do
ALL LINKS ARE ACTIVE IN THE ONLINE PDF.
Acknowledgements
This work was supported by the US National Institutes of Environmental Health Sciences grant ES014811. T.I. is a David and Lucille Packard Fellow.
Glossary
- Radiation hybrid mapping
High-resolution mapping of human markers using X-ray exposure to fragment human chromosomes and fusing the irradiated cells with rodent cells. The frequency of co-occurrence of markers on the same fragment relates to their genomic distance.
- Fluorescence in situ hybridization
Fluorescently labelled DNA probes are hybridized to chromosomal DNA. This allows genes (probes) to be assigned to chromosomes and provides a rough estimate of the chromosomal position of the cloned fragment.
- Reverse-genetic screening
Identifying the mutant phenotype(s) associated with a known genetic mutation or a panel of known mutations, such as a gene-deletion library. This term contrasts with forward-genetic screening, which involves identifying the mutations that affect a given phenotype.
- Regression
A statistical method for predicting a dependent variable on the basis of one or more independent variables.
- Likelihood function
A statistical method for predicting the likelihood of an outcome that is conditional (dependent) on other evidence.
- Petri network
A modelling approach that depicts a process on a bipartite graph. Nodes are either places or transitions that are connected by directed arcs. Tokens are transmitted from places to transitions or from transitions to places.
Footnotes
Competing interests statement The authors declare no competing financial interests.
References
- 1.Yu A, et al. Comparison of human genetic and sequence-based physical maps. Nature. 2001;409:951–953. doi: 10.1038/35057185. [DOI] [PubMed] [Google Scholar]
- 2.Sturtevant AH. The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J. Exp. Zool. 1913;14:43–59. [Google Scholar]
- 3.Goss SJ, Harris H. New method for mapping genes in human chromosomes. Nature. 1975;255:680–684. doi: 10.1038/255680a0. [DOI] [PubMed] [Google Scholar]
- 4.Cox DR, Burmeister M, Price ER, Kim S, Myers RM. Radiation hybrid mapping: a somatic cell genetic method for constructing high-resolution maps of mammalian chromosomes. Science. 1990;250:245–250. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- 5.Fauth C, Speicher MR. Classifying by colors: FISH-based genome analysis. Cytogenet. Cell Genet. 2001;93:1–10. doi: 10.1159/000056937. [DOI] [PubMed] [Google Scholar]
- 6.Rowen L, Mahairas G, Hood L. Sequencing the human genome. Science. 1997;278:605–607. doi: 10.1126/science.278.5338.605. [DOI] [PubMed] [Google Scholar]
- 7.Green P. Whole-genome disassembly. Proc. Natl Acad. Sci. USA. 2002;99:4143–4144. doi: 10.1073/pnas.082095999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Twyman RM, Primrose SB. Techniques patents for SNP genotyping. Pharmacogenomics. 2003;4:67–79. doi: 10.1517/phgs.4.1.67.22582. [DOI] [PubMed] [Google Scholar]
- 9.Stein L. Genome annotation: from sequence to biology. Nature Rev. Genet. 2001;2:493–503. doi: 10.1038/35080529. [DOI] [PubMed] [Google Scholar]
- 10.Sharan R, Ideker T. Modeling cellular machinery through biological network comparison. Nature Biotechnol. 2006;24:427–433. doi: 10.1038/nbt1196. [DOI] [PubMed] [Google Scholar]
- 11.Fields S. High-throughput two-hybrid analysis. The promise and the peril. FEBS J. 2005;272:5391–5399. doi: 10.1111/j.1742-4658.2005.04973.x. [DOI] [PubMed] [Google Scholar]
- 12.Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317.A landmark paper that explores a large genetic interaction network in yeast, and introduces the idea of genetic congruence — a second-degree genetic interaction.
- 13.Greenwald I. The C. elegans Research Community, editor. [4 August 2005];WormBook. [online], < http://www. wormbook.org>. (doi/10.1895/ wormbook.1.10.1)
- 14.Botstein D, et al. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology. Pringle J, Broach J, Jones E, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1997. [Google Scholar]
- 15.Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nature Rev. Genet. 2007;8:437–449. doi: 10.1038/nrg2085.A review of theory and approaches to mapping genetic interaction networks.
- 16.Bork P, et al. Protein interaction networks from yeast to human. Curr. Opin. Struct. Biol. 2004;14:292–299. doi: 10.1016/j.sbi.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 18.Jansen RC. Studying complex biological systems using multifactorial perturbation. Nature Rev. Genet. 2003;4:145–151. doi: 10.1038/nrg996. [DOI] [PubMed] [Google Scholar]
- 19.Sprinzak E, Altuvia Y, Margalit H. Characterization and prediction of protein–protein interactions within and between complexes. Proc. Natl Acad. Sci. USA. 2006;103:14718–14723. doi: 10.1073/pnas.0603352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suthram S, Shlomi T, Ruppin E, Sharan R, Ideker T. A direct comparison of protein interaction confidence assignment schemes. BMC Bioinformatics. 2006;7:360. doi: 10.1186/1471-2105-7-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee I, Date SV, Adai AT, Marcotte EM. A probabilistic functional network of yeast genes. Science. 2004;306:1555–1558. doi: 10.1126/science.1099511. [DOI] [PubMed] [Google Scholar]
- 22.Rhodes DR, et al. Probabilistic model of the human protein–protein interaction network. Nature Biotechnol. 2005;23:951–959. doi: 10.1038/nbt1103. [DOI] [PubMed] [Google Scholar]
- 23.Beyer A, et al. Integrated assessment and prediction of transcription factor binding. PLoS Comput. Biol. 2006;2:e70. doi: 10.1371/journal.pcbi.0020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollunder J, Beyer A, Wilhelm T. Identification and characterization of protein subcomplexes in yeast. Proteomics. 2005;5:2082–2089. doi: 10.1002/pmic.200401121. [DOI] [PubMed] [Google Scholar]
- 25.Collins SR, et al. Towards a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell. Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.von Mering C, et al. Comparative assessment of large-scale data sets of protein–protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750.The first comparison of the quality of various high-throughput physical interaction data sets.
- 27.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 29.Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Downard KM. Ions of the interactome: the role of MS in the study of protein interactions in proteomics and structural biology. Proteomics. 2006;6:5374–5384. doi: 10.1002/pmic.200600247. [DOI] [PubMed] [Google Scholar]
- 31.Legrain P, Wojcik J, Gauthier JM. Protein–protein interaction maps: a lead towards cellular functions. Trends Genet. 2001;17:346–352. doi: 10.1016/s0168-9525(01)02323-x. [DOI] [PubMed] [Google Scholar]
- 32.Myers CL, Barrett DR, Hibbs MA, Huttenhower C, Troyanskaya OG. Finding function: evaluation methods for functional genomic data. BMC Genomics. 2006;7:187. doi: 10.1186/1471-2164-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Estojak J, Brent R, Golemis EA. Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell. Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gunsalus KC, et al. Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature. 2005;436:861–865. doi: 10.1038/nature03876. [DOI] [PubMed] [Google Scholar]
- 35.Avery L, Wasserman S. Ordering gene function: the interpretation of epistasis in regulatory hierarchies. Trends Genet. 1992;8:312–316. doi: 10.1016/0168-9525(92)90263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ptacek J, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 37.Kelley R, Ideker T. Systematic interpretation of genetic interactions using protein networks. Nature Biotechnol. 2005;23:561–566. doi: 10.1038/nbt1096.The first large-scale identification of genetic interactions within and between pathways.
- 38.Ulitsky I, Shamir R. Pathway redundancy and protein essentiality revealed in the Saccharomyces cerevisiae interaction networks. Mol. Syst. Biol. 2007;3:104. doi: 10.1038/msb4100144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collins SR, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649.A large-scale identification of alleviating and aggravating genetic interactions and an interpretation of these interactions in the context of protein complexes.
- 40.Drees BL, et al. Derivation of genetic interaction networks from quantitative phenotype data. Genome Biol. 2005;6:R38. doi: 10.1186/gb-2005-6-4-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.St Onge RP, et al. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nature Genet. 2007;39:199–206. doi: 10.1038/ng1948.An example of using genetic interactions to order pathways involved in DNA damage.
- 42.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 43.Jana S. Simulation of quantitative characters from qualitatively acting genes. Theor. Appl. Genet. 1972;42:119–124. doi: 10.1007/BF00583413. [DOI] [PubMed] [Google Scholar]
- 44.Punnett RC. Mendelism. Macmillan; New York: 1913. [Google Scholar]
- 45.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 46.Ye P, et al. Gene function prediction from congruent synthetic lethal interactions in yeast. Mol. Syst. Biol. 2005;1:2005.0026. doi: 10.1038/msb4100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu H, Paccanaro A, Trifonov V, Gerstein M. Predicting interactions in protein networks by completing defective cliques. Bioinformatics. 2006;22:823–829. doi: 10.1093/bioinformatics/btl014. [DOI] [PubMed] [Google Scholar]
- 49.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 50.Goldberg DS, Roth FP. Assessing experimentally derived interactions in a small world. Proc. Natl Acad. Sci. USA. 2003;100:4372–4376. doi: 10.1073/pnas.0735871100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800.A large-scale analysis of the DNA binding patterns of most yeast transcription factors using ChIP–chip.
- 52.Workman CT, et al. A systems approach to mapping DNA damage response pathways. Science. 2006;312:1054–1059. doi: 10.1126/science.1122088.An example of the integration of physical ChIP–chip data with genetic knockout gene expression data to explore pathways involved in DNA damage.
- 53.Iyer VR, et al. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 54.Chiu R, et al. The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes. Cell. 1988;54:541–552. doi: 10.1016/0092-8674(88)90076-1. [DOI] [PubMed] [Google Scholar]
- 55.Vermeirssen V, et al. Transcription factor modularity in a gene-centered C. elegans core neuronal protein–DNA interaction network. Genome Res. 2007;17:1061–1071. doi: 10.1101/gr.6148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ideker T, et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 57.Yeang CH, et al. Validation and refinement of gene-regulatory pathways on a network of physical interactions. Genome Biol. 2005;6:R62. doi: 10.1186/gb-2005-6-7-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 59.Hu Z, Killion PJ, Iyer VR. Genetic reconstruction of a functional transcriptional regulatory network. Nature Genet. 2007;39:683–687. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- 60.Deplancke B, et al. A gene-centered C. elegans protein–DNA interaction network. Cell. 2006;125:1193–1205. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 61.Tu Z, Wang L, Arbeitman MN, Chen T, Sun F. An integrative approach for causal gene identification and gene regulatory pathway inference. Bioinformatics. 2006;22:e489–e496. doi: 10.1093/bioinformatics/btl234. [DOI] [PubMed] [Google Scholar]
- 62.Ott J. Analysis of Human Genetic Linkage. Johns Hopkins Univ. Press; Baltimore: 1999. [Google Scholar]
- 63.Zhao R, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the Hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 64.Tewari M, et al. Systematic interactome mapping and genetic perturbation analysis of a C. elegans TGFβ signaling network. Mol. Cell. 2004;13:469–482. doi: 10.1016/s1097-2765(04)00033-4. [DOI] [PubMed] [Google Scholar]
- 65.Pan X, et al. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 66.Zhang LV, et al. Motifs, themes and thematic maps of an integrated Saccharomyces cerevisiae interaction network. J. Biol. 2005;4:6. doi: 10.1186/jbiol23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nguyen DH, D’Haeseleer P. Deciphering principles of transcription regulation in eukaryotic genomes. Mol. Syst. Biol. 2006;2:2006.0012. doi: 10.1038/msb4100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yeang CH, Ideker T, Jaakkola T. Physical network models. J. Comput. Biol. 2004;11:243–262. doi: 10.1089/1066527041410382. [DOI] [PubMed] [Google Scholar]
- 69.Klipp E, Liebermeister W. Mathematical modeling of intracellular signaling pathways. BMC Neurosci. 2006;7:S10. doi: 10.1186/1471-2202-7-S1-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klipp E, Nordlander B, Kruger R, Gennemark P, Hohmann S. Integrative model of the response of yeast to osmotic shock. Nature Biotechnol. 2005;23:975–982. doi: 10.1038/nbt1114. [DOI] [PubMed] [Google Scholar]
- 71.Roberts L, Davenport RJ, Pennisi E, Marshall E. A history of the Human Genome Project. Science. 2001;291:1195. doi: 10.1126/science.291.5507.1195. [DOI] [PubMed] [Google Scholar]
- 72.Schadt EE, Lum PY. Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Reverse engineering gene networks to identify key drivers of complex disease phenotypes. J. Lipid Res. 2006;47:2601–2613. doi: 10.1194/jlr.R600026-JLR200. [DOI] [PubMed] [Google Scholar]
- 73.Lage K, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nature Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295.The first study to explain disease phenotypes by genome-wide mapping of genetic loci onto a human interaction network.
- 74.Bourgain C, Genin E, Cox N, Clerget-Darpoux F. Are genome-wide association studies all that we need to dissect the genetic component of complex human diseases? Eur. J. Hum. Genet. 2007;15:260–263. doi: 10.1038/sj.ejhg.5201753. [DOI] [PubMed] [Google Scholar]
- 75.Williams SM, et al. Problems with genome-wide association studies. Science. 2007;316:1840–1842. [PubMed] [Google Scholar]
- 76.Mathews CK. The cell: bag of enzymes or network of channels? J. Bacteriol. 1993;175:6377–6381. doi: 10.1128/jb.175.20.6377-6381.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Srere PA. Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 1987;56:89–124. doi: 10.1146/annurev.bi.56.070187.000513. [DOI] [PubMed] [Google Scholar]
- 78.Pollack G. Cells, Gels and the Engines of Life. Ebner & Sons; Seattle: 2001. [Google Scholar]
- 79.Pinney JW, Westhead DR, McConkey GA. Petri Net representations in systems biology. Biochem. Soc. Trans. 2003;31:1513–1515. doi: 10.1042/bst0311513. [DOI] [PubMed] [Google Scholar]
- 80.Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rual JF, et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 83.Stelzl U, et al. A human protein–protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 84.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 85.Suzuki H, et al. Protein–protein interaction panel using mouse full-length cDNAs. Genome Res. 2001;11:1758–1765. doi: 10.1101/gr.180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Uetz P, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 87.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 88.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 89.Pokholok DK, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 90.Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 91.Loh YH, et al. The OCT4 and NANOG transcription network regulates pluripotency in mouse embryonic stem cells. Nature Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 92.Wei CL, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 93.van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nature Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 94.Deplancke B, Dupuy D, Vidal M, Walhout AJ. A gateway-compatible yeast one-hybrid system. Genome Res. 2004;14:2093–2101. doi: 10.1101/gr.2445504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Walhout AJ. Unraveling transcription regulatory networks by protein–DNA and protein–protein interaction mapping. Genome Res. 2006;16:1445–1454. doi: 10.1101/gr.5321506. [DOI] [PubMed] [Google Scholar]
- 96.Berger MF, et al. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nature Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ooi SL, Shoemaker DD, Boeke JD. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nature Genet. 2003;35:277–286. doi: 10.1038/ng1258. [DOI] [PubMed] [Google Scholar]
- 98.Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nature Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 99.Lehner B, Tischler J, Fraser AG. RNAi screens in Caenorhabditis elegans in a 96-well liquid format and their application to the systematic identification of genetic interactions. Nature Protoc. 2006;1:1617–1620. doi: 10.1038/nprot.2006.245. [DOI] [PubMed] [Google Scholar]
- 100.Sahin O, et al. Combinatorial RNAi for quantitative protein network analysis. Proc. Natl Acad. Sci. USA. 2007;104:6579–6584. doi: 10.1073/pnas.0606827104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brem RB, Storey JD, Whittle J, Kruglyak L. Genetic interactions between polymorphisms that affect gene expression in yeast. Nature. 2005;436:701–703. doi: 10.1038/nature03865.A pioneering eQTL paper linking genetic variation in yeast to gene expression as a quantitative trait.
- 102.Bao L, et al. Combining gene expression QTL mapping and phenotypic spectrum analysis to uncover gene regulatory relationships. Mamm. Genome. 2006;17:575–583. doi: 10.1007/s00335-005-0172-2. [DOI] [PubMed] [Google Scholar]
- 103.Chesler EJ, Lu L, Wang J, Williams RW, Manly KF. WebQTL: rapid exploratory analysis of gene expression and genetic networks for brain and behavior. Nature Neurosci. 2004;7:485–486. doi: 10.1038/nn0504-485. [DOI] [PubMed] [Google Scholar]
- 104.Petretto E, et al. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet. 2006;2:e172. doi: 10.1371/journal.pgen.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 106.Phillips PC. The language of gene interaction. Genetics. 1998;149:1167–1171. doi: 10.1093/genetics/149.3.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Phillips PC, Otto SP, Whitlock MC. Beyond the Average: the Evolutionary Importance of Gene Interactions and Variability of Epistatic Effects in Epistasis and the Evolutionary Process. Oxford Univ. Press; New York: 2000. [Google Scholar]
- 108.Segre D, Deluna A, Church GM, Kishony R. Modular epistasis in yeast metabolism. Nature Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- 109.Tan K, Shlomi T, Feizi H, Ideker T, Sharan R. Transcriptional regulation of protein complexes within and across species. Proc. Natl Acad. Sci. USA. 2007;104:1283–1288. doi: 10.1073/pnas.0606914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Carter GW, et al. Prediction of phenotype and gene expression for combinations of mutations. Mol. Syst. Biol. 2007;3:96. doi: 10.1038/msb4100137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Carter GW, Rupp S, Fink GR, Galitski T. Disentangling information flow in the Ras-cAMP signaling network. Genome Res. 2006;16:520–526. doi: 10.1101/gr.4473506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Keene JD, Komisarow JM, Friedersdorf MB. RIP–Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nature Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- 113.Barrios-Rodiles M, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 114.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nature Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 116.Schadt EE, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nature Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 119.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Giaever G, et al. Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc. Natl Acad. Sci. USA. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]