Abstract
Living systems use RNA sequences known as riboswitches to detect the concentrations of small-molecule metabolites within cells, and to regulate the expression of genes that produce these metabolites. Like their natural counterparts, synthetic riboswitches also regulate gene expression in response to small molecules, however, because synthetic riboswitches can be engineered to respond to non-endogenous small molecules, they are powerful tools for chemical and synthetic biologists interested in understanding and reprogramming cellular behavior. In this review, we present an overview of natural riboswitches, highlight recent studies toward developing synthetic riboswitches, and provide an overview of emerging applications of these RNA switches in chemical biology.
The central dogma of molecular biology formulated by Crick was very simple. DNA was transcribed to mRNA, which was translated into proteins. However, it has become clear that biology is a bit more complicated. While the role of DNA as an information repository and the role of proteins as building blocks of the cell have remained largely intact, the role of RNA as a transient intermediate on the way from DNA to protein has been revised significantly.(1) We now know that RNA sequences are essential for the function of some enzymes,(2) can perform self-cleavage,(3) and can regulate gene expression either in cis(4, 5) or in trans.(6) Because RNA sequences can carry out diverse tasks and are amenable to engineering both in vitro and in vivo, they are particularly attractive for controlling cell behavior.
RATIONALE FOR REPROGRAMMING CELLS USING RNA
A major goal of synthetic biology is to program bacteria to autonomously perform a variety of tasks.(7) To operate autonomously, cells need to sense and respond to changes in their environment, evaluate their performance, and modify their behavior to accomplish the assigned tasks. Because most cellular behavior can be regulated at the genetic level, cells can be programmed to perform new duties by providing them with instructions in the form of new genetic material. By encoding genetic instructions that are executed by the cell only when it achieves a certain metabolic state, or when it detects a specific exogenous ligand, it is possible to program a cell to perform a desired task when it encounters specified conditions.
Traditionally, genetic engineering efforts have focused on introducing recombinant DNA into cells to direct the heterologous expression of proteins. However, for many applications in synthetic biology, RNA provides a powerful alternative to using proteins to regulate cell behavior. In addition to storing information, RNA sequences can fold into complex 3-dimensional structures that are capable of binding ligands and catalyzing chemical reactions, much in the way that proteins do. RNA sequences that can bind ligands are known as aptamers. Aptamers are particularly useful tools for reprogramming cellular behavior because in addition to recognizing ligands, appropriately placed aptamers can also regulate gene expression in a variety of organisms, and through a variety of mechanisms.(7–14) Such RNA switches, known as riboswitches, are ideal tools for modulating bacterial behavior because, as we will see, it is possible to create riboswitches that recognize new ligands, and to introduce these genetic switches into cells by well established techniques. Bacterial behavior can be controlled by using the riboswitch to directly regulate the expression of a single gene, or by incorporating the switch as a control element within a more elaborate genetic circuit. We begin this review by presenting an overview of naturally occurring riboswitches, move on to discuss the state-of-the-art in synthetic riboswitch engineering, and conclude by reviewing current and prospective applications of ligand-sensing RNA switches in chemical biology.
METABOLITE-SENSING CAN BE ACHIEVED WITH RNA
The elucidation of the cis-acting regulatory mechanism of the trp operon during the late 1970s demonstrated that the structure of an mRNA molecule may attenuate gene expression.(15) However, it was not yet apparent that gene expression could be regulated by a small-molecule binding directly to an mRNA. Over the past decade, it has become increasingly evident that small-molecules bind to mRNA molecules to regulate a variety of biochemical pathways across all domains of life through a mechanism known as riboswitch control.(16) Almost exclusively found in eubacteria, riboswitches are typically located in the 5´-untranslated regions (5´-UTRs) of metabolic genes, where they regulate expression of the genes in the pathway in response to the concentration of a specific metabolite. Notably, riboswitches perform their work without the need for protein cofactors.(17) Indeed, searches for riboswitches began because previous searches for protein mediators of several metabolite-regulated biochemical pathways proved fruitless.(18, 19) As an example, the discovery of an FMN riboswitch in Bacillus subtilis in 2002 ended a decade-long search for a putative repressor protein that might use FMN as a cofactor.(4)
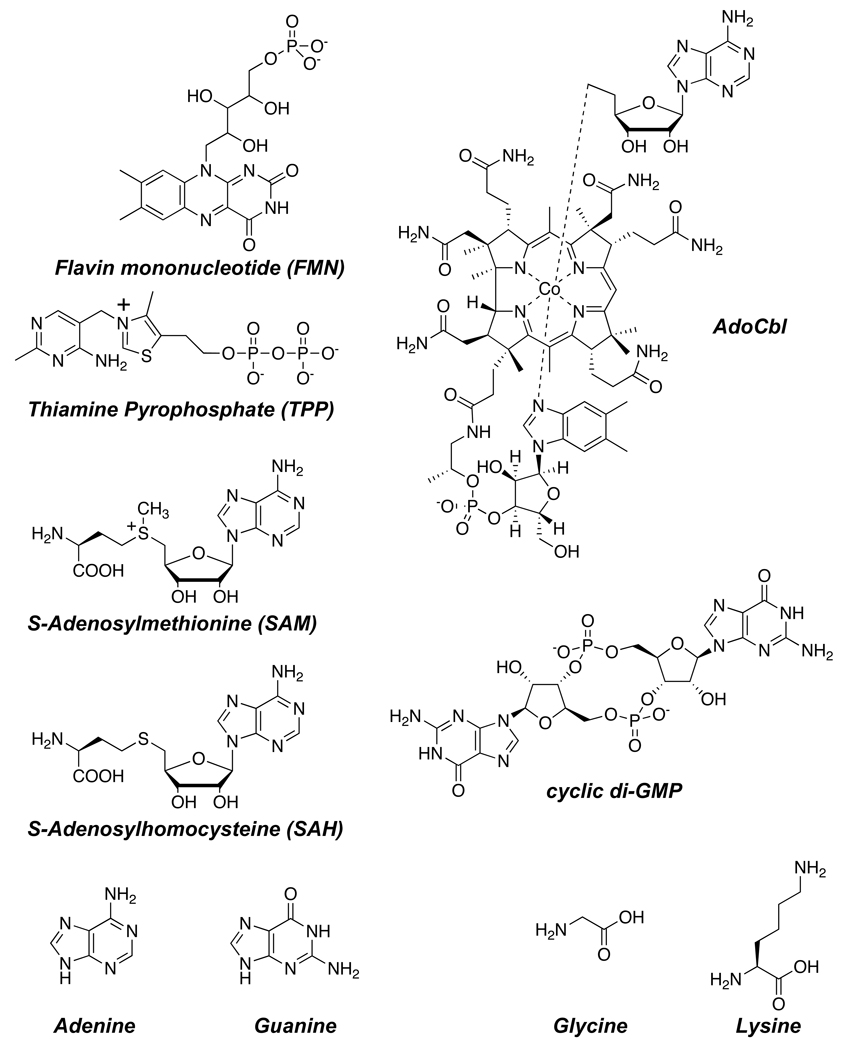
To date, at least a dozen riboswitch classes have since been discovered;(16) they respond to metabolites with diverse structures, such as flavin mononucleotide (FMN), thiamine pyrophosphate (TPP), adenosylcobalamin (AdoCbl), S-adenosylmethionine (SAM), S-adenosylhomocysteine (SAH), cyclic di-GMP, adenine, guanine, glycine, and lysine (Figure 1).(20–29) Each of these classes has been found, or is predicted to exist, within the genomes of many species of bacteria, leading some to postulate that riboswitches may be remnants of ancient metabolic ribozymes,(30) although others have speculated that the wide distribution of riboswitches is the result of horizontal gene transfer.(31) Regardless of their evolutionary origins, modern-day riboswitches are highly conserved genetic control elements that maintain metabolic homeostasis within cells.
Figure 1.
Examples of metabolites that bind natural riboswitches. These ligands vary substantially in size and structure, yet all are able to control gene expression by binding various RNA sequences in cells.
Riboswitches are composed of an aptamer domain, which binds the metabolite with high affinity and specificity, and an expression platform, which couples binding to a change in gene expression.(32) Riboswitches can operate by a variety of mechanisms to regulate the expression of a gene or set of genes in a metabolic pathway (Figure 2). Riboswitches have been discovered in diverse bacterial species,(20, 25–27, 33–35) a fungus,(36) and a plant.(37) Computational searches also indicate that riboswitches exist in archaea, suggesting that they are present in all kingdoms of life.(16) Barrick and Breaker provide an excellent overview of the distribution and mechanisms of confirmed and putative natural riboswitches.(16)
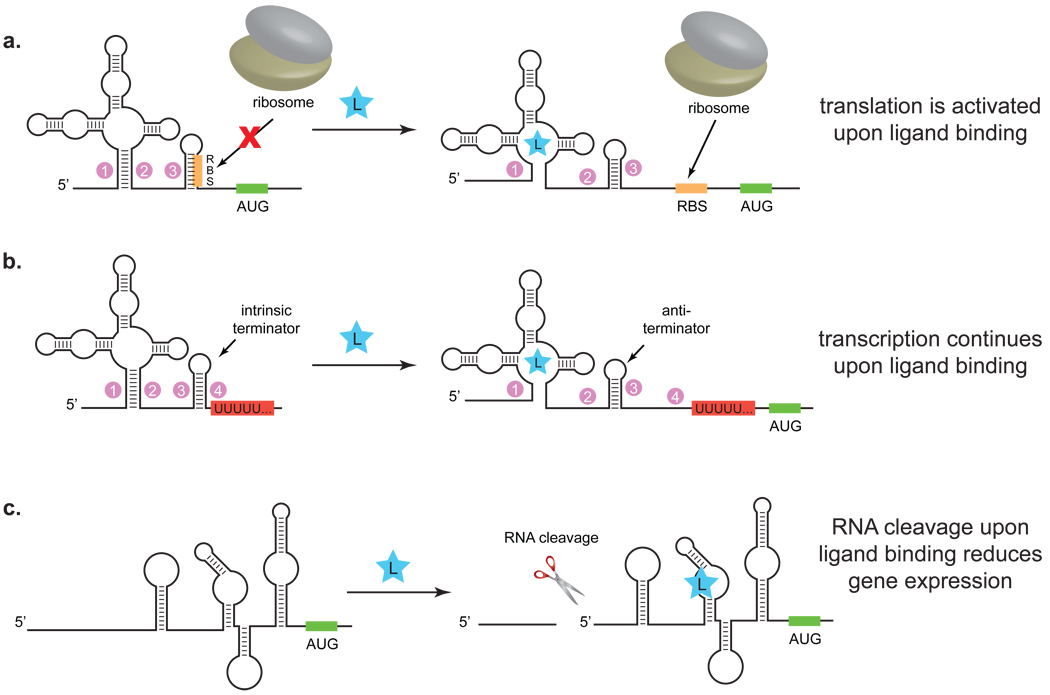
Figure 2.
Examples of riboswitch mechanisms. a) The ribosome binding site (RBS) is sequestered in the absence of ligand. Upon ligand binding, the RNA undergoes a conformational shift, revealing the RBS and enabling translation. b) In the absence of ligand, an intrinsic terminator stops transcription. Ligand binding induces a conformational shift that forms an anti-terminator, enabling expression of the downstream genes. c) The glmS riboswitch functions by a ligand-dependent splicing mechanism, whereby protein expression is diminished in the presence of the ligand.
Riboswitch-mediated changes in gene expression can occur either transcriptionally or translationally. The expression platform for a riboswitch that acts during transcription typically involves the ligand-dependent formation of an intrinsic terminator or anti-terminator structure. In contrast, riboswitches that operate at a translational level most often do so by masking or revealing the Shine-Dalgarno (SD) sequence (also known as the ribosome binding site; RBS) in a ligand-dependent fashion. When the Shine-Dalgarno sequence is revealed, the mRNA can bind to the ribosome and permit translation; masking the SD sequence represses translation. Transcriptionally regulated riboswitches are most often found in Gram-positive bacteria, while translationally regulated riboswitches are more common in Gram-negative bacteria.(16) While it is not yet clear why Gram-negative and Gram-positive bacteria appear to favor different riboswitch mechanisms, it is apparent that both expression platforms are functional across eubacteria. This flexibility is particularly attractive as researchers have become increasingly interested in developing synthetic riboswitches that function in diverse bacterial species.
SELECTIONS FOR SYNTHETIC APTAMERS
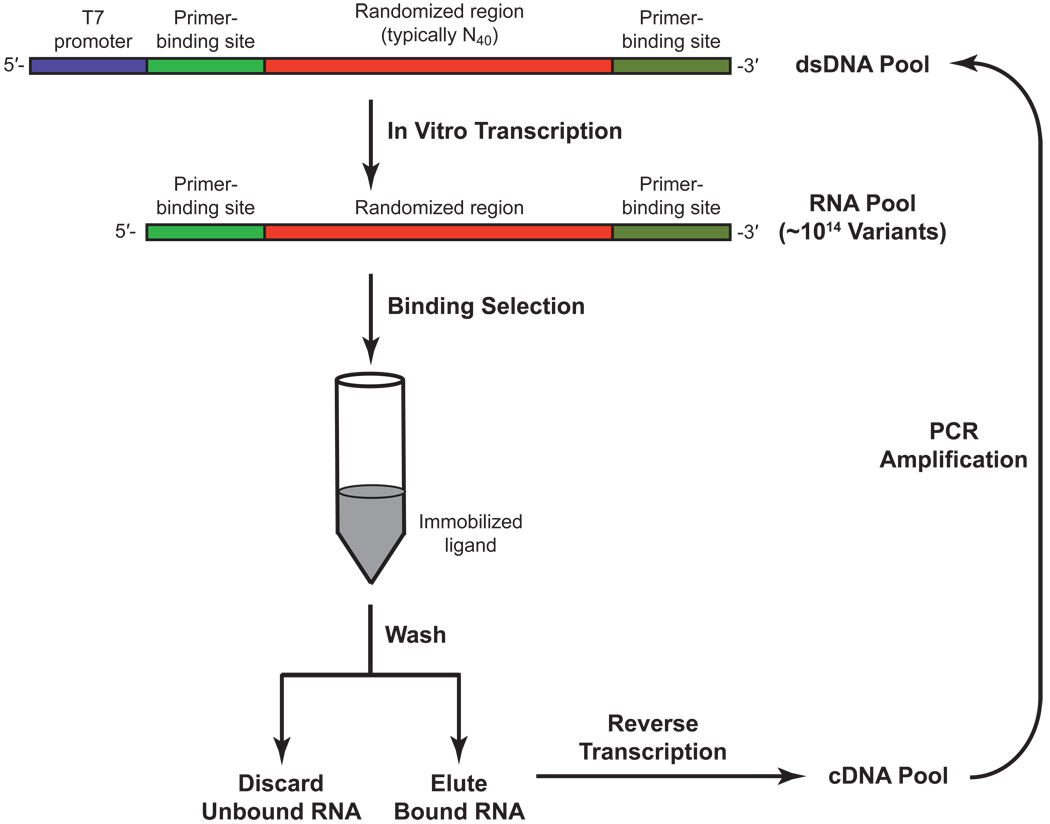
While the riboswitch mechanism was conclusively demonstrated in a natural system for the first time in 2002,(22) the idea that RNA could bind small molecules was already firmly established.(38, 39) In 1990, Ellington and Szostak reported an in vitro selection protocol to isolate RNA aptamers that could bind specific dyes.(38) In the same year, Tuerk and Gold independently reported a similar method to isolate protein-binding RNA sequences from large pools of randomized RNA sequences.(39) What is remarkable about these studies is that creation of an RNA aptamer did not require a pre-existing RNA scaffold. Compared to most protein engineering efforts, where an existing protein is subjected to computational design (40) or directed evolution methods,(41) RNA aptamers can be generated de novo by subjecting large pools of randomized oligonucleotides (~1014 unique sequences) to an in vitro selection process known as SELEX (Systematic Evolution of Ligands by EXponential enrichment).(39) Selections for RNA aptamers using SELEX require two alternating steps of partitioning and amplifying the desired sequences (Figure 3).
Figure 3.
Aptamer selection scheme. RNA aptamers may be isolated from large pools of randomized sequences using in vitro selection. The pool is enriched for sequences that bind an immobilized ligand and are eluted with free ligand. To obtain high-affinity binders, the selected sequences are reverse transcribed, amplified, and subjected to further rounds of selection.
To select aptamers, the starting pool of randomized RNA sequences is typically passed through a column that is covalently coupled to the desired target ligand. After the column is washed to remove unbound RNAs, the free ligand is used to competitively elute potential aptamers. This pool of initial binders is enriched by reverse transcription and PCR amplification, and the process is repeated to select for aptamers that bind the ligand with increased affinity. To select for aptamers that bind the ligand with high specificity, RNAs that bind to the column can be washed with structurally related ligands. After competitive elution, aptamers that remain bound to the column are expected to bind the desired ligand tightly, while discriminating against structurally related molecules. Typically, high-affinity RNA aptamers can be obtained with 10–15 rounds of in vitro selection.
As the tremendous potential of RNA aptamers became apparent, further studies were initiated to select for sequences that could bind new ligands tightly and with high specificity.(42) To enable more efficient identification of aptamers, many efforts have focused upon improving the SELEX strategy and have been reviewed extensively elsewhere.(43) As an alternative to SELEX-based selection strategies, allosteric selections based upon the directed evolution of ligand-dependent self-cleaving ribozymes have also enabled the isolation of RNA aptamers.(44, 45) Although allosteric selection can be particularly advantageous when it is undesirable or challenging to couple the target ligand to a solid support, this in vitro approach has not consistently led to the isolation of allosteric ribozymes that function well within the intracellular milieu.(46) The idea of selecting aptamers that recognize ligands free in solution, as opposed to bound to a solid support, has been pursued widely recently.(43, 47) In addition to obviating the need for synthesizing a solid-supported ligand, performing selections in the solution phase (ideally under conditions that mimic the environment in which the aptamer will be used), satisfies the maxim “you get what you screen for”.(48)
Many efforts toward more efficient aptamer selections have focused on using electrophoresis strategies to separate unbound RNAs from the desired aptamer complexes, and there is some evidence that this partitioning reduces the number of amplification cycles needed to discover aptamers.(49) However, while electrophoresis-based approaches show promise for identifying aptamers that bind protein or peptide targets, they have not (yet) proven generally effective for identifying aptamers that recognize small molecules.(49–51) This may be due, in part, to relatively small changes in charge and molecular weight upon ligand binding. In addition to classical separation technologies, a recent effort has implemented a magnetic-bead based selection on a chip-based microfluidic platform. While this technique may enable rapid and automatable selections for small-molecule binding aptamers, it does not overcome the disadvantage of having to couple the small-molecule to a solid support.(52)
While fundamentally new partitioning approaches may lead to more efficient aptamer selections, there are still many opportunities to improve current selection methods. For example, an automated buffer optimization strategy can identify an ideal buffer identity, buffer concentration, mono- and divalent salt concentrations, and pH before proceeding with selections for aptamers that bind a given target.(53) Additionally, a recent study aimed to improve the selection efficacy of traditional SELEX by decreasing the amplification bias introduced by the conventional PCR amplification step.(54) This approach, termed SELEX-T, replaces most of the PCR amplification cycles with T7 RNA polymerase amplification, and may be an important improvement for raising aptamers to both protein targets and small-molecule targets, alike. In our opinion, advances in selection technology are critical for maximizing the impact of aptamers and riboswitches in chemical biology.
Among the small-molecule binding aptamers discovered to date, perhaps the best-known example is the theophylline-binding aptamer, which was isolated by SELEX in 1994.(55) This aptamer binds theophylline tightly (KD = 320 nM) and with high specificity: The aptamer binds caffeine (which differs by an additional methyl group) 10,000-fold less tightly. The specificity of the RNA aptamer was 10-fold better than the performance of available antibodies, and the binding-affinity was 100-fold better than had been achieved with any previously identified small-molecule binding aptamer.(55) Furthermore, theophylline is an inexpensive FDA approved drug, whose activity and toxicity profiles have been characterized in a number of biological systems, making it a particularly attractive ligand choice for use in cells. Because of these desirable properties, many studies have drawn upon the theophylline aptamer to develop synthetic riboswitches, with the ultimate goal of pursuing more advanced applications in chemical biology.
FROM APTAMER TO RIBOSWITCH
Before riboswitches were identified in living systems, several groups anticipated that synthetic aptamers could be incorporated into mRNAs to regulate gene expression in living cells. Previous studies had shown that highly structured regions in the 5′-UTR of mRNAs could cause marked reductions in gene expression in both eukaryotic and prokaryotic cells.(56–58) Guided by these studies, Werstuck and Green incorporated a small-molecule binding RNA into the 5′-UTR of a gene, and showed that ligand-inducible gene expression could be accomplished in yeast.(14) Further studies showed that small-molecule binding aptamers could be incorporated within the 5′-UTR of genes in a variety of organisms to generate synthetic riboswitches that could respond to theophylline,(8, 12, 59) antibiotics,(60) or dyes.(9, 61) Since these initial observations, there has been increasing interest in the development of high-throughput screens or selections to isolate synthetic riboswitches that respond to a variety of new ligands.(11, 62, 63)
High-throughput screens and selections
In 2004, Desai and Gallivan reported the first example of a synthetic riboswitch that functioned in E. coli, while also providing proof-of-principle that a genetic screen could be used to isolate a rare functional riboswitch from a large pool of nonfunctional sequences.(8) In contrast to prior studies, in which ligand binding to the aptamer increased structure in the 5′-UTR of an mRNA and reduced translational efficiency,(10, 14, 58, 60) this study showed that ligand binding could result in an ~8-fold increase in gene expression. Although it was apparent that the spacing between the aptamer sequence and ribosome binding site (RBS) was an important determinant of switching activity, the mechanism by which this riboswitch functioned was not immediately clear.
To investigate the mechanism of switching in E. coli, Lynch et al. developed a high-throughput screen to isolate riboswitches from libraries in which the region separating the aptamer and the RBS was substituted with 4–8 randomized nucleotides.(64) With the help of a colony picking robot and an automated liquid handling system, they identified new riboswitch variants that could activate β-galactosidase expression by up to 36-fold in E. coli. More importantly, these studies demonstrated that these riboswitches functioned by sequestering the RBS in the ligand-free state and revealing the RBS in the theophylline-bound conformation. These results provided impetus for the development of a screen based upon fluorescence-activated cell sorting (FACS) to identify synthetic riboswitches from even larger genetic libraries (12 randomized bases; 412 = 16,777,216 sequences). While this library was not exhaustively screened, the FACS-assay identified theophylline-sensitive synthetic riboswitches that feature very low background expression levels, and activate protein translation up to 96-fold in E. coli. (65) In contrast, a recent FACS-based effort to identify riboswitches that function at the transcriptional level featured more modest (~8-fold) activation ratios.(66) It is possible that transcriptional riboswitches may be more challenging to obtain, or perhaps the sequence space of this library was not optimally designed. Additional rounds of FACS using more diverse genetic libraries may identify synthetic riboswitches with larger activation ratios.
As an alternative to these genetic screens, Yokobayashi and coworkers developed a genetic selection for synthetic riboswitches based on expression of the TetA protein.(67) Cells that express TetA are resistant to tetracycline but are sensitive to NiCl2, and cells lacking TetA are sensitive to tetracycline but are resistant to NiCl2. Such a selection allows gross control over dual phenotypes (live or dead), but may not allow the graded control afforded by a genetic screen. Nevertheless, Nomura and Yokobayashi were able to assay 75,000 unique clones to identify TPP riboswitches that activate, rather than repress, gene expression in E. coli. Moreover, they identified a riboswitch that could activate gene expression 11-fold in the presence of TPP, completely reversing the activity of the parent riboswitch, which represses gene expression 9-fold in the presence of the ligand.(67)
Finally, our group reported a high throughput selection for synthetic riboswitches based on ligand-dependent changes in cell motility.(68) This simple screen not only isolated several riboswitches that activate gene expression in the presence of ligand, but also simultaneously optimized the riboswitches for dynamic ranges that are conducive to cell migration on semi-solid agar. Because ligand-dependent changes in cell motility are a desirable phenotype for a variety of applications, including bioremediation and cell targeting, we anticipate that motility selections may provide a foundation for a variety of new applications in chemical and synthetic biology.
EMERGING APPLICATIONS OF RIBOSWITCHES
Riboswitches as tools for regulated gene expression
Ligand-inducible expression systems are important genetic tools for common laboratory organisms such as E. coli and B. subtilis. However, many of these inducers (such as IPTG) are too expensive to be useful on the industrial scale. Natural riboswitches that are activated by amino acids may therefore represent an affordable alternative for such applications. Toward this goal, a tandem glycine riboswitch from B. subtilis was used for glycine-inducible production of β-galactosidase in B. subtilis cells.(69) Although this system provided only 6-fold induction when glycine was added, it may be worthwhile to revisit this general strategy if riboswitches with more ideal induction parameters can be developed.
In a recent report, Jin et al. proposed that ligand-sensitive riboswitches may be useful genetic tools for the production of conditional hypermorphic mutants, particularly in cases where null mutants are lethal.(70) To test this hypothesis, a theophylline-sensitive synthetic riboswitch was designed and used to regulate the chromosomal copy of an essential E. coli gene that is known to modulate motility (csrA). The creation of a csrA-lacZ fusion at this chromosomal position verified that expression levels were very low in the absence of theophylline, but approached wild type expression levels in the presence of theophylline. The authors further showed that the cells were non-motile in the absence of the inducer, but regained their motility phenotype in the presence of theophylline. These experiments fortuitously revealed that CsrA is a negative regulator of autoaggregation in E. coli. The ability of a synthetic riboswitch to permit reversible and tunable ligand-dependent gene expression of a protein over its native expression range suggests that synthetic riboswitches may find broad use in studying microbial genetics.
Riboswitches as antimicrobial targets
RNA is a primary target of many antibacterial compounds. While it has been known for quite some time that ribosomal RNA is an important antimicrobial target, it was recently discovered that some antibiotics whose mechanisms of actions were previously unknown may act, in part, by targeting riboswitches.(71) Following up on observations that some roseoflavin-resistant strains of B. subtilis featured mutations within an FMN aptamer sequence, Lee et al. demonstrated that this naturally-occurring antimicrobial binds the FMN riboswitch as a major target.(72)
Additionally, it has been shown that two lysine analogs that repress the growth of some Gram-positive bacteria (l-aminoethylcysteine and dl-4-oxalysine) also bind the lysC riboswitch of B. subtilis.(73) Although the primary antimicrobial mechanism of these lysine analogs may not involve riboswitch-binding,(74) Breaker and coworkers were inspired by this discovery and asked if they could identify new antimicrobials that target natural riboswitches.(75) With the goal of identifying an antimicrobial compound that could specifically repress bacterial purine metabolism, they tested a panel of 16 guanine analogs for the ability to bind the B. subtilis guanine riboswitch.(75) By pairing in vitro in-line probing assays with in vivo growth inhibition and reporter gene expression assays, they identified a specific analog that may inhibit B. subtilis growth by the intended mechanism. Although B. subtilis is not pathogenic, these advancements serve as a guide for future efforts to screen for riboswitch-binding antimicrobial agents in pathogenic bacteria.
Riboswitches as Boolean logic gates
While most natural riboswitches consist of a single aptamer domain and an expression platform, some natural riboswitches are more complex.(76) Some riboswitches, such as the tandem TPP riboswitch(77) from B. anthracis and the tandem glycine riboswitch(28) from B. subtilis, have two aptamers that work together to produce a more digital response than can be achieved by riboswitches that employ only a single aptamer. Other complex riboswitches, such as the metE tandem riboswitches (SAM and AdoCbl) from B. clausii, function independently of one another to constitute a two-input Boolean NOR logic gate.(76) In this system, high concentrations of SAM repress the metE operon, as well as other related operons, in a widespread effort to prevent further SAM biosynthesis. Additionally, AdoCbl independently represses the metE operon because this cofactor enables MetH to synthesize methionine more efficiently than MetE. Thus, high concentrations of either SAM or AdoCbl cause transcriptional termination of metE RNA. The net result is a NOR logic gate, whereby MetE is produced at high levels only when both SAM and AdoCbl are present at low concentrations. These complex riboswitches have inspired others to develop synthetic logic gates to reprogram cell behavior and to engineer metabolic pathways.
Shortly before the first riboswitch was reported, Jose et al. recognized that the catalytic benefits gained by cooperative binding in allosteric proteins might be harnessed in the RNA realm by constructing binary allosteric ribozymes.(78) In an impressive demonstration of modular rational design, these researchers engineered a binary ribozyme that would self-cleave only in the presence of two effectors (theophylline and FMN). In vitro studies demonstrated that this binary ribozyme responds in a digital fashion, exhibiting little cleavage in the presence of a single effector, but providing a ~300-fold rate enhancement when both effectors are present.(78)
Building upon these earlier efforts, Win and Smolke recently engineered several versions of binary ribozymes to obtain genetic logic gates that operate by Boolean logic.(79) By incorporating the previously reported theophylline and tetracycline aptamers in various positions relative to a self-cleaving ribozyme, the researchers sought to construct AND, NOR, NAND, and OR genetic gates that might function within living cells. When tested in yeast, however, it was found that each of these allosteric ribozymes exhibited less than 3-fold modulation of reporter gene expression when the appropriate ligands were present.(79) The modest performance of these ribozymes in cells compared to their in vitro counterparts (78) might be improved by subjecting the ribozymes to an in vivo selection.(46)
Wieland et al. recently performed in vivo selections for allosteric ribozymes featuring improved modulation of reporter gene expression in bacteria,(80, 81) suggesting that this strategy might be extended to optimize binary riboswitches for in vivo applications. With a similar strategy in mind, Yokobayashi and coworkers used their TetA dual genetic selection system to perform in vivo selections for complex riboswitches, which function in E. coli as AND or NAND Boolean logic gates.(11) The AND gates exhibited particularly good properties, as gene expression remained low unless both theophylline and thiamine were present, which then enabled up to 18-fold induction of gene expression in vivo. These impressive results suggest that dual genetic selection may be a useful approach for creating Boolean logic gates in living cells.
Riboswitch-based control of bacterial behavior
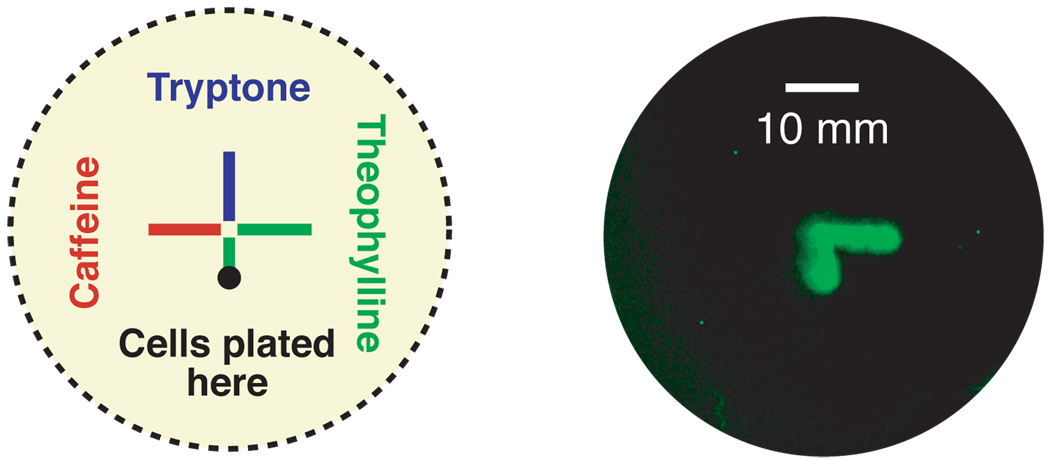
Because riboswitches are versatile tools for controlling gene expression, they can be used to reprogram a variety of bacterial behaviors. Bacterial chemotaxis has been studied extensively and is well understood at the genetic level. The ability to modulate bacterial motility in response to arbitrary chemical signals would provide new tools for bioremediation and drug delivery. We hypothesized that the E. coli chemotaxis system could be reprogrammed by placing a key chemotaxis signaling protein (cheZ) under the control of a theophylline-sensitive riboswitch.(82) Reprogrammed cells would then migrate up gradients of this ligand and autonomously localize to regions of high theophylline concentration, which is a behavior that cannot be accomplished by the natural E. coli chemotaxis system. To test this hypothesis, theophylline or caffeine was pipetted in a T-shape pattern on the surface of a semi-solid media plate. Reprogrammed cells were then spotted at the bottom of the theophylline path. As shown in Figure 4, the population of reprogrammed cells migrated up the first portion of the theophylline path, and then made a right turn to continue following this ligand, without migrating off the path. This precise localization to the ligand represents a sharp contrast to the behavior of wild type E. coli (which do not stop moving, and thus cannot localize to a chemical signal), and such behavior may prove useful for targeting cells.
Figure 4.
Controlling cell motility with a synthetic riboswitch. E. coli containing the cheZ gene under the control of a theophylline-sensitive synthetic riboswitch are plated at the bottom of a path containing theophylline and caffeine on semi-solid agar (the path is diagrammed to the left). Cells migrate exclusively along the theophylline path and disregard the path of caffeine, which is not recognized by the riboswitch.
Future Prospects
Just as natural riboswitches can regulate gene expression in response to small-molecule ligands during transcription or translation, synthetic riboswitches can be engineered to repress or activate gene expression in a ligand-dependent fashion. To maintain the optimal concentrations of critical cellular metabolites, natural riboswitches generally bind ligands to regulate the expression of metabolic genes. In contrast, synthetic riboswitches can be employed to regulate the expression of any gene (or genetic circuit) in response to any non-toxic molecule that is capable of being bound by RNA. This feature should enable RNA switches to play an increasingly important role as chemical biologists seek to modulate many types of cellular behavior in response to a broad range of chemical signals. We anticipate that as selection strategies improve, synthetic riboswitches will be easier to obtain, and that they will become a standard tool for the chemical biologist.
Acknowledgments
We thank Dennis Mishler for helpful comments. This work was supported by the NIH (GM074070).
Glossary
- RNA aptamer
An RNA sequence that binds a ligand with high affinity and specificity.
- Riboswitch
An RNA consisting of an aptamer and an expression platform that controls gene expression in a ligand-dependent fashion, without the need for protein cofactors.
- 5´-UTR
The 5´-untranslated region of a messenger RNA (mRNA).
- Ribozyme riboswitch
An RNA that performs self-cleavage in a ligand-dependent fashion.
- SELEX
“Systematic Evolution of Ligands by EXponential Enrichment”; an in vitro selection process to discover aptamers.
- Chemotaxis
The process by which an organism directs its motion in response to a chemical cue.
- NAND logic gate
An AND function with an inverted output. Output is low only when all inputs are high.
- NOR logic gate
An OR function with an inverted output. Output is high only when all inputs are low.
References
- 1.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stark BC, Kole R, Bowman EJ, Altman S. Ribonuclease P: an enzyme with an essential RNA component. Proc. Natl. Acad. Sci. 1978;75:3717–3721. doi: 10.1073/pnas.75.8.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass BL, Cech TR. Specific interaction between the self-splicing RNA of Tetrahymena and its guanosine substrate: implications for biological catalysis by RNA. Nature. 1984;308:820–826. doi: 10.1038/308820a0. [DOI] [PubMed] [Google Scholar]
- 4.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 5.Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. XIST RNA and the mechanism of X chromosome inactivation. Annu. Rev. Genet. 2003;36:233–278. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 6.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 7.Gallivan JP. Toward reprogramming bacteria with small molecules and RNA. Curr. Opin. Chem. Biol. 2007;11:612–619. doi: 10.1016/j.cbpa.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai SK, Gallivan JP. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J. Am. Chem. Soc. 2004;126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- 9.Grate D, Wilson C. Inducible regulation of the S. cerevisiae cell cycle mediated by an RNA aptamer-ligand complex. Bioorg. Med. Chem. 2001;9:2565–2570. doi: 10.1016/s0968-0896(01)00031-1. [DOI] [PubMed] [Google Scholar]
- 10.Harvey I, Garneau P, Pelletier J. Inhibition of translation by RNA-small molecule interactions. RNA. 2002;8:452–463. doi: 10.1017/s135583820202633x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma V, Nomura Y, Yokobayashi Y. Engineering complex riboswitch regulation by dual genetic selection. J. Am. Chem. Soc. 2008;130:16310–16315. doi: 10.1021/ja805203w. [DOI] [PubMed] [Google Scholar]
- 12.Suess B, Fink B, Berens C, Stentz R, Hillen W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucl. Acids Res. 2004;32:1610–1614. doi: 10.1093/nar/gkh321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Topp S, Gallivan JP. Riboswitches in unexpected places--a synthetic riboswitch in a protein coding region. RNA. 2008;14:2498–2503. doi: 10.1261/rna.1269008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werstuck G, Green MR. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 15.Lee F, Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc. Natl. Acad. Sci. 1977;74:4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends. Biochem. Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Gelfand MS, Mironov AA, Jomantas J, Kozlov YI, Perumov DA. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 1999;15:439–442. doi: 10.1016/s0168-9525(99)01856-9. [DOI] [PubMed] [Google Scholar]
- 19.Kil YV, Mironov VN, Gorishin I, Kreneva RA, Perumov DA. Riboflavin operon of Bacillus subtilis: unusual symmetric arrangement of the regulatory region. Mol. Gen. Genet. 1992;233:483–486. doi: 10.1007/BF00265448. [DOI] [PubMed] [Google Scholar]
- 20.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 22.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 23.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 24.Wang JX, Lee ER, Morales DR, Lim J, Breaker RR. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol. Cell. 2008;29:691–702. doi: 10.1016/j.molcel.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 27.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 28.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 29.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soukup JK, Soukup GA. Riboswitches exert genetic control through metabolite-induced conformational change. Curr. Opin. Struct. Biol. 2004;14:344–349. doi: 10.1016/j.sbi.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 31.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Riboswitches: the oldest mechanism for the regulation of gene expression? Trends Genet. 2004;20:44–50. doi: 10.1016/j.tig.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Nudler E. Flipping riboswitches. Cell. 2006;126:19–22. doi: 10.1016/j.cell.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 33.Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, Mandal M, Rudnick ND, Breaker RR. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer MM, Roth A, Chervin SM, Garcia GA, Breaker RR. Confirmation of a second natural preQ(1) aptamer class in Streptococcaceae bacteria. RNA. 2008;14:685–695. doi: 10.1261/rna.937308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warner DF, Savvi S, Mizrahi V, Dawes SS. A Riboswitch Regulates Expression of the Coenzyme B12-Independent Methionine Synthase in Mycobacterium tuberculosis: Implications for Differential Methionine Synthase Function in Strains H37Rv and CDC1551. J. Bacteriol. 2007;189:3655–3659. doi: 10.1128/JB.00040-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–U497. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 37.Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 39.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 40.Lippow SM, Tidor B. Progress in computational protein design. Curr. Opin. Biotechnol. 2007;18:305–311. doi: 10.1016/j.copbio.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bloom JD, Arnold FH. In the light of directed evolution: pathways of adaptive protein evolution. Proc. Natl. Acad. Sci. 2009;106 Suppl 1:9995–10000. doi: 10.1073/pnas.0901522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berens C, Thain A, Schroeder R. A tetracycline-binding RNA aptamer. Bioorg. Med. Chem. 2001;9:2549–2556. doi: 10.1016/s0968-0896(01)00063-3. [DOI] [PubMed] [Google Scholar]
- 43.Stoltenburg R, Reinemann C, Strehlitz B. SELEX--a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Koizumi M, Soukup GA, Kerr JN, Breaker RR. Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nat. Struct. Biol. 1999;6:1062–1071. doi: 10.1038/14947. [DOI] [PubMed] [Google Scholar]
- 45.Soukup GA, Emilsson GA, Breaker RR. Altering molecular recognition of RNA aptamers by allosteric selection. J. Mol. Biol. 2000;298:623–632. doi: 10.1006/jmbi.2000.3704. [DOI] [PubMed] [Google Scholar]
- 46.Link KH, Breaker RR. Engineering ligand-responsive gene-control elements: lessons learned from natural riboswitches. Gene Ther. 2009;16:1189–1201. doi: 10.1038/gt.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mosing RK, Bowser MT. Microfluidic selection and applications of aptamers. J. Sep. Sci. 2007;30:1420–1426. doi: 10.1002/jssc.200600483. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt-Dannert C, Arnold FH. Directed evolution of industrial enzymes. Trends Biotechnol. 1999;17:135–136. doi: 10.1016/s0167-7799(98)01283-9. [DOI] [PubMed] [Google Scholar]
- 49.Mosing RK, Bowser MT. Isolating aptamers using capillary electrophoresis-SELEX (CE-SELEX) Methods Mol. Biol. 2009;535:33–43. doi: 10.1007/978-1-59745-557-2_3. [DOI] [PubMed] [Google Scholar]
- 50.Berezovski M, Drabovich A, Krylova SM, Musheev M, Okhonin V, Petrov A, Krylov SN. Nonequilibrium Capillary Electrophoresis of Equilibrium Mixtures: A Universal Tool for Development of Aptamers. J. Am. Chem. Soc. 2005;127:3165–3171. doi: 10.1021/ja042394q. [DOI] [PubMed] [Google Scholar]
- 51.Drabovich A, Berezovski M, Krylov SN. Selection of Smart Aptamers by Equilibrium Capillary Electrophoresis of Equilibrium Mixtures (ECEEM) J. Am. Chem. Soc. 2005;127:11224–11225. doi: 10.1021/ja0530016. [DOI] [PubMed] [Google Scholar]
- 52.Lou X, Qian J, Xiao Y, Viel L, Gerdon AE, Lagally ET, Atzberger P, Tarasow TM, Heeger AJ, Soh HT. Micromagnetic selection of aptamers in microfluidic channels. Proc. Natl. Acad. Sci. 2009;106:2989–2994. doi: 10.1073/pnas.0813135106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stovall GM, Cox JC, Ellington AD. Automated optimization of aptamer selection buffer conditions. J. Assoc. Lab. Automation. 2004;9:117–122. [Google Scholar]
- 54.Tsuji S, Hirabayashi N, Kato S, Akitomi J, Egashira H, Tanaka T, Waga I, Ohtsu T. Effective isolation of RNA aptamer through suppression of PCR bias. Biochem. Bioph. Res. Co. 2009;386:223–226. doi: 10.1016/j.bbrc.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 55.Jenison R, Gill S, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 56.Paraskeva E, Atzberger A, Hentze MW. A translational repression assay procedure (TRAP) for RNA-protein interactions in vivo. Proc. Natl. Acad. Sci. USA. 1998;95:951–956. doi: 10.1073/pnas.95.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stripecke R, Oliveira CC, McCarthy JE, Hentze MW. Proteins binding to 5' untranslated region sites: a general mechanism for translational regulation of mRNAs in human and yeast cells. Mol. Cell. Biol. 1994;14:5898–5909. doi: 10.1128/mcb.14.9.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Smit MH, Van Duin J. Secondary Structure of the Ribosome Binding-Site Determines Translational Efficiency - a Quantitative-Analysis. Proc. Natl. Acad. Sci. USA. 1990;87:7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson K, Syrett H, Knudsen S, Ellington A. Group I aptazymes as genetic regulatory switches. BMC Biotechnology. 2002;2:21. doi: 10.1186/1472-6750-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suess B, Hanson S, Berens C, Fink B, Schroeder R, Hillen W. Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucl. Acids Res. 2003;31:1853–1858. doi: 10.1093/nar/gkg285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buskirk AR, Landrigan A, Liu DR. Engineering a ligand-dependent RNA transcriptional activator. Chem. Biol. 2004;11:1157–1163. doi: 10.1016/j.chembiol.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 62.Famulok M. Allosteric aptamers and aptazymes as probes for screening approaches. Curr. Opin. Mol. Ther. 2005;7:137–143. [PubMed] [Google Scholar]
- 63.Weigand JE, Sanchez M, Gunnesch EB, Zeiher S, Schroeder R, Suess B. Screening for engineered neomycin riboswitches that control translation initiation. RNA. 2008;14:89–97. doi: 10.1261/rna.772408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lynch SA, Desai SK, Sajja HK, Gallivan JP. A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function. Chem. Biol. 2007;14:173–184. doi: 10.1016/j.chembiol.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lynch SA, Gallivan JP. A flow cytometry-based screen for synthetic riboswitches. Nucl. Acids Res. 2009;37:184–192. doi: 10.1093/nar/gkn924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fowler CC, Brown ED, Li Y. A FACS-based approach to engineering artificial riboswitches. ChemBioChem. 2008;9:1906–1911. doi: 10.1002/cbic.200700713. [DOI] [PubMed] [Google Scholar]
- 67.Nomura Y, Yokobayashi Y. Reengineering a natural riboswitch by dual genetic selection. J. Am. Chem. Soc. 2007;129:13814–13815. doi: 10.1021/ja076298b. [DOI] [PubMed] [Google Scholar]
- 68.Topp S, Gallivan JP. Random walks to synthetic riboswitches--a high-throughput selection based on cell motility. ChemBioChem. 2008;9:210–213. doi: 10.1002/cbic.200700546. [DOI] [PubMed] [Google Scholar]
- 69.Phan TT, Schumann W. Development of a glycine-inducible expression system for Bacillus subtilis. J. Biotechnol. 2007;128:486–499. doi: 10.1016/j.jbiotec.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 70.Jin Y, Watt RM, Danchin A, Huang JD. Use of a riboswitch-controlled conditional hypomorphic mutation to uncover a role for the essential csrA gene in bacterial autoaggregation. J. Biol. Chem. 2009;284:28738–28745. doi: 10.1074/jbc.M109.028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat. Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- 72.Lee ER, Blount KF, Breaker RR. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 2009;6:187–194. doi: 10.4161/rna.6.2.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR. Antibacterial lysine analogs that target lysine riboswitches. Nat. Chem. Biol. 2007;3:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- 74.Ataide SF, Wilson SN, Dang S, Rogers TE, Roy B, Banerjee R, Henkin TM, Ibba M. Mechanisms of resistance to an amino acid antibiotic that targets translation. ACS Chem. Biol. 2007;2:819–827. doi: 10.1021/cb7002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim JN, Blount KF, Lim J, Link KH, Breaker R. Design and Antimicrobial Action of Purine Analogs that Bind Guanine Riboswitches. ACS Chem. Biol. 2009 doi: 10.1021/cb900146k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- 77.Welz R, Breaker RR. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA. 2007;13:573–582. doi: 10.1261/rna.407707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jose AM, Soukup GA, Breaker RR. Cooperative binding of effectors by an allosteric ribozyme. Nucl. Acids Res. 2001;29:1631–1637. doi: 10.1093/nar/29.7.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wieland M, Benz A, Klauser B, Hartig JS. Artificial ribozyme switches containing natural riboswitch aptamer domains. Angew. Chem. Int. Ed. Engl. 2009;48:2715–2718. doi: 10.1002/anie.200805311. [DOI] [PubMed] [Google Scholar]
- 81.Wieland M, Hartig JS. Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew. Chem. Int. Ed. Engl. 2008;47:2604–2607. doi: 10.1002/anie.200703700. [DOI] [PubMed] [Google Scholar]
- 82.Topp S, Gallivan JP. Guiding bacteria with small molecules and RNA. J. Am. Chem. Soc. 2007;129:6807–6811. doi: 10.1021/ja0692480. [DOI] [PMC free article] [PubMed] [Google Scholar]